Abstract
Previous work has shown that the C-terminal portion of BphA, especially two amino acid segments designated region III and region IV, influence the regiospecificity of the biphenyl dioxygenase (BPDO) toward 2,2′-dichlorobiphenyl (2,2′-CB). In this work, we evolved BPDO by shuffling bphA genes amplified from polychlorinated biphenyl-contaminated soil DNA. Sets of approximately 1-kb DNA fragments were amplified with degenerate primers designed to amplify the C-terminal portion of bphA. These fragments were shuffled, and the resulting library was used to replace the corresponding fragment of Burkholderia xenovorans LB400 bphA. Variants were screened for their ability to oxygenate 2,2′-CB onto carbons 5 and 6, which are positions that LB400 BPDO is unable to attack. Variants S100, S149, and S151 were obtained and exhibited this feature. Variant S100 BPDO produced exclusively cis-5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl from 2,2′-CB. Moreover, unlike LB400 BPDO, S100 BphA catalyzed the oxygenation of 2,2′,3,3′-tetrachlorobiphenyl onto carbons 5 and 6 exclusively and it was unable to oxygenate 2,2′,5,5′-tetrachlorobiphenyl. Based on oxygen consumption measurements, variant S100 oxygenated 2,2′-CB at a rate of 16 ± 1 nmol min−1 per nmol enzyme, which was similar to the value observed for LB400 BPDO. cis-5,6-Dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl was further oxidized by 2,3-dihydro-2,3-dihydroxybiphenyl dehydrogenase (BphB) and 2,3-dihydroxybiphenyl dioxygenase (BphC). Variant S100 was, in addition, able to oxygenate benzene, toluene, and ethyl benzene. Sequence analysis identified amino acid residues M237S238 and S283 outside regions III and IV that influence the activity toward doubly ortho-substituted chlorobiphenyls.
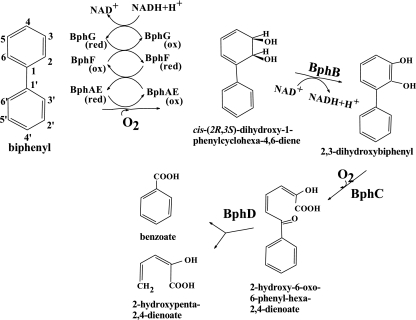
Biphenyl dioxygenase (BPDO) catalyzes the first reaction of the biphenyl catabolic pathway. BPDO is of particular interest because of its potential application as biocatalyst to oxygenate polychlorinated biphenyls (PCBs) and also to manufacture fine chemicals. BPDO comprises three components: the iron-sulfur oxygenase (BphAE) made up of α (BphA; Mr = 51,000) and β (BphE; Mr = 22,000) subunits, the ferredoxin (BphF; Mr = 12,000), and the ferredoxin reductase (BphG; Mr = 43,000) (15, 17). The encoding genes for Burkholderia xenovorans LB400 (10, 13), which is the best PCB degrader of natural origin, are bphA (BphAE α subunit), bphE (BphAE β subunit), bphF (BphF), and bphG (BphG). BPDO catalyzes a 2,3-dihydroxylation of biphenyl (15) (Fig. 1). The turnover rate of reaction as well as the specificity and regiospecificity toward substrate analogs are crucial for its application as biocatalyst. The C-terminal portion of BphAE α subunit is involved in substrate specificity and regiospecificity toward substrate analogs (11, 33). Identification of the amino acid residues that are critical to determine the substrate specificity and regiospecificity and their mechanism of action will help design strategies to engineer new, higher performing enzymes. Several investigations using site-directed mutagenesis, and molecular evolution approaches have allowed the identification of amino acids that are critical for the enzyme specificity and regiospecificity toward chlorobiphenyls (11, 33). Of particular interest is a segment of seven amino acid residues called region III (3, 5, 24). The sequence of amino acids for region III of B. xenovorans LB400 BphA is T335F336N337N338I339R340I341 (24). Besides region III amino acids, other residues, among which are residues Asn377Phe378 (also called region IV) of LB400 BphA (24, 30, 32) as well as Phe384 (36) and Gln255, Ile258, and Ala268 (29), were also found to influence the catalytic properties of the dioxygenase. Furthermore, structural analysis of Rhodococcus sp. strain RHA1 BphA1A2 (corresponding to BphAE of LB400) (12) and comparison with other aryl hydroxylating dioxygenases has confirmed the fact that some of the residues listed above are near the substrate inside the catalytic pocket and thus likely to interact with it to determine the specificity and regiospecificity. However, other residues that according to a model based on the structure of the naphthalene dioxygenase were not in direct contact with the substrate were also found to influence the enzyme specificity and regiospecificity (36).
FIG. 1.
The biphenyl catabolic pathway of B. xenovorans LB400, showing the three components of LB400 BPDO.
LB400 BPDO is among the few BPDOs that oxygenate the doubly ortho-substituted congener 2,2′-dichlorobiphenyl (2,2′-CB). The oxygenation occurs principally on the vicinal ortho-meta carbons 2 and 3 (3, 16). For many years, 5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl was believed to be the minor metabolite generated when LB400 BPDO catalyzed the oxygenation of 2,2′-CB. However, it was recently shown that LB400 BphAE produced cis-3,4-dihydro-3,4-dihydroxy-2,2′-dichlorobiphenyl as a minor metabolite and that 5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl was not generated by this reaction (3). Saturation mutagenesis of region III amino acids generated variant BphAEs with extended PCB-degrading potency (5). Many of these variants had changed their regiospecificity toward 2,2′-CB to oxygenate principally the vicinal meta-para carbons 3 and 4 instead of carbons 2 and 3 (3, 5), and none of them catalyzed vicinal ortho-meta carbons 5 and 6. Pseudomonas pseudoalcaligenes KF707 produced a BPDO presumed to catalyze a 5,6-dioxygenation of 2,2′-CB (28, 32). However, the catalytic activity of KF707 BPDO toward this congener and other ortho-substituted chlorobiphenyls is poor (32). Furthermore, the identity of the dihydrodiol metabolite produced from 2,2′-CB has only been deduced by gas chromatography-mass spectrometry (GC-MS) analysis (32) and further analyses are needed to assess its identity as 5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl.
Natural diversity of homologous genes retains principally the beneficial mutations through natural selection. It is therefore far more conservative, functionally and structurally, than random mutations. This explains why family shuffling of homologous genes generates libraries that comprise a high proportion of active clones covering a large array of the sequence space (8, 27). However, family shuffling is limited to the genes available in databanks that were cloned from cultivable bacteria. On the other hand, it has been estimated that only 1% of the soil microorganisms can be cultured (2). The size of the population that contributes to the diversity of natural genes in the environment is therefore considerably bigger than the one that is accessible from cultivated bacteria (34). Given these premises, in order to increase the diversity of the library of chimeric genes generated from family shuffling, we have designed a strategy that uses soil DNA to evolve bphA. Amplifying a whole gene from soil microbial populations is not feasible because conserved sequences are required to design the degenerate primers used for the polymerase reaction, and these are not usually located at the extremities of genes. Therefore, we chose to amplify from soil DNA a segment of the bphA C-terminal portion that, based on the crystal structure of Rhodococcus sp. strain RHA1 BphA1 (12), comprised the residues that are lining the BphAE catalytic site. The resulting bank of amplified DNA fragments was shuffled to generate a library of hybrid fragments that was used to replace the corresponding portion of LB400 bphA.
