Abstract
The mature 507-residue RgpB protein belongs to an important class of extracellular outer membrane-associated proteases, the gingipains, from the oral pathogen Porphyromonas gingivalis that has been shown to play a central role in the virulence of the organism. The C termini of these gingipains along with other outer membrane proteins from the organism share homologous sequences and have been suggested to function in attachment of these proteins to the outer membrane. In this report, we have created a series of truncated and site-directed mutants of the C terminus from a representative member of this class, the RgpB protease, to investigate its role in the maturation of these proteins. Truncation of the last two residues (valyl-lysine) from the C terminus is sufficient to create an inactive version of the protein that lacks the posttranslational glycosylation seen in the wild type, and the protein remains trapped behind the outer membrane. Alanine scanning of the last five residues revealed the importance of the C-terminal motif in mediating correct posttranslational modification of the protein. This result may have a wider implication in a novel secretory pathway in distinct members of the Cytophaga-Flavobacterium-Bacteroidetes phylum.
Porphyromonas gingivalis is an important organism in the pathogenesis of periodontal disease or “gum disease.” This common disease is characterized by a chronic inflammatory response by the host immune system against a polymicrobial biofilm which accumulates around the tooth surface in individuals with poor oral hygiene. In severe cases, the retreat of bone away from the area of inflammation results in the loss of the tooth. In individuals with weakened immune systems, such as individuals with poorly controlled diabetes and AIDS, the disease often progresses rapidly, leading to the loss of many teeth and its associated morbidity (16). More recently, mounting evidence has suggested a causative link between periodontal disease and cardiovascular disease (10, 18). In this respect, the association of P. gingivalis in patients with periodontal disease has been confirmed through many epidemiological, serological, in vitro, and animal studies (12, 22, 34).
RgpB belongs to an important family of clan CD cysteine proteases, the gingipains, exclusively from the bacterium P. gingivalis. Members of this family have been demonstrated to play a central role in the pathogenicity of the organism in periodontal disease (24). In addition to providing P. gingivalis with a general proteolytic tool for the degradation of proteinaceous nutrients for growth, gingipains have also been found to be essential for the processing of surface fimbrial proteins to facilitate bacterial adhesion to the host tissues (32, 55), to enable bacterial evasion of the host immune response through cytokine degradation and surface receptor cleavage (33), to encourage tissue inflammation and to enhance vascular permeability by dysregulating the coagulation and fibrinolytic pathways (23), to cause the exudation of plasma nutrients and erythrocytes (24), and to partake in the acquisition of iron and porphyrin from hemoglobin which are essential for growth of the organism (35, 46). Furthermore, numerous animal studies have shown attenuation of P. gingivalis virulence in inactivation mutants of these enzymes, underlining the important role they play in the pathogenesis of periodontal disease (14, 56).
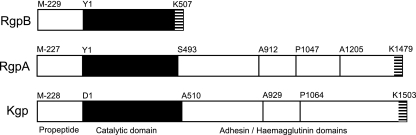
The gingipain family is comprised of three proteases: two highly homologous enzymes having specificity for cleavage after arginine residues, known as RgpA and RgpB, and one having specificity for lysine residues, named Kgp (39). They are comprised of a typical cleavable signal peptide, a propeptide, a catalytic domain, and with the exception of RgpB, several C-terminal adhesin/hemagglutination domains (Fig. 1). These proteins exist mainly as extracellular outer membrane (OM)-anchored entities (on the outer membrane or on released vesicles) in most strains of P. gingivalis (37). Presently, it is not known how gingipains are translocated across the outer membrane, since P. gingivalis is missing type II and IV secretory systems (5, 44) and the structures of the nascent translated gingipain polypeptides are not compatible with type I, III, and V systems operating in gram-negative bacteria (40). With the recent report of a type VI secretion system in a number of gram-negative pathogens (29, 41), homologues of the described secretion apparatus could not be found in the P. gingivalis genome (J. Potempa, personal communication). Hence, it has been suggested that P. gingivalis is using a novel