Abstract
The herpesvirus glycoprotein H (gH) and gL associate to form a heterodimer that plays a central role in virus-driven membrane fusion. When archetypal alpha- or betaherpesviruses lack gL, gH misfolds and progeny virions are noninfectious. In order to define the role that gL plays in gamma-2 herpesvirus infections, we disrupted its coding sequence in murine gammaherpesvirus-68 (MHV-68). MHV-68 lacking gL folded gH into a conformation antigenically distinct from the form that normally predominates on infected cells. gL-deficient virions bound less well than the wild type to epithelial cells and fibroblasts. However, they still incorporated gH and remained infectious. The cell-to-cell spread of gL-deficient viruses was remarkably normal, as was infection, dissemination, and latency establishment in vivo. Viral membrane fusion was therefore gL independent. The major function of gL appeared to be allowing gH to participate in cell binding prior to membrane fusion. This function was most important for the entry of MHV-68 virions into fibroblasts and epithelial cells.
Glycoprotein H (gH) and gL are conserved in all mammalian herpesviruses (8, 18, 20). They associate in the endoplasmic reticulums of infected cells to form a heterodimer that is generally essential for infectivity (11, 17, 26). The gH/gL heterodimer functions primarily to fuse virion and cell membranes during viral entry (11, 12, 26). gL lacks a transmembrane domain or an obvious fusion peptide, so gH is likely to be the key mediator of fusion, probably acting in conjunction with gB (7, 23, 32). The only known function for gL is to chaperone gH folding. Nevertheless, gL is essential for the infectivity of herpes simplex virus (33), pseudorabiesvirus (21), and cytomegalovirus (17).
Several herpesviruses also use gH/gL for cell type-specific binding or signaling, often by adding an accessory protein to the heterodimer. Thus, cytomegalovirus uses a gH/gL/gO complex to infect fibroblasts and a gH/gL/UL128/UL130 complex to infect epithelial cells (41). The human herpesvirus 6 gH/gL/gQ fusion complex binds to CD46, whereas its gH/gL/gO fusion complex does not (28). The herpes simplex virus gH/gL—alphaherpesviruses are not known to express more than one fusion complex—binds to αVβ3 integrins (31). Epstein-Barr virus (EBV) uses gH/gL to bind to (26) and penetrate epithelial cells (42). When gp42 is added to the fusion complex it binds to major histocompatibility complex class II glycoproteins, and this is essential for EBV to penetrate B cells (43). Whether gamma-2 herpesviruses express alternative fusion complexes or use gH/gL for cell binding is unknown. There has generally been less analysis of lytic functions for gammaherpesvirus than for alpha- or betaherpesviruses because the human gammaherpesviruses, EBV and the Kaposi's sarcoma-associated herpesvirus (KSHV), cause disease mainly when latent and establish predominantly latent infections in vitro. However, an inextricable link between lytic and latent infections is increasingly apparent in gammaherpesvirus pathogenesis (19). Continual exchange between latent and lytic infection pools is likely (13, 16), and the lytic gene expression found in sites of predominant latency may be an important source of viral immune evasion (37). Thus, the control of gammaherpesvirus infections is likely to require an understanding of their lytic functions.
EBV and KSHV both have narrow species tropisms. The in vivo significance of their in vitro-defined gene functions, therefore, has to remain largely speculative. However, the conservation of genes between different gammaherpesviruses means that it is not always necessary to study EBV and KSHV directly. The discovery of a murid gammaherpesvirus, MHV-68 (3), has consequently opened up gammaherpesviruses in general to experimental pathogenesis (30, 40). Like EBV and KSHV, MHV-68 colonizes epithelial cells and B cells. MHV-68 has only a limited capacity to tell us about the diseases caused by its human counterparts, since these tend to be pathogen specific, but it has enormous potential to tell us how gammaherpesviruses normally function. MHV-68 has been isolated several times from yellow-necked mice. It appears to infect a variety of small rodents, including house mice (25), and behaves like a natural pathogen of inbred, conventional mice, in that it persists without causing disease unless there is immune suppression (39). The information provided by MHV-68 is particularly relevant to our understanding of EBV and KSHV lytic functions, since 90% of MHV-68 lytic genes have obvious homologs in all three viruses. Even when molecular mechanisms differ, the functional parallels remain striking (36).
EBV, KSHV, and MHV-68 all encode gH and gL homologs, although MHV-68 and KSHV lack the EBV gp42. The KSHV gL appears to be required for the correct folding and transport of transfected gH (29). The MHV-68 gL influences gH conformation, but it is not required for transfected gH to reach the cell surface (14). In order to learn more about the role of gL in the context of infection, we disrupted its coding exon (ORF47) in the MHV-68 genome. Surprisingly, gL proved to be dispensable for both virus replication in vitro and host colonization in vivo. MHV-68 lacking gL did not fold gH into its usual virion conformation, and this was associated with reduced binding to some cell types. But once infection had been established, it spread with remarkably normal kinetics.
MATERIALS AND METHODS
Mice.
Female BALB/c and C57BL/6 mice were purchased from Harlan U.K. Ltd. (Bicester, United Kingdom), housed in the Cambridge University Department of Pathology, and infected intranasally with MHV-68 when 6 to 8 weeks old under Home Office Project License 80/1579.
Cell lines.
Baby hamster kidney (BHK-21) fibroblasts, CHO-K1 cells, NIH 3T3-CRE fibroblasts (38), Vero cells, NS0 cells, NMuMG epithelial cells, MCCD epithelial cells, and murine embryonic fibroblasts (MEFs) were grown in Dulbecco's modified Eagle medium (Invitrogen, Paisley, United Kingdom) supplemented with 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum (PAA Laboratories, Linz, Austria). Medium for MEFs was further supplemented with 50 μM 2-mercaptoethanol. CHO-gH cells were made by transfecting CHO-K1 cells with a glycosylphosphatidylinositol-linked form of the gH extracellular domain (14).
Viral mutagenesis.