As mentioned above, in previous investigations (3, 4, 5) we have examined the effect of altering the region III amino acid pattern on the catalytic activity towards ortho-substituted chlorobiphenyls. None of the numerous variants obtained in the course of these investigations was able to catalyze a 5,6-dioxygenation of ortho-chlorinated congeners. This suggested that other residues influence the capacity to bind and orient these congeners inside the catalytic pocket. In this regard, the purpose of this work was to isolate from the environment new sets of amino acid combinations of the C-terminal portion of BphA that would allow the resulting BphAE enzymes to catalyze an efficient dioxygenation of 2,2′-CB onto carbons 5 and 6. We anticipated that such variants would oxygenate efficiently onto carbons 5 and 6 and other congeners, such as 2,2′,3,3′-tetrachlorobiphenyl, which are oxygenated principally onto carbons 4 and 5 by LB400 BPDO (5). Therefore, we have also examined the catalytic properties of these variants towards 2,2′,3,3′-CB and towards other biphenyl analogs.
MATERIALS AND METHODS
Bacterial strains, plasmids, chemicals, and general protocols.
Escherichia coli DH11S (20) and C41(DE3) (22) (Statagene, La Jolla, CA) were used in this study. Plasmids pQE31[LB400 bphAE], pDB31[LB400 bphFG], and pDB31[LB400 bphFGBC] were described previously (3, 4, 5). DNA protocols were done according to Sambrook et al. (26). DNA was extracted from soil according to the protocol described by Cho et al. (7), except that the final DNA preparation was further purified by agarose gel electrophoresis. DNA was sequenced at the Genome Quebec DNA Sequencing Center (Montreal, Quebec, Canada). The chlorobiphenyls (>99% pure) used in this work were from AccuStandard (New Haven, CT). Benzene, toluene, and ethyl benzene were from Aldrich Chemicals (Milwaukee, WI).
Strategy to shuffle soil DNA.
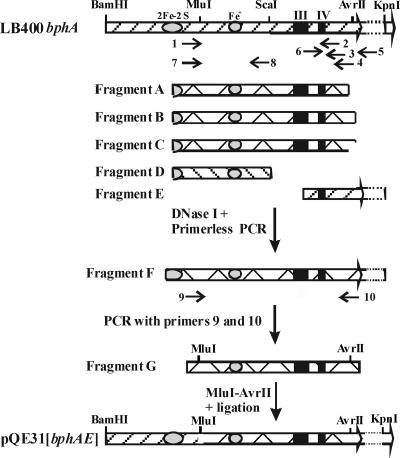
The strategy to evolve LB400 bphA is depicted in Fig. 2. DNA extracted from PCB-contaminated soils was PCR amplified with degenerate primers designed to amplify the C-terminal portion of bphA. The resulting DNA was evolved by one round of shuffling to generate a library of DNA fragments that was used to replace the corresponding portion of LB400 bphA. Contaminated soils G7, G8, and G9 used in this study were described previously (14).
FIG. 2.
Strategy used to shuffle soil DNA with LB400 bphA. LB400 bphA is drawn at the top, showing the positions of regions III and IV, the Rieske cluster, and the mononuclear iron. Degenerate primers (small arrows) 1, 2, 3, and 4 were used to amplify fragments A, B, and C from soil DNA. These fragments were shuffled with D and E amplified, respectively, from B-356 bphA with primers 7 and 8 and from LB400 bphAE with primers 5 and 6. The resulting library of 864-bp MluI/AvrII fragments was used to replace the corresponding fragment in pQE31[LB400 bphAE]. The sequences of the primers are indicated in the text. The right-end side limit of bphA in pQE31[LB400 bphAE] is indicated by the large arrowhead.
In a first series of experiments, sets of primers were designed to recover an optimal representation of bphA from soil DNA. Based on sequence alignment of bphA genes, four degenerate primers (one forward and three reverse) were designed. These four primers could potentially amplify three DNA fragments, A, B, and C, from bphA. Forward primer 1 (5′ GGC TGG GCC TAC GAC ANC GC 3′) started at a position corresponding to bp 370 of LB400 bphA; reverse primers 2 (5′ ADV CCS CGB GCC GCB TCH TCG 3′), 3 (5′-CRT CGT CCT GMT CRA AVA CRC C 3′), and 4 (5′ TT GAG SGY VKC CCA GYB VGG 3′) started at positions corresponding to bp 1322, 1165, and 1373, respectively, of LB400 bphA. These primers were tested to amplify ten homologous bphA genes from our collection plus the DNA extracted from each of the soils, G7, G8, and G9. As shown in Table 1, a fragment of expected size was amplified from all of the ten homologous genes tested with at least one of the three possible sets of primers. Furthermore, the three sets of primers amplified DNA fragments of expected sizes from each sample of soil DNA.
TABLE 1.
Ability of degenerate primers to PCR amplify a segment of bphA from indicated strain or soil DNA
| PCB-degrading strain or contaminated soil (reference) | Degenerate primersa
|
||
|---|---|---|---|
| 1 and 2 | 1 and 3 | 1 and 4 | |
| C. testosteroni B-356 (1) | + | + | ND |
| B. xenovorans | |||
| LB400 (1) | + | − | ND |
| B-206 (1) | − | + | + |
| Bacillus sp. strain B-257 (1) | |||
| − | + | ND | |
| Pseudomonas alcaligenes B-357 (1) | + | − | ND |
| Pseudomonas sp. | |||
| B-218 (1) | − | − | + |
| YT-01 (1) | + | − | + |
| Ralstonia eutropha H850 (1) | + | − | ND |
| Corynebacterium sp. strain MB-1 (1) | + | − | ND |
| Pseudomonas sp. strain P2 (25) | − | − | + |
| Soil G7 (14) | + | + | + |
| G8 (14) | + | + | + |
| G9 (14) | + | + | + |
See text for the bp sequence of each primer. +, presence of a DNA fragment of expected size; −, no DNA fragment of expected size; ND, not determined.
Based on these results, the DNA from each soil, G7, G8, and G9, was PCR amplified individually using a mixture of primers 1, 2, 3, and 4. Then, the DNA fragments amplified from each soil sample were combined. The resulting mixture of fragments was shuffled with fragments D and E, using the strategy shown in Fig. 2, which was designed to prevent self-hybridization of the parental LB400 bphA. Fragment D was amplified from Comamonas testosteroni B-356 bphA, and fragment E was amplified from LB400 bphA (Fig. 2). The 525-bp fragment D was amplified with the set of forward and reverse primers 7 (5′ GGC TGG GCC TAC GAC ATC GC 3) and 8 (5′ CTT GCG TCC AGT ACT GTG TGA 3). The 447-bp fragment E was amplified with primers 5 (5′ GTG GTC CTG CAA CTT TAT CC 3′) and 6 (5′-CAA TGA AAT CGA GGT GTG GG 3′). Neither fragments D nor E encoded the portion of BphA corresponding to the fragment comprised between residues E298 and P347 of LB400 BphA. Therefore, this segment of the library of evolved proteins could only be encoded by DNA amplified from soil. The combined DNA fragments amplified from soil plus fragments D and E were digested with DNase I to prepare the 50- to 100-bp fragments for DNA shuffling according to the method of Zhao and Arnold (35). Expand high-fidelity DNA polymerase was used for the primerless PCR step. Conditions were as recommended by Roche Diagnostics Canada except that the program was as follows for 60 cycles: 94°C for 45 s, 42°C for 1 min, and 72°C for 1 min followed by a 10-min period at 72°C. The approximately 1-kb fragment F obtained from the primerless PCR was amplified with the inner primers 9 (5′ CTG GGG GCC GTT GCA GGC AC 3′) and 10 (5′ TTG AGC GTG GCC CAC CTA 3′). Expand high-fidelity DNA polymerase was used under the conditions recommended by Roche Diagnostics Canada. The resulting 903-bp fragment G was digested with MluI and AvrII to replace the corresponding 864-bp fragment in pQE31[LB400 bphAE]. This library of plasmids was transformed into E. coli DH11S(pDB31[bphFGBC]) or E. coli DH11S(pDB31[bphFGB]), and the clones were screened for their capacity to transform 2,2′-CB into a yellow or pinkish metabolite as described previously (3).
