Abstract
The RNA genome of influenza A virus, which forms viral ribonucleoprotein complexes (vRNPs) with viral polymerase subunit proteins (PA, PB1, and PB2) and nucleoprotein (NP), is transcribed and replicated in the nucleus. NP, the major component of vRNPs, has at least two amino acid sequences that serve as nuclear localization signals (NLSs): an unconventional NLS (residues 3 to 13; NLS1) and a bipartite NLS (residues 198 to 216; NLS2). Although both NLSs are known to play a role in nuclear transport, their relative contributions to viral replication are poorly understood. We therefore investigated their contributions to NP subcellular/subnuclear localization, viral RNA (vRNA) transcription, and viral replication. Abolishing the unconventional NLS caused NP to localize predominantly to the cytoplasm and affected its activity in vRNA transcription. However, we were able to create a virus whose NP contained amino acid substitutions in NLS1 known to abolish its nuclear localization function, although this virus was highly attenuated. These results indicate that while the unconventional NLS is not essential for viral replication, it is necessary for efficient viral mRNA synthesis. On the other hand, the bipartite NLS, whose contribution to the nuclear transport of NP is limited, was essential for vRNA transcription and NP's nucleolar accumulation. A virus with nonfunctional NLS2 could not be generated. Thus, the bipartite NLS, but not the unconventional NLS, of NP is essential for influenza A virus replication.
Influenza A virus, a member of the family Orthomyxoviridae, is characterized by segmented RNA genomes of negative polarity (16). The viral genome encodes at least 11 proteins (4, 16) and consists of eight single-stranded RNA segments. These genomic RNAs are incorporated into virions as viral ribonucleoprotein complexes (vRNPs) that comprise viral RNA (vRNA), heterotrimeric viral polymerase subunit proteins (PA, PB1, and PB2), and nucleoprotein (NP). A unique property of influenza viruses among RNA viruses is that every step of vRNA synthesis take place in the nucleus by use of the nuclear machinery of the host cells (16). Therefore, newly synthesized proteins required for the vRNPs must be transported into the nuclei of the cells.
The transport of proteins (larger than 50 kDa) from the cytoplasm into the nucleus is regulated by signal-mediated processes. Peptide motifs that allow the proteins to be imported through the nuclear pore complex are referred to as nuclear localization signals (NLSs). They are rich in basic amino acids and bind to NLS receptors (e.g., karyopherin family members), which are responsible for the nuclear translocation of target proteins (12, 31). Of the influenza A virus proteins, NLSs have been found in three polymerase subunits, the matrix M1 protein (which associates with vRNP complexes), the nonstructural NS1 protein, and NP, the primary component of vRNPs (for a review, see reference 6).
NP has at least two NLS sequences. An unconventional NLS (termed here NLS1) is located between residues 3 and 13 and is responsible for NP binding to karyopherins α1 and α2 (27). The conserved basic residues at positions 7 and 8 are critical for NP import into the nucleus (19). Cros et al. (5) recently demonstrated that NLS1 is indispensable not only for the nuclear localization of NP but also for the nuclear import of vRNPs in digitonin-permeabilized cells. The second NLS is a bipartite signal located in the middle of NP (residues 198 to 216 [29]; termed here NLS2). This signal was identified as a counterpart of an NLS of Thogoto virus NP, which accumulates in the nucleus but lacks the amino acid sequence of the NLS1 of influenza A virus. NLS2 can function as an NLS to a limited extent when it is fused to a cytoplasmic reporter protein (29). Its contribution to NP nuclear import, however, is not as great as that of NLS1 (29). Furthermore, a mutant NP lacking both of these two NLSs is still transported to the nucleus to a limited extent, suggesting the existence of at least one additional NLS (3, 19). The data just described were obtained by expressing mutant NPs or proteins fused with the NLSs of NP, and the contribution of each NLS to viral replication was not clear.
Therefore, to understand the significance of the NLSs of influenza A virus NP for viral replication, we investigated the effect of mutations in these NLSs on the subcellular/subnuclear localization of NP and on vRNA transcription. Furthermore, to clarify their contributions to viral replication, we attempted to determine the nucleotide sequences required for efficient incorporation of NP segments into progeny virions (the so-called “packaging signal”). This experimental step was necessary since NLS1 was potentially located in the packaging signal of the NP segment, given the sequence information of other gene segments (10, 11, 18, 28). Using this knowledge of the NP packaging signal, we then generated recombinant viruses with mutations in NLS1. Our results show that NLS2, but not NLS1, is essential for viral replication.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney COS-7 cells and human embryonic kidney 293 and 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Madin-Darby canine kidney (MDCK) cells were maintained in minimal essential medium containing 5% newborn calf serum. All cells were maintained at 37°C in 5% CO2. A/WSN/33 (H1N1) (WSN) virus was generated by reverse genetics as previously described (20) and propagated in MDCK cells. A/Puerto Rico/8/34 (H1N1) (PR8) virus propagated in embryonated hen's eggs was kindly provided by Hiroshi Kida.
Reverse genetics.
Influenza viruses or virus-like particles (VLPs) were generated by using plasmid-based reverse genetics as previously described (20). Briefly, plasmids possessing the cDNA of WSN viral genes under the control of the human RNA polymerase I promoter and the mouse RNA polymerase I terminator (referred to as PolI plasmids) were used to produce of vRNAs of WSN virus. WSN viral proteins were expressed by using pCAGGS/MCS (15, 22) under the control of the chicken β-actin promoter. PolI plasmids and protein expression plasmids were mixed with a transfection reagent, Trans IT 293 (Mirus, Madison, WI) in Opti-MEM (GIBCO-BRL) and added to 293T cells. At 48 h posttransfection, viruses or VLPs in the culture supernatant were harvested.
Construction of plasmids.
To generate a plasmid that would produce NP vRNA containing mutations in the NLS1 coding sequence, we performed site-directed mutagenesis with PCR. In brief, PCR was performed with pPolI-WSN-NP (20) (termed here pPolI/NP) as the template and with complementary primers Pol-NP-NLS1mut-F (5′-GACCAAAGGCACCGCAGCATCTTACGAACA-3′; underlining indicates altered nucleotides) and Pol-NP-NLS1mut-R (5′-TGTTCGTAAGATGCTGCGGTGCCTTTGGTC-3′). The PCR products were introduced into Escherichia coli, yielding pPolI/NP-NLS1mut. A plasmid that would produce NP-NLS2mut vRNA (pPolI/NP-NLS2mut) was constructed in a similar fashion with primers Pol-NP-NLS2mut-F (5′-GGGGTGAGAATGGAGCGGCAACAGCGATTGCTTATGAAAG-3′) and Pol-NP-NLS2mut-R (5′-CTTTCATAAGCAATCGCTGTTGCCGCTCCATTCTCACCCC-3′).
