Abstract
Pradimicin A (PRM-A), an antifungal nonpeptidic benzonaphtacenequinone antibiotic, is a low-molecular-weight (molecular weight, 838) carbohydrate binding agent (CBA) endowed with a selective inhibitory activity against human immunodeficiency virus (HIV). It invariably inhibits representative virus strains of a variety of HIV-1 clades with X4 and R5 tropisms at nontoxic concentrations. Time-of-addition studies revealed that PRM-A acts as a true virus entry inhibitor. PRM-A specifically interacts with HIV-1 gp120 and efficiently prevents virus transmission in cocultures of HUT-78/HIV-1 and Sup T1 cells. Upon prolonged exposure of HIV-1-infected CEM cell cultures, PRM-A drug pressure selects for mutant HIV-1 strains containing N-glycosylation site deletions in gp120 but not gp41. A relatively long exposure time to PRM-A is required before drug-resistant virus strains emerge. PRM-A has a high genetic barrier, since more than five N-glycosylation site deletions in gp120 are required to afford moderate drug resistance. Such mutated virus strains keep full sensitivity to the other known clinically used anti-HIV drugs. PRM-A represents the first prototype compound of a nonpeptidic CBA lead and, together with peptide-based lectins, belongs to a conceptually novel type of potential therapeutics for which drug pressure results in the selection of glycan deletions in the HIV gp120 envelope.
Adsorption and fusion (entry) inhibitors of human immunodeficiency virus (HIV) are targeted to viral envelope glycoprotein gp120 or gp41 or to cellular (co)receptor CD4, CXCR4, or CCR5 (30). Enfuvirtide (T20; Fuzeon), which targets HIV gp41, is the first and so far only entry inhibitor officially approved for HIV treatment (13). Several carbohydrate-binding agents (CBA) derived from different prokaryotic, plant, invertebrate, or vertebrate species with specificity for mannose (Man), β-galactose, or N-acetylglucosamine (GlcNAc) were reported to be endowed with antiviral activity, in particular against HIV, in several cell culture systems (for an overview, see reference 3). Recently, several of these CBAs (Galanthus nivalis agglutinin [GNA], Hippeastrum hybrid agglutinin [HHA], and Urtica dioica agglutinin [UDA]) and the prokaryotic cyanovirin were shown to be potential drug candidates for microbicidal use in the prevention of HIV infection (4, 39). Due to their carbohydrate-binding properties, they interact with the viral envelope glycoprotein and efficiently prevent the HIV entry process. Interestingly, we have recently demonstrated that both Man- and GlcNAc-specific plant lectins select for mutant HIV strains that contain deletions of glycans in the gp120 envelope (5, 6). Such a resistance spectrum is unique among the known HIV entry inhibitors. The mutated virus strains kept full sensitivity to the inhibitory effects of other entry inhibitors (5, 6). A potential disadvantage of using natural products as HIV therapeutics, however, is their protein nature. It is indeed not technically easy and is relatively costly to produce and purify such protein CBAs on a large scale; they may have poor, if any, oral bioavailability, and they may also trigger an immune response when administered systemically on a frequent-application basis. Therefore, an intensive search for the discovery and study of low-molecular-weight CBAs is highly advisable (2).
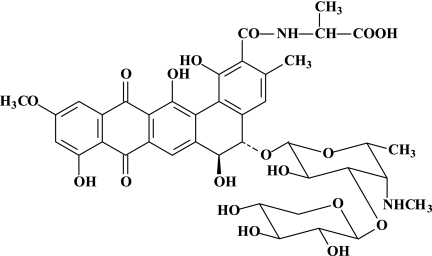
In the course of screening for new antibiotics active against fungi, an actinomycete strain (Actinomadura hibisca) was found to produce pradimicin A (PRM-A), which shows activity against systemic fungal infections (26). PRM-A has a unique structure containing the following moieties: the amino acid d-alanine, the carbohydrates d-xylose and 4,6-dideoxy-4-methylamino-d-galactose, and a substituted 5,6-dihydrobenzo[a]naphtacenequinone (Fig. 1). Interestingly, PRM-A was also shown to be inhibitory to HIV-1 (36, 37). The biological effect of PRM-A had been ascribed to binding to mannose residues (37, 38). This proposed mechanism of antiviral action of PRM-A is in agreement with the observation that the antibiotic binds to terminal d-mannopyranoside and Ca2+ to yield a ternary complex (40; for more-detailed information on the mechanisms of the antifungal activity and molecular interactions of PRM-A, see references 18, 19, 25, and 45).
FIG. 1.
Structural formula of pradimicin A.
Given the interesting antiviral properties and unique resistance profile of the mannose- and GlcNAc-specific plant lectins, we undertook an extensive study of the anti-HIV properties and antiviral resistance profile of PRM-A as the prototype compound among the nonpeptidic CBAs. Our study revealed that PRM-A behaves as an “artificial (nonpeptidic) lectin” endowed with an interesting antiviral (i.e., HIV) chemotherapeutic activity that can force the virus to follow a unique mutational pattern in its gp120 envelope to escape drug pressure.
MATERIALS AND METHODS
Test compounds.
PRM-A (molecular weight [MW], 838) (Fig. 1) was isolated and purified from the fermentation broth of Actinomyces sp. TP-A0016 (10). The mannose-specific plant lectins from GNA and HHA were derived and purified from these plants, as previously described (42, 43). AMD3100 was obtained from AnorMed (Langley, Canada), and dextran sulfate-5000 was obtained from Sigma (St. Louis, MO). The monoclonal antibody (MAb) 2G12 was obtained from the Medical Research Council, Potter Bar, Hertfordshire, United Kingdom, and from Polymun Scientific (Vienna, Austria). The MAbs B12, NEA9205, and F105 were obtained from DuPont NEN Research Products (Boston, MA) and provided by the NIH AIDS Research and Reference Reagent program (27, 28, 46).
Cells.
Human T-lymphocytic CEM and Sup T1 cells were cultivated in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) (BioWittaker Europe, Verviers, Belgium), 2 mM l-glutamine, and 0.075 M NaHCO3. C8166 cells were grown and maintained in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, 0.1% sodium bicarbonate, and 20 μg gentamicin per ml.
Viruses.
HIV-1(IIIB) was provided by R. C. Gallo and M. Popovic (at that time of the National Cancer Institute, National Institutes of Health, Bethesda, MD). The HIV-1/NL4-3.GFP11 strain that expresses an enhanced version of green fluorescent protein (GFP) (35) as a replacement for Nef has been described previously (41). For this molecular clone, the expression of GFP in infected cells is a measurement of virus production (11). For all tests described, HIV-1/NL4-3.GFP11 was obtained from the transfection of 293T cells. One milliliter of virus-containing 293T cell supernatant was used to infect 8 × 106 MT-4 cells in 40 ml of culture medium. Three days after being infected, the supernatant was collected and used as the virus input in our assays.
Generation of VSV-G-pseudotyped HIV-1.
The vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped HIV-1(with env deleted) was generated by transient cotransfection of recombinant pHIV-1/NL4-3-ΔE-GFP with env deleted (48) (a kind gift of R. Siliciano, Johns Hopkins University, Baltimore MD) and pVSV-G in 293T cells, using calcium phosphate. The supernatants containing VSV-G-pseudotyped HIV-1 were collected 60 h after transfection. The cell debris was removed from the supernatant by centrifugation (450 × g for 10 min), and the supernatants were used for infection experiments or stored at −80°C.
Production of GFP-labeled HIV-1 virions.
Fluorescent virions were produced by transient cotransfection of pHIV-1/NL4-3 (48) and pGFP-Vpr, a plasmid expressing the GFP-Vpr fusion protein (a kind gift from G. N. Pavlakis, National Cancer Institute, Frederick MD). The supernatants holding GFP-Vpr-containing HIV-1 were collected 60 h after transfection. The cell debris was removed from the supernatant by centrifugation (450 × g for 10 min), and the supernatants were used for infection experiments or stored at −80°C.
Antiretrovirus assays.
The methodology of the anti-HIV assays has been described previously. Briefly, CEM cells (4.5 × 105 cells/ml) were suspended in fresh culture media and infected with HIV-1 at 100 50% cell culture infective dose per ml of cell suspension in the presence of appropriate dilutions of the test compounds. After 4 to 5 days of incubation at 37°C, giant cell formation was recorded microscopically in the CEM cell cultures. The 50% effective concentration (EC50) corresponds to the compound concentrations required to prevent syncytium formation by 50% in the virus-infected CEM cell cultures. Several test compounds were also tested against HIV-1 in CEM cell culture media containing 2, 5, 10, 20, or 40% FBS.
In the cocultivation assays, 5 × 104 persistently HIV-1-infected HUT-78 cells (designated HUT-78/HIV-1) were mixed with 5 × 104 Sup T1 cells, along with appropriate concentrations of the test compounds. After 16 to 20 h, marked syncytium formation was noted in the control cell cultures, and the number of syncytia was determined under a microscope.