We disrupted the gL coding sequence (genomic coordinates 65437 to 65024) by deleting its rapid amplification of cDNA ends (RACE)-mapped start codon and signal sequence (gL-DEL), by combining this deletion with premature stop codons (gL-DEL-STOP), or by introducing stop codons into the coding sequence for the gL signal peptide (gL-STOP). We first PCR amplified genomic coordinates 63873 to 65361 of the MHV-68 genomes as a SacI/KpnI-restricted fragment and coordinates 65438 to 66946 as an XbaI/SphI-restricted fragment (Phusion DNA polymerase; New England Biolabs, Hitchin, United Kingdom). These were cloned into the corresponding restriction sites of pSP73 (Promega Corporation, Southampton, United Kingdom). The genomic segments were then subcloned as a SacI/SphI fragment into the SacI/SphI sites of the pST76K-SR shuttle vector and recombined into an MHV-68 genomic bacterial artificial chromosome (BAC) (1). This deleted the RACE-mapped gL AUG (14), its signal sequence, and the next five codons (gL-DEL). A diagnostic BamHI restriction site from the pSP73 polylinker was left in their place. A second mutant was made by ligating the self-complementary oligonucleotide 5′-CTAGCTAGCTAGGATCCGAATTCGGATCCTAGCTAGCTAG-3′ into a SmaI restriction site between the same genomic flanks (gL-DEL-STOP). Again, the mutated gL locus was subcloned as a SacI/SphI fragment into pST76K-SR and recombined into the MHV-68 BAC. A third mutant (gL-STOP) had the same oligonucleotide inserted into a blunted AccI site (genomic coordinate 65380). This was done by subcloning a BamHI/KpnI genomic fragment (coordinates 64765 to 66120) from the BamHI-N genomic clone (10) into the BamHI/KpnI sites of pSP73, digesting it with AccI, blunting with Klenow fragment DNA polymerase, and dephosphorylating with Antarctic alkaline phosphatase (New England Biolabs). The phosphorylated oligonucleotide was then ligated in to terminate gL translation 2 amino acids before its predicted signal sequence cleavage site (gL-STOP). When the mutated genomic fragment was subcloned into pST76K-SR, we were unable to obtain sucrose-sensitive colonies. We therefore first subcloned the gL-STOP mutation into the pSP73-gL-DEL construct as a BstEII/KpnI fragment (genomic coordinates 65022 to 66120) and then transferred it to pST76K-SR using SphI and SacI as before. We also isolated revertants of the gL-DEL-STOP and gL-STOP BACs in which the disrupted gL locus was restored to its wild-type form. Fluorescence-tagged MHV-68 was made by attaching enhanced green fluorescent protein (eGFP) to the C terminus of gM (24). This virus is described in detail in reference 15. Briefly, the eGFP coding sequence from pEGFP-N3 (Clontech, Palo Alto, CA) was inserted between PCR-amplified genomic flanks so as to place it just upstream of the gM stop codon. The eGFP coding sequence plus its MHV-68 genomic flanks were then subcloned into pST76K-SR and recombined into the MHV-68 BAC, thereby fusing eGFP to the endogenous gM. An eGFP-tagged gL mutant was made by serially combining the gM-eGFP and gL-DEL-STOP mutations. Infectious virus was reconstituted by transfecting each BAC into BHK-21 cells using Fugene-6 (Roche Diagnostics Ltd., Lewes, United Kingdom). The loxP-flanked BAC/eGFP cassette was removed by passaging viruses through NIH 3T3-CRE cells.
Antibodies.
B-cell hybridomas were generated by fusing splenocytes from MHV-68-immune mice with NS0 cells using polyethylene glycol 1500 (14), and selected with azaserine (1 μg/ml)-hypoxanthine (100 μM). Monoclonal antibodies (MAbs) were concentrated from hybridoma supernatants by ammonium sulfate precipitation and quantitated by enzyme-linked immunosorbent assay against mouse immunoglobulin G (IgG) standards. The MAbs used in this study are listed in Table 1.
TABLE 1.
MHV-68-specific MAbs used in this study
| MAb | Isotype | Target | Source or reference |
|---|---|---|---|
| 1A2 | IgG2a | gH | This study |
| 2E6 | IgG2a | gH | This study |
| 9A3 | IgG2a | gH | This study |
| 9B10 | IgG2a | gH | This study |
| T2C12 | IgG2a | gH/gL | 14 |
| T7G1 | IgG2a | gH/gL | 14 |
| 7E5 | IgG2a | gH/gL | 14 |
| 8C1 | IgG2b | gH or gH/gL | 14 |
| T6D11 | IgG2b | gH or gH/gL | 14 |
| T1A1 | IgG2a | gp150 | 9 |
| T7H9 | IgG2a | gB | 21 |
| 3F7 | IgG2a | gN | 23 |
| 16D2 | IgG2a | ORF4 | 14 |
| 7D1-C12 | IgG2a | ORF17 (capsid) | This study |
| LSB11 | IgG1 | gp150 | 15 |
Virus growth and titration.
Virus stocks were grown and titered by plaque assay using BHK-21 cells or MEFs. Monolayers were fixed in 10% formaldehyde after 5 days and stained with 0.1% toluidine blue. Plaques were counted with a plate microscope (9). To titer infectious virus in lungs, the lungs were homogenized in complete medium, frozen, thawed, and sonicated. Tissue debris was pelleted by brief centrifugation (1,000 × g, 1 min). Infectious virus in homogenate supernatants was then measured by plaque assay. Latent virus in spleens was measured by infectious-center assay (9). Single spleen cell or mediastinal lymph node cell suspensions were cultured on MEF monolayers, which were fixed and stained for plaque counting after 6 days. Preformed infectious virus—that forming plaques even after freeze-thawing—contributes <1% of the infectivity recoverable from lymphoid tissue, so the infectious-center assay essentially measures reactivating latent virus.
Viral DNA quantitation.
Viral genome loads were measured by real-time PCR (2). Genomic coordinates 24832 to 25071 were amplified by PCR (Rotor Gene 3000; Corbett Research, Cambridge, United Kingdom) from 10 ng of DNA extracted (Wizard genomic DNA purification kit; Promega Corporation) from spleens or lymph nodes. The PCR products were quantitated with Sybr green (Invitrogen), the copy number was calculated by comparison with a standard curve of cloned MK3 template, and the products were serially diluted in control spleen DNA and amplified in parallel. Amplified products were distinguished from paired primers by melting curve analysis and the correct sizes of the amplified products confirmed by electrophoresis and staining with ethidium bromide.
Southern blotting.
Viral DNA was extracted by alkaline lysis (9), digested with restriction endonucleases, electrophoresed through 0.8% agarose in Tris-acetate buffer, and transferred to positively charged nylon membranes (Roche Diagnostics Ltd., Lewes, United Kingdom). A [32P]dCTP-labeled probe (APBiotech, Little Chalfont, United Kingdom) was generated by random primer extension (Nonaprimer kit; Qbiogene, Bingham, United Kingdom). Membranes were hybridized with the probe (65°C, 18 h), washed to a stringency of 0.2% SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS), and exposed to X-ray film.
Metabolic labeling and immunoprecipitation.
MHV-68-infected cells were washed twice in phosphate-buffered saline (PBS), cultured for 30 min in cysteine- and methionine-free medium with dialyzed fetal calf serum, and labeled for 6 h with 35S-labeled cysteine-methionine (4). The cells were then cultured in normal medium for a further 18 h and virions recovered from supernatants by ultracentrifugation (20,000 × g, 2 h). The virions were lysed (30 min, 4°C) in 1% Triton X-100, 50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, plus Complete protease inhibitors (Roche Diagnostics). Insoluble debris was removed by centrifugation (13,000 × g, 15 min). The supernatants were precleared with normal rabbit serum plus protein A-Sepharose. gH was then immunoprecipitated with gH-specific, gH/gL-specific, or pan-gH MAbs followed by protein A-Sepharose. The Sepharose beads were washed five times in lysis buffer. The precipitated proteins were denatured by heating (95°C, 5 min) in Laemmli's buffer and resolved by SDS-polyacrylamide gel electrophoresis (PAGE). Dried gels were exposed to X-ray film.
Immunoblotting.
Virions were lysed and denatured by heating (95°C, 5 min) in Laemmli's buffer. Virion proteins were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes (5). The membranes were probed with MHV-68-specific MAbs plus horseradish peroxidase-conjugated rabbit anti-mouse IgG polyclonal antibody (PAb; Dako Cytomation, Ely, United Kingdom), followed by enhanced chemiluminescence substrate development (APBiotech).
Flow cytometry.