To construct a plasmid for the expression of NP-NLS2mut, pPolI/NP-NLS2mut was amplified by PCR with primers CA-NP-F (5′-CACACAGAATTCATGGCGACCAAAGGCACCAAACGATCTTAC-3′) and CA-NP-R (5′-CACACAGCTAGCTTAATTGTCGTACTCCTCTGCATTGTCTCCGAAG-3′). The PCR products were digested with NheI and EcoRI and cloned into pCAGGS/MCS, resulting in pCAGGS/NP-NLS2mut. To construct plasmids for the expression of NP-NLS1mut and NP-NLS1+2mut, pPolI/NP or pPolI/NP-NLS2mut was amplified by PCR with primers CA-NP-NLS1mut-F (5′-CACACAGAATTCATGGCGACCAAAGGCACCGCAGCATCTTACGAAC-3′) and CA-NP-R. The PCR products were digested with NheI and EcoRI and cloned into pCAGGS/MCS, resulting in pCAGGS/NP-NLS1mut and pCAGGS/NP-NLS1+2mut, respectively.
To produce plasmids for the expression of c-Myc-tagged NP, we prepared the DNA fragment corresponding to the sequence of the c-Myc tag with a linker by annealing oligonucleotides N-myc(+) (5′-AATTCACCATGGAACAAAAACTCATCTCAGAAGAGGATCTGGC-3′) and C-myc(−) (5′-GGCCGCCAGATCCTCTTCTGAGATGAGTTTTTGTTCCATGGTG-3′). They were digested with EcoRI and NotI and cloned into pCAGGS/MCS, resulting in pCAGGS/N-Myc. The open reading frame of NP or NP-NLS1mut, amplified by PCR with primer set CA-NP(NotI)-F (5′-CACACAGCGGCCGCGATGGCGACCAAAGGCACCAAACGATCTTAC-3′) or CA-NP-NLS1mut(NotI)-F (5′-CACACAGCGGCCGCGATGGCGACCAAAGGCACCGCAGCATCTTACGAAC-3′) and CA-NP-R, were digested with NotI and NheI and cloned into pCAGGS/N-Myc, resulting in pCAGGS/Myc-NP and pCAGGS/Myc-NP-NLS1mut.
Plasmids for the expression of yellow fluorescent protein (YFP) fused with NP (NP-YFP) and NP-NLS2mut (NP-NLS2mut-YFP) were constructed as follows. To prepare the DNA fragment of YFP, the open reading frame of YFP (Clontech, Palo Alto, CA) was amplified by PCR with primers CXFP-NotI-F (5′-CACACACAGCGGCCGCGGGAGGAGTGAGCAAGGGCGAGGAGCTGTT C-3′) and CXFP-BglII-R (5′-CACACAAGATCTTACTTGTACAGCTCGTCCATGCCGAGAG-3′). The PCR products were digested with NotI and BglII and cloned into pCAGGS/MCS, resulting in pCAGGS/C-YFP. NP and NP-NLS2mut gene fragments were amplified by PCR with pPolI/NP and pPolI/NP-NLS2mut as the templates and with primers CA-NP-F and CA-NP(NotI)-R (5′-CACACAGCGGCCGCTTAATTGTCGTACTCCTCTGCATTGTCTCCG AAG-3′). The PCR products were digested with EcoRI and NotI and cloned into pCAGGS/C-YFP, resulting in pCAGGS/NP-YFP and pCAGGS/NP-NLS2mut-YFP, respectively.
For construction of pCAGGS/NLS2-NP-NLS2mut and pCAGGS/NLS2mut-NP-NLS2mut, pPolI/NP-NLS2mut was amplified by PCR with primers NLS2-add-F (5′-CACACAGAATTCATGAAACGTGGGATCAATGATCGGAACTTCTGGAGGGGTGAGAATGGACGGAGAACAAGGATGGCGACCAAA GGCACC-3′) or NLS2mut-add-F (5′-CACACAGAATTCATGAAACGTGGGATCAATGATCGGAACTTCTGGAGGGGTGAGAATGGAGCGGCAACA GCGATGGCGACCAAAGGCACC-3′) and CA-NP-R, respectively. The PCR products were digested with NheI and EcoRI and cloned into pCAGGS/MCS. Construction of pCAGGS/NP-KR1 and pCAGGS/NP-KR2 was performed similarly with primers CA-KR1-F (5′-CACACAGAATTCATGGCGACCAAAGGCACCAAAAGATCTTACGAAC-3′) and CA-KR2-F (5′-CACACAGAATTCATGGCGACCAAAGGCACCAAGAGGTCTTACGAAC-3′), respectively.
To prepare a series of plasmids that produce RNAs containing the complementary sequence for the open reading frame of enhanced green fluorescent protein (GFP; Clontech) flanked by different lengths of the coding and/or noncoding regions of the NP segment, the sequence of NP vRNA was cloned into pUC19, resulting in pUC19/NP. pPolI/NP(300)GFP(300) was used to produce negative-sense RNA containing the 3′ noncoding ends of NP vRNA, a complementary sequence encoding 100 amino-terminal NP codons and the GFP open reading frame, two consecutive stop codons (TAA-TAG) in the negative sense, and 300 bases of the 5′ coding region and the 5′ noncoding region of NP vRNA. This plasmid was produced by amplifying pUC19/NP by inverse PCR with back-to-back primers NP-300-F (5′-CACACAGGTCTCAATAAAGGGCTTCCTCGGGCC-3′) and NP-300-R (5′-CACACAGGTCTCACCATTACTCTCCTGTATATAGGTCCTCCAG-3′). The PCR products were digested with BsaI, and the GFP gene was cloned into the BsaI sites, resulting in pUC19/NP(300)GFP(300). The DNA fragment obtained from pUC19/NP(300)GFP(300) by digestion with BsmBI was inserted between the BsmBI sites of a PolI plasmid. A series of NP coding deletion mutants was similarly produced by inverse PCR. These mutants were designated according to the number of nucleotides derived from the NP coding region; for example, the NP(120)GFP(30) RNA segment contains the 3′ noncoding region, 120 nucleotides of the coding region of NP vRNA, the GFP sequence, and 30 nucleotides of the 3′ coding and the 5′ noncoding regions of NP vRNA. The NP(0)GFP(0) RNA segment contains the 3′ noncoding region of NP vRNA, the GFP sequence, and the 5′ noncoding region of NP vRNA.