Site-directed mutagenesis.
Site-directed mutagenesis of variant S100 bphA to change positions M237S238 → T237T238, L283→ S283, F370 → Y370, and T377 → N377 were performed using a two-step mutagenesis protocol similar to the one described for saturation mutagenesis of bphA (23). For each mutant, as the first step, the right-end side of S100 bphA was amplified using forward primers F100-237-38 (5′ GGCACCACGACGCACCTGTCC 3′), F100-283 (5′ ATGAGCCGGGCTCACTCATGGCGGTAATG 3′), F100-370 (5′ ATCAAGGAAGAATATCGTCGGCAC 3′), and F100-377 (5′ CACAACATCCGCAACTTCTCTGCAGG 3′) individually, in conjunction with the reverse primer 2059R (5′ GGTACCCCCTAGAAGAACATGCT 3′) (23); for the second step, the left-end side of S100 bphA was amplified using reverse primers R100-237-238 (5′ GGACAGGTGCGTCGTGGTGCC 3′), R100-283 (5′ TTACCGCCATGAGTGAGCCCGGCTCATC 3′), R100-370 (5′ GTGCCGACGATATTCTTCCTTG 3′), and R100-377 (5′ CCTGCAGAGAAGTTGCGGATGTTGTG 3′), in conjunction with the forward primer 1F (5′ CGGGATCCGATGAGTTCAGCAATCA 3′) (23). All other steps to assemble the two fragments and to subclone the 864-bp MluI-AvrII fragment into pQE31[LB400 bphAE] were performed exactly as described previously (23).
Assays to examine the catalytic activity of E. coli cells with BphAE expression variants.
The identities and amounts of metabolites produced from chlorobiphenyls or other biphenyl analogs by resting cell suspensions of isopropyl-β-d-thiogalactopyranoside (IPTG)-induced E. coli(pDB31[bphFG])(pQE31[LB400 bphAE]), E. coli(pDB31[bphFG])(pQE31[S100 bphAE]), E. coli(pDB31[bphFG])(pQE31[S149 bphAE]), or E. coli(pDB31[bphFG])(pQE31[S151 bphAE]) and by E. coli carrying pDB31[bphFG] plus one of the variants M237S238 → T237T238, L283→ S283, F370 → Y370, or T377 → N377 of pQE31[S100 bphAE] were evaluated using previously described procedures (3, 4) to grow the cell and to quantify and analyze the metabolites by GC-MS after derivatization with n-butyl boronate (nBuB) or N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) to produce the trimethylsilyl (TMS) derivatives.
Assays to examine the catalytic activity of His tag-purified BphAE variants.
The catalytic activity of variant BphAEs toward chlorobiphenyls or benzene, toluene, or ethyl benzene was evaluated using BPDOs reconstituted from His-tagged purified components. Each enzyme component was produced in E. coli C41(DE3) expressing the encoding gene from pET-14b (Novagen, Madison, WI), following previously published procedures (23). Plasmids pET-14b bphAE S100, S149, S151, and II-9 were constructed by subcloning the 2,079-bp BamHI-HindIII DNA fragment from each of the pQE31 S100, S149, S151, and II-9 bphAE plasmids. The enzyme purification was performed according to previously described protocols for LB400 and RR41 BphAE (23).
The enzyme assays to examine the catalytic activities toward chlorobiphenyls were performed in 50 mM morpholineethanesulfonic (MES) buffer (pH 6.1, 37°C) as described previously (17). The metabolites were extracted from the reaction vial and analyzed by GC-MS after derivatization with nBuB or BSTFA according to previously described protocols (3, 23). Catalytic activities were quantified according to protocols described previously from the measurement of substrate depletion (18) or from the sum of the area of the peaks of metabolites (3) recorded by GC-MS analysis. In some cases, the catalytic activity was evaluated by recording the oxygen consumption rates using a Clarke-type Hansatech model DW1 oxygraph (19). In this case, the assay conditions were as above, except that the pH of the 50 mM MES buffer was adjusted to 5.5.
The activities toward benzene, toluene, and ethyl benzene were evaluated in a coupled reaction by measuring the production of their meta cleavage metabolite at the optimal wavelengths of absorption, which were found to be, respectively, 375 nm, 389 nm, and 398 nm for the meta cleavage metabolites derived from benzene, toluene, and ethyl benzene.
Isolation and characterization of 5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl.
5,6-Dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl was produced from a resting cell suspension of IPTG-induced E. coli(pDB31[bphFG])(pQE31[S100 bphAE]). It was extracted with ethyl acetate and purified by high-performance liquid chromatography (HPLC). The ethyl acetate extract was evaporated under a stream of nitrogen and the residues were dissolved in a mixture of water-methanol-acetonitrile (50:25:25, vol/vol). The solution was injected onto an analytical Eclipse XDB-C8 reverse-phase column (4.6 mm by 150 mm). The column was equilibrated with water-methanol-acetonitrile (50:25:25). The compound was eluted using the same solvent mixture at 1 ml/min. The Agilent model 1314 variable wavelength detector was set at 276 nm, which was previously determined to be the maximal wavelength of the dihydro-dihydroxy-dichlorobiphenyl metabolite of 2,2′-CB (16). Under these conditions, 5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl eluted as a single peak. The peak of the metabolite was collected, the solvent phase was evaporated under a stream of nitrogen, and the residual aqueous phase was extracted with ethyl acetate. The identity and purity of the metabolite was confirmed by GC-MS analyses of its butylboronate derivative (5, 21). Nuclear magnetic resonance (NMR) spectra were obtained at the NMR spectrometry center of INRS-Institut Armand-Frappier with a Brucker 500-mHz spectrometer. The analyses were carried out in deuterated acetonitrile at room temperature.
Metabolism of 5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl by biphenyl catabolic enzymes.
Purified preparations of B. xenovorans LB400 2,3-dihydro-2,3-dihydroxybiphenyl dehydrogenase (BphB) and 2,3-dihydroxybiphenyl dioxygenase (BphC) were used to assess their ability to metabolize the 5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl. The procedures to produce these enzymes as His-tagged proteins by recombinant E. coli cells and to purify them have been described previously (3, 6). BphB assays using as substrate HPLC-purified 5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl were performed in 400-μl 50 mM MES buffer, pH 7.0. The catalytic activity was measured by recording the substrate depletion by GC-MS analysis. The metabolites were extracted from the reaction vial with ethyl acetate at neutral pH and derivatized with nBuB or BSTFA for GC-MS analysis (3).
The ability of BphC to oxidize 5,6-dihydroxy-2,2′-dichlorobiphenyl was verified in a coupled reaction using His-tagged purified BphB plus His-tagged purified BphC and HPLC-purified 5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl as substrate. The activity was measured in 50 mM MES buffer, pH 7.0, containing 0.6 nmol BphB, 0.6 nmol His-tagged purified LB400 BphC, and 200 μM NAD in a final volume of 400 μl. The enzyme activity was monitored by measuring substrate depletion by GC-MS analysis. For GC-MS analysis of the meta cleavage metabolites, the assay medium was acidified to pH 3.5, extracted with ethyl acetate, and treated with BSTFA.
Structural modeling of variant BphAEs.
The interactive 3D-JIGSAW structural modeling software of the UK Cancer Research Institute (http://www.bmm.icnet.uk/servers/3djigsaw/) was used to model the structure of S100, S149, and S151 BphAEs using as template the crystal structure of Rhodococcus sp. strain RHA1 BphA1.
Nucleotide sequence accession numbers.
The accession numbers for DNA sequences of S100, S149 and S151 bphA are respectively EF090937, EF090938 and EF090939.
RESULTS
Screening of BphAE variants able to oxygenate 2,2′-CB onto carbons 5 and 6.
