Abstract
Following acute infection, bovine herpesvirus 1 establishes latency in sensory neurons of trigeminal ganglia (TG). Reactivation from latency occurs periodically, resulting in the shedding of infectious virus. The latency-related (LR) RNA is abundantly expressed in TG of latently infected calves, and the expression of LR proteins is necessary for dexamethasone-induced reactivation from latency. Previously published studies also identified an alternatively spliced LR transcript which is abundantly expressed in TG at 7 days after infection and has the potential to encode a novel LR fusion protein. Seven days after infection is when extensive viral gene expression is extinguished in TG and latency is established, suggesting that LR gene products influence the establishment of latency. In this study, we used a bacterial two-hybrid assay to identify cellular proteins that interact with the novel LR fusion protein. The LR fusion protein interacts with two proteins that can induce apoptosis (Bid and Cdc42) and with CCAAT enhancer binding protein alpha (C/EBP-α). Additional studies confirmed that the LR fusion protein interacts with human or insect C/EBP-α. C/EBP-α protein expression is induced in TG neurons of infected calves and after dexamethasone-induced reactivation from latency. Wild-type C/EBP-α, but not a DNA binding mutant of C/EBP-α, enhances plaque formation in bovine cells. We hypothesize that interactions between the LR fusion protein and C/EBP-α promote the establishment of latency.
Bovine herpesvirus-1 (BHV-1) is an Alphaherpesvirinae subfamily member (35) that establishes lifelong latency in ganglionic neurons of the peripheral nervous system following acute replication in the mucosal epithelium. Infection of permissive cells (13) or calves (65) leads to rapid cell death, in part due to apoptosis. Infection can also cause conjunctivitis, pneumonia, genital disorders, abortions, and a complex upper respiratory tract infection referred to as shipping fever (bovine respiratory complex) (59). BHV-1 transiently depresses cell-mediated immunity (8, 21-23), resulting in secondary bacterial infections and pneumonia (shipping fever). BHV-1 infection and shipping fever cost the United States cattle industry at least $500 million/year (5, 46). Although modified live vaccines are available, they can cause disease in young calves or abortion in pregnant cows.
Viral gene expression is regulated in three distinct phases: immediate early (IE), early (E), or late (L). IE gene expression is stimulated by a virion component, alpha gene trans-inducting factor (44), and bICP0. The bICP0 protein is constitutively expressed throughout productive infection because the gene has an IE and E promoter. The bICP0 protein trans-activates all viral promoters and thus promotes efficient productive infection (reviewed in references 36 and 38). IE proteins activate E gene expression, and viral DNA replication ensues. L gene expression then occurs, culminating in virion production.
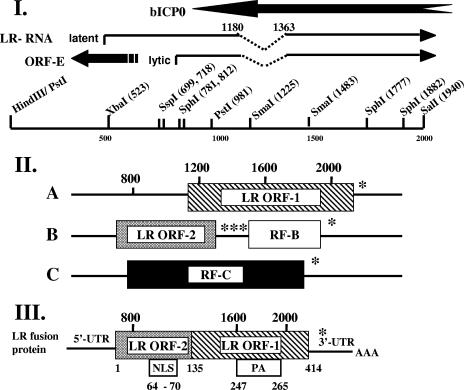
The BHV-1 latency-related (LR) RNA is abundantly transcribed in latently infected neurons (40, 54, 55) and is antisense to the bICP0 gene. The LR gene has two open reading frames (ORF-1 and ORF-2) and two reading frames that lack an initiating ATG (RF-B and RF-C) (Fig. 1, panel II). A fraction of LR RNA is polyadenylated and alternatively spliced in trigeminal ganglia (TG), suggesting that different LR proteins are expressed at specific times during the latency-reactivation cycle (13, 27) (Fig. 1, panel I). One such transcript is expressed in TG at 7 days after infection (7d cDNA) and has the potential to encode an ORF-2/ORF-1 fusion protein (Fig. 1, panel III, LR fusion protein) (11, 12). When the 7d cDNA is expressed in insect cells from a recombinant baculovirus, a 40-kDa protein is expressed (33).
FIG. 1.
Schematic of LR gene and major ORF encoded by the 7d cDNA. (I) Map of the PstI-SalI fragment that contains the LR promoter, 5′ terminus of the LR transcript, ORF-E, and part of the bICP0 protein coding sequences. The 5′ end of the LR transcript in TG is located upstream of the start site mapped during productive infection of cultured bovine cells (11, 12, 27). The positions of the 5′ and 3′ splice sites of the 7d cDNA are shown above the latent LR transcript (12). These splice sites have not been detected during productive infection, and thus the splice sites shown here are in the general region where splicing occurs. ORF-E is antisense relative to the LR gene (29). (II) The organization of the major ORFs in the LR gene was described previously (40). Numbers on top indicate the genomic nucleotide positions, and those on the bottom refer to amino acid positions. The hatched box denotes LR-ORF-1. RF-B (white box) and RF-C (black box) that follow LR-ORF-2 (stippled box) do not contain a methionine at their respective amino termini. Stars denote the positions of in-frame stop codons. (III) Organization of the major ORFs encoded by the 7d cDNA. NLS refers to the nuclear localization sequence and PA to proline/alanine-rich sequences present in the LR fusion protein. Nucleotide numbers refer to the genomic sequence that was initially reported for the LR gene (40), and the numbers below the LR fusion protein are amino acid numbers of the ORF.
A mutant BHV-1 strain with three stop codons at the N terminus of ORF-2 (LR mutant virus) was constructed (30). ORF-2 and RF-C are expressed in bovine cells infected with wild-type (wt) or the LR-rescued virus but not in cells infected with the LR mutant virus (33). Calves infected with the LR mutant exhibit diminished clinical symptoms and reduced shedding of infectious virus from the eye, TG, or tonsil (30, 31, 51). The LR mutant virus does not reactivate from latency following treatment with dexamethasone (DEX) (31), whereas all calves latently infected with wt virus or the LR-rescued virus shed infectious virus following DEX treatment (31). Thus, disruption of LR protein expression is necessary for DEX-induced reactivation from latency in calves. LR gene products inhibit cell proliferation, bICP0 RNA expression (6, 18, 56), and apoptosis (10). LR protein expression is necessary for inhibiting apoptosis but not for cell growth or bICP0 expression (6, 16, 18, 56). It is not clear whether LR protein expression enhances the establishment or maintenance of latency and/or reactivation from latency.
In this study, we performed a two-hybrid screening to identify cellular proteins that interact with the LR fusion protein. Several such cellular proteins were identified. Two of these proteins, Bid and Cdc42, can induce apoptosis. Since the LR gene inhibits apoptosis (10, 43), the ability of an LR protein to interact with proapoptotic cellular proteins was expected. The LR fusion protein also interacts with CCAAT enhancer binding protein alpha (C/EBP-α) in the two-hybrid assay. Independent studies confirmed that the LR fusion protein interacts with insect or human C/EBP-α. Induction of C/EBP-α protein expression is detected in TG neurons of acutely infected calves and DEX-induced reactivation from latency. The overexpression of wt C/EBP-α, but not a DNA binding mutant, also enhances productive infection. Collectively, these studies suggest that interactions between the LR fusion protein and C/EBP-α repress productive infection in TG neurons, thus enhancing the establishment of latency.