To produce reporter NP vRNA encoding firefly luciferase, an amplified DNA fragment encoding firefly luciferase was cloned into pPolI/NP(0)GFP(0) instead of GFP, resulting in pPolI/NP(0)Fluc(0).
Plasmids to produce the NP(60)NP, NP(60)NP-NLS1mut, NP(60)NP-NLS2mut, and NP(60)NP-NLS1+2mut vRNAs were constructed by inverse PCR (23) with the back-to-back primers, followed by oligonucleotide insertion as follows. First, two ATG codons (nucleotides 82 to 84 and 92 to 94) were converted to ATC in the NP vRNA of pPolI/NP by site-directed mutagenesis with PCR primers ATC-F (5′-CGATCTTACGAACAGATCGAGACTGATCGAGAACGCCAGAATGC-3′) and ATC-R (5′-GCATTCTGGCGTTCTCGATCAGTCTCGATCTGTTCGTAAGATCG-3′). An insert fragment corresponding to the first 60 nucleotides of the NP coding region, which contains a GCG mutation at the initiation codon of the NP open reading frame (nucleotides 37 to 39) and a partial Kozak consensus sequence (GCCGCC) was then created by PCR with the mutated pPolI/NP produced above as a template and primers NPInsertF (5′-CACACACGTCTCAGCGGCGACCAAAGGCACCAAACGATCTTACGAACAGA-3′) and NPInsertR (5′-CACACACGTCTCACCATGGCGGCCTGGCGTTCTCGATCAGTCTCGATCTGTTCGTAAGATCGT-3′). This insert fragment was cloned into pPolI/NP, pPolI/NP-NLS1mut, pPolI/NP-NLS2mut, and pPolI/NP-NLS1+2mut, resulting in pPolI/NP(60)NP, pPolI/NP(60)NP-NLS1mut, pPolI/NP(60)NP-NLS2mut, and pPolI/NP(60)NP-NLS1+2mut, respectively.
All plasmid constructs were sequenced to ensure that no undesirable mutations were introduced.
Immunofluorescent staining.
At 12 h posttransfection, cells on 35-mm glass base dishes (IWAKI; Asahi Techno Glass, Chiba, Japan) were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min and permeabilized with 0.5% Triton X-100 for 10 min. NP was detected by incubating cells with an anti-NP monoclonal antibody (347/3; prepared in our laboratory) for 30 min and with Texas Red-labeled secondary antibody (Calbiochem-Novabiochem, La Jolla, CA) for 30 min. Nuclei were visualized with Hoechst 33342 (Invitrogen, Carlsbad, CA). c-Myc-tagged NP and fibrillarin were detected by incubating cells with an anti-c-Myc polyclonal rabbit antibody (C3956; Sigma, St. Louis, MO) or an antifibrillarin monoclonal antibody (38F3; Abcam, Cambridge, United Kingdom), respectively, for 30 min and with Alexa Fluor 594-labeled anti-rabbit secondary antibody (Molecular Probes, Eugene, OR) or Alexa Fluor 488-labeled anti-mouse secondary antibody (Molecular Probes), respectively, for 30 min. Samples were observed under a confocal laser microscope (LSM510META; Carl Zeiss, Jena, Germany).
Time-lapse imaging.
Transfected cells on glass base dishes were placed on the stage of an LSM510META that was warmed at 37°C in 5% CO2 with an Incubator XL-3 (PeCon, Jena, Germany). Images were acquired with C-APOCHROMAT 40x/1.2 W Corr (Carl Zeiss).
Luciferase assay.
Luciferase assay was performed with a dual-luciferase reporter assay system (Promega, Madison, WI) on a microplate luminometer (Veritas; Turner Biosystems, Sunnyvale, CA) according to the manufacturer's instructions. As an internal control for the dual-luciferase assay, pGL4.74[hRluc/TK] (Promega) was used.
Identification of the nucleotide sequence in the NP coding region that contributes to efficient incorporation of NP vRNA into virions.
We identified the packaging signal of the NP segment by using reverse genetics as previously described (18). In brief, we constructed plasmids to produce RNA possessing the complementary sequence of the enhanced GFP open reading frame flanked by various lengths of the coding and/or noncoding regions of the NP segment (see Fig. 6). One of these plasmids and seven plasmids to produce the remaining vRNA segments and four plasmids for the expression of NP, PA, PB1, and PB2, which are necessary and sufficient for vRNA transcription and replication, were cotransfected into 293T cells to generate VLPs possessing mutant NP vRNA. Since the transfectant VLPs do not possess intact NP, a functional NP must be supplied in trans for reporter GFP expression. To provide NP in trans, the PR8 strain was used as helper influenza virus. The supernatants containing VLPs were mixed with PR8 virus at a multiplicity of infection of 0.1 and transferred to MDCK cells. At 12 h postinfection, the cells were dissociated with trypsin and spun down for 5 min at 3,000 × g at 4°C. Cell pellets were resuspended in PBS containing 2% fetal calf serum and 0.1% sodium azide. The Cells were then incubated on ice with an anti-WSN-HA monoclonal antibody (967/8), which specifically binds to the HA protein of WSN but not PR8 virus, and with a rhodamine-labeled secondary antibody (CHEMICON, Hampshire, United Kingdom). We then performed fluorescence-activated cell sorter analysis with a FACScalibur (Becton Dickinson, Heidelberg, Germany) and the CellQuest software (Becton Dickinson). The efficiency of mutant NP vRNAs incorporation into infectious VLPs was calculated by dividing the number of GFP-expressing cells by the total number of GFP- and/or HA-expressing cells.
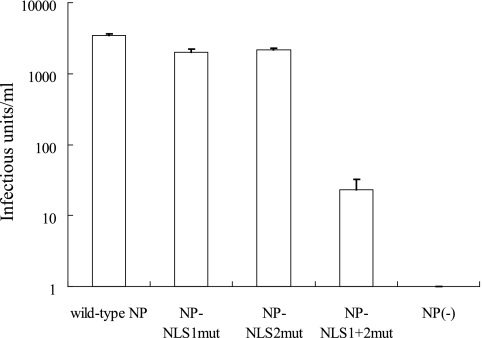
FIG. 6.
Comparison of mutant NPs' ability to support the generation of infectious VLPs. VLPs were generated with the plasmids for the expression of wild-type NP and of mutant NPs by using the plasmid-based VLP generation system previously described (21). Plasmids for the expression of wild-type NP and the indicated mutant NPs were transfected into 293T cells together with the protein expression plasmids for PB2, PB1, PA, HA, NA, M1, M2, NS1, and NS2 and pPolI-GFP (21). The culture supernatants were harvested 48 h posttransfection and mixed with helper virus (wild-type WSN virus at a multiplicity of infection of 1). MDCK cells were infected with these mixtures, and GFP-expressing cells were counted 24 h later. Error bars indicate the standard error of three independent experiments.