Cells exposed to eGFP+ viruses were washed in PBS and analyzed directly for green channel fluorescence (35). For surface staining, cells were incubated (1 h, 4°C) with MHV-68 glycoprotein-specific MAbs followed by fluorescein isothiocyanate-conjugated rabbit anti-mouse IgG PAb (Dako Cytomation) or Alexa 633-conjugated or Alexa 488-conjugated goat anti-mouse PAb (Invitrogen). For intracellular staining, cells were fixed in 1% paraformaldehyde (30 min at room temperature) and then permeabilized with 0.1% saponin. Cells were then washed after antibody staining and analyzed on a FACSort using Cellquest software (Becton Dickinson, Oxford, United Kingdom).
Immunofluorescence.
Adherent cells were washed in PBS, fixed in 2% paraformaldehyde, and then permeabilized with 0.1% NP-40. The MHV-68 gp150 was detected with MAb LSB11 plus Alexa 488-conjugated goat anti-mouse IgG1 (Invitrogen). Nuclei were counterstained with DAPI (4′,6′-diamidino-2-phenylindole). Fluorescence was visualized with an Olympus IX70 microscope plus a Retiga 2000R camera line (QImaging).
RESULTS
Identification of gH-specific MAbs.
We have previously used MAbs derived from MHV-68-infected mice to identify two types of gH epitope (14). These MAbs were all selected for strong staining of MHV-68-infected BHK-21 cells. Most (45/48 MAbs tested) required both gH and gL for recognition (gH/gL specific). Three recognized gH and gH/gL equally well (pan-gH). Because gL is small and probably lies close to the virion membrane in the gH/gL heterodimer (14), we hypothesized that most gH/gL-specific MAbs recognize gL-dependent gH epitopes rather than compound epitopes or gL itself. The lack of these epitopes on gH when it was expressed alone therefore implied that other epitopes would be displayed in their place. As gH and gL are unlikely to remain always associated in vivo, gH-only epitopes should also elicit antibody responses in MHV-68-infected mice. We therefore looked for gH-only specificities among MAbs from MHV-68-infected mice by testing their recognition of a glycosylphosphatidylinositol-linked form of the gH extracellular domain (14) expressed without gL.
Fourteen MAbs that recognized cells expressing gH alone, corresponding to a similar abundance in mice to gH/gL-specific MAbs, were obtained. (The gB-specific MAb frequency was similar in each fusion and therefore provided an internal control of fusion efficiency.) Pan-gH MAbs such as 8C1 stained CHO-gH cells and MHV-68-infected BHK-21 cells comparably (Fig. 1). One of the gH-selected MAbs matched this pattern. gH/gL-specific MAbs such as T7G1 stained infected cells but not CHO-gH cells. No gH-selected MAbs matched this pattern. The 13 remaining gH-selected MAbs (represented by 2E6, 9A3, and 9B10 in Fig. 1) all stained CHO-gH cells more strongly than MHV-68-infected cells. Thus, their epitopes were not simply independent of gL like those of the pan-gH MAbs but were expressed better when gL was absent (gH only). None of the gH-specific MAbs detected denatured gH on immunoblots (data not shown). They therefore recognized a distinct gH conformation rather than the unfolded polypeptide. The fact that gH/gL-specific and gH-only MAbs were both considerably more numerous than pan-gH MAbs implied that gL substantially changed gH antigenicity.
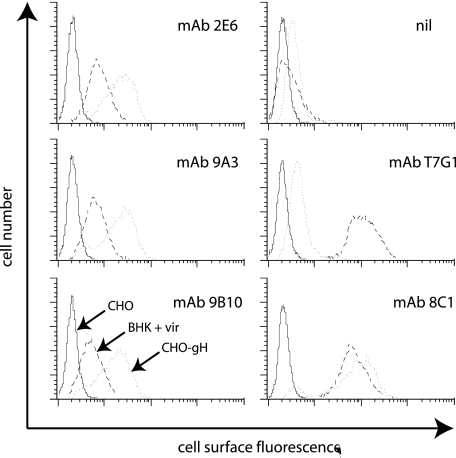
FIG. 1.
Flow-cytometric identification of gL-dependent and gL-independent MAbs recognizing the MHV-68 gH. BHK-21 cells were infected (18 h, 2 PFU/cell) with wild-type MHV-68 (BHK + vir). CHO-gH cells stably express a glycosylphosphatidylinositol-linked form of the gH extracellular domain without gL. MAbs 2E6, 9A3, and 9B10 were selected for their recognition of CHO-gH cells. MAb T7G1 recognizes gH on MHV-68-infected cells but not on cells expressing only gH. MAb 8C1 recognizes gH in either context. nil, secondary antibody only.
The stronger staining of CHO-gH cells by pan-gH MAbs than by gH-only MAbs suggested that gH-only epitopes expressed in isolation might have limited stability or accessibility. Even so, gH-only MAbs stained CHO-gH cells more strongly than infected cells. Their relatively weak staining of infected cells explained why we had not identified these MAbs before, when strong infected-cell recognition had been our primary selection criterion. gH-only MAbs still gave some staining, so not all the gH molecules on cell surfaces were associated with gL, but the large majority appeared to be so.
Generation of gL-deficient MHV-68 mutants.
We tested the functional importance of gL for MHV-68 replication by disrupting ORF47. We previously mapped the major start site of the MHV-68 gL mRNA to genomic coordinate 65489 (14). The first AUG of this mRNA corresponds to the fifth ATG of ORF47, and the mRNA encodes a predicted 137-amino-acid protein with a 21-amino-acid leader sequence (Fig. 2A). We first disrupted this coding sequence by deleting its AUG, its signal sequence, and the next five codons (gL-DEL). It was still theoretically possible that some form of gL could be produced from one of the upstream ORF47 AUGs, which remained in frame. We therefore generated a second mutant, with the same deletion but also multiple stop codons in its place (gL-DEL-STOP). We made a third mutant by inserting stop codons without any associated deletion into an AccI site at genomic coordinate 65380 (gL-STOP). This terminated gL translation 2 amino acids before the end of its predicted signal sequence. Restriction digests of BAC DNA (data not shown) and Southern blots of viral DNA (Fig. 2) confirmed the expected genomic structure for all three mutants, plus revertants of the gL-DEL-STOP and gL-STOP mutants.
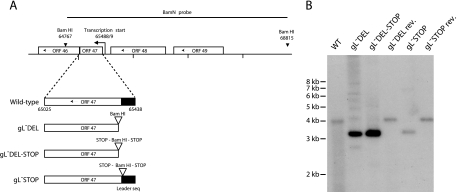
FIG. 2.
Generation of gL-deficient MHV-68 mutants. A. The gL coding sequence within ORF47 was disrupted by deleting its 5′ end including its start codon (gL-DEL), by combining the deletion with premature stop codons (gL-DEL-STOP), or by inserting stop codons near the end of the coding sequence for its predicted signal peptide (gL-STOP). Each mutation incorporated a new BamHI restriction site. B. Viral DNA was digested with BamHI, resolved by agarose gel electrophoresis, and hybridized with a 32P-labeled BamHI-N probe, as shown in panel A. The 4,048-bp wild-type (WT) band becomes 3,375 bp for the gL-DEL and gL-DEL-STOP mutants and 3,433-bp for the gL-STOP mutant. The remaining 600-bp fragments are too small to appear on this gel.