MATERIALS AND METHODS
Cells and virus.
Bovine kidney (MDBK) cells were grown in Earle's modified Eagle's medium supplemented with 5% fetal calf serum (FCS). Human neuroblastoma cells (SK-N-SH) and bovine testicle cells (9.1.3) were grown in Earle's modified Eagle's medium supplemented with 10% FCS. Spodoptera frugiperda (SF9) insect cells were maintained in Grace's supplemented insect medium (Gibco) with 10% FCS. All media contained penicillin (10 U/ml) and streptomycin (100 μg/ml).
The wt Cooper strain of BHV-1 was obtained from the National Veterinary Services Laboratory, Animal and Plant Health Inspection Services (Ames, IA). The BHV-1 gC-blue virus contains the β-galactosidase gene inserted into the glycoprotein C gene, which was provided by S. Chowdhury (Kansas State University). A recombinant baculovirus expressing the 7d cDNA sequence (His66) was described previously (33), and His66 was used to infect SF9 insect cells.
Plasmids.
pBT7day was generated by cloning the SphI fragment of the 7d cDNA (33) into the bait plasmid pBT. Cloning was done in such a way that the LR fusion protein is in frame with the Escherichia coli λcI repressor coding sequences. The target plasmid (pTRG) contains a rat brain cDNA library fused in frame to the RNA polymerase alpha subunit.
Plasmid pC/EBP-α contains the wt mouse C/EBP-α cDNA in an adenoviral vector (pAdTrack) with corresponding mammalian expression promoter/enhancer sequences (61). Plasmid pC/EBP-α 290mts contains a mutation that produces a single amino acid change (R290A) in C/EBP-α, which results in loss of DNA binding activity (61). This mutant is referred to as C/EBP-αΔDB in this study.
The bICP0 plasmid encodes bICP0 coding sequences cloned into a human cytomegalovirus expression vector.
Bacterial two-hybrid assay.
All procedures were performed according to recommendations of the manufacturers of the BacterioMatch II two-hybrid system (Stratagene). An E. coli reporter strain was cotransformed with pBT7day and pTRG containing the rat brain cDNA library. Cotransformants were screened by using medium lacking histidine and containing 3-aminotriazole, as recommended by the manufacturer. Positive interactions were further confirmed by activation of the streptomycin resistance gene on selective plates. The pTRG plasmid derived from positive interactions was isolated by standard methods. DNA sequencing reactions of the respective inserts in pTRG were performed at the University of Nebraska DNA sequencing core facility. A BLAST search at the NCBI website was performed to identify proteins encoded by the respective cDNAs.
Binding assays.
The His66 recombinant baculovirus, containing the 7d cDNA expressed as a histidine-tagged protein (33), was used to infect SF9 cells. Cells were collected when the cytopathic effect was 70 to 80%, lysed, and then loaded on a 2-ml Ni+ column under (i) native or (ii) denaturing conditions, according to the manufacturer's recommendations (Probound, K850-01; Invitrogen).
(i) SF9 cells (infected or uninfected) were suspended in native binding buffer (50 mM NaPO4, 500 mM NaCl, 10 mM imidazole [pH 8.0]), followed by three freeze-thaw cycles and a final 10-min spin at 3,000 × g. Eleven milligrams of protein was loaded onto the Ni+ column equilibrated with native binding buffer and allowed to bind for 1 h at 4°C. Three washes of 10 ml each were performed using native wash buffer (50 mM NaPO4, 500 mM NaCl, 20 mM imidazole [pH 8]).
(ii) Lysis of SF9 cells (infected or uninfected) was conducted under denaturing conditions by suspending cells in a solution of 6 M guanidine-HCl, 20 mM NaPO4 (pH 7.8), and 500 mM NaCl. The cell lysate was passed through an 18-gauge needle to ensure total lysis. Eleven milligrams of total protein was loaded onto the Ni+ column equilibrated with denaturing binding buffer (8 M urea, 20 mM NaPO4, 500 mM NaCl [pH 7.8]) and allowed to bind for 1 h at 4°C. The column was washed with denaturing binding buffer (pH 7.8 or 6 or 5.6) (two washes for each pH; 4 ml in each wash).
After SF9 proteins were bound under denaturing conditions, the Ni+ column was equilibrated in native binding buffer (see above) and blocked for 30 min with 0.01% bovine serum albumin (BSA) in native binding buffer at 4°C. SK-N-SH cells were lysed in NP-40 lysis buffer, and 3 mg of protein was applied to the column, which was incubated for 1 h at 4°C. Finally, native wash buffer was applied to the column (three 10-ml washes).
In all cases, the contents of the column were analyzed by boiling the beads in sodium dodecyl sulfate (SDS) reducing sample buffer, and the released proteins were loaded on a 12% SDS-polyacrylamide gel electrophoresis (PAGE) gel. After electrophoresis, proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore), and Western blotting was performed using an anti-C/EBP-α specific antibody (14AA; Santa Cruz Biotechnology).
SDS-PAGE and Western blotting.
Standard 12% SDS-PAGE gels were prepared and used for these studies. Proteins were transferred to PVDF membrane (Millipore), blocked for 1 h by using 5% nonfat dry milk with phosphate-buffered saline-0.05% Tween 20 (PBS-T), and incubated with primary antibody overnight at 4°C. The C/EBP-α antibody was diluted 1:500 in the blocking solution. After 30 min of washing with PBS-T, the blots were incubated with donkey anti-rabbit horseradish peroxidase-conjugated immunoglobulin G (Amersham Biosciences), which was diluted 1:1,000 in nonfat dry milk with PBS-T. Blots were washed for 30 min with PBS-T and exposed to Amersham ECL reagents, and then autoradiography was performed.
Immunohistochemistry.
wt-infected calves were euthanized, and their TG were fixed in neutral buffered formalin and then embedded in paraffin. Thin sections (4 to 5 um) were cut and mounted onto slides. Tissue sections were incubated for 20 min at 65°C, followed by two incubations of 10 min in xylene and rehydration in graded alcohols. Tissue sections were then incubated with 3% hydrogen peroxide in PBS (pH 7.4) for 20 min at room temperature to block endogenous peroxidase. After three washes in Tris-buffered saline (TBS; 5 min each) at room temperature, tissue sections were digested with 0.05% protease XIV (P-5147; Sigma) in Tris buffer (0.05 M Tris, pH 7.6) for 20 min at 37°C. Tissue sections were then blocked with 5% normal goat serum diluted in TBS containing 0.25% BSA for 45 min at room temperature in a humidified chamber.
Primary anti-C/EBP-α antibody (sc-14AA; Santa Cruz Biotechnology) was used at a 1:250 dilution, incubated overnight in a humidified chamber at 4°C, and washed in TBS (pH 7.6) the next day. Biotinylated goat anti-rabbit immunoglobulin G (Vector Labs) was then incubated with the section for 30 min at room temperature in a humidified chamber. Next, the avidin biotinylated enzyme complex was added to slides for 30 min at room temperature in a humidified chamber. After three washes in TBS, slides were incubated with freshly prepared substrate (Vector NovaRed substrate kit for peroxidase; Vector Labs), rinsed with distilled water, and counterstained with hematoxylin. Thin sections from mock-infected calves were used as a negative control.