Immunostaining of plaques.
At 48 h postinfection, virus-infected MDCK cells were fixed with 4% paraformaldehyde in PBS for 30 min and permeabilized with 0.1% Triton X-100 for 30 min. After incubation with an anti-WSN virus polyclonal antibody for 30 min and with a biotinylated secondary antibody (VECTASTAIN ABC kit; Vector Laboratories, Burlingame, CA) for 30 min, the cells were treated with ABC reagent (Vector Laboratories) according to the procedures provided by the manufacturer. The samples were then reacted with diaminobenzidine tetrahydrochloride (Sigma).
Replicative properties of transfectant viruses.
The culture supernatants of MDCK cells infected with viruses at a multiplicity of infection of 0.001 were harvested at different times postinfection. The viral titers in the supernatants were determined by plaque assay on MDCK cells.
RESULTS
NLS1 plays the central role in NP nuclear localization.
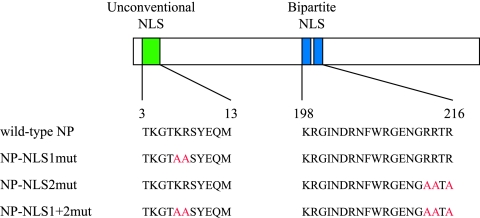
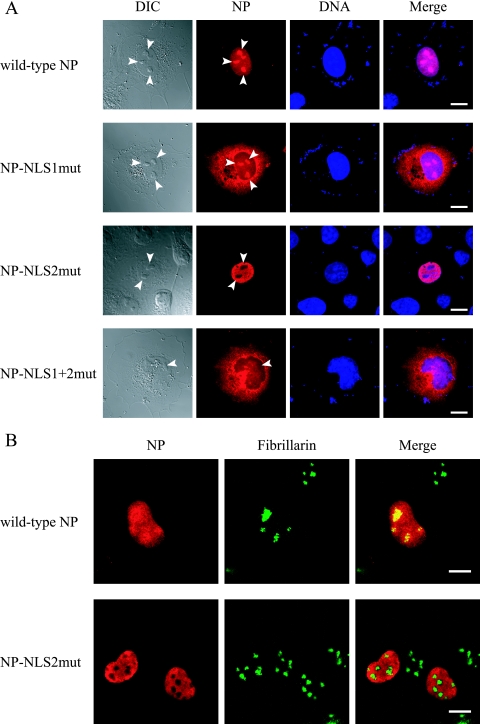
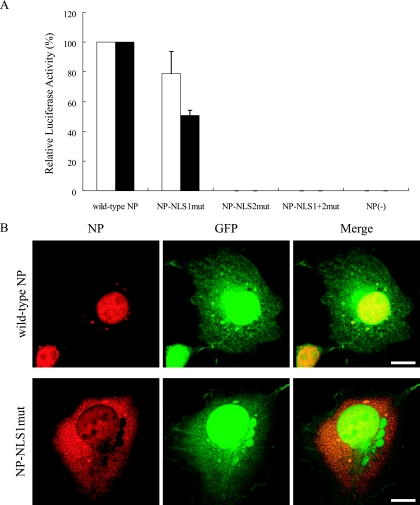
The basic residues in NP NLSs are important for its nuclear localization (19, 29). To confirm this previous finding, we constructed plasmids expressing NP in which alanine residues replaced the basic residues in NLS1 (Fig. 1, NP-NLS1mut), in NLS2 (Fig. 1, NP-NLS2mut), and in both (Fig. 1, NP-NLS1+2mut). We transfected COS-7 cells with each plasmid and assessed NP localization by immunofluorescent staining with an anti-NP monoclonal antibody. At 12 h posttransfection, wild-type NP and NP-NLS2mut accumulated in nuclei whereas NP-NLS1mut and NP-NLS1+2mut predominantly localized to the cytoplasm (Fig. 2A). These differences in subcellular localization indicate that NP nuclear localization is regulated mainly by NLS1 with a limited contribution from NLS2, in accordance with previous reports (5, 19, 27, 29).
FIG. 1.
Schematic diagrams of wild-type and mutant NPs. The unconventional NLS (residues 3 to 13) and the bipartite NLS (residues 198 to 216) are represented in green and blue, respectively. The alanine replacements known to abolish their nuclear transport activity (5) are in red.
FIG. 2.
Alteration of the subcellular/subnuclear localization of NP following alanine replacements in its NLSs. (A) Subcellular localization of mutant NPs. Plasmids for the expression of wild-type or mutant NP were transfected into COS-7 cells. At 12 h posttransfection, the cells were fixed and incubated with an anti-NP antibody (red) and with Hoechst dye (blue). The far left column shows differential interference contrast images. The far right column shows merged fluorescent images (Merge). Arrowheads point to nucleoli. Scale bars, 10 μm. (B) Subnuclear localization of wild-type NP and NP-NLS2mut. Plasmids for the expression of c-Myc-tagged wild-type NP or NP-NLS2mut were transfected into COS-7 cells. At 12 h posttransfection, cells were fixed and incubated with an anti-c-Myc antibody (red) and with an antibody against the nucleolar protein fibrillarin (green). The right column shows merged fluorescent images.
NLS2 plays the central role in NP nucleolar localization.
We also observed that wild-type NP, but not NP-NLS2mut, localized to the nucleolus at 12 h posttransfection (Fig. 2A). To prove that this nucleolus localization was dependent on NLS2, we examined the colocalization of c-Myc-tagged wild-type and mutant NPs with fibrillarin, a marker protein for the nucleolus. We found that NP did indeed localize to the nucleolus and that this localization was NLS2 dependent (Fig. 2B).
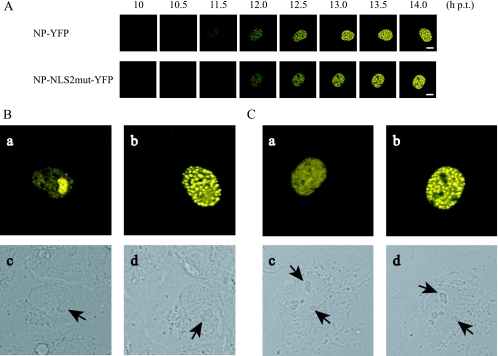
To understand the temporal change in NP subnuclear localization, we fused YFP to the C terminus of wild-type NP (NP-YFP) and to that of NP-NLS2mut (NP-NLS2mut-YFP) and examined their expression in COS-7 cells by time-lapse analysis. Both of the fusion proteins showed localization patterns similar to those shown in Fig. 2A. Figure 3A shows time-lapse images of a single cell expressing NP-YFP or NP-NLS2mut-YFP at 30-min intervals. Both fusion proteins accumulated in the nuclei and eventually exhibited punctate staining (e.g., at 14 h posttransfection). By using software to enhance image contrast (Fig. 3Ba and Ca), we found that NP-YFP appeared extensively in the nucleolus at the same time as NP began to be detected (e.g., at 10 h posttransfection), whereas NP-NLS2mut-YFP failed to accumulate in the nucleolus. These data indicate that NP nucleolar accumulation is a transient event in the early phase of NP expression and dependent on NLS2.