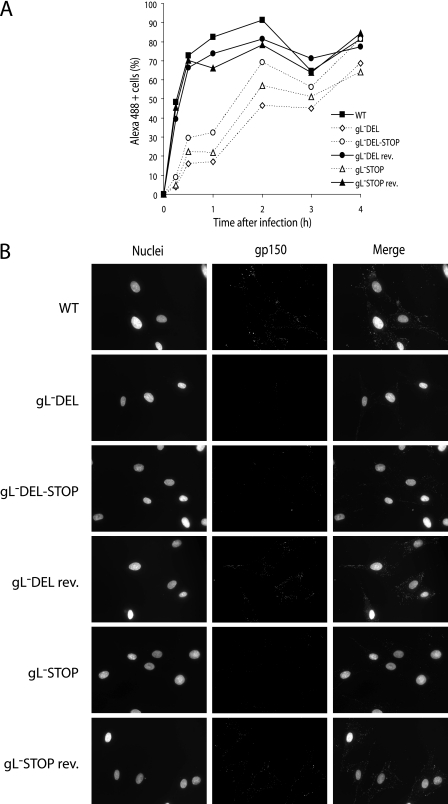
All three gL mutants produced infectious virus after BAC DNA transfection into BHK-21 cells. Virus propagation was noticeably slower than with parallel transfections of wild-type BAC DNA: extensive cytopathic effects were typically evident after 7 days rather than 4. This finding was consistent between at least two independent BAC clones for each mutant. Nevertheless, we could readily grow high-titer gL-deficient virus stocks. Flow cytometry of infected cells showed no evidence of gH/gL heterodimer expression by any of the gL mutants (Fig. 3A). Instead, it showed enhanced presentation of gH-only epitopes, indicating that the folding of gH in infected cells mirrored that seen in transfected cells (14). The gL-DEL and gL-DEL-STOP mutants also showed lower cell surface expression of other viral glycoproteins (Fig. 3A), particularly for gN (24) and gB (22), which are likely to reach the cell surface only in virions. The gB and gN content of gL− virions themselves was not reduced (Fig. 3B). Thus, there may have been fewer gL− virions attached to infected-cell surfaces. An alternative explanation is that the gL knockouts were less infectious than the wild type. Although all the cells were infected based on viral eGFP expression (data not shown), fewer may have been supporting late lytic gene expression. The gL-STOP mutant similarly showed no gH/gL expression and reduced levels of the ORF4 complement control protein and gp150, although its expression of gN and gB was normal.
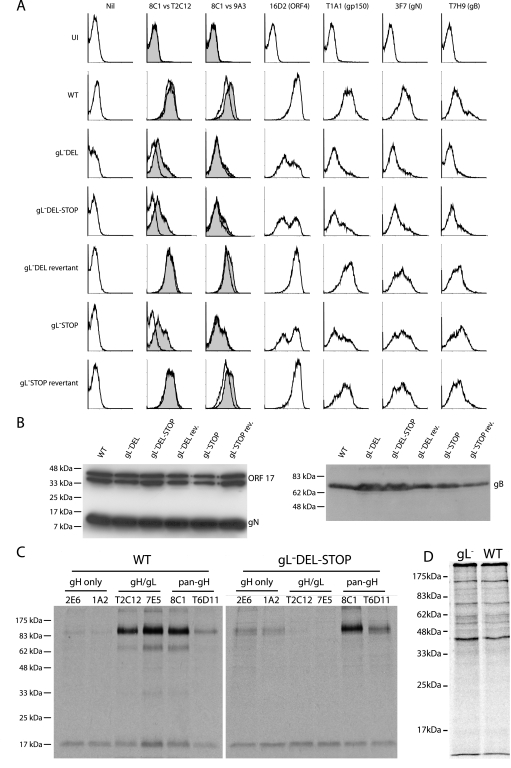
FIG. 3.
Glycoprotein expression by gL-deficient MHV-68. A. BHK-21 cells were left uninfected (UI) or infected (1 PFU, 24 to 48 h) with gL− or gL+ viruses as indicated. The cells were then trypsinized and stained for viral glycoproteins. nil, secondary antibody only. For gH expression, gH/gL-specific (T2C12) and gH-only (9A3) MAbs (open histograms) are shown in comparison to a pan-gH MAb (8C1) (filled histograms). eGFP-expressing viruses were used, and each histogram shows an equivalent eGFP+-gated population to ensure equivalent levels of infection. WT, wild type. B. Viral proteins (equivalent to 5 × 104 PFU/lane for each virus) were denatured, resolved by SDS-PAGE, and immunoblotted for gB and gN as indicated. MAb 7D1-C12 specifically detects ORF17, either in cells transfected with an ORF17 expression plasmid or as a glutathione S-transferase-ORF17 fusion protein expressed in Escherichia coli (data not shown). It provides a loading control. The doublet band reflects the fact that ORF17 is autoproteolytically cleaved. C. gH was immunoprecipitated from 35S-labeled virions. The MAbs used were gH only (2E6, 1A2), gH/gL specific (T2C12, 7E5), or pan-gH (8C1, T6D11). gH migrates at an apparent molecular mass of 85 kDa. The nonspecifically immunoprecipitated 19-kDa band probably corresponds to the abundant ORF65 capsid component. D. Input labeled virion proteins used for immunoprecipitation in panel C.
We examined the conformation of virion gH by labeling the proteins made in BHK-21 cells infected with either wild-type MHV-68 or the gL-DEL-STOP mutant (Fig. 3C and D). Labeled virions were recovered from infected-cell supernatants. gH was then immunoprecipitated from virion lysates with gH/gL-specific, gH-only, or pan-gH MAbs. Levels of total gH recovery were the same. However, gH/gL-specific MAbs recovered no gH from the gL-deficient virions and gH-only MAbs recovered gH poorly from the wild-type virions. In agreement with the flow cytometry data of Fig. 3A, therefore, virion gH failed to adopt its normal conformation when gL was not present. Instead it favored an otherwise minority conformation.
Growth in vitro of gL-deficient MHV-68 mutants.
The relatively slow spread of cytopathic effects after transfecting gL-deficient BACs into BHK-21 cells suggested that gL contributed to in vitro virus propagation. We assayed the replication of the gL-DEL mutant more formally by incubating BHK-21 cells with virus at low multiplicity for 1 h at 37°C, washing off unbound virus with PBS, and then assaying infectious virus titers with time by plaque assay (Fig. 4A). A substantial, gL-dependent growth deficit was evident.
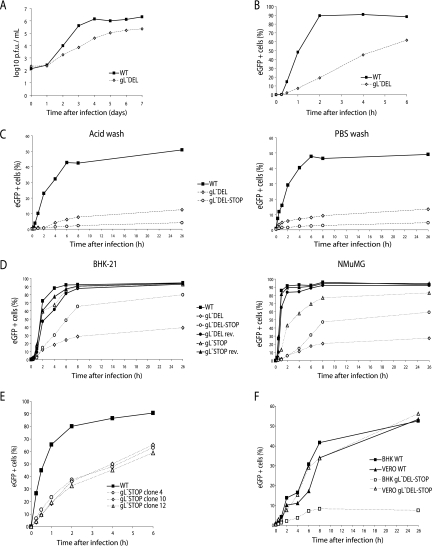
FIG. 4.