Antiviral activity of test compounds against HIV-1 clade isolates in PBMCs.
Primary clinical isolates representing different HIV-1 clades and an HIV-2 isolate (see Table 2) were all kindly provided by L. Lathey of BBI Biotech Research Laboratories, Inc., Gaithersburg, MD, and their coreceptor (R5 or X4) use was determined (33). Antiviral testing of these isolates in peripheral blood mononuclear cells (PBMCs) was done as previously described (6). Briefly, PBMCs from healthy donors were stimulated with phytohemagglutinin (PHA) at 2 μg/ml (Sigma, Bornem, Belgium) for 3 days at 37°C. The PHA-stimulated blasts were then seeded at 0.5 × 106 cells per well into a 48-well plate containing various concentrations of the compound in cell culture medium (RPMI 1640) containing 10% fetal calf serum and interleukin-2 (25 U/ml, R&D Systems Europe, Abingdon, United Kingdom). The virus stocks were added at a final dose of 250 pg of p24 or p27/ml. The cell supernatant was collected at day 12, and the HIV-1 core antigen (Ag) in the culture supernatant was analyzed by a p24 Ag enzyme-linked immunosorbent assay kit (Perkin Elmer, Boston, MA). For HIV-2 p27 Ag detection, the INNOTEST from Innogenetics (Temse, Belgium) was used.
TABLE 2.
Inhibitory activity of CBAs on anti-HIV-1 activity in CEM cell cultures in the presence of different concentrations of serum
| Compound | EC50a (μg/ml) for indicated conc of newborn bovine serum
|
||||
|---|---|---|---|---|---|
| 2% | 5% | 10% | 20% | 40% | |
| PRM-A | 4.0 ± 0.0 | 3.5 ± 0.7 | 4.0 ± 0.0 | 3.5 ± 0.7 | 4.0 ± 0.0 |
| GNA | 1.7 ± 1.2 | 0.90 ± 0.14 | 1.0 ± 0.28 | 1.0 ± 0.28 | 1.6 ± 0.85 |
| HHA | 0.55 ± 0.35 | 0.58 ± 0.59 | 0.65 ± 0.49 | 0.75 ± 0.35 | 1.0 ± 0.28 |
EC50 for inhibition of HIV-1-induced cytopathic effect in CEM cell cultures.
Time-of-drug-addition experiment with GFP-expressing HIV-1/NL4-3.GFP11.
The time-of-drug-addition experiments have been previously described (11). Briefly, C8166 cells were infected with HIV-1/NL4-3.GFP11. Following a 1-h adsorption period, the cells were distributed in a 96-well plate at 45,000 cells/well and incubated at 37°C. Test compounds or culture medium (control) was added at different time points (0, 1, 2, 3, 4, 5, 6, 7, 8, 24, and 25 h) after virus infection. Dextran sulfate (Sigma, St. Louis, MO) was used at 100 μg/ml, AMD3100 (Sigma) at 5 μg/ml, nevirapine (Boehringer Ingelheim) at 2 μg/ml, HHA (kind gift of E. Van Damme, Ghent University, Belgium) at 100 μg/ml, and PRM-A at 50 μg/ml. No cytotoxicity was observed at these compound concentrations. The number of GFP-expressing cells was monitored by fluorescence-activated cell sorter (FACS) analysis at 30 h after infection. The value for the number of GFP-expressing cells in the positive control was set at 100%, and for each time point the percentage of infected cells was calculated relative to this value. Flow cytometric analysis of the virus-exposed cell cultures was performed on a FACSCalibur flow cytometer equipped with a 488-nm argon-ion laser and a 530/30-nm bandpass filter (FL1; for detection of GFP-associated fluorescence; Becton Dickinson, San Jose, CA). Before the acquisition process was done, the cells were pelleted at 1,000 rpm for 10 min and fixed in a 3% paraformaldehyde solution. Acquisition was stopped when 10,000 events were counted. Data analysis was carried out with CellQuest software (BD Biosciences). The cell debris was excluded from the analysis by gating on forward- versus side-scatter dot plots.
Selection and isolation of PRM-A-resistant HIV-1 strains.
The procedure followed for the selection of drug-resistant virus mutants was essentially as recently described (5, 6). Briefly, HIV-1(IIIB) was added to CEM cell cultures in 48-well plates in the presence of PRM-A at a concentration equal to one- to twofold the EC50. For the generation of drug-resistant virus mutants, an increased drug concentration was administered when the previous cell culture became fully cytopathogenic. The drug resistance selection schedule is depicted in Fig. 2. Ten virus isolates from four independent series of drug resistance selections were taken at different time points during the drug resistance selection process.
FIG. 2.
Time-of-drug-addition experiment. C8166 cells were infected with HIV-1/NL4-3.GFP11, and test compounds were added at different times after infection. Virus-associated GFP expression was measured by flow cytometry at 30 h postinfection. Depending on the target of the drug action, addition of the compounds could be delayed for a certain number of hours without loss of antiviral activity. The later the drug target occurs in the virus infection process, the longer the drug addition can be delayed before antiviral activity is lost.
Genotyping of the HIV-1 env region.
Proviral DNA was extracted from cell pellets using the QIAamp Blood mini kit (QIAGEN, Hilden, Germany). The genotypes of both the gp120 and gp41 genes were determined by this assay, as recently described (44).
Antibody staining and flow cytometry.
Human T-lymphoid MT-4 and CEM cells were washed once with phosphate-buffered saline (PBS) containing 2% FBS and preincubated with the CBAs GNA, HHA, and PRM-A or the specific CXCR4 antagonist AMD3100 at the indicated concentrations for 30 min at room temperature. After being centrifugated and washed with PBS containing 2% FBS, the cells were incubated for 30 min on ice with a panel of different anti-human CXCR4 monoclonal antibodies (i.e., clones 12G5, 2B11 [BD Pharmingen)], and 44717.111 [R&D Systems Europe]), all phycoerythrin [PE] conjugated) and anti-human CD4 MAbs [i.e., clones SK3(Leu3a) and L120(CD4 v4), both PE-conjugated (BD Pharmingen), and OKT4 and OKT4A, both fluorescein isothiocyanate (FITC)-conjugated (Ortho Diagnostic Systems, Raritan, NJ)] in PBS containing 2% FBS. Thereafter, the cells were washed twice with PBS, fixed in 1% paraformaldehyde in PBS and analyzed on a FACSCalibur flow cytometer equipped with CellQuest software (Becton Dickinson, San Jose, CA). As a negative control for a specific background staining, the cells were stained in parallel with Simultest isotype control MAb (Becton Dickinson).
Evaluation of the immunosuppressive activity of PRM-A.
The immunosuppressive activity of PRM-A was evaluated in PHA-stimulated PBMCs and in mixed leukocyte cultures (MLC) with mitomycin-C-treated PBMCs from donor number 1 as stimulators and untreated PBMCs from another donor (donor number 2) as responders. The PBMCs were stimulated with PHA (2 μg/ml (Sigma), and 105 PBMCs/well were set up in triplicate in a 96-well microtiter plate in the presence of a dilution series of PRM-A. For the MLC cocultures, 105 PBMCs from each donor were set up in triplicate in a 96-well microtiter plate in the presence of a dilution series of PRM-A. At day 3 for PHA-stimulated PBMCs and day 5 for the MLC, 0.4 μCi of [3H]thymidine (Amersham Pharmacia, Buckinghamshire, United Kingdom) with a radiospecific activity of 5 Ci/mmol was added to each well. The plates were harvested 12 h later, and [3H]thymidine incorporation, which is a measure of lymphocyte proliferation, was measured in a scintillation counter (Canberra-Packard, Zellik, Belgium) and expressed as counts per minute.
Surface plasmon resonance binding assay.
A BIAcore X apparatus (BIAcore Inc, Piscataway, NJ) was used. Surface plasmon resonance (SPR) was exploited to measure changes in the refractive index caused by the ability of PRM-A and HHA to bind to gp120 immobilized to a BIAcore sensor chip. Seven and a half micrograms per milliliter of recombinant gp120 protein (HIV-1 MN strain), produced and purified from baculovirus (Immunodiagnostics Inc., Woburn, MA), was allowed to react with a flow cell of a CM3 sensor chip that was previously activated with 50 μl of a mixture of 0.2 M N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide hydrochloride and 0.05 M N-hydroxysuccinimide. These experimental conditions allowed the immobilization of 3,000 resonance units (RU; ∼0.025 pmol of gp120) (14, 15). Similar results were obtained for the immobilization of bovine serum albumin (BSA), used as a negative control and for Blanco subtraction. Increasing concentrations of PRM-A or HHA in HBS buffer (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20 [pH 7.4]) were then injected over the GST-Tat or BSA surfaces for 4 min (to allow their association with immobilized proteins) and then washed until dissociation was observed. For PRM-A only, the injections were also made in HBS with EDTA omitted and 10 mM Ca2+ added. After every run, the sensor chip was regenerated with an injection of 10 mM glycine buffer (pH 1.5). The SPR signal was expressed in terms of RU.