FIG. 3.
Contribution of NLS2 to NP nucleolar localization. (A) Time-lapse imaging of NP-YFP and NP-NLS2mut-YFP expression. COS-7 cells were transfected with the plasmids for the expression of NP-YFP and NP-NLS2mut-YFP and examined with a confocal laser microscope. Time frames at 30-min intervals are shown. Scale bars, 10 μm. (B) A change in the subnuclear localization of NP-YFP in a cell. High-magnification views of upper images in panel A at 10 (a) and 14 (b) h posttransfection (h p.t.) and corresponding differential interference contrast images (c and d, respectively) are shown. Contrast of images in parts a and b were enhanced by using Adobe Photoshop Elements 2.0 software. Arrows point to nucleoli. (C) Under the conditions identical to those described for panel B, NP-NLS2mut-YFP displays a change in subnuclear localization.
NLS2 is important for vRNA transcription.
To investigate whether the alanine substitutions in NLS1 and/or NLS2 affect vRNA transcription, we prepared pPolI/NP(0)Fluc(0), which expresses a reporter vRNA containing the open reading frame of firefly luciferase flanked by 3′ and 5′ NP noncoding sequences. Protein expression plasmids for the viral polymerase subunits and for wild-type NP or each of the mutant NPs were cotransfected into cells with pPolI/NP(0)Fluc(0) and pGL4.74[hRluc/TK], which expresses the Renilla luciferase and was used as an internal control. In this experiment, 293 cells were used because, unlike 293T and COS-7 cells, they do not express simian virus 40 T antigen, which would amplify transfected plasmids and affect the accurate quantification of the reporter protein.
At 12 and 24 h posttransfection, the cells were harvested to measure luciferase activities. NP-NLS1mut exhibited substantial luciferase activity (more than 50% of the activity of wild-type NP at both time points), whereas NP-NLS2mut and NP-NLS1+2mut showed almost no activity (Fig. 4A), suggesting that NLS2, but not NLS1, is crucial for vRNA transcription.
FIG. 4.
Comparison of NP NLS mutants' support of vRNA transcription. (A) NP activity to support vRNA transcription as measured by reporter luciferase activity. Plasmids for the expression of wild-type or mutant NP were transfected into 293 cells together with protein expression plasmids for viral polymerase subunits and pPolI/NP(0)Fluc(0), which encodes vRNA possessing a reporter firefly luciferase gene flanked by 3′ and 5′ noncoding regions. At 12 h (white bars) and 24 h (black bars) posttransfection, cells were subjected to the dual-luciferase assay. Relative firefly luciferase activity, normalized to the Renilla luciferase activity used as an internal control (see Materials and Methods), is shown. Error bars indicate the standard error of the mean of three independent experiments. (B) Simultaneous visual analysis of the mutant NP's ability to support vRNA transcription and NP subcellular localization. Plasmids for the expression of wild-type NP and NP-NLS1mut were transfected into COS-7 cells together with protein expression plasmids for viral polymerase subunits (i.e., PA, PB1, and PB2) and pPolI/NP(0)GFP(0), which encodes vRNA possessing a reporter GFP gene flanked by 3′ and 5′ noncoding regions. At 12 h posttransfection, cells were fixed and incubated with an anti-NP antibody (red). The right column shows merged fluorescence images. Scale bars, 10 μm.
Since NP-NLS1mut, which is not efficiently transported to the nucleus, supported a substantial level of reporter gene expression, we investigated whether the other components of vRNP (i.e., viral polymerase subunits and/or reporter vRNA) compensated for the deficiency of NP-NLS1mut for nuclear localization. To achieve this, we prepared pPolI/NP(0)GFP(0), which encodes the GFP open reading frame flanked by NP noncoding sequences. This reporter plasmid and expression plasmids for viral polymerase subunits were cotransfected into COS-7 cells with plasmids for the expression of wild-type NP or NP-NLS1mut. At 12 h posttransfection, the cells were subjected to an immunofluorescence analysis with an anti-NP antibody, which allowed us to visually inspect individual cells expressing both NP and the reporter GFP. As shown in Fig. 4B, GFP expression was observed in cells transfected with a plasmid for wild-type NP or NP-NLS1mut, and a large proportion of NP-NLS1mut still localized to the cytoplasm, indicating that the other components of the vRNP do not alter the subcellular localization of NP-NLS1mut by complex formation. Therefore, a small but sufficient amount of NP-NLS1mut must be in the nucleus to support the level of the reporter protein expression observed (Fig. 4).
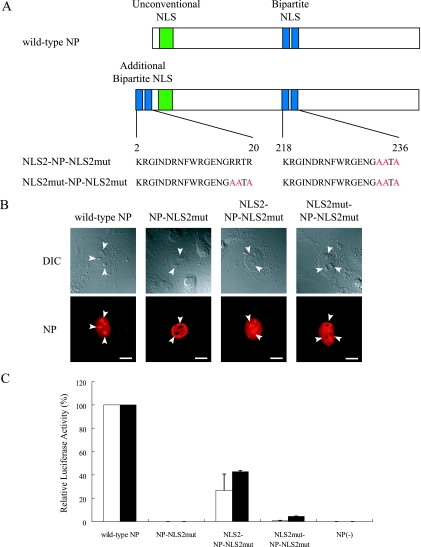
To further test the importance of NLS2 for vRNA transcription, we constructed plasmids for the expression of NP-NLS2mut fused with an additional intact NLS2 (NLS2-NP-NLS2mut) or with NLS2mut (NLS2mut-NP-NLS2mut, as a negative control) at the amino terminus (Fig. 5A). The addition of NLS2, but not NLS2mut, at the amino terminus made NP-NLS2mut localize to the nucleoli, confirming that NLS2 functions as a nucleolus localization signal (Fig. 5B). In addition, NLS2, but not NLS2mut, added to the amino terminus of NP-NLS2mut recovered the function of NP-NLS2mut to support vRNA transcription, although to a limited extent (Fig. 5C, NLS2-NP-NLS2mut), indicating that the NLS2 sequence has a role in vRNA transcription and can function in a position other than its original location in NP.