Infectivity assays with gL-deficient MHV-68 mutants. A. BHK-21 cells were infected (0.01 PFU/cell, 1 h, 37°C) with wild-type (WT) or gL-DEL MHV-68. Infectious virus in replicate cultures was determined thereafter by plaque assay. B. BHK-21 cells were exposed to eGFP-expressing wild-type or gL-DEL MHV-68 (1 PFU/cell) for the times indicated and then washed with PBS and cultured overnight. Viral infection was assayed by flow cytometry for eGFP expression. C. MEFs were exposed to eGFP-expressing wild-type, gL-DEL, or gL-DEL-STOP MHV-68 (1 PFU/cell) for the times indicated and then washed either with PBS (pH 7.4) or with isotonic (pH 3) buffer (acid wash). Viral infection was assayed by measuring eGFP expression 18 h later as for panel B. D. BHK-21 fibroblasts or NMuMG epithelial cells were infected with eGFP-expressing gL− or gL+ viruses as for panel B. The cells were washed with PBS after the time indicated, and eGFP expression was assayed the next day. E. Three different BAC isolates of the gL-STOP mutant were reconstituted into infectious virus and then compared with the wild type for their capacity to infect BHK-21 cells. F. BHK-21 and Vero cells were exposed to gL− or gL+ MHV-68 for different times before being washed with PBS as for panel D.
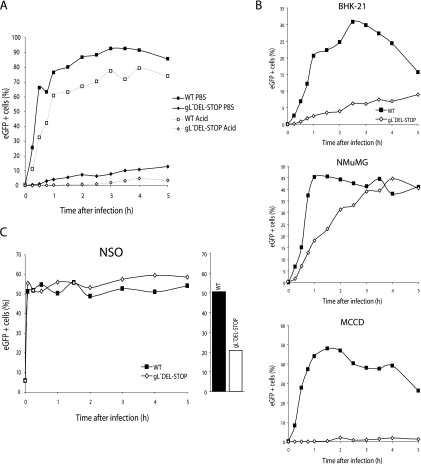
Since gH/gL is likely to be involved in virion entry, we next tested the first process in entry, attachment, by incubating BHK-21 cells with eGFP-expressing wild-type or gL-DEL MHV-68 (0.5 PFU/cell) for various times before washing the cells with PBS. The number of infected cells was determined by flow-cytometric assay of viral eGFP expression 18 h later (Fig. 4B). It should be noted that in plaque assays of virus stocks the virus inoculum was not removed so as to minimize any under-titering of gL− viruses, and immunoblots had established that the protein content of gL− stocks was comparable to the wild type for an equivalent number of PFU (Fig. 3B).
The gL-DEL mutant appeared slow to attach, in that wild-type infection became PBS wash resistant within 2 h, whereas gL-DEL infection continued to increase from 6 to 24 h of exposure to virus. The gL-DEL and gL-DEL-STOP mutants also showed profound single-cycle infection deficits in MEFs (Fig. 4C). These deficits were equivalent whether the cells were washed with PBS (pH 7.4) to remove unbound virions or in acid (pH 3) to inactivate any virions that had not penetrated the plasma membrane. Thus, gL− mutants appeared to be impaired in cell binding rather than cell penetration. (MHV-68 penetration of fibroblasts equates to endocytosis [14].)
The single-cycle infection deficit was quite variable between different gL mutants (Fig. 4D) and even between different stocks of the same mutant. The reason for this variability remains unclear. In the experiment shown, the gL-STOP mutant, which had shown the least reduction in cell surface glycoprotein staining (Fig. 3A), had a minimal BHK-21 infection deficit (Fig. 4D) but was appreciably reduced in NMuMG epithelial cell infection (Fig. 4D). In other experiments (Fig. 4E), gL-STOP mutants showed a more substantial deficit in BHK-21 cell infection. Interestingly, gL did not seem to contribute appreciably to the infection Vero cells (Fig. 4F).
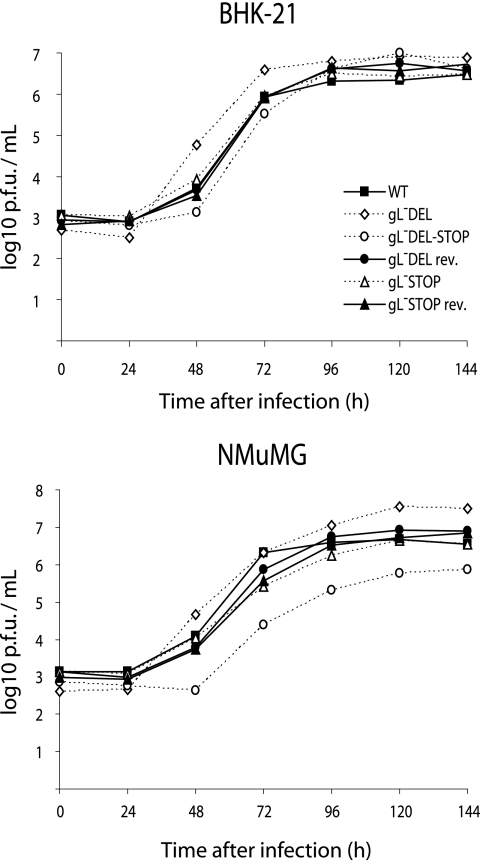
We then retested in vitro virus growth while allowing for reduced cell binding by not washing off the input virus, as in our standard plaque assay protocol. Viral replication was again quite variable between different gL mutants, but overall there was now no significant growth deficit in either BHK-21 or NMuMG cells (Fig. 5).
FIG. 5.
Growth of gL-deficient MHV-68 with allowance made for reduced cell binding. BHK-21 fibroblasts or NMuMG fibroblasts were infected at low multiplicity (0.01 PFU/cell) with gL− or gL+ MHV-68 and then cultured without removing the virus inoculum. The infectious virus in replicate cultures was determined thereafter by plaque assay. WT, wild type.
Impaired cell binding of gL-deficient MHV-68 mutants.
We visualized cell binding directly by either staining the cells exposed to virus for a known virion glycoprotein or tracking the transfer of fluorescence to cells from eGFP-tagged virions. All three gL mutants showed reduced binding to BHK-21 cells as measured by flow-cytometric staining for virion gN (Fig. 6A). Equivalent cells visualized by immunofluorescence are shown in Fig. 6B. gp150 was much more efficiently transferred to cell membranes by wild-type MHV-68.
FIG. 6.
Decreased cell binding by gL-deficient MHV-68 mutants. A. BHK-21 cells were exposed to gL− or gL+ viruses (2 PFU/cell) at 37°C for the times indicated and then fixed, permeabilized, and stained with the gN-specific MAb 3F7 plus an Alexa 488-conjugated mouse IgG-specific secondary antibody. Cells were analyzed for positive staining by flow cytometry. WT, wild type. B. BHK-21 cells were exposed to gL− or gL+ viruses for 30 min as in panel A and then washed with PBS and stained for gp150 with MAb LSB11 plus an Alexa 488-conjugated secondary antibody. Nuclei were counterstained with DAPI.