RNA extraction.
Extraction of RNA was performed as described by Chomczynski and Sacchi (9). Briefly, TG were minced into small pieces, placed into 10 ml of solution D (4 M guanidine thiocyanate, 25 mM sodium citrate [pH 7.0], 0.5% sarcosyl, 14 mM β-mercaptoethanol), and homogenized. After two phenol extractions were performed, RNA concentrations were determined spectrophotometrically (260 nm) and RNA was reprecipitated in 3 volumes of ethanol.
RNA was prepared from MDBK cells using TRIzol reagent as described previously (49, 50).
DNase I treatment and reverse transcription.
Three micrograms of total RNA was treated with 1 U of DNase I (Invitrogen) for 15 min at room temperature in the presence of a RNase inhibitor (RNAsin; Promega, Madison, WI). After DNase I treatment, samples were incubated at 65°C for 7.5 min in the presence of 2 mM EDTA to eliminate DNase I activity. One microgram of total RNA (DNase I treated) plus 1 μg of random hexamers were incubated at 65°C for 7.5 min and then chilled on ice. Sixteen microliters of ice-cold reverse transcription mix (20 mM Tris-HCl [pH 8.3], 50 mM KCl, 2.5 mM MgCl2), 100 μg BSA/ml, 1 mM dithiothreitol, 0.5 mM (each) deoxynucleoside triphosphates, 10 U of RNasin, and 100 U of reverse transcriptase (Invitrogen) was added. The reaction mixture was incubated for 10 min at 25°C and then for 50 min at 42°C. As a control for DNA contamination in RNA samples, 2 μg of RNA (DNase I treated) was mixed with ice-cold reverse transcription mix lacking RT.
PCRs.
An aliquot (2 μl) of the reverse transcription reaction mixture was used for each PCR using primers specific for bovine C/EBP-α (+AGTCCGTGGACAAGAACAGC and −GGTCATTGTCACTGGTCAGC). Bovine growth hormone (GH) primers (+GCTTTCGCCCTGCTCTGCC and −TCCTGCCTCCCCACCCCTA) were used as an internal control. PCRs were carried out in 50-μl reactions containing 5 mM MgCl2, 200 μM concentrations of each deoxynucleoside triphosphate, 1 μM concentrations of each primer, and 1 U Taq polymerase. Five microliters of GC melt (Advantage GC-2; BD Biosciences) was then added to the reaction mixture. Amplification was carried out with 37 cycles of denaturing at 95°C for 45 s, annealing at 53°C for 45 s, and extending at 72°C for 1 min. Upon completion of the last cycle, reactions were further incubated at 72°C for 10 min to ensure complete extension of the amplified product. PCR products were electrophoresed by using a 2% agarose gel.
Cell lysis and protein quantitation.
MDBK or SK-N-SH whole-cell lysate was prepared using NP-40 lysis buffer (1% NP-40, 50 mM Tris-HCl [pH 8], 150 mM NaCl). The NP-40 lysis buffer also contained 1 tablet of protease inhibitor cocktail (11 8361 53001; Roche) for each 10 ml of buffer. Protein concentrations were measured using a Bradford reagent kit (500-0006; Bio-Rad).
Analysis of C/EBP-α expression on plaque formation.
The gCblue virus grows to titers similar to those for wt BHV-1 and was grown in MDBK cells. Procedures for preparing BHV-1 genomic DNA have been described previously (18, 30-32). 9.1.3 cells were cotransfected with the gCblue viral DNA and one of the plasmids encoding bICP0, C/EBP-α, C/EBP-αΔDB, or empty expression vector (pcDNA3.1−) using Lipofectamine 2000 (Invitrogen). β-Galactosidase-positive (β-Gal+) cells were counted at 24 h after transfection as described previously (18, 30-32). The number of β-Gal+ cells in cultures transfected with pcDNA3.1− and gCblue genomic DNA was set at a value of 1 for each experiment. All other transfections were examined for β-Gal+ cells and are expressed as induction (n-fold) relative to the control. This representation of the data minimizes differences in cell density, variation in Lipofectamine 2000 lots, and transfection efficiency.
RESULTS
Identification of cellular proteins that interact with the LR fusion protein.
The bacterial two-hybrid system was used to identify cellular proteins that bind to the LR fusion protein. A fragment containing the 7d cDNA (11) (Fig. 1) was cloned into the pBT vector such that the major ORF was in frame with λcI coding sequences (pBT7day). Since BHV-1 can replicate in cottontail rats (48), a commercially available rat brain cDNA library was used as target sequences. Cotransformation of E. coli with the pBT7day and pTRG plasmids resulted in several positive colonies. Rat cDNA inserts were sequenced, and BLAST analysis was performed to identify cellular proteins that interacted with the LR fusion protein (Table 1). As expected, we did not obtain positive colonies when bacteria were transformed with pTRG or pBT7day alone.
TABLE 1.
Rat proteins that interact with the LR fusion protein encoded by 7d cDNAa
| Description of protein | GenBank accession no. |
|---|---|
| Apoptosis regulator | |
| BH3 domain from Bid | BC023963 |
| Cdc42 (GTPase) | NM009861 |
| BCR-related gene | BC043673 |
| NADH dehydrogenase 4, mitochondrial | BC020382 |
| Transcription factor | |
| CCAAT enhancer binding protein | BC011319 |
| High mobility group nuclear protein 2 (Hmgn2) | NM016957 |
| Topoisomerase 1 binding protein (Btbd2) | XM354550 |
| Other | |
| Elongation factor 1-alpha 1 homologue | AK088041 |
| Ribosomal phosphoprotein P0 | BC003833 |
| Ubiquitin C | NM019639 |
| B-cell and monocyte-activating chemokine precursor | AF144754 |
| RIKEN cDNA 2610510H01 gene | BC005803 |
| RIKEN cDNA D130012J15 gene | NM172271 |
Procedures for performing the BacterioMatch II two-hybrid screening are described in Materials and Methods.
Two cellular genes that encode proapoptotic proteins (the BH3 domain of Bid and Cdc42) and two other genes that encode proteins with potential roles in apoptosis (NADH dehydrogenase 4 and a BCR-related gene) were identified in the two-hybrid screening (Table 1). The Bcr protein is a GTPase-activating protein (14), suggesting that it activates the GTPase activity of Cdc42. Since the LR gene can inhibit apoptosis in cultured cells (10, 25) and TG of infected calves (43), we expected that LR proteins would interact with proapoptotic proteins. Determining the significance of the other binding proteins will require additional studies. We began by examining the significance of the LR fusion protein interacting with C/EBP-α.
The LR fusion protein interacts with human and insect C/EBP-α.