FIG. 5.
Contribution of NLS2 to vRNA transcription. (A) Schematic diagram of NLS2-NP-NLS2mut and NLS2mut-NP-NLS2mut. The NLS2 or NLS2mut motif was added to the amino terminus of NP-NLS2mut. The alanine replacements introduced to abolish the nuclear transport activity of NLS2 are in red. (B) Subcellular localization of mutant NPs. Plasmids for the expression of wild-type and mutant NP were transfected into COS-7 cells. At 12 h posttransfection, cells were fixed and incubated with an anti-NP antibody. The lower parts are differential interference contrast images. Arrowheads point to nucleoli. Scale bars, 10 μm. (C) NP support of vRNA transcription as measured by reporter luciferase activity. Plasmids for the expression of wild-type NP and the indicated mutant NPs were transfected into 293 cells together with protein expression plasmids for viral polymerase subunits and pPolI/NP(0)Fluc(0). At 12 h (white bar) and 24 h (black bar) posttransfection, cells were subjected to a dual-luciferase assay. Relative firefly luciferase activity, normalized to Renilla luciferase activity, is shown. Error bars indicate the standard error of three independent experiments.
Effects of alanine substitutions in NP NLSs on the formation of functional RNPs and their incorporation into virus particles.
To investigate whether the alanine substitutions in NLS1 and/or NLS2 influenced the formation of functional RNPs and their incorporation into virus particles, we generated VLPs containing mutant NPs by using a plasmid-based VLP generation system (21) in which all of the structural proteins of the virus-like particles are derived from expression plasmids and in which a reporter vRNA is produced by cellular RNA polymerase I. In this system, we found no substantial reduction in the titers of infectious VLPs when alanine substitutions were introduced into one of the NLSs (Fig. 6), indicating that the abolition of one NLS does not abrogate the formation of functional RNPs or their incorporation into virus particles. However, a substantial reduction in the number of infectious VLPs was observed upon the introduction of alanine substitutions into both NLSs (i.e., NP-NLS1+2mut), which indicates that the NLSs can compensate for each other with respect to the function tested here.
Identification of the nucleotide sequence in the NP coding region that contributes to the efficient incorporation of NP vRNA into virions.
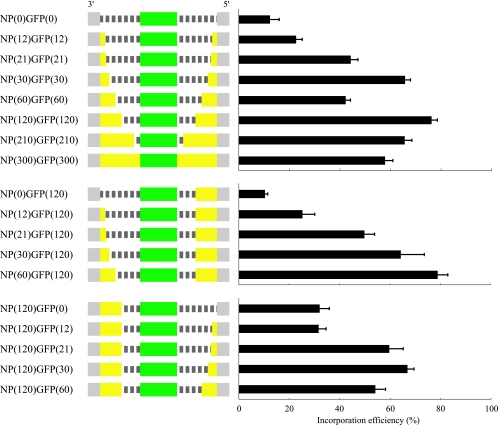
Given that the NP mutant NP-NLS1mut was functional for vRNA transcription and for the formation of functional RNPs and their incorporation into virus particles, we asked whether NLS1 is essential for viral replication. To answer this question, however, we needed to introduce alanine substitutions into NLS1. But NLS1 is located at the amino terminus of the coding region of the NP, the location of the packaging signal for the other six segments of influenza A viruses (i.e., HA [28], NA [11], NS [10], PA, PB1, and PB2 [9, 17, 18] segments). Since the exact site of the packaging signal for the NP segment has not been identified, we needed to obtain this information. We therefore prepared a series of plasmids for test NP vRNAs (Fig. 7). With these plasmids, we generated VLPs, counted the VLP-infected cells by fluorescence-activated cell sorter analysis, and calculated the efficiency of mutant NP vRNA incorporation into infectious VLPs as previously described (18). The vRNA incorporation efficiency gradually diminished, as the length of the 3′ or the 5′ NP coding region was reduced (Fig. 7), indicating that the coding region of the NP vRNA is required for efficient packaging of the NP segment. Under these conditions, 60 nucleotides at the 3′ end and 120 nucleotides at the 5′ end of the coding region were sufficient for efficient NP vRNA incorporation into virions (Fig. 7).
FIG. 7.
Schematic diagram of mutant NP vRNAs and their incorporation efficiency into virions. The noncoding and coding regions are represented by gray and yellow bars, respectively, while the dashed lines indicate nucleotides deleted from the NP coding regions. GFP open reading frames were inserted in frame with the NP open reading frames, as indicated by green bars. All mutants are shown in the negative-sense orientation. The incorporation efficiencies of these test NP vRNAs were calculated as described in Materials and Methods. Error bars indicate the standard error of three independent experiments.
Production of a mutant NP vRNA that is efficiently incorporated into virions.
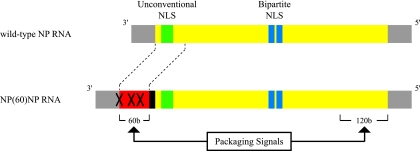
To determine whether NLS1 is required for viral replication, we constructed a plasmid for the production of a mutant NP vRNA (Fig. 8). NP vRNA derived from this plasmid would contain the additional 60 nucleotides that correspond to the coding sequence of the 3′ end of the NP open reading frame, i.e., the minimum sequence required at this end of the NP segment for virion incorporation. To eliminate expression of truncated NP from the inserted nucleotides, the NP start codon and downstream ATG codons in the 60-nucleotide insert were changed to a GCG codon and ATC codons, respectively [× in NP(60)NP RNA in Fig. 8]. Further, for optimal translation of the authentic coding region, a partial Kozak consensus sequence (GCCGCC) in the negative sense (black box) was placed between the 60-nucleotide insert and the downstream NP open reading frame. Thus, this recombinant NP vRNA, NP(60)NP RNA, should encode intact NP. With plasmid pPolI/NP(60)NP, nucleotide substitutions that would generate alanine in NLS1 and/or NLS2 were then introduced. A series of plasmids were created for the production of mutant NP vRNAs that contain the inserted 60 nucleotides and the mutations in NLS1, NLS2, or both [i.e., pPolI/NP(60)NP-NLS1mut, pPolI/NP(60)NP-NLS2mut, or pPolI/NP(60)NP-NLS1+2mut]. We also made a plasmid that lacked the 60-nucleotide insert but contained mutations in the region encoding NLS1 in pPolI/NP (designated pPolI/NP-NLS1mut).
FIG. 8.