We tagged wild-type MHV-68 and the gL-DEL-STOP mutant by modifying gM to include eGFP at its C-terminal end. The eGFP-tagged gL-DEL-STOP mutant showed profound binding deficits that were equivalent whether the cells were washed at neutral or at low pH (Fig. 7A). NMuMG epithelial cells showed—surprisingly in view of their greater single-cycle infection deficit—somewhat less binding deficit than BHK-21 cells (Fig. 7B). The deficit for MCCD epithelial cells was very marked (Fig. 7B). Overall, there was evidence of gL-dependent MHV-68 binding to both epithelial cells and fibroblasts. In contrast, NS0 cells, a myeloma cell line known to be permissive for MHV-68 infection (39), showed rapid binding with no difference between gL+ and gL− viruses (Fig. 7C).
FIG. 7.
Tracking cell binding with eGFP-tagged MHV-68. A. BHK-21 cells were exposed to either wild-type (WT) or gL-deficient versions of MHV-68 with eGFP-tagged gM (2 PFU/cell) and then cultured at 37°C. The cells were washed with PBS or at low pH after the time indicated and analyzed by flow cytometry for green fluorescence. B. BHK-21 fibroblasts, NMuMG epithelial cells, and MCCD epithelial cells were exposed to gL− or gL+ versions of the gM-eGFP virus as for panel A. The cells were washed with PBS at the time indicated and then analyzed by flow cytometry for green fluorescence. The decline from maximum fluorescence with time may reflect the destruction of gM-eGFP in lysosomes following endocytosis and fusion. C. NS0 cells were analyzed for green fluorescence after exposure to gL− or gL+ gM-eGFP MHV-68 as for panel C. A 2-h parallel infection of BHK-21 cells at the same multiplicity (2 PFU/cell) is shown.
Dissemination of gL-deficient MHV-68 mutants in vivo.
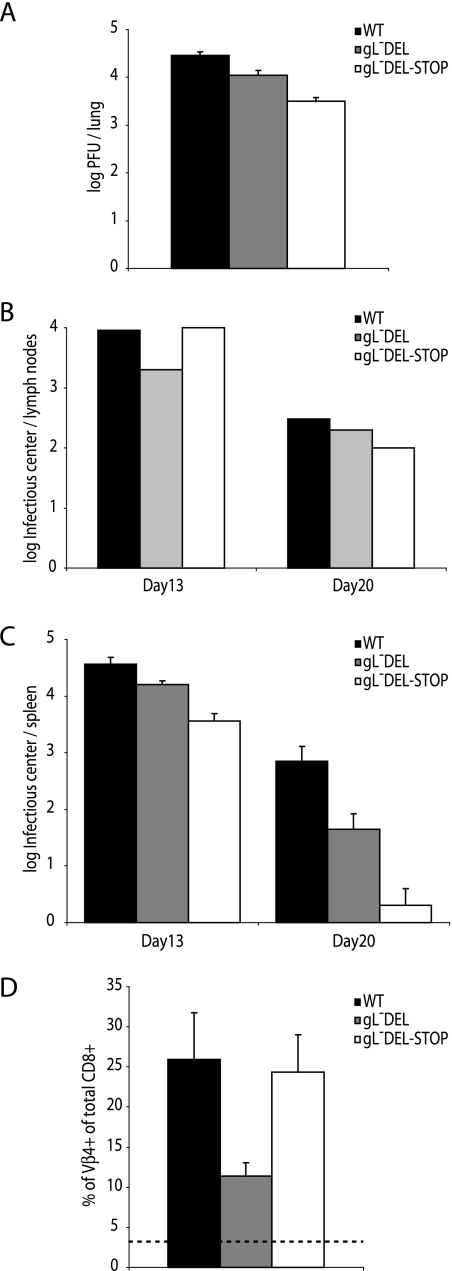
We used intranasal infection to compare gL− and gL+ viruses in host colonization. We first infected C57BL/6 mice with 2 × 104 PFU of the gL-DEL mutant, the gL-DEL-STOP mutant, or wild-type MHV-68. The gL− mutants showed some reduction in lytic replication in mouse lungs (Fig. 8A). However, the deficit was small. The colonization of draining lymph nodes was essentially normal (Fig. 8B). Somewhat less virus was recovered from spleens in infectious-center assays, but the gL− mutants clearly had no major problem colonizing this site (Fig. 8C). All the viruses stimulated the expansion of Vβ4+ CD8+ T cells, whose activation provides an immunological marker of MHV-68 latency establishment. Thus, a lack of gL had relatively little effect on host colonization.
FIG. 8.
Replication of gL− and gL+ MHV-68 in C57BL/6 mice. A. C57BL/6 mice were infected intranasally with gL+ or gL− viruses. Seven days later, the infectious virus titer in lungs was determined by plaque assay. The titers of gL− viruses were significantly reduced relative to the wild type (WT) (P < 0.02 by Student's t test). B. Mediastinal lymph nodes were removed at the times indicated after infection with gL+ or gL− viruses, pooled from five mice, and assayed for reactivable MHV-68 by infectious-center assay. C. Spleens from the same mice were analyzed individually by infectious-center assay. Each bar shows the mean ± standard error of the mean (SEM) for each group of five. The gL-DEL titers were not significantly different from the wild type at day 13. The other gL− virus titers did show a significant reduction (P < 0.03). D. Splenic CD8+ T cells were analyzed for Vβ4+ subset expansion at 20 days postinfection. Each bar shows the mean ± SEM for five mice per group. The dashed line shows the value for naive mice.
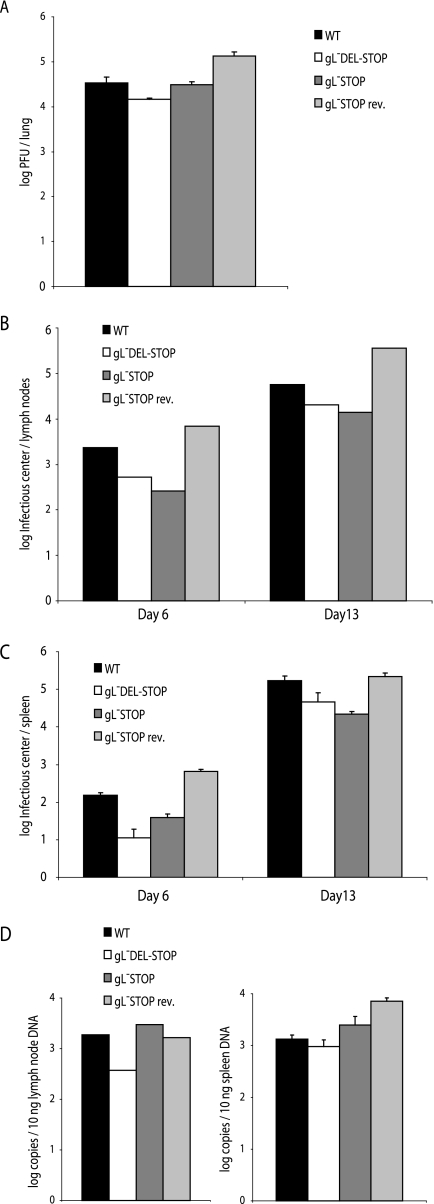
We then tested the colonization of BALB/c mice by the gL-DEL-STOP and gL-STOP mutants, comparing them with wild-type virus and a revertant of the gL-STOP mutant. In these experiments we used a 10-fold-lower inoculum (2,000 PFU/mouse). The small lytic replication deficit of the gL-DEL-STOP mutant was maintained, but the gL-STOP mutant showed no deficit relative to the wild type (Fig. 9A). Both mutants showed slightly lower infectious-center assay titers in mediastinal lymph nodes (Fig. 9B) and spleens (Fig. 9C), suggesting a reduction in latency amplification. However, viral DNA loads in these sites were normal (Fig. 9D), so the lower infectious-center assay titers may have reflected simply less-efficient in vitro detection of reactivation from latency. Overall, gL-deficient mutants appeared to be fully capable of in vivo host colonization.