RESULTS
Antiviral activity of CBAs in cell culture.
PRM-A inhibited the cytopathic effects induced by the virus in CEM and C8166 cell cultures at an EC50 of 2.8 and 4.0 μg/ml (i.e., 3.3 and 4.8 μM), respectively (Table 1). It was not toxic to the cell cultures at 50 μg/ml (i.e., 60 μM), which is the highest soluble compound concentration that could be obtained in cell culture. The mannose-specific plant lectins GNA and HHA had EC50 values of 0.48 and 0.29 μg/ml (i.e., 0.01 and 0.006 μM), respectively, in CEM and 1.3 and 0.77 μg/ml (i.e., 0.02 and 0.015 μM), respectively, in C8166 cell cultures, and were not toxic at 100 μg/ml (i.e., 2 μM). DS-5000 showed antiviral activity comparable to that of GNA and HHA when it was measured in micrograms per milliliter but lower antiviral activity when it was measured in micromolars (∼0.07 μM). Whereas the antiviral activity of PRM-A could be reversed by 10-fold in the presence of mannan, with the plant lectins it lost antiviral activity by 50- to 100-fold. In contrast, DS-5000 was not affected in its antiviral potential by mannan (Table 1).
TABLE 1.
Inhibitory activity of CBAs on anti-HIV-1 activity in CEM cell cultures
| Compound | EC50a (μg/ml) for indicated cell culture:
|
IC50b (μg/ml) for HUT-78/HIV-1 plus Sup T1 | CC50c (μg/ml) for C8166 and CEM cells | |||
|---|---|---|---|---|---|---|
| C8166 cells | CEM cells | CEM cells plus 2.5 mg mannan/ml | ||||
| PRM-A | 4.0 ± 0.0 | 2.8 ± 1.0 | 32 ± 11 | 3.4 ± 1.3 | >50 | |
| GNA | 1.3 ± 0.66 | 0.48 ± 0.28 | 27 ± 11 | 6.0 ± 2.8 | >100 | |
| HHA | 0.77 ± 0.21 | 0.29 ± 0.19 | 25 ± 8.7 | 4.3 ± 2.5 | >100 | |
| DS-5000 | 0.37 ± 0.15 | 0.33 ± 0.12 | 0.30 ± 0.1 | 2.5 ± 0.1 | >100 | |
EC50 for inhibition of HIV-1-induced cytopathic effect in CEM cell cultures.
IC50 for inhibition of syncytium formation between HUT-78/HIV-1 and Sup T1 cells.
CC50, 50% cytostatic concentration for inhibition of CEM or C8166 cell proliferation. Due to the limited solubility of PRM-A, a concentration higher than 50 μg/ml could not be reliably tested.
Several CBAs, including pradimicin A, have also been evaluated for their anti-HIV-1 activity in CEM cell cultures in the presence of various amounts of bovine serum (2% to up to 40%). The antiviral activities of the CBAs were not affected by increasing amounts of serum (Table 2).
PRM-A efficiently prevented giant cell formation between persistently HIV-1(IIIB)-infected HUT-78/HIV-1 and uninfected Sup T1 cells at an EC50 of 3.4 μg/ml, that is, at a drug concentration that is equally effective in preventing cytopathic effects induced by HIV in CEM cell cultures (Table 1). In contrast, the peptidic CBAs such as GNA and HHA and the polyanionic compound DS-5000 lost ∼10-fold of their inhibitory potential in the cocultivation assay compared with that for the infection assay with free virus (Table 1).
PRM-A was also endowed with a consistent suppressive activity against a wide variety of X4- and R5-tropic HIV-1 clade isolates in PBMCs (Table 3). In contrast, the mannose-specific plant lectins showed much higher variability in their virus-suppressive potential (i.e., 1.2 μg/ml [group O] to ≥100 μg/ml [clade C]).
TABLE 3.
Inhibitory activity of CBAs on HIV strains and clinical HIV-1 clade isolates in PBMC cultures
| CBA | EC50a (μg/ml) for indicated HIV strain of indicated group or clade and with indicated tropism:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clade A | Clade B | Clade C | Clade D | Clade E | Clade F | Clade G | Group O | NL4.3 | IIIB | BaL | HIV-2 | |
| UG273 | US2 | ETH2220 | UG270 | ID12 | BZ163 | BCF-DIOUM | BCF06 | BV-5061W | ||||
| R5 tropic | R5 tropic | R5 tropic | X4 tropic | R5 tropic | R5 tropic | R5 tropic | X4 tropic | X4 tropic | X4 tropic | R5 tropic | X4 tropic | |
| PRM-A | 2.2 | 2.3 | 8.7 | 3.1 | 3.4 | 11 | 2.2 | NDb | 0.69 | 2.8 | 2.2 | NDb |
| GNA | 27 | 17 | ≥100 | >20 | 19 | 25 | ≥50 | 1.9 | 1.4 | 0.33 | 6.5 | 9.5 |
| HHA | 29 | 5.4 | 44 | 4.9 | 12 | 4.6 | 41 | 1.2 | 0.35 | 1.1 | 6.0 | 8.9 |
EC50 for inhibition of HIV replication in cell cultures.
ND, not determined.
Mechanism of antiviral action of PRM-A.
Three separate experiments were designed to reveal the target of PRM-A interaction with the virus. In a time-of-drug-addition experiment, C8166 cell cultures were infected with a HIV-1/NL4.3-GFP strain that contains an optimized version of a GFP gene as a replacement for the nef gene and the compounds were added at 1, 2, 3, … 9 h after infection, as indicated in Fig. 2. The extent of GFP expression at 30 h postinfection was used as a parameter of virus infection (Fig. 2). Depending on the target of drug action, addition of the compounds could be delayed for a certain number of hours characteristic for each compound without loss of antiviral activity. The presence of dextran sulfate-5000 (an adsorption inhibitor) and AMD3100, a CXCR4 coreceptor antagonist of HIV-1, at the time of virus infection was required to display full antiviral potential. A delay in drug addition of as short as 1 to 2 h postinfection resulted in significantly decreased suppression of virus replication by dextran sulfate-5000 and AMD3100. Also, the mannose-specific plant lectin HHA and PRM-A needed to be present from the very beginning of the virus infection to fully suppress the infection, suggesting an early target of interaction. In contrast, addition of the reverse transcriptase inhibitors UC-781 and nevirapine could be markedly delayed (up to 5 h) after virus infection before losing their antiviral potential (Fig. 2).
To further investigate the effect of PRM-A on the HIV-1 entry process, VSV-G-pseudotyped (env-deficient) GFP-encoding HIV-1 was exposed to C8166 cells in the absence (control) or presence of different concentrations of the test compounds (Fig. 3). Forty-eight hours postinfection, the supernatants were analyzed for GFP production. The reverse transcriptase inhibitors UC-781 and nevirapine dose dependently suppressed GFP expression by the pseudotyped virus-infected cells, while the CXCR4 antagonist AMD3100, HHA, and PRM-A completely lost their inhibitory effect, pointing to the specific interaction of the HIV-1 envelope with susceptible cells as the target of antiviral intervention of PRM-A and HHA (Fig. 3).
FIG. 3.
Effect of test compounds against VSV-G-pseudotyped HIV-1. C8166 cells were infected with GFP-encoding, env-deleted HIV-1 pseudotyped with envelope VSV-G and treated with different compounds at different concentrations. The letters on the x axis represent different concentrations of the respective compounds (in μg/ml, for AMD3100, A = 0, B = 0.008, C = 0.04, D = 0.2, E = 1, and F = 5; for PRM-A, A = 0, B = 0.08, C = 0.4, D = 2, E = 10, and F = 50; for HHA, A = 0, B = 0.16, C = 0.8, D = 4, E = 20, and F = 100; for UC781, A = 0, B = 0.0016, C = 0.008, D = 0.04, E = 0.2, and F = 1; for nevirapine, A = 0, B = 0.0008, C = 0.004, D = 0.02, E = 0.1, and F = 0.5). The infected cells were assayed for GFP expression at 48 h postinfection. One hundred percent of GFP expression for each compound was defined as that for the control culture without the compound added.