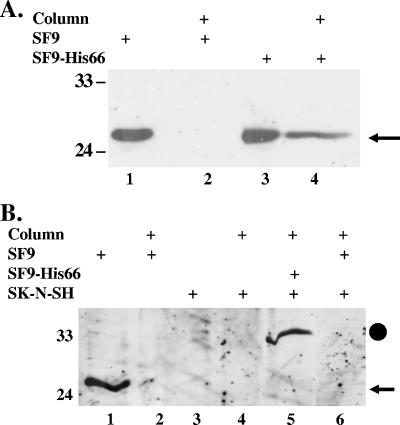
The interaction between C/EBP-α and the BHV-1 LR fusion protein (Table 1) was of particular interest because Epstein-Barr virus (EBV), human herpesvirus 8 (HHV-8), and herpesvirus saimiri (HVS) productive infections are stimulated by C/EBP-α (28, 60, 62, 67). To confirm the interaction between the LR fusion protein and C/EBP-α, a recombinant baculovirus that expresses the LR fusion protein as a histidine-tagged protein (recombinant baculovirus His66) (33) was used. Because mammalian C/EBP-α shares significant homology with the Drosophila melanogaster homolog (64), we tested whether the insect C/EBP-α homolog was bound to the His-tagged LR fusion protein. SF9 insect cells were infected with the recombinant baculovirus His66, and infected cells were lysed and then loaded onto a Ni+ affinity column using native conditions to preserve protein-protein interactions. A Western blot analysis of proteins bound to the column demonstrated that insect C/EBP-α was retained in the Ni+ column only when SF9 cells were infected with the recombinant baculovirus His66 (Fig. 2A, lane 4). We also noted that the insect C/EBP-α (approximately 27 kDa) (Fig. 2A) was smaller than the human homolog (approximately 33 kDa) (Fig. 2B).
FIG. 2.
The LR fusion protein binds to the C/EBP-α protein expressed in SF9 cells and human neuroblastoma cells (SK-N-SH). (A) Eleven milligrams of cell lysate prepared from uninfected SF9 cells (SF9) or SF9 cells infected with the His66 baculovirus (SF9-His66) were bound to the Ni+ column using native conditions. After the column was washed, the column beads were boiled in SDS reducing buffer and the bound proteins electrophoresed by 12% SDS-PAGE. Lanes 1 and 3 contain 250 μg of the indicated whole-cell lysate. Proteins were transferred to a PVDF membrane and probed with anti-C/EBP-α antibody that was diluted 1:500. The arrow denotes the position of the insect C/EBP-α homolog (approximately 27 kDa). Lanes 2 and 4 contain proteins bound to the Ni+ column. (B) Eleven milligrams of uninfected (SF9) or infected (SF9-His66) insect cell lysate was prepared using denaturing conditions (see Materials and Methods), and the protein was then applied to a Ni+ column. Unbound proteins were removed by washing the column. Three milligrams of cell lysate prepared from SK-N-SH cells was then incubated for 1 h with the Ni+ column. Unbound protein was removed by extensive washing as described in Materials and Methods. Column beads were boiled in SDS reducing buffer, and proteins were electrophoresed by 12% SDS-PAGE, transferred to a PVDF membrane, and then probed with the anti-C/EBP-α antibody diluted 1:500. The closed circle denotes the ∼33-kDa C/EBP-α detected with SK-N-SH cells, and the arrow denotes the position of the insect C/EBP-α homolog. Lanes 1 and 3 contain 250 μg of the indicated cell lysate. Lanes 2, 4, 5, and 6 contain proteins that were bound to the Ni+ column. Molecular weight markers are in kilodaltons.
To test whether the LR fusion protein interacted with mammalian C/EBP-α, the recombinant LR fusion protein was bound to the Ni+ column under denaturing conditions. Whole-cell extract prepared from SK-N-SH cells was subsequently incubated with the LR fusion protein that was immobilized on the Ni+ column. Under denaturing conditions, the insect C/EBP-α protein was not retained in the Ni+ column (Fig. 2B, lane 2). When SK-N-SH cell extract was incubated with the column containing the LR fusion protein, a band of the expected size of human C/EBP-α (approximately 33 kDa) was retained in the Ni+ column and was recognized by the C/EBP-α antibody (Fig. 2B, lane 5). In contrast, when only SF9 cell extract was incubated with the column, human C/EBP-α was not bound to the column (Fig. 2B, lane 6). As expected, the human C/EBP-α protein was not retained on the Ni+ column lacking the LR fusion protein (Fig. 2B, lane 4). In summary, these results demonstrated that the LR fusion protein interacted with human or insect C/EBP-α protein.
Analysis of C/EBP-α protein expression following infection of bovine cells.
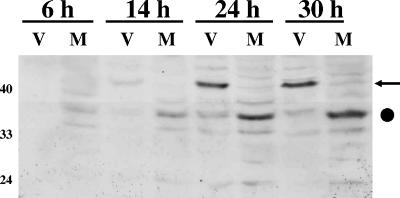
Since the LR fusion protein interacted with the C/EBP-α protein, we tested whether C/EBP-α protein levels were altered during viral infection. During the course of productive infection, a prominent 41-kDa protein in infected cells was induced; it was recognized by the C/EBP-α antibody (Fig. 3, lanes V). This 41-kDa protein is the approximate size of the long translational form of C/EBP-α (41, 47). The levels of a 33-kDa band recognized by the C/EBP-α antibody increased in mock-infected MDBK cells as a function of time after plating (Fig. 3, lanes M). Induction of the 41-kDa protein recognized by the C/EBP-α antibody was detected at 24 h after infection when cells were infected at a high (5) or low (0.01) multiplicity of infection (Fig. 3 and data not shown). In summary, these studies suggested that BHV-1 infection of cultured cells led to induction of the long form of C/EBP-α.
FIG. 3.
Expression of C/EBP-α during productive infection. MDBK cells were infected with wt BHV-1 (V) using a multiplicity of infection of 5, and whole-cell lysate was collected at 6, 14, 24, or 30 h after infection. M, mock-infected cells. Cells were lysed with NP-40 lysis buffer, and 250 μg of protein was electrophoresed by 12% SDS-PAGE. Proteins in the gel were transferred onto a PVDF membrane and probed with anti-C/EBP-α that was diluted 1:500. The arrow denotes the 42-kDa isoform of C/EBP-α, and the closed circle denotes a 35-kDa band that was recognized by the antibody. Molecular weight markers are in kilodaltons.
Analysis of C/EBP-α expression in TG of infected calves.
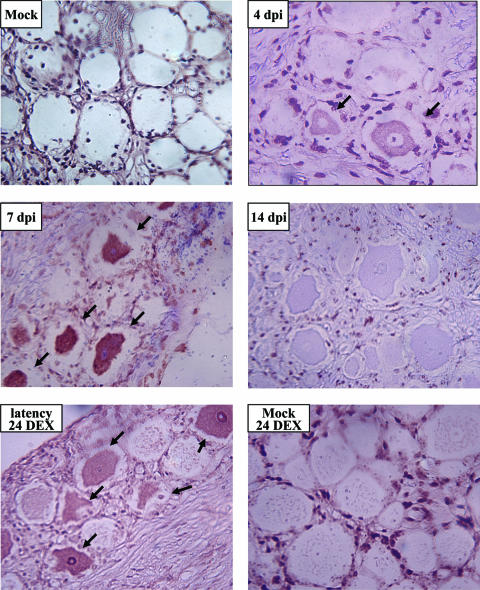
To examine C/EBP-α protein expression in TG of infected calves, immunohistochemistry was performed using the C/EBP-α specific antibody. C/EBP-α protein expression was readily detected in TG neurons of calves that were infected for 4 or 7 days (Fig. 4). At 4 days after infection, C/EBP-α protein expression was primarily localized to the perinuclear region, whereas at 7 days after infection, C/EBP-α was detected in the nucleus of certain neurons. In contrast, C/EBP-α protein expression was not detected in TG neurons of an uninfected calf (Fig. 4). However, a few nonneuronal cells in all calves, including mock-infected calves, expressed C/EBP-α. At 14 days after infection, there appeared to be slight neuronal staining (Fig. 4), which was not observed in mock-infected calves or at 60 days after infection (latency) (data not shown). Calves latently infected with BHV-1 undergo reactivation from latency following administration of DEX, a synthetic corticosteroid (54). At 24 (Fig. 4) or 48 (data not shown) h after a single dose of DEX, C/EBP-α protein expression was readily detected in a subset of TG neurons. In contrast, when a mock-infected calf was administered the same dose of DEX for 24 h, C/EBP-α protein expression was not detected in TG neurons (Fig. 4).