FIG. 9.
Replication of gL− and gL+ MHV-68 in BALB/c mice. A. BALB/c mice were infected intranasally with gL+ or gL− viruses. Six days later, the infectious virus titer in lungs was determined by plaque assay. There was no significant difference between gL− and wild-type (WT) virus titers. B. Mediastinal lymph nodes were removed at the times indicated after infection with gL+ or gL− viruses, pooled from five mice, and assayed for reactivable MHV-68 by infectious-center assay. C. Spleens from the same mice were analyzed individually by infectious-center assay. Each bar shows the mean ± standard error of the mean (SEM) for each group of five. At day 6, the gL− titers were significantly lower than wild type (P < 0.01 by Student's t test). By day 13, only the gL-DEL-STOP titers were lower (P < 0.03). D. DNA was extracted from individual spleens. The viral genome copy number was then determined by real-time PCR amplification of genomic coordinates 24832 to 25071 and comparison with parallel amplifications of plasmid template dilutions. Mediastinal lymph nodes were assayed as pools of five mice, spleens were assayed individually, and each bar shows the mean ± SEM. The gL− viruses were not significantly different from the wild type.
DISCUSSION
Many viruses devote just one protein to cell binding and penetration. Herpesviruses, by contrast, engage cells via multiple glycoproteins, not all of which are always required. Such flexibility allows for more-complex viral behavior, mediated, for example, through cell type-specific receptor signaling. However, membrane fusion remains an obligate step, and the conserved herpesvirus proteins that drive it, gB, gH, and gL, have, where tested, proved to be essential for virion entry. MHV-68 provides the first example of gL being nonessential and in doing so suggests how gH might function in gamma-2 herpesviruses.
We have previously shown that the transfected MHV-68 gL associates with gH to create a heterodimer that is antigenically distinct from gH expressed alone. We have shown here that gL similarly sets the gH conformation in infected cells. Infectivity assays established that gH does not simply misfold without gL but rather adopts a legitimate, albeit distinct, functional form. The MHV-68 gL is therefore a nonobligate chaperone for gH, conferring on it a cell binding function that is separable from its presumed role in membrane fusion. A lack of gL reduced the binding of MHV-68 to fibroblasts and epithelial cells, but not to Vero cells (Fig. 5) or NS0 cells (Fig. 6). MHV-68 almost certainly binds to glycosaminoglycans (9), and glycosaminoglycans on Vero cells may compensate for a gL-related attachment deficit. But NS0 cells are relatively low in surface glycosaminoglycans (9), so here gL-independent binding suggested the use of a receptor different from those on epithelial cells and fibroblasts, perhaps analogous to the different B-cell and epithelial cell receptors used by EBV (6). However, the functional significance of NS0 cell binding by gL-deficient MHV-68 must remain speculative, as the normal route of MHV-68 entry into the B-cell pool is unknown. For example, the A20 B-cell line is essentially uninfectable by MHV-68 unless its glycosaminoglycan expression is increased (2). Only a small fraction of ex vivo spleen cells bound eGFP-tagged MHV-68 as strongly as NS0 cells did (data not shown), and MHV-68 does not appear to infect NS0 cells efficiently unless gp150 is removed (9). Thus, all we can say at present is that MHV-68 attachment to NS0 cells was gL independent whereas its attachment to fibroblast or epithelial cell lines was not.
Reduced cell binding should compromise infection by cell-free virions more than infection by direct spread between adjacent cells, and this was observed (Fig. 5). Efficient virion entry may also be less important for single-exposure experimental infections than for natural transmission; the latter presumably involves limiting virion numbers and requires on average many contacts. This explains, perhaps, the surprisingly mild in vivo phenotype of the gL knockouts. As yet there is no transmission model for MHV-68. Thus, our data do not suggest that gL is functionally redundant, only that its contribution to host colonization is small once an initial infection has been established.
Cell-free virions also face more difficulty than those passing from cell to cell in establishing a suitable setting for subsequent virus replication, because infected cells have more opportunities to communicate with their neighbors. Receptor cross-linking represents the first opportunity for virions to signal, and other herpesviruses provide abundant precedent for this being functionally important. Some such function for the MHV-68 gH/gL might explain the failure of gL− virions to reach wild-type levels of NMuMG cell infection (Fig. 4D), even though the binding deficit here was quite mild (Fig. 7B). However, there was no evidence for the membrane fusion deficit that might have been predicted from previous herpesvirus gL descriptions. In this regard, the viability of the gL mutants contrasted completely with our (24) and others' (27, 34) inability to propagate MHV-68 gH mutants. It seems unlikely that gH alone and gH/gL participate independently in fusion, as they appear to be conformationally quite different (<10% of gH-specific MAbs recognized both). More likely is that gH-only infection represents a downstream entry step, reached via gL dissociation. Still further steps are likely, since gL− infection, like that of wild-type MHV-68 (14), was inhibited by ammonium chloride, bafilomycin, or chlorpromazine (data not shown), implying a continued requirement for endocytosis and low pH.
The variability of the gL knockout phenotype, plus some discrepancy between the slow virus recovery after BAC transfection (data not shown) and relatively normal growth curves (Fig. 5), suggested that compensatory mutations might have partially rescued a gL-dependent replication deficit (21). None of the mutants showed gH/gL epitope expression (Fig. 3A), and none was found to have secondary DNA sequence changes in gH (data not shown), but other compensatory changes are possible, for example, an increase in gB expression. Such questions require a more complete understanding of MHV-68 glycoprotein function. They do not alter our main conclusions: the MHV-68 gL altered gH folding and promoted attachment to epithelial cells and fibroblasts but was inessential for viral propagation either in vitro or in vivo.
Acknowledgments
Laurent Gillet is a Postdoctoral Researcher of the Fonds National Belge de la Recherche Scientifique. Philip Stevenson is a Wellcome Trust Senior Clinical Fellow (GR076956MA). This work was also supported by Medical Research Council grants G0400427 and G9800903.
Footnotes
Published ahead of print on 18 October 2006.