A library of BphAE variants was prepared using the protocol shown in Fig. 2 and screened for their ability to transform 2,2′-CB to a yellow- or pinkish-colored metabolite. Among 1,200 E. coli DH11S(pDB31[bphFGBC])(pQE31[bphAE]) transformants, 20 were able to transform 2,2′-CB. Ten of those were sequenced (not shown). None of them were identical. Their MluI-AvrII fragment exhibited between 70 and 95% homology with the corresponding fragment of LB400 bphA. One of the variants (S100) produced a single dihydro-dihydroxy-2,2′-CB metabolite that exhibited a gas chromatographic retention time clearly distinct from that of cis-3,4-dihydro-3,4-dihydroxy-2,2′-CB (Table 2), which was previously identified as a major metabolite produced by the evolved p4 BphAE (5). Two other variants (S149 and S151) that produced 2,3-dihydroxy-2′-dichlorobiphenyl as a major metabolite plus a small amount of this new metabolite were retained for further characterization.
TABLE 2.
Metabolites produced from ortho-substituted PCB congeners by variant dioxygenases
| Congener | Metabolite | RTe | Activity relative to LB400a
|
Ratio of metabolites producedb
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| S100 | S149 | S151 | LB400 | S100 | S149 | S151 | |||
| 2,2′-CB | 2,3-Dihydroxy-2′-chlorobiphenyl | 14.9 | 1 | 1 | 1 | 92 (6) | 0.2 (0.2) | 60 (1) | 63 (5) |
| 5,6-Dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl | 16.3 | — | 99.8 (0.2) | 23 (1) | 18 (2) | ||||
| 3,4-Dihydro-3,4-dihydroxy-2,2′-dichlorobiphenyl | 17.5 | 8 (1) | — | 17 (1) | 19 (2) | ||||
| 2,6-CB | 3,4-Dihydro-3,4-dihydroxy-2′,6′(or 2,6)-dichlorobiphenylc | 17.2 | 1,500 | 1 | 500 | ND | 24 (1) | 0.5 (1) | 10 (1) |
| 3,4-Dihydro-3,4-dihydroxy-2,6(or 2′,6′)-dichlorobiphenylc | 17.5 | ND | 1 (1) | 99 (1) | 74 (1) | ||||
| 2,3-Dihydro-2,3-dihydroxy-2′,6′-dichlorobiphenylc | 18.5 | ND | 70 (5) | 0.5 (1) | 16 (1) | ||||
| 2,4′,5-CBd | 2′,3′-Dihydro-2′,3′-dihydroxy-2,5,4′-trichlorobiphenyl | 22.7 | 1 | 1 | 0.5 | 1 (1) | 100 | 4 (1) | 1 (1) |
| 3,4-Dihydro-3,4-dihydroxy-2,5,4′-trichlorobiphenyl | 24.3 | 99 (1) | 0 | 96 (1) | 99 (1) | ||||
| 2,2′,3,3′-CB | 5,6-Dihydro-5,6-dihydroxy-2,2′,3,3′-tetrachlorobiphenyl | 25.3 | 1 | 1 | 1 | 5 (1) | 100 | 95 (2) | 93 (5) |
| 4,5-Dihydro-4,5-dihydroxy-2,2′,3,3′-tetrachlorobiphenyl | 26.7 | 95 (1) | — | 5 (2) | 7 (4) | ||||
| 2,2′,5,5′-CBd | 3,4-Dihydro-3,4-dihydroxy-2,2′,5,5′-tetrachlorobiphenyl | 24.4 | 0 | 1 | 1 | 100 | — | 100 | 100 |
Activity of each BphAE relative to LB400 BphAE was calculated from the ratio of the sum of the area under the peaks of all metabolites produced from each congener after 2 min of incubation with 1 nmol of enzyme following the protocol described in Materials and Methods. The variance of the ratio of the sum of metabolites was <15% in all cases.
Metabolite ratio based on the area of the peaks of metabolites detected by GC-MS analysis of butylboronate-derived metabolites produced by purified enzyme preparations. —, no metabolite detected; ND, not determined.
The exact position of dioxygenation has not been determined precisely.
Data were obtained with IPTG-induced E. coli cells expressing the indicated BphAE following the protocol described in Materials and Methods. SDS-PAGE gels of cell total proteins indicate similar levels of expression of BphAEFG for all clones used in this experiment.
RT is retention time in min of each metabolite under the chromatographic conditions used.
The specific activity of the His-tagged purified preparations of S100, S149, and S151 BphAEs toward biphenyl was evaluated at 37°C and pH 5.5. Under these conditions, the initial rates of oxygen consumption were, respectively, 30.5 ± 5, 84.9 ± 2, 48.7 ± 2, and 74.8 ± 8 nmol−1 min−1 per nmol LB400, S100, S149, and S151 BphAE. Under the same conditions, when 2,2′-CB was the substrate, the oxygen consumption rates were, respectively, 16 ± 1, 16 ± 1, 16 ± 1, and 13 ± 1 nmol min−1 per nmol of LB400, S100, S149, and S151 BphAE. Therefore, these variants oxygenated 2,2′-CB at rates comparable to that of LB400 BphAE.
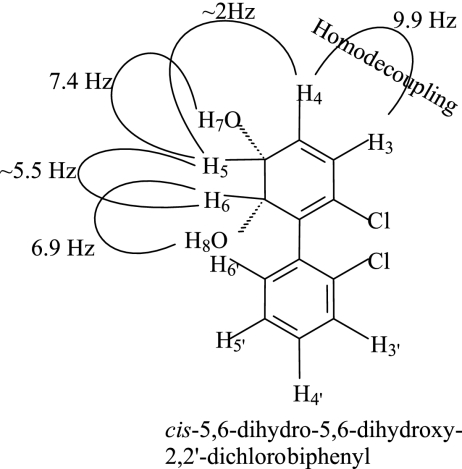
The single metabolite produced from 2,2′-CB by S100 BphAE eluted as a single peak at 9.4 min in the HPLC system described in Materials and Methods. It exhibited a λmax at 262 nm. The compound could only be either 5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl or 4,5-dihydro-4,5-dihydroxy-2,2′-dichlorobiphenyl. When the NMR spectrum was taken in deuterated acetonitrile, signals were detected at values between 7.30 and 7.40 ppm, which correspond to the four aromatic protons (m, H6′), (m, H5′-4′), and (m, H3′) (Fig. 3). In the higher portion of the field, signals characteristic of aliphatic protons were recorded at 6.04 ppm (dd, H3), 6.02 ppm (dd, H4), 4.56 ppm (m, H5), and 4.27 ppm (dd, H6), in addition to the two hydroxylic protons at 3.25 ppm (d, H7) and 3.33 ppm (d, H8), respectively. H5 was weakly coupled to H4 (J∼2 Hz). The absence of correlation between the other vinyl proton (H3) and the methine protons (H5 and H6) indicated that the two vinyl protons were vicinal. This excludes 4,5-dihydro-4,5-dihydroxy-2,2′-dichlorobiphenyl as metabolite. H5 was coupled to H7 (J = 7.4) and H6 was coupled to H8 (J = 6.9 Hz), and H5 and H6 were coupled to each other (J∼5.5). These latter values are consistent with the theoretical values predicted for the cis configuration. When H5 was irradiated at 4.56 ppm, two doublets for H3 and H4 were observed with a coupling constant of 9.9 Hz, characteristic of a Z-configuration of this system (Fig. 3). This confirmed that the two vinyl protons H3-4 were vicinal. Together, these data confirmed the identity of the metabolite as cis-5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl.
FIG. 3.
Structure of cis-5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl produced from catalytic oxygenation of 2,2′-CB by S100 BphAE and their NMR coupling constants. The coupling constant of 9.9 Hz shown between H3 and H4 was obtained after homodecoupling by irradiating H5 at 4.56 ppm.
Metabolism of cis-5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl.