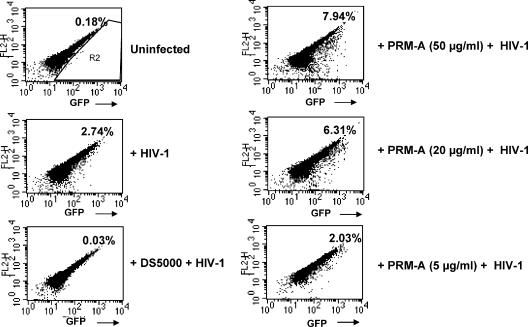
To study in more detail the mechanisms of action of PRM-A on HIV-1-specific entry, C8166 cell cultures were exposed to HIV-1/NL4.3 virions labeled with GFP-Vpr fusion proteins in the intact virion particles to monitor virion attachment to target cells in the presence of different concentrations of PRM-A and DS-5000. Interestingly, the presence of PRM-A showed a dose-dependent increase in cell-associated GFP fluorescence, pointing to enhanced binding of HIV-1 to the C8166 cells. In contrast, DS-5000 completely prevented appearance of cell-associated GFP fluorescence, pointing to an efficient inhibition of virus adsorption to the cells (Fig. 4).
FIG. 4.
Flow cytometric analysis of human C8166 cells incubated with GFP-Vpr-labeled HIV-1 virions. C8166 cells were incubated in the presence or absence of different compounds for 30 min at 37°C. (Panels 1 and 2) Medium (control); (panel 3) 100 μg/ml DS5000; (panels 4, 5, and 6) different concentrations of PRM-A. The cells were then inoculated with GFP-Vpr-labeled HIV-1 (15 ng of p24) for 6 h at 37°C. The cells were subsequently washed with PBS, fixed with 3% paraformaldehyde, and analyzed by flow cytometry for GFP fluorescence. The toxicity of the compounds was assessed by comparing the forward- versus the side-scatter dot plots. Cell debris was excluded from the analysis by gating on forward- versus side-scatter dot plots. The percentage of GFP-positive cells (R2) is indicated in the upper right hand corner of each panel.
Selection and genotypic characterization of PRM-A-resistant HIV-1(IIIB) strains.
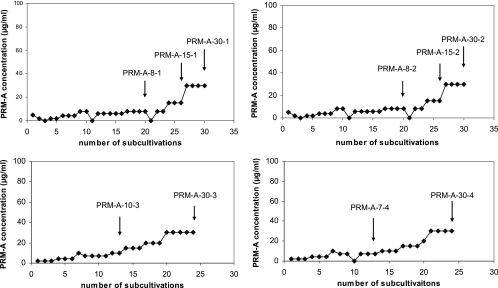
HIV-1(IIIB)-infected CEM cell cultures were exposed to PRM-A at 2 to 5 μg/ml, that is at an approximately one- to twofold-higher concentration than its EC50 (Fig. 5A -D). Four independent series of subcultivations were performed. Only when giant cell formation was abundantly visible in the drug-exposed virus-infected cell cultures was the PRM-A concentration increased stepwise. At several time points during the selection process, virus isolates were stored at −80°C and used for genotypic and phenotypic characterization (Fig. 5). A substantial number of cultivations (at least 15 to 20 subcultivations; 4 or 5 days/subcultivation) were required before the drug concentration could be increased without losing the virus from the infected cell cultures. Indeed, when PRM-A was added to the virus-infected cell cultures during the selection process at concentrations two- to threefold higher than the previous PRM-A concentration, the virus was easily lost upon further subcultivations. During the PRM-A-resistance selection process, a total of ten virus strains were isolated and characterized (Fig. 5A to D).
FIG. 5.
Four independent selection pathways for PRM-A-resistant HIV-1 strains. The drug concentrations were increased when a marked viral cytopathic effect was observed in the cell cultures. The time points at which virus isolates were propagated are indicated by arrows and a designation that corresponds with the virus strains depicted in Table 3.
A variety of amino acid changes occurred in gp120 but not gp41 of the drug-exposed virus strains (Table 4). Importantly, the mutations found in gp120 were located at N-glycosylation sites or at T/S sites that are part of the glycosylation motif NXT/S. Each isolate contained a variety of multiple amino acid mutations at glycosylation sites in gp120 (Table 4). In a number of cases, there was still a mixture of wild-type and mutated amino acids present in the virus preparation. The virus isolates taken early during the selection process already contained 4 to 5 different glycan deletions in their gp120s (i.e., PRM-A-8-1, PRM-A-8-2, and PRM-A-7-4). Further selection, up to the highest PRM-A concentration, resulted in the addition of several more glycan deletions in gp120 (i.e., PRM-A-15-1, PRM-A-30-1, PRM-A-30-3, and PRM-A-30-4). The virus isolates at the end of the selection could contain up to eight different glycan deletions. Interestingly, in the first series of virus strain isolates, an insertion of five amino acids containing an extra putative glycosylation site was observed in the different isolates (Table 4). However, it should be mentioned that this glycosylation site is present in most HIV-1 strains but appears to be deleted in the HIV-1(IIIB) strain that was used in our drug resistance selection experiments. When all mutated glycosylation sites (with deleted glycans) were spotted on the three-dimensional structure of HIV-1 gp120 (the amino acid numbering is according to Kwong et al., 21), none of the six glycans in the V1/V2 area were deleted. In the major remaining part of gp120, no clustered mutational pattern could be detected. The glycan deletions had obviously appeared in a rather disperse manner (Fig. 6). However, most of the glycan deletions occurred at glycosylation sites that were reported to carry a high-mannose type of glycan (23) (Table 4).
TABLE 4.
Genotypic characterization of gp120 of mutant HIV-1(IIIB) strains that emerged under escalating PRM-A exposure in CEM cell cultures
| N-glycan sitea | Genotypic characteristics of indicated strain:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PRM-A-8-1 | PRM-A-15-1 | PRM-A-30-1 | PRM-A-8-2 | PRM-A-15-2 | PRM-A-30-2 | PRM-A-10-3 | PRM-A-30-3 | PRM-A-7-4 | PRM-A-30-4 | |
| 88NVT90 | 90T/I | |||||||||
| 136NDT138 | ||||||||||
| 141NSS143 | ||||||||||
| 156NCS158 | ||||||||||
| 160NIS162 | ||||||||||
| 186NDT188 | ||||||||||
| 197NTS199 | ||||||||||
| 230NKT232* | 232T/M | 232T/M | 230N/S | 230N/S/D/G | 230N/S/D/G | 232T/M | 232M | 232T/M | ||
| 234NGT236* | 234N/I; 236T/I | 234N/I; 236T/I | 234N/I; 236T/I | 236T/I | 236T/I | |||||
| 241NVS243* | ||||||||||
| 262NGS264* | ||||||||||
| 276NFT278 | ||||||||||
| 289NQS291* | 291F | 291F | 291F | 291F | 291F | 291F | 291S/F | |||
| 295NCT297* | 297T/I | 297I | 297I | 295K | 295K | 295K | 295N/Y; 297T/I | 295N/Y; 297T/I | 297I | 297I |
| 301NNT303 | 303I | 303I | 303I | 301N/Y; 303T/I | 301N/Y; 303T/I | 301Y | ||||
| 332NIS334* | 334N/S | 334N | 334N | 334N/S | 334N | |||||
| 339NNT341* | 341A | 341A | 341A | 341A | 341A | 341T/I | 341I | |||
| 356NKT358 | ||||||||||
| 386NST388* | ||||||||||
| 392NST394* | ||||||||||
| 395b | FNSTW395-396ins | FNSTW395-396ins | FNSTW395-396ins | |||||||
| 397NST399 | ||||||||||
| 406NNT408 | ||||||||||
| 448NIT450* | 450N/Y | 450N | ||||||||
| 463NES465c | ||||||||||
Amino acid mutations according to Kwong et al. (21). An asterisk denotes the presence of a high-mannose-type glycan according to Leonard et al. (23). Boldface represents amino acid changes.
In the HIV-1/IIIB strain used in the resistance selection experiments, a glycosylation site (present in most other HIV-1 strains [i.e., NL4.3]) is absent. It reappeared during the series-1 selection experiments, downstream of amino acid position 395, as an insertion of five amino acids containing glycosylation motif NST.
This glycosylation site is absent in the HIV-1(IIIB) strain used in the PRM-A selection experiments due to the presence of NGP instead of the NES glycosylation motif.
FIG. 6.
Mapping of the deleted glycosylation sites in gp120 of HIV-1 strains isolated under escalating PRM-A concentrations in CEM cell cultures. HIV-1 (HXB-2) gp120 contains 24 numbered N-glycosylation sites (circles) (21, 23). The N-glycosylation sites that were found to be deleted in the ten PRM-A-exposed virus strains are in red. The N-glycosylation sites that were not found to be mutated are in green. The N-glycosylation site that was created during the drug selection process (series-1) is indicated in blue.
Phenotypic characterization of PRM-A-exposed HIV-1(IIIB) strains.