Schematic diagram of NP(60)NP RNA. The noncoding and coding regions are represented by gray and yellow bars, respectively. NP(60)NP RNA contains the 60 nucleotides that correspond to the coding sequence of the packaging signal at the 3′ end of the NP segment (red bar) and a partial Kozak consensus sequence (black bar). The × symbols indicate the GCG mutation at the NP start codon and the ATC mutations at the downstream ATG codons, which abolish expression of the truncated NP derived from the inserted nucleotides. Both RNAs are shown in the negative-sense orientation.
NLS2, not NLS1, is essential for viral replication.
To test the role of NLS1 in viral replication, we attempted to generate recombinant viruses possessing mutant NPs by reverse genetics. During this experiment, we found that recombination occurred between plasmids designed to express wild-type NP and mutant NP-vRNA, resulting in the emergence of revertant viruses (data not shown). Hence, for subsequent experiments, we performed reverse genetics with plasmids for the expression of mutant NPs, whose nucleotide sequences were identical to the open reading frames of the plasmids for the production of corresponding mutant NP vRNAs.
With this system, we were able to generate viruses possessing NP-NLS1mut, but not NP-NLS2mut or NP-NLS1+2mut, as well as viruses possessing the wild-type NP, including one with the additional 60 nucleotides in its NP segment [NP(60)NP]. The inserted 60 nucleotides were maintained in the NP segments of both the NP(60)NP and NP(60)NP-NLS1mut viruses (data not shown). The recombinant virus, NP(60)NP-NLS1mut virus, formed plaques substantially smaller than those formed by wild-type WSN virus or by NP(60)NP virus (Fig. 9A). The growth of the NP(60)NP-NLS1mut virus in cell culture was substantially attenuated (Fig. 9B, >4-log reduction in virus titer at 36 h postinfection). These results indicate that the NLS1 of NP is not essential for viral replication, although it supports efficient viral replication by promoting NP nuclear localization. The results also show that NLS2 is essential for viral replication, likely because of its role in vRNA transcription.
FIG. 9.
Characterization of the recombinant virus expressing NP-NLS1mut. (A) Plaque phenotype of wild-type WSN and recombinant viruses. MDCK cells were infected with wild-type WSN virus, NP(60)NP virus, or NP(60)NP-NLS1mut virus and overlaid with agarose. At 48 h postinfection, cells were subjected to immune staining with an anti-WSN virus polyclonal antibody. Scale bars, 1 mm. (B) Growth comparison of wild-type WSN, NP(60)NP, and NP(60)NP-NLS1mut viruses. MDCK cells were infected with wild-type WSN, NP(60)NP, or NP(60)NP-NLS1mut virus at a multiplicity of infection of 0.001. The culture supernatants were subjected to plaque assay on MDCK cells at the indicated times postinfection. Error bars indicate the standard error of triplicate infections. Symbols: ○, wild-type WSN virus; •, NP(60)NP virus; ▴, NP(60)NP-NLS1mut virus.
DISCUSSION
In this report, we have demonstrated that the unconventional NLS of influenza A virus NP (i.e., NLS1) is not essential for viral replication, whereas the bipartite NLS (NLS2) is required for this function. NLS2 does not interact with karyopherin α1 or α2 (27), and its contribution to NP nuclear localization is limited (Fig. 2A) (5, 19). However, NLS2 does serve as a signal for NP nucleolar localization (Fig. 2B and 3A). The nucleolus contains complexes of proteins and various types of RNA but is not separated by membranes from the surrounding nucleoplasm. Although nucleolar localization of NP was reported more than 20 years ago (7, 8), the peptide motif for NP nucleolar localization remained unknown. Weber et al. (30) proposed that the amino acid sequence (R/KR/K)X(R/K) functions as a nucleolar localization signal. The amino acid sequence at positions 213 to 216 of NP (RRTR) matches perfectly with this signal motif. Recent large-scale proteomics analyses failed to identify a simple motif conserved among nucleolar proteins (1, 25); however, given that the 19 amino acids encompassing these four amino acids (RRTR) function as a nucleolar signal even when moved to the amino terminus of NP (Fig. 5B), this region (likely the RRTR sequence) certainly seems to serve as a nucleolar localization signal. In most of the previous reports describing NP localization, NP appears to be absent from the nucleoli (e.g., see figures in reference 19). This is not surprising since our data show that NP nucleolar localization is a transient event in the early phase of protein expression (Fig. 3A and B). The nucleolus is a dynamic nuclear domain and known as the site of ribosome biogenesis (i.e., rRNA transcription, pre-rRNA processing, and ribosome subunit assembly). Recent studies have demonstrated that the nucleolus also plays a significant role in several other cellular functions, including cell cycle control and mRNA transport (2, 14, 24, 26). It is also known that various viral proteins are targeted to the nucleolus (13). Although further studies are needed to reveal a mechanism and the biological significance of NP nucleolar localization, this event may be an important aspect of vRNA transcription, which is known to require NP.
A mutant NP containing substitutions in NLS1 supported vRNA transcription at a substantial level (Fig. 4A and 5C). Since vRNA synthesis takes place in the nucleus (16) and NP nuclear localization is mainly regulated by NLS1 (Fig. 2A), this result was unexpected. It may be that a limited amount of NP was transported to the nucleus despite the alanine mutations in NLS1 and that this amount of NP in the nucleus was sufficient for vRNA transcription. Cros et al. (5) reported that NLS1 plays a crucial role in the nuclear localization of NP and vRNP and in viral replication. Our results are consistent with those reported by Cros et al. (5) in that alanine substitutions in NLS1 severely attenuated the virus (Fig. 9). It is, however, not clear whether the limited growth of the virus is the result of incomplete abolition of NLS1 function or of an unidentified NLS compensating for the lack of NLS1 function.
Recent studies have revealed that both ends of the coding regions, in addition to the noncoding regions, are required for efficient packaging of vRNAs into virions for all of the influenza A virus segments examined to data (i.e., every segment except the M and NP segments) (9-11, 17, 18, 28). Here we found that this concept extends to the NP segment (Fig. 6). However, the specific role(s) of these coding regions in viral genome packaging remains to be elucidated.
In summary, we have demonstrated that the unconventional NLS of influenza A virus NP, even with alanine substitutions for key basic residues in this signal, supported virus replication, albeit to a limited extent. We have further shown that the bipartite NLS, located in the middle of NP, functions as a nucleolar localization signal and is essential for vRNA synthesis and, therefore, viral replication (Table 1). Further studies on the significance of NP nucleolar localization should provide information essential to our understanding of influenza virus replication and could potentially identify a nucleolar transport pathway.
TABLE 1.