When 0.6 nmol His-tagged purified BphB, 100 nmol NAD, and 75 nmol cis-5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl were added to the enzyme assay, approximately 10 nmol of substrate was consumed when the reaction was run for 5 min at 37°C. The BSTFA-derivatized cis-5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl decomposed under the chromatographic settings we used. However, a peak corresponding to TMS-5,6-dihydroxy-2,2′-dichlorobiphenyl was well resolved and could be analyzed by mass spectrometry. The spectrum included ions at m/z = 398 M+, m/z = 383 (M-15), m/z = 363 (M-Cl), m/z = 310 (M-Me4Si), m/z = 275 (M-Cl-Me4Si), and m/z = 260 (M-Cl-Me4Si-Me), which are characteristic features of hydroxy-chlorobiphenyls (21).
cis-5,6-Dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl was transformed to a yellow-colored metabolite by a coupled reaction involving BphB plus BphC. The metabolite exhibited a λmax at 395 nm. When the reaction was initiated with 75 nmol cis-5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl, 10 nmol of substrate was consumed after 5 min and no 5,6-dihydroxy-2,2′-dichlorobiphenyl was detected. This indicates that BphC is not the limiting step in this reaction. The yellow metabolite was extracted with ethyl acetate and derivatized with BSTFA for GC-MS analysis. A single peak of metabolite was detected. Its MS spectrum comprised diagnostically important ions at m/z = 430 (M+), m/z = 415 (M-CH3), m/z = 395 (M-Cl), and m/z = 313 (M-COOTMS). Based on a previous report (21), these features identified the resulting metabolite as 2-hydroxy-5-chloro-6-oxo-6-(2-chlorophenyl)-hexa-2,4-dienoic acid, the meta cleavage metabolite produced from catalytic dioxygenation of 5,6-dihydroxy-2,2′-dichlorobiphenyl.
Catalytic properties of evolved BphAEs toward ortho-substituted chlorobiphenyls and other biphenyl analogs.
The apparent ratio of metabolites generated from 2,2′-CB, 2,2′,3,3′-CB, and 2,2′,5,5′-CB by purified enzyme preparations of LB400, S100, S149, and S151 BphAEs were compared (Table 2). As indicated above, all three variants oxygenated 2,2′-CB at a rate similar to that of LB400 BphAE. When 2,2′-CB was the substrate, S100 BphAE produced almost exclusively cis-5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl. On the other hand, similar to LB400 BphAE, both S149 and S151 produced 2,3-dihydroxy-2′-chlorobiphenyl as a major metabolite, but in addition both enzymes generated cis-5,6-dihydro-5,6-dihydroxy-2,2′-dichlorobiphenyl. In a previous report, LB400 BphAE was found to oxygenate 2,2′,3,3′-CB principally onto carbons 4 and 5 to generate cis-4,5-dihydro-4,5-dihydroxy-2,2′,3,3′-tetrachlorobiphenyl as the major metabolite and cis-5,6-dihydro-5,6-dihydroxy-2,2′,3,3′-tetrachlorobiphenyl resulting from a 5,6-dioxygenation reaction was produced in a much lesser amount (3). Based on the area of GC-MS peaks of metabolites, purified preparations of S100, S149, and S151 BphAE oxygenated 2,2′,3,3′-CB at rates similar to that of LB400 BphAE. However, S100 BphAE oxygenated this substrate onto carbons 5 and 6 exclusively to produce cis-5,6-dihydro-5,6-dihydroxy-2,2′,3,3′-tetrachlorobiphenyl, which was also the major metabolite produced by S149 and S151 BphAEs (Table 2). Unlike purified preparations of S149 and S151 BphAEs that produce 3,4-dihydro-3,4-dihydroxy-2,2′,5,5′-CB from 2,2′,5,5′-CB, S100 BphAE was unable to oxygenate this substrate. Therefore, S100 is unable to effect a 3,4-dioxygenation of 2,2′,5,5′-CB. This explains why S100 BphAE exhibited a much different pattern of metabolites toward 2,4′,5-CB than LB400, S149, and S151 BphAEs. Two dihydro-dihydroxy metabolites are likely to be produced from 2,4′,5-CB, 2′,3′-dihydro-2′,3′-dihydroxy-2, and 4′,5-trichlorobiphenyl resulting from a 2,3-dioxygenation of the para-substituted ring and 3,4-dihydro-3,4-dihydroxy-2,4′,5-trichlorobiphenyl resulting from a 3,4-dioxygenation of the disubstituted ring. Suenaga et al. (28) have shown that LB400 BphAE produced 3,4-dihydro-3,4-dihydroxy-2,4′,5-trichlorobiphenyl from 2,4′,5-CB. S149 and S151 BphAEs metabolized 2,4′,5-CB very similarly to LB400 BphAE. On the other hand, S100 metabolized 2,4′,5-CB similarly to P. pseudoalcaligenes KF707 BPDO, which was found to generate 2′,3′-dihydro-2′,3′-dihydroxy-2,4′,5-trichlorobiphenyl exclusively from 2,4′,5-CB (28). This is consistent with the inability of these two dioxygenases to effect a 3,4-dioxygenation of the 2,5-substituted ring. Finally, when IPTG-induced resting cells of recombinant E. coli expressing S100, S149, or 151 BphAE were incubated with 2,2′,4,4′-CB for 18 h, a small amount (approximately 1%) of the substrate was converted into a single metabolite whose GC-MS features corresponded to a dihydro-dihydroxy-tetrachlorobiphenyl which can only be 5,6-dihydro-5,6-dihydroxy-2,2′,4,4′-tetrachlorobiphenyl (data not shown in Table 2). This metabolite was not produced when recombinant E. coli cells expressing LB400 BphAE were incubated in identical conditions. Interestingly, based on the sum of metabolites produced, when 1 nmol of enzyme was reacted with 2,6-CB for 1 min, S100 BphAE oxygenated this substrate 1,500 times better than LB400 BphAE (Table 2). Three metabolites were produced. Assuming the major metabolite was the one derived from a 2,3-dioxygenation of the nonchlorinated ring, the two others must have been produced from a 3,4-dioxygenation of the chlorinated or unchlorinated ring. It is noteworthy that the ratio of metabolites produced from 2,6-CB by S151 and S149 BphAE differed significantly from the one produced by S100 BphAE. At this time, the exact position of 3,4-dioxygenation (on the chlorinated or nonchlorinated ring) is not known, but data show that although S100 BphAE cannot catalyze a 3,4-dioxygenation of 2,2′-CB or 2,2′,5,5′-CB, it certainly catalyzes a 3,4-dioxygenation of 2,6-CB.
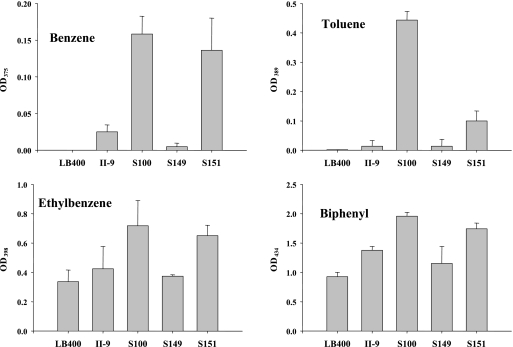
It has been reported that purified LB400 BphAE cannot oxygenate benzene and toluene (15). E. coli cells expressing LB400 BphAE poorly oxygenated ethyl benzene (29). Unlike LB400 BphAE, purified preparations of S100 BphAE oxygenated benzene and toluene efficiently and they oxygenated ethyl benzene more efficiently than LB400 BphAE (Fig. 4). To a lesser degree, S149 and S151 BphAEs were also able to oxygenate benzene, toluene, and ethyl benzene. Therefore, the catalytic pocket of these variants was able to bind monocyclic aromatic compounds efficiently enough to allow a catalytic reaction to occur.
FIG. 4.