PRM-A showed a gradually decreasing inhibitory potential against those virus strains that were isolated at a later stage during the drug resistance selection process (Table 5). It lost antiviral potential against the virus strains at most by 3- to 10-fold except for strain HIV-1/PRM-A-30-3, for which the decrease in antiviral efficacy was 18-fold. GNA and HHA lost 20-fold to up to 50-fold of their inhibitory activity. In contrast, DS-5000 and AMD3100 kept their full suppressive effects to all evaluated virus strains (Table 4). The monoclonal antibody 2G12, which showed specificity against a well-defined glycan-containing epitope of HIV-1 gp120 (31, 32), completely lost its antiviral potential against all mutant virus isolates [EC50 for wild-type HIV-1(IIIB), 1.1 μg/ml versus >50 μg/ml for all mutant viruses] (Table 5). Also, the MAb B12 that recognizes peptide segments of gp120 that lie at the periphery of the CD4 binding site (9), the MAb F105 that recognizes the CD4 binding site on gp120 (27, 28, 46), and the MAb NEA-9205 that is directed to the gp120 V3 loop (34) have been evaluated against several PRM-A-exposed mutant HIV-1 strains. It was found that some of the MAbs (i.e., B12) keep their inhibitory potential against the different mutant virus strains, whereas other MAbs, such as F105 and NEA-9205, show antiviral efficacies that differ depending on the nature of the glycan deletions in the viral gp120 (Table 5).
TABLE 5.
Inhibitory activity of CBAs to pradimicin A-exposed HIV-1(IIIB) isolates in CEM cell culturesa
| Compound | EC50 (μg/ml) for indicated isolate:
|
||||||
|---|---|---|---|---|---|---|---|
| HIV-1(IIIB) | PRM-A-8-1 | PRM-A-30-1 | PRM-A-8-2 | PRM-A-30-2 | PRM-A-30-3 | PRM-A-30-4 | |
| PRM-A | 2.8 ± 1.0 | 7.5 ± 0.7 | 32 ± 25 | 8.0 ± 0.0 | 15 ± 7.1 | 50 | 32.5 ± 24.7 |
| GNA | 0.48 ± 0.28 | 2.5 ± 0.0 | 4.0 ± 0.0 | 0.90 ± 0.14 | 4.85 ± 3.04 | 19.0 ± 15.6 | 13.0 ± 9.9 |
| HHA | 0.29 ± 0.19 | 1.85 ± 0.49 | 5.5 ± 2.1 | 0.55 ± 0.35 | 5.0 ± 1.4 | 16.0 ± 5.7 | 8.0 ± 2.8 |
| DS-5000 | 0.33 ± 0.12 | 0.30 ± 0.14 | 0.8 ± 0.0 | 0.3 ± 0.0 | 0.28 ± 0.17 | 1.25 ± 0.35 | 1.25 ± 0.35 |
| AMD3100 | 0.05 ± 0.03 | 0.019 ± 0.002 | 0.045 ± 0.007 | 0.036 ± 0.006 | 0.070 ± 0.028 | 0.06 | 0.08 |
| MAb 2G12 | 1.1 ± 0.12 | >50 | >50 | >50 | >50 | >50 | >50 |
| MAb B-12 | 0.012 ± 0.011 | 0.020 ± 0.017 | 0.021 ± 0.009 | 0.017 ± 0.005 | |||
| MAb NEA-9205 | 0.0003 ± 0.00007 | 0.0003 ± 0.0002 | 0.025 ± 0.006 | 0.0004 ± 0.0003 | |||
| MAb F105 | 0.17 ± 0.05 | 0.68 ± 0.50 | ≥1 | 0.80 ± 0.56 | |||
Data are expressed as the EC50 (μg/ml) for inhibition of HIV-1-induced cytopathic effect in CEM cell cultures.
Interaction of PRM-A with the HIV (co)receptors CD4 and CXCR4 and with HIV-1 envelope gp120.
To reveal whether PRM-A exerts its antiviral activity solely by interaction with the viral gp120 or whether potential interaction with the cell membrane also plays a role in the eventual antiviral activity of the drug, PRM-A, the mannose-specific plant lectins GNA and HHA, and the CXCR4 antagonist AMD3100 (as a control inhibitory agent) were evaluated for their interference with a variety of anti-CD4 (i.e., SK3, OKT4, and OKT4A) and anti-CXCR4 (i.e., 12G5, 2B11, and 44717.111) MAbs for binding to CD4 and CXCR4 in MT-4 cell cultures. Data were obtained by flow cytometric analysis. Neither PRM-A (250 μg/ml) nor the other CBAs (GNA and HHA at 20 μg/ml) measurably affected the anti-CD4 and anti-CXCR4 MAb binding to CD4 and CXCR4 abundantly present on T-lymphocyte (i.e., MT-4 and CEM) cell cultures (data not shown). The drug concentrations used in these experiments were at least two orders of magnitude higher than their effective antiviral concentrations. As a control, the CXCR4 antagonist AMD3100 strongly prevented anti-CXCR4 MAb binding to the MT-4 cells but did not prevent anti-CD4 MAb binding.
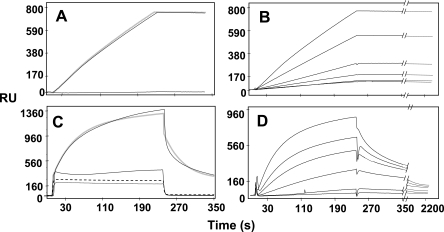
PRM-A and HHA were also investigated for their binding capacity for HIV-1 gp120, using the BIAcore technology. When tested in HBS buffer, HHA interacted with gp120 immobilized on the BIAcore sensor chip (Fig. 7A), while PRM-A did not show any binding capacity (Fig. 7C). However, the gp120 binding capacity of PRM-A became evident when EDTA was omitted and Ca2+ (10 mM) was added to the HBS buffer (Fig. 7C). The same buffer only slightly increased the binding of HHA to gp120 (less than 20%) without altering the binding parameters (data not shown). When tested in the appropriate buffers (unmodified HBS for HHA and HBS without EDTA and containing 10 mM Ca2+ for PRM-A), HHA and PRM-A showed a similar binding capacity (800 to 960 RU bound at the end of the injection phase). The specificity of the interactions was demonstrated by the lack of significant binding for the compounds when tested at the maximal doses on a BSA-coated sensor chip (Fig. 7A and C). In a second series of experiments, increasing concentrations of the two compounds were injected over the gp120 surface to evaluate the binding parameters. HHA, once bound, hardly detached from the gp120 surface, even when the dissociation phase was prolonged to 2,200 s (Fig. 7B), that is, at experimental conditions in which PRM-A almost completely dissociated (Fig. 7D). Accordingly, the dissociation rate of PRM-A is significantly higher than that of the plant lectin (Table 6). The dissociation constant (KD) for the interaction of HHA with gp120 was in the low nanomolar range, while that calculated for PRM-A proved significantly higher, being equal to ∼2.7 μM (Table 6). Interestingly, while the molar ratio of gp120 versus the drug could be calculated to be approximately 1:1.5 for HHA, a value of 1:33 was found for PRM-A (Table 6).
FIG. 7.
Interaction of HHA and PRM-A with gp120. In the first series of experiments, HHA (A) was injected at 5 nM in HBS over BIAcore sensor chips containing p120 (upper sensor grams) or BSA (lower sensor grams). PRM-A (C) was injected at 700 nM in HBS over gp120 (dotted sensor gram) or BSA (hacked sensor gram) surfaces. Alternatively, PRM-A was injected into HBS without EDTA but in the presence of 10 mM Ca2+ over gp120 (upper straight sensor gram) or BSA (lower straight sensor gram) surfaces (C). The solid black lines represent the experimental data. The gray lines represent the fit (A and C). In a second series of experiments, HHA (B) and PRM-A (D) were injected into the appropriate buffers at increasing concentrations over the gp120 surface (HHA at 0.3, 0.75, 0.5, 1.3, and 5 nM; PRM-A at 100, 150, 200, 350, 500, and 700 nM). In panels B and D, the sensor grams shown are those obtained after blank subtraction.
TABLE 6.
Binding parameters of the interactions of HHA and PRM-A to gp120 immobilized to a BIAcore sensor chipa
| Compound | Association rate (1/Ms) | Dissociation rate (1/s) | Dissociation constant (M) | Molecular ratio of gp120 to compound |
|---|---|---|---|---|
| HHA | 4.15 × 105 | 2.1 × 10−6 | 7.7 × 10−9 | 1:1.5 |
| PRM-A | 7.93 × 103 | 4.01 × 10−3 | 2.7 × 10−6 | 1:33 |
Binding parameters were calculated by the nonlinear-curve-fitting software package BIAevaluation 3.2, using a single-site model with a drifting baseline. Only sensorgrams whose fitting gave values of x2 close to 10 were used (20). Data for equilibrium binding between the compounds and gp120 were used to calculate an affinity value (KD) independently of the kinetics of binding. The correlation coefficient of the linear regression of the equilibrium binding data was always higher than 0.8.
Effect of PRM-A on immunological parameters.