FIG. 4.
Analysis of C/EBP-α protein expression in TG of BHV-1-infected calves. At the indicated times after infection, thin sections of TG were prepared and stained with the anti-C/EBP-α antibody as described in Materials and Methods. Arrows denote C/EBP-α positive neurons. Magnification, ×400. dpi, days postinfection; 24 DEX, 24 h after DEX treatment.
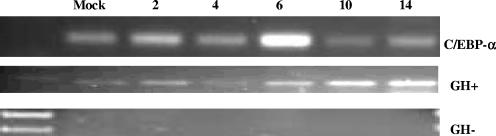
RT-PCR was performed on TG samples of calves infected with wt BHV-1 or left uninfected. C/EBP-α, but not GH, RNA levels were higher at 6 days after infection than at 10 or 14 days after infection (Fig. 5). Although C/EBP-α RNA was detected in mock-infected calves (Fig. 5), we assume expression occurred in nonneuronal cells or C/EBP-α was unstable in neurons, because it was not detected in TG neurons prepared from mock-infected calves (Fig. 4). In summary, these studies indicated that C/EBP-α protein expression was detected in TG neurons of acutely infected calves or following DEX-induced reactivation from latency.
FIG. 5.
Detection of C/EBP-α RNA in TG of BHV-1-infected cattle. RT-PCR for C/EBP-α was performed using RNA prepared from TG on the indicated days after infection or from a mock-infected calf (Mock). RNA extraction, RT-PCR, and PCR were performed as described in Materials and Methods. The cDNA for the respective samples was generated with random primers. Bovine GH was used as an internal control for cDNA amplification and DNA contamination. Positive (GH+) and negative (GH−) RT-PCRs (without reverse transcriptase) are shown. The first lane contains a 100-bp DNA ladder (New England BioLabs).
wt C/EBP-α, but not a DNA binding mutant, increases plaque formation.
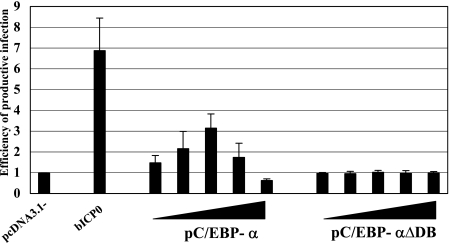
Since C/EBP-α stimulates HHV-8, HVS, or EBV gene expression and productive infection (28, 60, 62, 67), we hypothesized that C/EBP-α may enhance BHV-1 productive infection. To test whether the overexpression of C/EBP-α enhanced plaque formation, bovine cells were cotransfected with gCblue genomic DNA and increasing concentrations of wt pC/EBP-α or a DNA binding mutant (pC/EBP-αΔDB) (Fig. 6). Transfection of 9.1.3 cells with BHV-1 genomic DNA yields low levels of infectious virus. Cotransfection of BHV-1 DNA with a plasmid expressing bICP0 enhances productive infection and virus yield (Fig. 6) (32). At 24 h after transfection, cells were fixed and β-Gal+ cells identified. This time point was used as the time to count β-Gal+ cells to minimize the number of virus-positive cells that result from virus spread. Furthermore, at later times, many of the β-Gal+ cells lift off the dish, making it difficult to count virus-positive cells (18, 32). The number of β-Gal+ cells directly correlates with the number of plaques produced following transfection with the BHV-1 blue virus (17, 18, 32). When a 16:1 molar ratio of pC/EBP-α to genomic gCblue viral DNA was cotransfected into 9.1.3 cells, a threefold increase in plaques was observed relative to pC/EBP-αΔDB or the blank expression vector (Fig. 6). At high concentrations of pC/EBP-α, the number of plaques was slightly lower than the numbers obtained with the empty expression vector or with equivalent amounts of pC/EBP-αΔDB. pC/EBP-α was less efficient than bICP0 at activating productive infection, even when transfected at four times the level of bICP0.
FIG. 6.
The DNA binding domain of C/EBP-α is necessary to enhance productive infection. Bovine cells (9.1.3) were cotransfected with increasing amounts of pC/EBP-α or pC/EBP-αΔDB (13 ng, 52 ng, 0.21 μg, 0.83 μg, or 3.32 μg) or bICP0 (52 ng), and the gCblue virus genome (0.83 μg DNA). Ratios of pC/EBP-α or pC/EBP-αΔDB DNA molecules to viral genomic DNA molecules are 1:1, 4:1, 16:1, 64:1, or 256:1, respectively. The ratio between bICP0 molecules and viral DNA molecules is 4:1. An empty expression vector (pcDNA3.1−) was used to maintain equivalent amounts of DNA. At 24 h after transfection, cells were fixed and stained, and the number of β-Gal+ cells was counted. The number of β-Gal+ cells in the vector control (pcDNA3.1−) was set at onefold. The number of β-Gal+ cells in each well was calculated as the change (n-fold) compared to the vector control. The results are the averages from three independent experiments.
DISCUSSION
In this study, we identified cellular proteins that interact with the major ORF encoded by an alternatively spliced polyadenylated LR transcript that is abundantly expressed in TG of calves at 7 days after infection (11, 12). Seven days after infection is important with respect to establishing latency in calves, because after this time most viral gene expression is extinguished in TG (57). Only the LR gene (40) and ORF-E (29) are abundantly expressed after 7 days postinfection. Therefore, we predicted that LR proteins, in part, regulate the transition between productive infection and establishment of latency.
The ability of the LR gene (10, 25) and products encoded by the 7d cDNA (data not shown) to inhibit apoptosis correlates with the LR gene being necessary for reactivation from latency after DEX treatment (31). During the transition from acute infection to establishment of latency, an LR mutant virus induces higher levels of apoptosis in TG of infected calves, which reduces establishment from latency (43). Furthermore, insertion of the wt LR gene (52), but not the LR gene containing stop codons at the 5′ terminus of ORF-2 (45), into a herpes simplex virus type 1 latency-associated transcript minus mutant virus restores high levels of spontaneous reactivation in the rabbit eye model and explant-induced reactivation. Finally, the ability of herpes simplex virus type 1 latency-associated transcript to inhibit apoptosis is crucial for wt levels of reactivation from latency (34). Taken together, these observations suggest that the ability of the LR gene to inhibit apoptosis is one function that promotes the establishment of latency in a pool of neurons that can subsequently reactivate from latency.