Production of yellow meta cleavage metabolites from benzene, toluene, and ethyl benzene by a coupled reaction involving the indicated His-tagged purified BphAEFGs plus His-tagged purified LB400 BphB and LB400 BphC. The formation of the yellow metabolite was quantified at the corresponding maximum wavelength of absorption. Results are expressed as average values ± the standard deviations.
Sequence analysis of S100, S149, and S151 bphA and structure modeling of S100 BphAE.
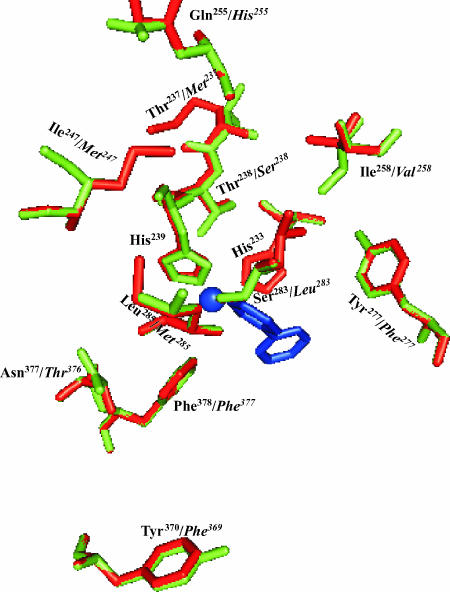
The amino acid sequence patterns of S100, S149, and S151 are shown in Fig. 5. Over the entire protein, the amino acid sequence of variant S100 BphA exhibits, respectively, 95% and 94% identity with those of variants S149 and S151. S100, S149, and S151 exhibit, respectively, 93%, 94%, and 96% identity with LB400 BphA. Based on the sequence alignment, aside from region III amino acids that differ from those of LB400 BphA, residues that are common to the three variants and differ from LB400 BphA are E179, M237, S238, M247, H255, V258, S268, F277, and M285, and in addition residue S283 of LB400 was replaced by an aliphatic hydrophobic amino acid in all three variants. According to a model of S100 BphAE obtained using the 3D-JIGSAW software based on the crystal structure (1ULJ.pdb) of Rhodococcus sp. strain RHA1 BphA1 (12), none these nine residues are within four Å from the substrate (Fig. 6). However, residues M237S238 are part of a small segment of amino acids that are ended on each side by H233 and H239, two residues that coordinate the mononuclear iron. Residue L283 (corresponding to L274 of RHA1 BphA1) is a residue that according to the RHA1 BphA1 structure shifted significantly after substrate binding (12). In addition, although in our model of S100 BphAE, L283 was at 6.2 Å from the substrate, based on the Pseudomonas fluorescens IP01 cumene dioxygenase crystal structure, the corresponding residue L284 is part of a loop involved in the formation of the active site of this oxygenase (9).
FIG. 5.
Alignment of sequences of LB400, S100, S149, and S151 BphAs. Only the residues of the C-terminal portion that differ in at least one BphA are shown in the alignment. Residues that are identical in S100, S149, and S151 BphAs are shown in bold. III and IV refer to BphA regions III and IV. The amino acid numbering at the top is for LB400, II-9, and S151 BphAs; the bottom (italic) numbering is for S100 and S149 BphAs.
FIG. 6.
Superposition of a three-dimensional model, using the Rasmol software, of selected amino acid residues of LB400 BphAE (green) and S100 BphAE (red) (names in italics) that differ in both proteins. The figure shows their conformation and their position relative to the active iron (shown as a blue sphere) and the substrate and relative to His233 and His239 that coordinate the active iron. None of the residues are within 4 Å of the active site in S100 BphAE.
Site-directed mutagenesis of residues likely to influence specificity and regiospecificity.
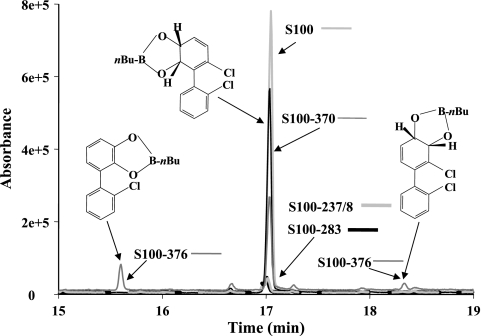
Because our analysis indicated that M237S238 and L283 of S100 BphA were likely candidates to influence substrate specificity or regiospecificity, we exchanged them to, respectively, T237T238 and S283 as in LB400 BphA. Both S100 (M237S238 → T237T238) and S100 (L283→ S283) variants retained their capacity to catalyze a 5,6-dioxygenation of 2,2′-CB, but interestingly, based on the amount of metabolite produced by recombinant E. coli cells expressing these enzymes, they exhibited considerably lesser activity toward this substrate (Fig. 7). In this case, biphenyl was oxygenated at a similar rate for control cells incubated under the same conditions (not shown) and all clones produced similar amounts of BphAEFG as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of cell extracts (not shown), showing that the difference in activity toward 2,2′-CB was not the result of differential enzyme expression.
FIG. 7.
Total ion chromatogram showing the metabolites produced from 2,2′-CB by E. coli expressing either S100 BphAE or the indicated S100 variants. IPTG-induced cells were incubated for 18 h with 100 nmol 2,2′-CB and then extracted with ethyl acetate, and the metabolites were derivatized with butylboronate for GC-MS analysis. Controls of E. coli cells incubated in identical conditions produced similar amounts of 2,3-dihdyro-2,3-dihydroxybiphenyl when biphenyl was the substrate. Furthermore, SDS-PAGE of the cell extracts showed similar levels of production of BphAE for all strains.
Previous reports (3, 5, 24, 28, 30, 32) showed that region III and region IV of BphA strongly influence the turnover rate of reaction and regiospecificity toward ortho-chlorinated congeners; residue N377 of LB400 BphA was found to be especially critical in determining the capacity of the enzyme to catalyze a meta-para dioxygenation of the biphenyl ring (28, 30). Unlike S100 BphAE, S149 and S151 BphAEs produced metabolites resulting from 2,3- and 3,4-dioxygenation (Table 2). Since T376 of S100 BphA was replaced by N in S149 and S151 BphAs we have exchanged T376 of S100 BphA with N. When a resting cell suspension of recombinant E. coli expressing S100 (T376→N) BphAE was incubated with 2,2′-CB, based on the area of GC-MS peaks of metabolites, this enzyme was less efficient than the parent S100 BphAE in converting the substrate (Fig. 7). In addition, S100 (T376→N) BphAE produced 2,3-dihydroxy-2′-chlorobiphenyl and 3,4-dihydro-3,4-dihydroxy-2,2′-chlorobiphenyl in an approximately 50-50 ratio. This confirms the importance of residue N377 of LB400 BphA in determining its capacity to oxygenate the biphenyl ring onto carbons 3 and 4, although in our model T377 of S100 BphA is at 8.2 Å from the substrate, thus far from it.
In previous work, variant II-9 BphA was described with an amino acid sequence that is identical to that of LB400 BphA except that the sequence pattern of region III of II-9 BphA is G335I336N337T338I339R340T341 instead of T335F336N337N338I339R340I341 (4). Therefore, the amino acid sequence pattern of region III of variant II-9 is identical to that of S100 BphA. However, II-9 BphAE is unable to oxygenate 2,2′-CB onto carbons 5 and 6 (Table 2). The fact that S100, the T376→N mutant of S100, S149, and S151 BphAEs were able to catalyze a 5,6-dioxygenation of 2,2′-CB but II-9 BphAE was not shows that amino acid residues other than those of region III influence the regiospecificity of BphA toward di-ortho-substituted biphenyl analogs and their capacity to catalyze the oxygenation onto the ortho-meta carbons 5 and 6.