Many lectins are endowed with mitogenic activity. We recently found mannose-specific cyanovirin mitogenic in cell culture and also stimulatory to several differentiation markers (7). Therefore, the effect of PRM-A was examined on several immunological parameters. Whereas PHA caused a marked stimulation of [3H]thymidine incorporation into PBMC DNA, PRM-A had no such stimulatory (mitogenic) effect at any of the concentrations tested (20, 4, and 0.8 μg/ml). However, PRM-A inhibited PHA-induced stimulation at its highest tested concentration (20 μg/ml) (data not shown). Lower PRM-A concentrations had no effect at all on the PHA stimulatory effect. Finally, in mixed lymphocyte reaction assays, PRM-A was devoid of any significant inhibitory activity on the stimulatory T-lymphocytic response at the tested concentrations (20 and 4 μg/ml). Thus, PRM-A seems not to interfere with these particular immunological parameters at its antiviral concentrations.
DISCUSSION
Pradimicin A represents the first prototype drug of a class of nonpeptidic low-molecular-weight (MW, 838) CBAs that show consistent antiviral (i.e., HIV) properties in cell cultures. Time-of-drug-addition studies pointed to an early event in the infection cycle of HIV as the target of therapeutic intervention by PRM-A (Fig. 2). The lack of inhibitory activity against VSV-G-pseudotyped HIV-1 infection indicates that the entry process is inhibited by the drug, pointing to a specific inhibition of the interaction of the HIV-1 gp120 envelope with the target cells (Fig. 3). The molecular mechanism of action of PRM-A likely requires binding to the mannose moieties that are part of the glycans on HIV gp120, because mannan can reverse the antiviral activity of the drug. The interaction of PRM-A with Man is Ca2+-dependent, as evidenced by our BIAcore studies that showed the strict requirement of Ca2+ for binding to HIV gp120. These observations are in agreement with the early findings on the Ca2+ requirement of PRM-A for antifungal activity and the yielding of a ternary complex among PRM-A, mannopyranoside, and Ca2+ (40). Also, evidence is provided that such interaction eventually yields a ternary complex consisting of two molecules of PRM-A, four molecules of d-mannopyranoside, and one atom of Ca2+ (40). In this respect, PRM-A behaves as a C-type “artificial lectin” in its interaction with glycans (such as DC-SIGN, present on the dendritic cell membrane), in contrast to the mannose-specific GNA and HHA plant lectins that do not require Ca2+ for efficient interaction with gp120, as is also demonstrated by the BIAcore studies (Fig. 7). Given the requirements of calcium for the binding of PRM-A to carbohydrates, the antiviral efficacies of PRM-A, and GNA and HHA, which do not require Ca2+, were examined in the presence of a broad variety (2% to 40%) of serum concentrations. Similar antiviral efficacies were observed.
Although virus infection/replication could be completely blocked by PRM-A, the drug seems to enhance virus interactions with its target cells in a dose-dependent manner (Fig. 4). In this respect, the enhancing effects of PRM-A were more pronounced than those of the mannose-specific HHA (data not shown). Since DS-5000 clearly showed the opposite effect (Fig. 4), we hypothesize that CBAs such as plant lectins, as well as the nonpeptidic low-molecular-weight PRM-A, may be endowed with a cross-linking ability that creates a network of cross-linked and immobilized surface glycoproteins between the virus particle gp120 envelope and the target cell membrane. Given the evidence of the presence of two PRM-A molecules in the ternary complex that have mannose oligomers and Ca2+ and a total of two mannose binding sites for each PRM-A molecule, this cross-linking hypothesis may be viable. PRM-A may then inhibit virus entry by cross-linking glycans, affecting their mobility and preventing the virus particle gp120 from adopting the conformations necessary for efficient virus-cell fusion. This proposed mechanism of action, however, has still to be proven. However, such a cross-linking activity on surface glycoproteins has also been proposed for the antiviral action of the θ-defensin retrocyclin 2 (a multivalent lectin), which inhibits influenza virus infection by blocking membrane fusion mediated by the viral hemagglutinin (22). It was also shown that human β-defensin 3 and mannan-binding C-type lectin block viral fusion by creating a protective barricade of immobilized surface glycoproteins. This general binding mechanism may explain the broad-spectrum antiviral activity of many multivalent lectins of the immune system (22).
The KD of PRM-A/gp120 interaction (in the presence of Ca2+) was markedly higher than that of the mannose-specific plant lectin HHA. This correlates with our observation that PRM-A is 5- to 10-fold less inhibitory to HIV in cell culture than the HHA plant lectin. However, PRM-A has a much lower variability of antiviral activity against a broad variety of X4- and R5-tropic HIV-1 clades than the plant lectins HHA and GNA. Whereas the plant lectins HHA and GNA are reported to recognize α(1-3) and/or α(1-6) mannose oligomers (43), it has been suggested that PRM-A predominantly recognizes α(1-2) mannose residues (38) that are predominantly present on high-mannose-type glycans of HIV-1 gp120. Cyanovirin N, which also selectively recognizes α(1-2) mannose residues (8), is also more consistently active against all different HIV-1 clades than HHA and GNA (7).
Since CBAs, in particular PRM-A, may simultaneously bind to multiple sites on HIV gp120, this drug differs from the currently available anti-HIV drugs that stoichiometrically bind to their target molecules (at an obligatory 1:1 ratio). Indeed, one reverse transcriptase or protease or gp41 molecule can bind to only one interacting drug molecule (nonnucleoside reverse transcriptase inhibitor, protease inhibitor, or fusion inhibitor) at the same moment. As a consequence, one or a few mutations that appear in a target, like reverse transcriptase, protease, or gp41, usually result in a marked degree of drug resistance. Instead, the data obtained from the BIAcore studies revealed a molar ratio for gp120 to drug of 1:33 (Table 6), which means that a high number of PRM-A molecules may bind to one gp120 molecule. Data in the literature has revealed a stoichiometry ratio of 1:5 for the α1,2-mannose-specific cyanovirin (24), which is still higher than the 1:1.5 ratio found for HHA (Table 6). These striking differences in stoichiometry may be directly correlated with the size of the drug molecules: 50,000 D for HHA, 11,000 D for cyanovirin, and 838 D for PRM-A. Obviously, the smaller the size of the molecule, the easier the carbohydrates on gp120 can be reached by the CBAs without steric hindrance. Therefore, the multiple binding sites on gp120 for CBAs make these drugs much less susceptible to sensitivity loss by the deletion of a few glycosylation sites in gp120, resulting in a high genetic barrier for the CBAs. Moreover, in particular for high-molecular-weight CBAs such as HHA, one single glycan deletion in HIV gp120 may immediately make space for an efficient interaction of one of the other (remaining) glycans with the CBA. As a consequence, multiple glycans need concomitantly to be deleted before CBAs will show a decreased antiviral efficacy. These properties are indeed observed in our drug resistance selection experiments. Interestingly, whereas PRM-A also seems to be able to induce apoptosis-like cell death in Saccharomyces cerevisiae, a mutant that lacks a putative N-glycosylation site in the extracellular domain of a surface protein of the yeast became resistant to the PRM-A derivative (18), showing a molecular mechanism of resistance development (deletion of glycosylation sites) comparable to that found in our studies for HIV.
The very slow (and moderate) resistance development of HIV-1 against the plant lectins (5, 6) and PRM-A (this study) is in full agreement with the mechanisms of action and interaction of PRM-A with its target molecule. Given the uniqueness of the drug resistance profile (i.e., glycan deletions in HIV gp120) of the CBAs, in casu PRM-A, mutant HIV strains that emerge under CBA pressure are not expected to loose their susceptibilities to other anti-HIV drugs, including the variety of available entry inhibitors as also shown in our drug-susceptibility studies. Instead, HIV-1 strains resistant to other drug targets such as RT, protease, and gp41 are not expected to loose susceptibility to CBAs, including PRM-A. However, it was previously shown that mannose (i.e., HHA, GNA, and cyanovirin)- and GlcNAc (i.e., UDA)-specific plant lectins also select for glycan deletions in the HIV-1 gp120 (5-7, 47). Such deletions predominantly occurred at similar glycosylation sites, as observed for PRM-A. Consequently, PRM-A also showed decreased antiviral activity against such GNA-, HHA-, and UDA-resistant virus strains (data not shown) and vice versa: GNA, HHA, and UDA partially lost antiviral activity against the PRM-A-resistant virus strains (Table 5). Although no glycan deletions have been observed so far in gp41 under PRM-A pressure, it cannot be excluded that such mutations might appear at higher concentrations (with a more-soluble PRM-A derivative) or longer selection times for compensatory purposes.