The results from this study suggested that the antiapoptotic functions of the LR gene rely on the ability of LR proteins to interact with Bid or Cdc42. Bid links the extrinsic pathway of apoptosis to the intrinsic pathway; following cleavage by caspase 8, Bid interacts with mitochondria (42), inducing cytochrome C and Smac/Diablo release (63). Bid is also specifically cleaved by granzyme B (3, 4, 53), suggesting that the ability of LR proteins to interact with Bid impairs cytotoxic T-lymphocyte-induced death of infected neurons. Neuronal stress, including removal of nerve growth factor from neuronal cultures (39, 68), induces activity of Cdc42, a Rho GTPase family member (24). When PC12 cells (rat neuron-like cell line) are treated with nerve growth factor, they differentiate and behave like mature sympathetic neurons (19, 20). If PC12 cells or primary sympathetic neurons are deprived of nerve growth factor, Cdc42 is activated, resulting in apoptosis (39, 68). Since DEX reproducibly induces BHV-1 reactivation from latency (1, 2, 7, 26, 37, 54, 58, 66) and DEX is a potent stressor, Cdc42 may be activated in TG.
C/EBPs belong to the bZIP (basic and leucine zipper domain) family of transcription factors (64). The D. melanogaster genome contains 27 bZIP genes, of which 21 have a mammalian counterpart (15). Two of these bZIP proteins have homology with human C/EBP-α outside the bZIP region, showing 76% and 36% identity in the basic region and 23% and 28% identity in the leucine zipper (15). The C/EBP-α mRNA is translated into a 30- or 42-kDa protein, because there is an in-frame ATG codon downstream of the first ATG and both ATGs can be used as start codons. The shorter form of C/EBP-α retains dimerization and DNA binding domains but possesses altered trans-activation potential (41), because the N-terminal domain contains trans-activating domains (41, 47). C/EBP-α regulates cell cycle regulation and differentiation (41), suggesting that interactions with the LR fusion protein helps neurons recover from infection.
The interaction between the LR fusion protein and C/EBP-α may also influence viral gene expression, because C/EBP-α stimulates lytic gene transcription of EBV, HHV-8, and HVS (28, 60, 62, 67). C/EBP-α binds to the major lytic viral transactivators of EBV and HHV-8, Zta and Rta proteins, respectively. Binding of viral activators stabilizes C/EBP-α, and the protein complex directly binds to viral IE promoters to activate viral transcription (28, 62, 67). Several lines of evidence suggested that C/EBP-α enhanced BHV-1 productive infection. First, a BHV-1-encoded or -induced product led to the induction of the full-length C/EBP-α isoform, or C/EBP-α was posttranslationally modified (Fig. 3). Secondly, there was a positive correlation between viral gene expression in TG and C/EBP-α protein expression (Fig. 4) because C/EBP-α was detected in TG neurons at times (4 and 7 days after infection) when viral gene expression (57) and infectious virus (31) are detected. Finally, wt C/EBP-α, but not a DNA binding mutant, enhanced BHV-1 plaque formation (Fig. 6). These studies suggested that C/EBP-α activates certain key BHV-1 promoters, directly or indirectly, which is consistent with the role that C/EBP-α plays in EBV, HHV8, or HVS productive infection. We hypothesize that, in addition to inhibiting apoptosis, interactions between the LR fusion protein and C/EBP-α repress productive infection, thus promoting the establishment of latency in neurons that can support reactivation from latency. Studies designed to test these predictions are in progress.
Acknowledgments
This work was supported by USDA grants (2002-35204, 2006-01627, and 2005-01554) and a Public Health Service grant (1P20RR15635) (C.J.). F.M. was partially supported by a Maude Hammond Fling fellowship from the University of Nebraska. V.G. was partially supported by a NIH Ruth L. Kirschstein fellowship (T32 AI060547).
Footnotes
Published ahead of print on 20 September 2006.
REFERENCES
- 1.Ackermann, M., E. Peterhans, and R. Wyler. 1982. DNA of bovine herpesvirus type 1 in the trigeminal ganglia of latently infected calves. Am. J. Vet. Res. 43:36-40. [PubMed] [Google Scholar]
- 2.Ackermann, M., and R. Wyler. 1984. The DNA of an IPV strain of bovid herpesvirus 1 in sacral ganglia during latency after intravaginal infection. Vet. Microbiol. 9:53-63. [DOI] [PubMed] [Google Scholar]
- 3.Alimonti, J. B., L. Shi, P. K. Baijal, and A. H. Greenberg. 2001. Granzyme B induces BID-mediated cytochrome c release and mitochondrial permeability transition. J. Biol. Chem. 276:6974-6982. [DOI] [PubMed] [Google Scholar]
- 4.Barry, M., J. A. Heibein, M. J. Pinkoski, S. F. Lee, R. W. Moyer, D. R. Green, and R. C. Bleackley. 2000. Granzyme B short-circuits the need for caspase 8 activity during granule-mediated cytotoxic T-lymphocyte killing by directly cleaving Bid. Mol. Cell. Biol. 20:3781-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowland, S. L., and P. E. Shewen. 2000. Bovine respiratory disease: commercial vaccines currently available in Canada. Can. Vet. J. 41:33-48. [PMC free article] [PubMed] [Google Scholar]
- 6.Bratanich, A. C., N. D. Hanson, and C. Jones. 1992. The latency-related gene of bovine herpesvirus 1 inhibits the activity of immediate-early transcription unit 1. Virology 191:988-991. [DOI] [PubMed] [Google Scholar]
- 7.Brown, G. A., and H. J. Field. 1990. Experimental reactivation of bovine herpesvirus 1 (BHV-1) by means of corticosteroids in an intranasal rabbit model. Arch. Virol. 112:81-101. [DOI] [PubMed] [Google Scholar]
- 8.Carter, J. J., A. D. Weinberg, A. Pollard, R. Reeves, J. A. Magnuson, and N. S. Magnuson. 1989. Inhibition of T-lymphocyte mitogenic responses and effects on cell functions by bovine herpesvirus 1. J. Virol. 63:1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski, P., and N. Sacchi. 1987. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 10.Ciacci-Zanella, J., M. Stone, G. Henderson, and C. Jones. 1999. The latency-related gene of bovine herpesvirus 1 inhibits programmed cell death. J. Virol. 73:9734-9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devireddy, L., Zhang, Y., and C. Jones. 2003. Cloning and initial characterization of an alternatively spliced transcript encoded by the bovine herpes virus 1 latency-related gene. J. Neurovirol. 9:612-622. [DOI] [PubMed] [Google Scholar]
- 12.Devireddy, L. R., and C. Jones. 1998. Alternative splicing of the latency-related transcript of bovine herpesvirus 1 yields RNAs containing unique open reading frames. J. Virol. 72:7294-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devireddy, L. R., and C. J. Jones. 1999. Activation of caspases and p53 by bovine herpesvirus 1 infection results in programmed cell death and efficient virus release. J. Virol. 73:3778-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diekmann, D., S. Brill, M. D. Garrett, N. Totty, J. Hsuan, C. Monfries, C. Hall, L. Lim, and A. Hall. 1991. Bcr encodes a GTPase-activating protein for p21rac. Nature 351:400-402. [DOI] [PubMed] [Google Scholar]
- 15.Fassler, J., D. Landsman, A. Acharya, J. R. Moll, M. Bonovich, and C. Vinson. 2002. B-ZIP proteins encoded by the Drosophila genome: evaluation of potential dimerization partners. Genome Res. 12:1190-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geiser, V., and C. Jones. 2005. Localization of sequences within the latency-related gene of bovine herpesvirus 1 that inhibit mammalian cell growth. J. Neurovirol. 11:563-570. [DOI] [PubMed] [Google Scholar]