Several other residues that are not shared by all three variants and that were far from the active center on our S100 BphA model were also exchanged individually for the residue of LB400 BphA. None of them affected the specificity or regiospecificity toward 2,2′-CB. Residue F369 of S100 BphA which is located at 13.5 Å is an example. As shown in Fig. 7, recombinant E. coli cells expressing S100 (F369→Y369) or parental S100 BphA produced similar amounts of 5,6-dihydro,5,6-dihydroxy-2,2′-dichlorobiphenyl.
DISCUSSION
Shuffling of soil DNA to evolve LB400 bphA generated variants S100, S149, and S151 that exhibited novel catalytic properties toward di-ortho-substituted chlorobiphenyls. All three variants have acquired the capacity to oxygenate 2,2′-CB onto the ortho-meta carbons 5 and 6. In addition, unlike variants S149 and S151, variant S100 was unable to oxygenate the meta-para carbons of any of the ortho-substituted congeners tested (2,2′-CB, 2,5,4′-CB, 2,2′,3,3′-CB, and 2,2′,5,5′-CB) but it was able to effect a 3,4-dioxygenation of 2,6-CB. Unlike LB400 BphAE that preferentially oxygenates carbons 2 and 3 of 2,2′-CB to catalyze an oxygenolytic dehalogenation (3, 16), P. pseudoalcaligenes KF707 BPDO was reported to catalyze the oxygenation of 2,2′-CB exclusively on carbons 5 and 6 (28). However, the latter performed poorly on ortho-substituted chlorobiphenyls. Interestingly, variant S100 BphAE performed as well as LB400 BphAE on 2,2′-CB and 2,2′-3,3′-CB and it performed much better than LB400 BphAE on 2,6-CB. Although the inability of S100 BphAE to effect an oxygenolytic dehalogenation appears to be a disadvantage compared to LB400 BphAE, data show that 5,6-dihydro-5,6-dihydroxy-2,2′-chlorobiphenyl can be further metabolized by BphB and BphC. Furthermore, the ability of S100 BphAE to catalyze a 5,6-dihydroxylation of 2,2′,3,3′-CB represents an advantageous feature compared to LB400 BphAE that preferentially oxygenates this congener onto carbons 4 and 5 to produce a dead end dihydrodiol metabolite (3).
Data obtained with the T376→N mutant of S100 BphA confirm previous reports (28, 30), showing that this residue is critical to confer the capacity to catalyze a 3,4-dioxygenation of the biphenyl ring. Yet, according to our model of S100 BphAE, this residue does not interact directly with the substrate. It therefore might play its role indirectly by modulating the positions of residues that interact with the substrate. For example, residue F377 of S100 BphA is lining the catalytic center (located at 3.9 Å from the substrate). Using the Rasmol software, we have superposed the S100 BphAE model created by the 3D-JIGSAW software with that of the S100 (T376→N) mutant. However, this software was not precise enough to show the influence of this mutation on the conformation of the residues lining the catalytic pocket. In fact, as shown in Fig. 6, Phe378 of LB400 BphA aligns perfectly with Phe377 of S100 BphA.
On the other hand, based on previous reports (3, 5, 24) that showed that region III of BphA strongly influences the turnover rate of reaction and regiospecificity toward ortho-chlorinated congeners, it would have been possible that the different sequence patterns at positions 335 to 340 of variants S100, S149, and S151 were responsible for at least part of their phenotypic differences. However, it is noteworthy that the amino acid sequence pattern of region III of the previously described II-9 BphA (4) is identical to the one of S100 BphA, and yet, this variant is unable to oxygenate 2,2′-CB onto carbons 5 and 6. Therefore, residues other than those of region III and of region IV influence the regiospecificity toward di-ortho-substituted chlorobiphenyls.
According to the sequence alignment shown on Fig. 5, at least nine residues are common to S100, S149, and S151 and could be responsible for the capacity to effect a 5,6-dioxygenation of 2,2′-CB. We have examined the effect of exchanging three of the residues that based on our model were likely to influence either the substrate activity or the regiospecificity. Interestingly, exchanging M237S238 for T237T238 and L283 for S283 of S100 BphA as in LB400 BphA created variants that exhibited lower reactivity toward 2,2′-CB but did not change their regiospecificity toward this substrate. Therefore, these positions appear to influence the distance between the reactants involved in the catalytic reaction, but they do not seem to influence the orientation of the substrate inside the catalytic pocket. Therefore, based on the data provided in this work, it is impossible at this time to identify specific residues outside regions III and IV that are responsible for the capacity of S149, S151, and especially S100 to orient the substrate in the proper position to allow catalytic dioxygenation onto carbons 5 and 6 of the chlorobiphenyl carrying an ortho substituent on both rings. However, data show that regiospecificity is a complex process that depends upon interactions that occur between amino acid residues that line the catalytic pocket and the substrate, but in addition it involves other residues that might act by modulating the conformation of the residues lining the catalytic pocket.
In a previous work, changing H255V258G268F277 of P. pseudoalcaligenes KF707 BphA1 to Q255I258A268Y277 as in LB400 BphA created variant 1072 that, unlike both parents, had acquired the capacity to oxygenate benzene, toluene, and ethyl benzene efficiently (29). Individually these residues did not significantly affect the specificity toward monocyclic compounds. The combination of the four mutations was required to change the specificity. Surprisingly, although the sequence pattern of variant S100 BphA for these positions (H255V258S268F277) is very similar to that of KF707 BphA1, like variant 1072, S100 BphAE can oxygenate these compounds efficiently. None of these residues are located in the vicinity of the catalytic center of our model, which confirms data obtained by modeling KF707 BphA1A2 (31). Based on their model structures, Suenaga et al. (31) found that changing H255V258S268F277 of KF707 BphA to Q255I258A268Y277 caused a slight enlargement of the enzyme substrate binding cavity. Our model was not precise enough to reveal such changes, but our results show that the same residues contribute differently to the substrate specificity, depending on the overall structure of the catalytic pocket.
Altogether, the observation made regarding the catalytic features of the variant BphAEs obtained in this investigation brings to mind that changing substrate specificity is a complex process; as pointed out recently by Zielinski et al. (37), it not only involves amino acid residues that interact directly with the substrate, but it also involves residues that are not part of the pocket lining and that are not predictable through structural modeling. These residues might indirectly influence the shape of the catalytic pocket or the conformation of the residues that are involved in substrate binding and positioning inside the catalytic pocket. Furthermore, we cannot exclude the possibility that these residues modulate the conformational changes induced during substrate binding which might be required to accommodate substrate analogs.
Acknowledgments
This work was supported by the Natural Science and Engineering Research Council of Canada grant numbers STPGP 257566-02 and 0039579.
We thank Nicolas Leberre for assistance in NMR analyses and their interpretation.
Footnotes
Published ahead of print on 1 December 2006.