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, N. J., J. S. May, and P. G. Stevenson. 2005. Gamma-herpesvirus latency requires T cell evasion during episome maintenance. PLoS Biol. 3:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaskovic, D., M. Stancekova, J. Svobodova, and J. Mistrikova. 1980. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 24:468. [PubMed] [Google Scholar]
- 4.Boname, J. M., and P. G. Stevenson. 2001. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15:627-636. [DOI] [PubMed] [Google Scholar]
- 5.Boname, J. M., B. D. de Lima, P. J. Lehner, and P. G. Stevenson. 2004. Viral degradation of the MHC class I peptide loading complex. Immunity 20:305-317. [DOI] [PubMed] [Google Scholar]
- 6.Borza, C. M., and L. M. Hutt-Fletcher. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8:594-599. [DOI] [PubMed] [Google Scholar]
- 7.Browne, H., B. Bruun, and T. Minson. 2001. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. J. Gen. Virol. 82:1419-1422. [DOI] [PubMed] [Google Scholar]
- 8.Cranage, M. P., G. L. Smith, S. E. Bell, H. Hart, C. Brown, A. T. Bankier, P. Tomlinson, B. G. Barrell, and T. C. Minson. 1988. Identification and expression of a human cytomegalovirus glycoprotein with homology to the Epstein-Barr virus BXLF2 product, varicella-zoster virus gpIII, and herpes simplex virus type 1 glycoprotein H. J. Virol. 62:1416-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lima, B. D., J. S. May, and P. G. Stevenson. 2004. Murine gammaherpesvirus 68 lacking gp150 shows defective virion release but establishes normal latency in vivo. J. Virol. 78:5103-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efstathiou, S., Y. M. Ho, and A. C. Minson. 1990. Cloning and molecular characterization of the murine herpesvirus 68 genome. J. Gen. Virol. 71:1355-1364. [DOI] [PubMed] [Google Scholar]
- 11.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis-Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuller, A. O., R. E. Santos, and P. G. Spear. 1989. Neutralizing antibodies specific for glycoprotein H of herpes simplex virus permit viral attachment to cells but prevent penetration. J. Virol. 63:3435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gangappa, S., S. B. Kapadia, S. H. Speck, and H. W. Virgin. 2002. Antibody to a lytic cycle viral protein decreases gammaherpesvirus latency in B-cell-deficient mice. J. Virol. 76:11460-11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gill, M. B., L. Gillet, S. Colaco, J. S. May, B. D. de Lima, and P. G. Stevenson. 2006. Murine gammaherpesvirus-68 glycoprotein H-glycoprotein L complex is a major target for neutralizing monoclonal antibodies. J. Gen. Virol. 87:1465-1475. [DOI] [PubMed] [Google Scholar]
- 15.Gillet, L., M. B. Gill, S. Colaco, C. M. Smith, and P. G. Stevenson. 2006. The murine gammaherpesvirus-68 glycoprotein B presents a difficult neutralization target to monoclonal antibodies derived from infected mice. J. Gen. Virol. 87:3515-3538. [DOI] [PMC free article] [PubMed]
- 16.Grundhoff, A., and D. Ganem. 2004. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J. Clin. Investig. 113:124-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutchinson, L., H. Browne, V. Wargent, N. Davis-Poynter, S. Primorac, K. Goldsmith, A. C. Minson, and D. C. Johnson. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenner, R. G., and C. Boshoff. 2002. The molecular pathology of Kaposi's sarcoma-associated herpesvirus. Biochim. Biophys. Acta 1602:1-22. [DOI] [PubMed] [Google Scholar]
- 20.Kaye, J. F., U. A. Gompels, and A. C. Minson. 1992. Glycoprotein H of human cytomegalovirus (HCMV) forms a stable complex with the HCMV UL115 gene product. J. Gen. Virol. 73:2693-2698. [DOI] [PubMed] [Google Scholar]
- 21.Klupp, B. G., and T. C. Mettenleiter. 1999. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J. Virol. 73:3014-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopes, F. B., S. Colaco, J. S. May, and P. G. Stevenson. 2004. Characterization of the MHV-68 glycoprotein B. J. Virol. 78:13370-13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopper, M., and T. Compton. 2004. Coiled-coil domains in glycoproteins B and H are involved in human cytomegalovirus membrane fusion. J. Virol. 78:8333-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.May, J. S., S. Colaco, and P. G. Stevenson. 2005. Glycoprotein M is an essential lytic replication protein of the murine gammaherpesvirus 68. J. Virol. 79:3459-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mistrikova, J., and D. Blaskovic. 1985. Ecology of the murine alphaherpesvirus and its isolation from lungs of rodents in cell culture. Acta Virol. 29:312-317. [PubMed] [Google Scholar]
- 26.Molesworth, S. J., C. M. Lake, C. M. Borza, S. M. Turk, and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 74:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moorman, N. J., C. Y. Lin, and S. H. Speck. 2004. Identification of candidate gammaherpesvirus 68 genes required for virus replication by signature-tagged transposon mutagenesis. J. Virol. 78:10282-10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mori, Y., P. Akkapaiboon, S. Yonemoto, M. Koike, M. Takemoto, T. Sadaoka, Y. Sasamoto, S. Konishi, Y. Uchiyama, and K. Yamanishi. 2004. Discovery of a second form of tripartite complex containing gH-gL of human herpesvirus 6 and observations on CD46. J. Virol. 78:4609-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naranatt, P. P., S. M. Akula, and B. Chandran. 2002. Characterization of gamma2-human herpesvirus-8 glycoproteins gH and gL. Arch. Virol. 147:1349-1370. [DOI] [PubMed] [Google Scholar]
- 30.Nash, A. A., and N. P. Sunil-Chandra. 1994. Interactions of the murine gammaherpesvirus with the immune system. Curr. Opin. Immunol. 6:560-563. [DOI] [PubMed] [Google Scholar]
- 31.Parry, C., S. Bell, T. Minson, and H. Browne. 2005. Herpes simplex virus type 1 glycoprotein H binds to αvβ3 integrins. J. Gen. Virol. 86:7-10. [DOI] [PubMed] [Google Scholar]
- 32.Pertel, P. E. 2002. Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J. Virol. 76:4390-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roop, C., L. Hutchinson, and D. C. Johnson. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 67:2285-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song, M. J., S. Hwang, W. H. Wong, T. T. Wu, S. Lee, H. I. Liao, and R. Sun. 2005. Identification of viral genes essential for replication of murine gamma-herpesvirus 68 using signature-tagged mutagenesis. Proc. Natl. Acad. Sci. USA 102:3805-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevenson, P. G., S. Efstathiou, P. C. Doherty, and P. J. Lehner. 2000. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc. Natl. Acad. Sci. USA 97:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevenson, P. G. 2004. Immune evasion by gamma-herpesviruses. Curr. Opin. Immunol. 16:456-462. [DOI] [PubMed] [Google Scholar]
- 37.Stevenson, P. G., J. S. May, X. G. Smith, S. Marques, H. Adler, U. H. Koszinowski, J. P. Simas, and S. Efstathiou. 2002. K3-mediated evasion of CD8+ T cells aids amplification of a latent gamma-herpesvirus. Nat. Immunol. 3:733-740. [DOI] [PubMed] [Google Scholar]
- 38.Stevenson, P. G., J. M. Boname, B. de Lima, and S. Efstathiou. 2002. A battle for survival: immune control and immune evasion in murine gamma-herpesvirus-68 infection. Microbes Infect. 4:1177-1182. [DOI] [PubMed] [Google Scholar]
- 39.Stevenson, P. G., and S. Efstathiou. 2005. Immune mechanisms in murine gammaherpesvirus-68 infection. Viral Immunol. 18:445-456. [DOI] [PubMed] [Google Scholar]
- 40.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1993. Interactions of murine gammaherpesvirus 68 with B and T cell lines. Virology 193:825-833. [DOI] [PubMed] [Google Scholar]
- 41.Wang, D., and T. Shenk. 2005. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl. Acad. Sci. USA 102:18153-18158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, X., and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus lacking glycoprotein gp42 can bind to B cells but is not able to infect. J. Virol. 72:158-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, X., W. J. Kenyon, Q. Li, J. Mullberg, and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72:5552-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]