- 17.Geiser, V., and C. Jones. 2003. Stimulation of bovine herpesvirus 1 productive infection by the adenovirus E1A gene and a cell cycle regulatory gene, E2F-4. J. Gen. Virol. 84:929-938. [DOI] [PubMed] [Google Scholar]
- 18.Geiser, V., M. Inman, Y. Zhang, and C. Jones. 2002. The latency related gene of bovine herpes virus-1 can inhibit the ability of bICP0 to activate productive infection. J. Gen. Virol. 83:2965-2971. [DOI] [PubMed] [Google Scholar]
- 19.Greene, L. A., J. M. Aletta, A. Rukenstein, and S. H. Green. 1987. PC12 pheochromocytoma cells: culture, nerve growth factor treatment, and experimental exploitation. Methods Enzymol. 147:207-216. [DOI] [PubMed] [Google Scholar]
- 20.Greene, L. A., and A. S. Tischler. 1976. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. USA 73:2424-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griebel, P. J., H. B. Ohmann, M. J. Lawman, and L. A. Babiuk. 1990. The interaction between bovine herpesvirus type 1 and activated bovine T lymphocytes. J. Gen. Virol. 71:369-377. [DOI] [PubMed] [Google Scholar]
- 22.Griebel, P. J., L. Qualtiere, W. C. Davis, A. Gee, H. Bielefeldt Ohmann, M. J. Lawman, and L. A. Babiuk. 1987. T lymphocyte population dynamics and function following a primary bovine herpesvirus type-1 infection. Viral Immunol. 1:287-304. [DOI] [PubMed] [Google Scholar]
- 23.Griebel, P. J., L. Qualtiere, W. C. Davis, M. J. Lawman, and L. A. Babiuk. 1988 1987. Bovine peripheral blood leukocyte subpopulation dynamics following a primary bovine herpesvirus-1 infection. Viral. Immunol. 1:267-286. [DOI] [PubMed] [Google Scholar]
- 24.Harwood, A., and V. M. M. Brage. 2003. Cdc42 and GSK-3: signals at the crossroads. Nat. Cell Biol. 5:275-277. [DOI] [PubMed] [Google Scholar]
- 25.Henderson, G., G.-C. Perng, A. Nesburn, S. Wechsler, and C. Jones. 2004. The latency-related gene encoded by bovine herpesvirus 1 can suppress caspase 3 and caspase 9 during productive infection. J. Neurovirol. 10:64-70. [DOI] [PubMed] [Google Scholar]
- 26.Homan, E. J., and B. C. Easterday. 1983. Experimental latent and recrudescent bovine herpesvirus-1 infections in calves. Am. J. Vet. Res. 44:309-313. [PubMed] [Google Scholar]
- 27.Hossain, A., L. M. Schang, and C. Jones. 1995. Identification of gene products encoded by the latency-related gene of bovine herpesvirus 1. J. Virol. 69:5345-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, J., G. Liao, H. Chen, F. Y. Wu, L. Hutt-Fletcher, G. S. Hayward, and S. D. Hayward. 2006. Contribution of C/EBP proteins to Epstein-Barr virus lytic gene expression and replication in epithelial cells. J. Virol. 80:1098-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inman, M., J. Zhou, H. Webb, and C. Jones. 2004. Identification of a novel bovine herpesvirus 1 transcript containing a small open reading frame that is expressed in trigeminal ganglia of latently infected cattle. J. Virol. 78:5438-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inman, M., L. Lovato, A. Doster, and C. Jones. 2001. A mutation in the latency-related gene of bovine herpesvirus 1 leads to impaired ocular shedding in acutely infected calves. J. Virol. 75:8507-8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inman, M., L. Lovato, A. Doster, C. Jones. 2002. A mutation in the latency-related gene of bovine herpesvirus 1 disrupts the latency-reactivation cycle in calves. J. Virol. 76:6771-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inman, M., Y. Zhang, V. Geiser, and C. Jones. 2001. The zinc ring finger in the bICP0 protein encoded by bovine herpesvirus-1 mediates toxicity and activates productive infection. J. Gen. Virol. 82:483-492. [DOI] [PubMed] [Google Scholar]
- 33.Jiang, Y., M. Inman, Y. Zhang, N. A. Posadas, and C. Jones. 2004. A mutation in the latency-related gene of bovine herpesvirus 1 inhibits protein expression of a protein from open reading frame 2 and an adjacent reading frame during productive infection. J. Virol. 78:3184-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin, L., G.-C. Perng, A. B. Nesburn, C. Jones, and S. L. Wechsler. 2005. A herpes simplex virus type 1 mutant expressing a baculovirus inhibitor of apoptosis gene in place of latency-associated transcript has a wild-type reactivation phenotype in the mouse. J. Virol. 79:12286-12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones, C. 1998. Alphaherpesvirus latency: its role in disease and survival of the virus in nature. Adv. Virus Res. 51:81-133. [DOI] [PubMed] [Google Scholar]
- 36.Jones, C. 2003. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin. Microbiol. Rev. 16:79-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones, C., T. J. Newby, T. Holt, A. Doster, M. Stone, J. Ciacci-Zanella, C. J. Webster, and M. W. Jackwood. 2000. Analysis of latency in cattle after inoculation with a temperature sensitive mutant of bovine herpesvirus 1 (RLB106). Vaccine 18:3185-3195. [DOI] [PubMed] [Google Scholar]
- 38.Jones, C., V. Geiser, G. Henderson, Y. Jiang, F. Meyer, S. Perez, and Y. Zhang. 2006. Functional analysis of bovine herpesvirus 1 (BHV-1) genes expressed during latency. Vet. Microbiol. 113:199-210. [DOI] [PubMed] [Google Scholar]
- 39.Kanamoto, T., M. Mota, K. Takeda, L. L. Rubin, K. Miyazono, H. Ichijo, and C. E. Bazenet. 2000. Role of apoptosis signal-regulating kinase in regulation of the c-Jun N-terminal kinase pathway and apoptosis in sympathetic neurons. Mol. Cell. Biol. 20:196-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kutish, G., T. Mainprize, and D. Rock. 1990. Characterization of the latency-related transcriptionally active region of the bovine herpesvirus 1 genome. J. Virol. 64:5730-5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lekstrom-Himes, J., and K. G. Xanthopoulos. 1998. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biol. Chem. 273:28545-28548. [DOI] [PubMed] [Google Scholar]
- 42.Li, H., H. Zhu, C. J. Xu, and J. Yuan. 1998. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94:491-501. [DOI] [PubMed] [Google Scholar]
- 43.Lovato, L., M. Inman, G. Henderson, A. Doster, and C. Jones. 2003. Infection of cattle with a bovine herpesvirus 1 strain that contains a mutation in the latency related gene leads to increased apoptosis in trigeminal ganglia during the transition from acute infection to latency. J. Virol. 77:4848-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Misra, V., S. Walker, S. Hayes, and P. O'Hare. 1995. The bovine herpesvirus alpha gene trans-inducing factor activates transcription by mechanisms different from those of its herpes simplex virus type 1 counterpart VP16. J. Virol. 69:5209-5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mott, K., N. Osorio, L. Jin, D. Brick, J. Naito, J. Cooper, G. Henderson, M. Inman, C. Jones, S. L. Wechsler, and G.-C. Perng. 2003. The bovine herpesvirus-1 LR ORF2 is crucial for this gene's ability to restore the high reactivation phenotype to a herpes simplex virus-1 LAT null mutant. J. Gen. Virol. 84:2975-2985. [DOI] [PubMed] [Google Scholar]
- 46.National Agricultural Statistics Service. 1996. Effect of bovine respiratory disease complex on the beef cattle industry. Agricultural Statistics Board, U.S. Department of Agriculture, Washington, DC.