REFERENCES
- 1.Ahmad, D., R. Massé, and M. Sylvestre. 1990. Cloning and expression of genes involved in 4-chlorobiphenyl transformation by Pseudomonas testosteroni: homology to polychlorobiphenyl-degrading genes in other bacteria. Gene 86:53-61. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barriault, D., F. Lépine, M. Mohammadi, S. Milot, N. Leberre, and M. Sylvestre. 2004. Revisiting the regiospecificity of Burkholderia xenovorans LB400 biphenyl dioxygenase toward 2,2′-dichlorobiphenyl and 2,3,2′,3′-tetrachlorobiphenyl. J. Biol. Chem. 279:47489-47496. [DOI] [PubMed] [Google Scholar]
- 4.Barriault, D., M. M. Plante, and M. Sylvestre. 2002. Family shuffling of a targeted bphA region to engineer biphenyl dioxygenase. J. Bacteriol. 184:3794-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barriault, D., and M. Sylvestre. 2004. Evolution of the biphenyl dioxygenase BphA from Burkholderia xenovorans LB400 by random mutagenesis of multiple sites in region III. J. Biol. Chem. 279:47480-47488. [DOI] [PubMed] [Google Scholar]
- 6.Barriault, D., M. Vedadi, J. Powlowski, and M. Sylvestre. 1999. cis-2,3-Dihydro-2,3-dihydroxybiphenyl dehydrogenase and cis-1,2-dihydro-1,2-dihydroxynaphathalene dehydrogenase catalyze dehydrogenation of the same range of substrates. Biochem. Biophys. Res. Commun. 260:181-187. [DOI] [PubMed] [Google Scholar]
- 7.Cho, J. C., D. H. Lee, Y. C. Cho, J. C. Cho, and S. J. Kim. 1996. Direct extraction of DNA from soil for amplification of 16S RNA gene sequences by polymerase chain reaction. J. Microbiol. 34:229-235. [Google Scholar]
- 8.Crameri, A., S. A. Raillard, E. Bermudez, and W. P. Stemmer. 1998. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391:288-291. [DOI] [PubMed] [Google Scholar]
- 9.Dong, X. S., S. Fushinobu, E. Fukuda, T. Terada, S. Nakamura, K. Shimizu, H. Nojiri, T. Omori, H. Shoun, and T. Wakagi. 2005. Crystal structure of the terminal oxygenase component of cumene dioxygenase from Pseudomonas fluorescens IP01. J. Bacteriol. 187:2483-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson, B. D., and F. J. Mondello. 1992. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J. Bacteriol. 174:2903-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furukawa, K., H. Suenaga, and M. Goto. 2004. Biphenyl dioxygenases: functional versatilities and directed evolution. J. Bacteriol. 186:5189-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furusawa, Y., V. Nagarajan, M. Tanokura, E. Masai, M. Fukuda, and T. Senda. 2004. Crystal structure of the terminal oxygenase component of biphenyl dioxygenase derived from Rhodococcus sp. strain RHA1. J. Mol. Biol. 342:1041-1052. [DOI] [PubMed] [Google Scholar]
- 13.Goris, J., P. De Vos, J. Caballero-Mellado, J. Park, E. Falsen, J. F. Quensen III, J. M. Tiedje, and P. Vandamme. 2004. Classification of the biphenyl- and polychlorinated biphenyl-degrading strain LB400T and relatives as Burkholderia xenovorans sp. nov. Int. J. Syst. Evol. Microbiol. 54:1677-1681. [DOI] [PubMed] [Google Scholar]
- 14.Guilbeault, B., M. Sondossi, D. Ahmad, and M. Sylvestre. 1994. Factors affecting the enhancement of PCB degradative ability of soil microbial populations. Int. Biodeterior. Biodegradation 33:73-91. [Google Scholar]
- 15.Haddock, J. D., and D. T. Gibson. 1995. Purification and characterization of the oxygenase component of biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. J. Bacteriol. 177:5834-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haddock, J. D., J. R. Horton, and D. T. Gibson. 1995. Dihydroxylation and dechlorination of chlorinated biphenyls by purified biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. J. Bacteriol. 177:20-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurtubise, Y., D. Barriault, and M. Sylvestre. 1996. Characterization of active recombinant his-tagged oxygenase component of Comamonas testosteroni B-356 biphenyl dioxygenase. J. Biol. Chem. 271:8152-8156. [DOI] [PubMed] [Google Scholar]
- 18.Hurtubise, Y., D. Barriault, and M. Sylvestre. 1998. Involvement of the terminal oxygenase β subunit in the biphenyl dioxygenase reactivity pattern toward chlorobiphenyls. J. Bacteriol. 180:5828-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imbeault, N. Y., J. B. Powlowski, C. L. Colbert, J. T. Bolin, and L. D. Eltis. 2000. Steady-state kinetic characterization and crystallization of a polychlorinated biphenyl-transforming dioxygenase. J. Biol. Chem. 275:12430-12437. [DOI] [PubMed] [Google Scholar]
- 20.Lin, J. J., M. Smith, J. Jessee, and F. Bloom. 1992. DH11S: an E. coli strain for preparation of single-stranded DNA from phagemid vectors. BioTechniques 12:718-7721. [PubMed] [Google Scholar]
- 21.Massé, R., F. Messier, C. Ayotte, M.-F. Lévesque, and M. Sylvestre. 1989. A comprehensive gas chromatographic/mass spectrometric analysis of 4-chlorobiphenyl bacterial degradation products. Biomed. Environ. Mass Spectrom. 18:27-47. [Google Scholar]
- 22.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 23.Mohammadi, M., and M. Sylvestre. 2005. Resolving the profile of metabolites generated during oxidation of dibenzofuran and chlorodibenzofurans by the biphenyl catabolic pathway enzymes. Chem. Biol. 12:835-846. [DOI] [PubMed] [Google Scholar]
- 24.Mondello, F. J., M. P. Turcich, J. H. Lobos, and B. D. Erickson. 1997. Identification and modification of biphenyl dioxygenase sequences that determine the specificity of polychlorinated biphenyl degradation. Appl. Environ. Microbiol. 63:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novakova, H., M. Vosahlikova, J. Pazlarova, M. Mackova, J. Burkhard, and K. Demnerova. 2002. PCB metabolism by Pseudomonas sp. P2. Int. Biodeterior. Biodegradation 50:47-54. [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 27.Stemmer, W. P. C. 2002. Molecular breeding of genes, pathways and genomes by DNA shuffling. J. Mol. Catal. B 19:3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suenaga, H., M. Goto, and K. Furukawa. 2006. Active-site engineering of biphenyl dioxygenase: effect of substituted amino acids on substrate specificity and regiospecificity. Appl. Microbiol. Biotechnol. 71:168-176. [DOI] [PubMed] [Google Scholar]
- 29.Suenaga, H., M. Mitsuoka, Y. Ura, T. Watanabe, and K. Furukawa. 2001. Directed evolution of biphenyl dioxygenase: emergence of enhanced degradation capacity for benzene, toluene, and alkylbenzenes. J. Bacteriol. 183:5441-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suenaga, H., A. Nishi, T. Watanabe, M. Sakai, and K. Furukawa. 1999. Engineering a hybrid pseudomonad to acquire 3,4-dioxygenase activity for polychlorinated biphenyls. J. Biosci. Bioeng. 87:430-435. [DOI] [PubMed] [Google Scholar]
- 31.Suenaga, H., M. Sato, M. Goto, M. Takeshita, and K. Furukawa. 2006. Steady-state kinetic characterization of evolved biphenyl dioxygenase, which acquired novel degradation ability for benzene and toluene. Biosci. Biotechnol. Biochem. 70:1021-1025. [DOI] [PubMed] [Google Scholar]
- 32.Suenaga, H., T. Watanabe, M. Sato, Ngadiman, and K. Furukawa. 2002. Alteration of regiospecificity in biphenyl dioxygenase by active-site engineering. J. Bacteriol. 184:3682-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sylvestre, M. 2004. Genetically modified organisms to remediate polychlorinated biphenyls. Where do we stand? Int. Biodeterior. Biodegradation 54:153-162. [Google Scholar]
- 34.Torsvik, V., L. Ovreas, and T. F. Thingstad. 2002. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science 296:1064-1066. [DOI] [PubMed] [Google Scholar]
- 35.Zhao, H., and F. H. Arnold. 1997. Optimization of DNA shuffling for high fidelity recombination. Nucleic Acids Res. 25:1307-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zielinski, M., S. Kahl, H. J. Hecht, and B. Hofer. 2003. Pinpointing biphenyl dioxygenase residues that are crucial for substrate interaction. J. Bacteriol. 185:6976-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zielinski, M., S. Kahl, C. Standfuss-Gabisch, B. Camara, M. Seeger, and B. Hofer. 2006. Generation of novel-substrate-accepting biphenyl dioxygenases through segmental random mutagenesis and identification of residues involved in enzyme specificity. Appl. Environ Microbiol. 72:2191-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]