Biological functions of wild-type and mutant NPs
| NP variant | Main intracellular locationa | vRNA transcription | Function to support infectivity of VLPb | Viral replication |
|---|---|---|---|---|
| Wild type | Nucleus including nucleolus | + | + | + |
| NP-NLS1mut | Cytoplasm | + | + | + |
| NP-NLS2mut | Nucleus | − | + | − |
| NP-NLS1+2mut | Cytoplasm | − | − | − |
Intracellular location examined 12 h posttransfection.
As measured in the experiments shown in Fig. 6.
Acknowledgments
We thank Susan Watson and Krisna Wells for editing the manuscript.
This work was supported by CREST (Japan Science and Technology Agency) and by grants-in-aid from the Ministries of Education, Culture, Sports, Science, and Technology and of Health, Labor, and Welfare of Japan and by National Institute of Allergy and Infectious Diseases Public Health Service research grants.
Footnotes
Published ahead of print on 18 October 2006.
REFERENCES
- 1.Andersen, J. S., C. E. Lyon, A. H. Fox, A. K. Leung, Y. W. Lam, H. Steen, M. Mann, and A. I. Lamond. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12:1-11. [DOI] [PubMed] [Google Scholar]
- 2.Bond, V. C., and B. Wold. 1993. Nucleolar localization of myc transcripts. Mol. Cell. Biol. 13:3221-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullido, R., P. Gomez-Puertas, C. Albo, and A. Portela. 2000. Several protein regions contribute to determine the nuclear and cytoplasmic localization of the influenza A virus nucleoprotein. J. Gen. Virol. 81:135-142. [DOI] [PubMed] [Google Scholar]
- 4.Chen, W., P. A. Calvo, D. Malide, J. Gibbs, U. Schubert, I. Bacik, S. Basta, R. O'Neill, J. Schickli, P. Palese, P. Henklein, J. R. Bennink, and J. W. Yewdell. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306-1312. [DOI] [PubMed] [Google Scholar]
- 5.Cros, J. F., A. Garcia-Sastre, and P. Palese. 2005. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic 6:205-213. [DOI] [PubMed] [Google Scholar]
- 6.Cros, J. F., and P. Palese. 2003. Trafficking of viral genomic RNA into and out of the nucleus: influenza, Thogoto and Borna disease viruses. Virus Res. 95:3-12. [DOI] [PubMed] [Google Scholar]
- 7.Davey, J., A. Colman, and N. J. Dimmock. 1985. Location of influenza virus M, NP and NS1 proteins in microinjected cells. J. Gen. Virol. 66(Pt. 11):2319-2334. [DOI] [PubMed] [Google Scholar]
- 8.Dimmock, N. J. 1969. New virus-specific antigens in cells infected with influenza virus. Virology 39:224-234. [DOI] [PubMed] [Google Scholar]
- 9.Dos Santos Afonso, E., N. Escriou, I. Leclercq, S. van der Werf, and N. Naffakh. 2005. The generation of recombinant influenza A viruses expressing a PB2 fusion protein requires the conservation of a packaging signal overlapping the coding and noncoding regions at the 5′ end of the PB2 segment. Virology 341:34-46. [DOI] [PubMed] [Google Scholar]
- 10.Fujii, K., Y. Fujii, T. Noda, Y. Muramoto, T. Watanabe, A. Takada, H. Goto, T. Horimoto, and Y. Kawaoka. 2005. Importance of both the coding and the segment-specific noncoding regions of the influenza A virus NS segment for its efficient incorporation into virions. J. Virol. 79:3766-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujii, Y., H. Goto, T. Watanabe, T. Yoshida, and Y. Kawaoka. 2003. Selective incorporation of influenza virus RNA segments into virions. Proc. Natl. Acad. Sci. USA 100:2002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Görlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 13.Hiscox, J. A. 2002. The nucleolus—a gateway to viral infection? Arch. Virol. 147:1077-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadowaki, T., M. Hitomi, S. Chen, and A. M. Tartakoff. 1994. Nuclear mRNA accumulation causes nucleolar fragmentation in yeast mtr2 mutant. Mol. Biol. Cell 5:1253-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobasa, D., M. E. Rodgers, K. Wells, and Y. Kawaoka. 1997. Neuraminidase hemadsorption activity, conserved in avian influenza A viruses, does not influence viral replication in ducks. J. Virol. 71:6706-6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1531. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 17.Liang, Y., Y. Hong, and T. G. Parslow. 2005. cis-acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. J. Virol. 79:10348-10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muramoto, Y., A. Takada, K. Fujii, T. Noda, K. Iwatsuki-Horimoto, S. Watanabe, T. Horimoto, H. Kida, and Y. Kawaoka. 2006. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J. Virol. 80:2318-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neumann, G., M. R. Castrucci, and Y. Kawaoka. 1997. Nuclear import and export of influenza virus nucleoprotein. J. Virol. 71:9690-9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neumann, G., T. Watanabe, and Y. Kawaoka. 2000. Plasmid-driven formation of influenza virus-like particles. J. Virol. 74:547-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 23.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pederson, T., and J. C. Politz. 2000. The nucleolus and the four ribonucleoproteins of translation. J. Cell Biol. 148:1091-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scherl, A., Y. Coute, C. Deon, A. Calle, K. Kindbeiter, J. C. Sanchez, A. Greco, D. Hochstrasser, and J. J. Diaz. 2002. Functional proteomic analysis of human nucleolus. Mol. Biol. Cell 13:4100-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visintin, R., and A. Amon. 2000. The nucleolus: the magician's hat for cell cycle tricks. Curr. Opin. Cell Biol. 12:372-377. [DOI] [PubMed] [Google Scholar]
- 27.Wang, P., P. Palese, and R. E. O'Neill. 1997. The NPI-1/NPI-3 (karyopherin α) binding site on the influenza A virus nucleoprotein NP is a nonconventional nuclear localization signal. J. Virol. 71:1850-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe, T., S. Watanabe, T. Noda, Y. Fujii, and Y. Kawaoka. 2003. Exploitation of nucleic acid packaging signals to generate a novel influenza virus-based vector stably expressing two foreign genes. J. Virol. 77:10575-10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber, F., G. Kochs, S. Gruber, and O. Haller. 1998. A classical bipartite nuclear localization signal on Thogoto and influenza A virus nucleoproteins. Virology 250:9-18. [DOI] [PubMed] [Google Scholar]
- 30.Weber, J. D., M. L. Kuo, B. Bothner, E. L. DiGiammarino, R. W. Kriwacki, M. F. Roussel, and C. J. Sherr. 2000. Cooperative signals governing ARF-Mdm2 interaction and nucleolar localization of the complex. Mol. Cell. Biol. 20:2517-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whittaker, G., M. Bui, and A. Helenius. 1996. Nuclear trafficking of influenza virus ribonucleoproteins in heterokaryons. J. Virol. 70:2743-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]