- 47.Nerlov, C., and E. B. Ziff. 1995. CCAAT/enhancer binding protein-α amino acid motifs with dual TBP and TFIIE binding ability co-operate to activate transcription in both yeast and mammalian cells. EMBO J. 14:4318-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Papp, Z., D. M. Middleton, S. K. Mittal, L. A. Babiuk, and M. E. Baca-Estrada. 1997. Mucosal immunization with recombinant adenoviruses: induction of immunity and protection of cotton rats against respiratory bovine herpesvirus type 1 infection. J. Gen. Virol. 78:2933-2943. [DOI] [PubMed] [Google Scholar]
- 49.Peng, W., G. Henderson, G.-C. Perng, A. B. Nesburn, S. L. Wechsler, and C. Jones. 2003. The gene that encodes the herpes simplex virus type 1 latency-associated transcript influences the accumulation of transcripts (Bcl-xL and Bcl-xS) that encode apoptotic regulatory proteins. J. Virol. 77:10714-10718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng, W., M. Inman, G. Henderson, S. L. Wechsler, L. BenMohamed, G.-C. Perng, and C. Jones. 2005. The locus encompassing the latency-associated transcript of herpes simplex virus type 1 interferes with and delays interferon expression in productively infected neuroblastoma cells and trigeminal ganglia of acutely infected mice. J. Virol. 79:6162-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perez, S., M. Inman, A. Doster, and C. Jones. 2005. Latency-related gene encoded by bovine herpesvirus 1 promotes virus growth and reactivation from latency in tonsils of infected calves. J. Clin. Microbiol. 43:393-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perng, G.-C., B. Maguen, L. Jin, K. R. Mott, N. Osorio, S. M. Slanina, A. Yukht, H. Ghiasi, A. B. Nesburn, M. Inman, G. Henderson, C. Jones, and S. L. Wechsler. 2002. A gene capable of blocking apoptosis can substitute for the herpes simplex virus type 1 latency-associated transcript gene and restore wild-type reactivation levels. J. Virol. 76:1224-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pinkoski, M. J., N. J. Waterhouse, J. A. Heibein, B. B. Wolf, T. Kuwana, J. C. Goldstein, D. D. Newmeyer, R. C. Bleackley, and D. R. Green. 2001. Granzyme B-mediated apoptosis proceeds predominantly through a Bcl-2-inhibitable mitochondrial pathway. J. Biol. Chem. 276:12060-12067. [DOI] [PubMed] [Google Scholar]
- 54.Rock, D., J. Lokensgard, T. Lewis, and G. Kutish. 1992. Characterization of dexamethasone-induced reactivation of latent bovine herpesvirus 1. J. Virol. 66:2484-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rock, D. L., S. L. Beam, and J. E. Mayfield. 1987. Mapping bovine herpesvirus type 1 latency-related RNA in trigeminal ganglia of latently infected rabbits. J. Virol. 61:3827-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schang, L. M., A. Hossain, and C. Jones. 1996. The latency-related gene of bovine herpesvirus 1 encodes a product which inhibits cell cycle progression. J. Virol. 70:3807-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schang, L. M., and C. Jones. 1997. Analysis of bovine herpesvirus 1 transcripts during a primary infection of trigeminal ganglia of cattle. J. Virol. 71:6786-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sheffy, B. E., and D. H. Davies. 1972. Reactivation of a bovine herpesvirus after corticosteroid treatment. Proc. Soc. Exp. Biol. Med. 140:974-976. [DOI] [PubMed] [Google Scholar]
- 59.Tikoo, S. K., M. Campos, and L. A. Babiuk. 1995. Bovine herpesvirus 1 (BHV-1): biology, pathogenesis, and control. Adv. Virus Res. 45:191-223. [DOI] [PubMed] [Google Scholar]
- 60.Wakenshaw, L., M. S. Walters, and A. Whitehouse. 2005. The herpesvirus saimiri replication and transcription activator acts synergistically with CAAT enhancer binding protein alpha to activate the DNA polymerase promoter. J. Virol. 79:13548-13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, G.-L., and N. A. Timchenko. 2005. Dephosphorylated C/EBP-a accelerates cell proliferation through sequestering retinoblastoma protein. Mol. Cell Biol. 25:1325-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, S. E., F. Y. Wu, Y. Yu, and G. S. Hayward. 2003. CCAAT/enhancer-binding protein-α is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters. J. Virol. 17:9590-9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, X. 2001. The expanding role of mitochondria in apoptosis. Genes Dev. 15:2922-2933.11711427 [Google Scholar]
- 64.Wedel, A., and H. W. LomsZeigler-Heitbrock. 1995. The C/EBP family of transcription factors. Immunobiology 193:171-185. [DOI] [PubMed] [Google Scholar]
- 65.Winkler, M. T., A. Doster, and C. Jones. 1999. Bovine herpesvirus 1 can infect CD4+ T lymphocytes and induce programmed cell death during acute infection of cattle. J. Virol. 73:8657-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winkler, M. T., A. Doster, J. H. Sur, and C. Jones. 2002. Analysis of bovine trigeminal ganglia following infection with bovine herpesvirus 1. Vet. Microbiol. 86:139-155. [DOI] [PubMed] [Google Scholar]
- 67.Wu, F. Y., S. E. Wang, H. Chen, L. Wang, S. D. Hayward, and G. S. Hayward. 2004. CCAAT/enhancer binding protein a binds to the Epstein-Barr Virus (EBV) ZTA protein through oligomeric interactions and contributes to cooperative transcriptional activation of the ZTA promoter through direct binding to the ZII and ZIIB motifs during induction of the EBV lytic cycle. J. Virol. 78:4847-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu, Z., A. C. Maroney, P. Dobrzanski, N. V. Kukekov, and L. A. Greene. 2001. The MLK family mediates c-Jun N-terminal kinase activation in neuronal apoptosis. Mol. Cell. Biol. 21:4713-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]