The antiviral properties of PRM-A and the mutational pattern the virus follows in an attempt to escape PRM-A drug pressure led us to consider that the application of CBAs may represent a novel therapeutic approach to HIV treatment that is entirely different in concept from currently existing therapeutic approaches. Indeed, exposure of HIV to CBAs such as PRM-A may put the virus in a dilemma: it must either eventually be eliminated from its host by continual suppression by the CBA, or it must escape the drug pressure by progressively deleting the glycosylation sites in its envelope, thereby becoming highly susceptible to neutralization and elimination by the immune system (2). The proposed therapeutic concept may display a thus far unprecedented concerted action between drug chemotherapy and the immune system. Indeed, besides a direct antiviral activity of the CBAs, CBA-induced deletions of gp120 glycans may have the potential to trigger the immune system against previously hidden highly immunogenic and antigenic epitopes on gp120, resulting in “therapeutic self-vaccination” (2). The observation that a number of known monoclonal neutralizing antibodies (i.e., B12, NEA-9205, and F105) did not display a more pronounced inhibitory effect and in some cases even displayed a decreased antiviral efficacy against the PRM-A-exposed mutant HIV-1 strains is not necessarily against this hypothesis. In fact, these antibodies have been generated against the wild-type virus but not the mutated virus (with glycan deleted) and thus may never have encountered the epitopes that became uncovered after the glycan deletions appeared in the gp120 envelope. Although a dual mechanism of antiviral action with involvement of the immune system has to be shown in the in vivo setting, the earlier findings of Reitter et al. (29) may be highly relevant in this context. It was indeed demonstrated that infection of monkeys with mutant SIV strains that contain as few as two deleted glycosylation sites in their envelope glycoproteins induced a much stronger production of neutralizing antibodies (to the uncovered, exposed, immunogenic envelope epitopes) and eventually resulted in markedly lower viral levels in the plasma (up to three orders of magnitude lower) (29). It now seems imperative to examine CBAs such as PRM-A in SIV-infected monkeys to determine whether a similar phenomenon would occur. Also, other viruses with a glycosylated envelope, such as para- and orthomyxoviruses (i.e., influenza virus), flaviviruses (i.e., hepatitis C virus), and coronaviruses (i.e., severe acute respiratory syndrome coronavirus), may be susceptible to the inhibitory activity of CBAs as well, putting the proposed therapeutic concept in a broader (antiviral) context.
Although general in vivo toxicity should be a realistic concern with the CBA therapeutic concept, it should be mentioned that in previous studies intravenous injection of plant lectins (i.e., HHA, GNA, and UDA) into mice did not result in acute animal toxicity (4, 6). Also, a PRM-A derivative was shown to have in vivo antifungal activity in the treatment of experimental pulmonary aspergillosis in persistently neutropenic rabbits. In this study, the drug was well tolerated at all dosing schedules (up to 150 mg/kg/day) and no toxicity was noted (16). Moreover, it was shown that the PRM-A derivative can bind only to terminal d-mannopyranosides of glycans (40), which are abundantly present on the HIV gp120 envelope, whereas normal animal cells show no binding of the drug, probably because d-mannopyranoside terminals which are necessary for PRM-A binding are very scarce on the cell surface (4, 40). Interestingly, large doses of PRM-A derivative have been administered to animals in the past with minimal toxicity (45), and one (more-soluble) compound derivative (BMS 181184) has already been subjected to phase I/II clinical trials to investigate its antifungal activity. In the clinical study, hepatic transaminase was significantly induced in the majority of volunteers (17). It would be important to reveal whether this side effect is entirely the result of, or not related to, the carbohydrate-binding property of the compound.
In conclusion, nonpeptidic CBAs such as PRM-A represent a novel therapeutic concept for HIV therapy. PRM-A represents the first nonpeptidic (low-molecular-weight) drug with an interesting anti-HIV activity, unique mutational pattern and resistance profile, and high genetic barrier. Moreover, the drug's lead may have the potential to act in vivo through a dual mechanism of action involving a “direct” antiviral activity (entry inhibition) and the triggering of the immune system against previously hidden immunogenic epitopes on HIV gp120 (“self-vaccination”). Therefore, CBAs in general, and the nonpeptidic PRM-A lead compound and its derivatives in particular, should be given prompt consideration for further (pre)clinical development as a potential new conceptual generation of anti-HIV agents.
Acknowledgments
This work was supported by the European Commission (René Descartes Prize-2001, Krediet nr. HPAW-2002-90001; and EMPRO no. 503558 of the 6th Frame Work Programme), the ANRS (France), the Fonds voor Wetenschappelijk Onderzoek (FWO) Krediet nr. G-0267-04, the Centers of Excellence of the Katholieke Universiteit at Leuven (Krediet nr. EF/05/015), and the Istituto Superiore di Sanità, AIDS project (Italy).
We are grateful to Ann Absillis, Lizette van Berckelaer, Yoeri Schrooten, Sandra Claes, Rebecca Provinciael, and Eric Fonteyn for excellent technical assistance, Mathy Froeyen for valuable help in drawing Fig. 6, and Christiane Callebaut for dedicated editorial help.
Footnotes
Published ahead of print on 18 October 2006.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balzarini, J. 2005. Targeting the glycans of gp120: a novel approach aimed at the Achilles heel of HIV. Lancet Infect. Dis. 5:726-731. [DOI] [PubMed] [Google Scholar]
- 3.Balzarini, J. 2006. Inhibition of HIV entry by carbohydrate-binding proteins. Antiviral Res. 71:237-247. [DOI] [PubMed] [Google Scholar]
- 4.Balzarini, J., S. Hatse, K. Vermeire, K. Princen, S. Aquaro, C.-F. Perno, E. De Clercq, H. Egberink, G. Vanden Mooter, W. Peumans, E. Vandamme, and D. Schols. 2004. Mannose-specific plant lectins from the Amaryllidaceae family qualify as efficient microbicides for prevention of human immunodeficiency virus infection. Antimicrob. Agents Chemother. 48:3858-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balzarini, J., K. Van Laethem, S. Hatse, K. Vermeire, E. De Clercq, W. Peumans, E. Van Damme, A. M. Vandamme, A. Bolmstedt, and D. Schols. 2004. Profile of resistance of human immunodeficiency virus to mannose-specific plant lectins. J. Virol. 78:10617-10627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balzarini, J., K. Van Laethem, S. Hatse, M. Froeyen, W. Peumans, E. Van Damme, and D. Schols. 2005. Carbohydrate-binding agents cause deletions of highly conserved glycosylation sites in HIV gp120. J. Biol. Chem. 280:41005-41014. [DOI] [PubMed] [Google Scholar]
- 7.Balzarini, J., K. Van Laethem, W. J. Peumans, E. J. M. Van Damme, A. Bolmstedt, F. Gago, and D. Schols. 2006. Mutational pathways, resistance profile, and side effects of cyanovirin relative to human immunodeficiency virus type 1 strains with N-glycan deletions in their gp120 envelopes. J. Virol. 80:8411-8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Botos, I., and A. Wlodawer. 2005. Proteins that bind high-mannose sugars of the HIV envelope. Prog. Biophys. Mol. Biol. 88:233-282. [DOI] [PubMed] [Google Scholar]
- 9.Bublik, E. M., S. Yeger-Azuz, and J. M. Gershoni. 2006. Computational prediction of the cross-reactive neutralizing epitope corresponding to the monoclonal antibody b12 specific for HIV-1 gp120. FASEB J. 20:1762-1774. [DOI] [PubMed] [Google Scholar]
- 10.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, D. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065-2071. [DOI] [PubMed] [Google Scholar]
- 11.Daelemans, D., C. Pannecouque, G. N. Pavlakis, O. Tabarrini, and E. De Clercq. 2005. A novel and efficient approach to discriminate between pre- and post-transcription HIV inhibitors. Mol. Pharmacol. 67:1574-1580. [DOI] [PubMed] [Google Scholar]
- 12.Dairi, T., Y. Hamano, Y. Igarashi, T. Furumai, and T. Oki. 1997. Cloning and nucleotide sequence of the putative polyketide synthase genes for pradimicin biosynthesis from Actinomadura hibisca. Biosci. Biotech. Biochem. 61:1445-1453. [DOI] [PubMed] [Google Scholar]
- 13.De Clercq, E. 2004. New anti-HIV agents in preclinical or clinical development. Front. Medic. Chem. 1:543-579. [Google Scholar]
- 14.Dey, A. K., M. Khati, M. Tang, R. Wyatt, S. M. Lea, and W. James. 2005. An aptamer that neutralizes R5 strains of human immunodeficiency virus type 1 blocks gp120-CCR5 interaction. J. Virol. 79:13806-13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer, P. B., M. Collin, G. B. Karlsson, W. James, T. D. Butters, S. J. Davis, S. Gordon, R. A. Dwek, and F. M. Platt. 1995. The α-glucosidase inhibitor N-butyldeoxynojirimycin inhibits human immunodeficiency virus entry at the level of post-CD4 binding. J. Virol. 69:5791-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez, C. E., A. H. Groll, N. Giri, D. Shetty, I. Al-Mohsen, T. Sein, E. Feuerstein, J. Bacher, S. Piscitelli, and T. J. Walsh. 1998. Antifungal activity of the pradimicin derivative BMS 18114 in the treatment of experimental pulmonary aspergillosis in persistently neutropenic rabbits. Antimicrob. Agents Chemother. 42:2399-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groll, A. H., T. Sein, V. Petraitis, R. Petriaitiene, D. Callender, C. E. Gonzalez, N. Giri, J. Bacher, S. Piscitelli, and T. J. Walsh. 1998. Compartmental pharmacokinetics and tissue drug distribution of the pradimicin derivative BMS 181184 in rabbits. Antimicrob. Agents Chemother. 42:2700-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiramoto, F., N. Nomura, T. Furumai, Y. Igarashi, and T. Oki. 2005. Pradimicin resistance of yeast is caused by a mutation of the putative N-glycosylation sites of osmosensor protein Sln1. Biosci. Biotechnol. Biochem. 69:238-241. [DOI] [PubMed] [Google Scholar]
- 19.Igarashi, Y., and T. Oki. 2004. Mannose-binding quinone glycoside MBQ: potential utility and action mechanisms. Adv. Appl. Microbiol. 54:147-166. [DOI] [PubMed] [Google Scholar]
- 20.Khalifa, M. B., L. Choulier, H. Lortat-Jacob, D. Altschuh, and T. Vernet. 2001. BIACORE data processing: an evaluation of the global fitting procedure. Anal. Biochem. 293:193-203. [DOI] [PubMed] [Google Scholar]
- 21.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leikina, E., H. Delanoe-Ayari, K. Melikov, M. S. Cho, A. Chen, A. J. Waring, W. Wang, Y. Xie, J. A. Loo, R. I. Lehrer, and L. V. Chernomordik. 2005. Carbohydrate-binding molecules inhibit viral fusion and entry by crosslinking membrane glycoproteins. Nature Immunol. 6:995-1001. [DOI] [PubMed] [Google Scholar]
- 23.Leonard, C. K., M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas, and T. J. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265:10373-10382. [PubMed] [Google Scholar]
- 24.O'Keefe, B. R., S. R. Shenoy, D. Xie, W. Zhang, J. M. Muschik, M. J. Currens, I. Chaiken, and M. R. Boyd. 2000. Analysis of the interaction between the HIV-inactivating protein cyanovirin-N and soluble forms of the envelope glycoproteins gp120 and gp41. Mol. Pharmacol. 58:982-992. [DOI] [PubMed] [Google Scholar]
- 25.Oki, T., and T. Dairi. 1994. Pradimicins: potential antifungal and anti-HIV agents. Exp. Opin. Ther. Patents 4:1483-1491. [Google Scholar]
- 26.Oki, T., M. Konishi, K. Tomatsu, K. Tomita, K. I. Saitoh, M. Tsunakawa, M. Nishio, T. Miyaki, and H. Kawaguchi. 1988. Pradimicin, a novel class of potent antifungal antibiotics. J. Antibiotics 41:1701-1704. [DOI] [PubMed] [Google Scholar]
- 27.Posner, M. R., T. Hideshima, T. Cannon, M. Mukherjee, K. H. Mayer, and R. A. Byrn. 1991. An IgG human monoclonal antibody that reacts with HIV-1/gp120, inhibits virus binding to cells, and neutralizes infection. J. Immunol. 146:4325-4332. [PubMed] [Google Scholar]
- 28.Posner, M. R., L. A. Cavacini, C. L. Emes, J. Power, and R. Byrn. 1993. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J. Acquir. Immune Defic. Syndr. 6:7-14. [PubMed] [Google Scholar]
- 29.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 30.Rusconi, S., E. Bulgheroni, and P. Citterio. 2004. Entry and fusion inhibitors of HIV. Expert Opin. Ther. Patents 14:733-748. [Google Scholar]
- 31.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollman Saphire, R. Stanfield, J. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. O. Saphire, D. Calarese, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, D. R. Burton, and P. M. Rudd. 2003. The carbohydrate epitope of the neutralizing anti-HIV-1 antibody 2G12. Adv. Exp. Med. Biol. 535:205-218. [DOI] [PubMed] [Google Scholar]
- 33.Schols, D., S. Struyf, J. Van Damme, J. A. Esté, G. Henson, and E. De Clercq. 1997. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J. Exp. Med. 186:1383-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schonning, K., B. Jansson, S. Olofsson, J. O. Nielsen, and J. S. Hansen. 1996. Resistance to V3-directed neutralization caused by the N-linked oligosaccharide depends on the quaternary structure of the HIV-1 envelope oligomer. Virology 218:134-140. [DOI] [PubMed] [Google Scholar]
- 35.Stauber, R. H., K. Horie, P. Carney, E. A. Hudson, N. I. Tarasova, G. A. Gaitanaris, and G. N. Pavlakis. 1998. Development and applications of enhanced green fluorescent protein mutants. BioTechniques 24:462-466, 468-471. [DOI] [PubMed] [Google Scholar]
- 36.Tanabe, A., H. Nakashima, O. Yoshida, N. Yamamoto, O. Tenmyo, and T. Oki. 1988. Inhibitory effect of new antibiotic, pradimicin A on infectivity, cytopathic effect and replication of human immunodeficiency virus in vitro. J. Antibiotics 41:1708-1710. [DOI] [PubMed] [Google Scholar]
- 37.Tanabe-Tochikura, A., T. S. Tochikura, J. R. Blakeslee, Jr., R. G. Olsen, and L. E. Mathes. 1992. Anti-human immunodeficiency virus (HIV) agents are also potent and selective inhibitors of feline immunodeficiency virus (FIV)-induced cytopathic effect: development of a new method for screening of anti-FIV substances in vitro. Antiviral Res. 19:161-172. [DOI] [PubMed] [Google Scholar]
- 38.Tanabe-Tochikura, A., T. S. Tochikura, O. Yoshida, T. Oki, and N. Yamamoto. 1990. Pradimicin A inhibition of human immunodeficiency virus: attenuation by mannan. Virology 176:467-473. [DOI] [PubMed] [Google Scholar]
- 39.Tsai, C. C., P. Emau, Y. Jiang, B. Tian, W. R. Morton, K. R. Gustafson, and M. R. Boyd. 2003. Cyanovirin-N as a topical microbicide prevents rectal transmission of SHIV89.6P in macaques. AIDS Res. Hum. Retrovir. 19:535-541. [DOI] [PubMed] [Google Scholar]
- 40.Ueki, T., M. Oka, Y. Fukagawa, and T. Oki. 1993. Studies on the mode of antifungal action of pradimicin antibiotics. III. Spectrophotometric sequence analysis of the ternary complex formation of BMY-28864 with D-mannopyranoside and calcium. J. Antibiotics 46:465-477. [DOI] [PubMed] [Google Scholar]
- 41.Valentin, A., W. Lu, M. Rosati, R. Schneider, J. Albert, A. Karlsson, and G. N. Pavlakis. 1998. Dual effect of interleukin 4 on HIV-1 expression: implications for viral phenotypic switch and disease progression. Proc. Natl. Acad. Sci. USA 95:8886-8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Damme, E. J. M., A. K. Allen, and W. J. Peumans. 1988. Related mannose-specific lectins from different species of the family Amaryllidaceae. Physiol. Plant 73:52-57. [Google Scholar]
- 43.Van Damme, E. J. M., W. J. Peumans, A. Pusztai, and S. Barocz (ed.). 1998. Handbook of plant lectins: properties and biomedical applications. John Wiley & Sons, Chichester, N.Y.
- 44.Van Laethem, K., Y. Schrooten, P. Lemey, E. Van Wijngaerden, S. De Wit, M. Van Ranst, and A.-M. Vandamme. 2005. A genotypic resistance assay for the detection of drug resistance in the human immunodeficiency virus type 1 envelope gene. J. Virol. Methods 123:25-34. [DOI] [PubMed] [Google Scholar]
- 45.Walsh, T. J., and N. Giri. 1997. Pradimicins: a novel class of broad-spectrum antifungal compounds. Eur. J. Clin. Microbiol. Infect. Dis. 16:93-97. [DOI] [PubMed] [Google Scholar]
- 46.Wilkinson, R. A., C. Piscitelli, M. Teintze, L. A. Cavacini, M. R. Posner, and C. M. Lawrence. 2005. Structure of the Fab fragment of F105, a broadly reactive anti-human immunodeficiency virus (HIV) antibody that recognizes the CD4 binding site of HIV type 1 gp120. J. Virol. 79:13060-13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Witvrouw, M., V. Fikkert, A. Hantson, C. Pannecouque, B. R. O'Keefe, J. McMahon, L. Stamatatos, E. De Clercq, and A. Bolmstedt. 2005. Resistance of human immunodeficiency virus type 1 to the high-mannose binding agents cyanovirin N and concanavalin A. J. Virol. 79:7777-7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, H., Y. Zhou, C. Alcock, T. Kiefer, D. Monie, J. Siliciano, Q. Li, P. Pham, J. Cofrancesco, D. Persaud, and R. F. Siliciano. 2004. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J. Virol. 78:1718-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]