secretion system which may be associated with gingipain maturation or posttranslational modification (44). It has been postulated that once the proteases are outside the cell, they are associated to the cell surface via various mechanisms: (i) through putative membrane-anchoring motifs in the highly conserved residues at the C terminus (53); (ii) through glycolipid/glycosylated modifications of the mature protein (15); or (iii) strong association with lipopolysaccharide (LPS) in the outer membrane of the organism through LPS/carbohydrate binding motifs (42). The crystal structure of the soluble form of RgpB has been reported to be missing the last 72 residues from the carboxyl terminus, providing evidence suggesting that this region is important for surface anchorage in the membrane-associated forms of the enzyme but not for enzymatic activity (11). Furthermore, Veith et al. (53) have reported that together with 11 other outer membrane proteins from P. gingivalis, the gingipains share highly conserved residues within the C terminus that may act as anchorage points for the protein to the outer membrane. Very recently, a study describing the buildup of inactive, partially processed enzymes in the culture medium and in the periplasmic space of P. gingivalis mutants carrying a recombinant form of RgpB that is missing the C-terminal 72 residues, suggesting a role of this domain in export and anchorage of the protein, was published (45). On the other hand, recombinantly expressed RgpB without the C terminus in a Saccharomyces cerevisiae host has been reported to have impaired functionality compared to the wild type, suggesting that the domain has more of an intramolecular chaperone function (28). To provide further experimental evidence to show the function of this C-terminal region in the maturation, processing, outer membrane translocation or anchorage of the gingipains, we embark on a series of truncations and site-directed mutagenesis of the terminal region in RgpB in order to shed light on the role of this tail.
FIG. 1.
Processed domains of the gingipains from P. gingivalis strain W83. Domain regions are labeled below the diagram, and the initial residue in each domain and the C-terminal residue positions are indicated above each protein. The C-terminal regions of these proteins are highly conserved (striped).
MATERIALS AND METHODS
Materials and reagents.
All bacterial media were sourced from Difco Laboratories (MD), while enzymes for molecular biology work were from Promega Inc. (WI) unless stated otherwise. QIAGEN (CA) kits were used in nucleic acid purifications including the QIAprep spin miniprep kit (for plasmid extraction), DNeasy tissue kit (for genomic DNA purification), QIAquick gel extraction kit (for PCR cleanup and gel extractions), and the RNeasy mini kit (for RNA purification). All general chemicals used were bought from Sigma-Aldrich Inc. (MO) unless noted otherwise. DNA oligonucleotide primers were synthesized by IDT Inc. (IA) and ethidium bromide was obtained from Bio-Rad Laboratories (CA). Anti-RgpB (18E6) monoclonal antibody (MAb) was produced on-site in a monoclonal facility at the University of Georgia, and the P. gingivalis antiglycan (1B5) MAb was a kind gift from Mike Curtis.
Bacterial strains and general growth conditions.
Porphyromonas gingivalis wild-type strain W83 and mutant derivatives were grown in enriched tryptic soy broth medium (eTSB; 30 g Trypticase soy broth, 5 g yeast extract, 5 mg hemin per liter, pH 7.5, and supplemented with 5 mM l-cysteine and 2 mg menadione) or blood eTSB agar (eTSB medium plus 15 g agar per liter and supplemented with 3% defibrinated sheep blood) at 37°C in an anaerobic chamber (Bactron IV; Sheldon Manufacturing Inc., OR) with an atmosphere of 90% N2, 5% CO2, and 5% H2. Escherichia coli strain DH5α was used for all plasmid construction work and was grown in Luria-Bertani medium and agar. For antibiotic selection in E. coli, ampicillin was used at 100 μg/ml and erythromycin at 300 μg/ml. For P. gingivalis growth selection on solid media, all antibiotics were used at 5 μg/ml and the concentration of antibiotics was doubled for selective P. gingivalis mutant growth in liquid culture.
Growth curve experiments.
Initial starter cultures were grown under antibiotic selection as appropriate, but subsequent passage culture for growth kinetics was carried out without antibiotic supplementation. Colony counting after four passages without antibiotic selection did not show any loss of antibiotic resistance in the clones, confirming the stable integration of the constructs into the host genome (data not shown). Cell densities of 3-day-old starter cultures were adjusted to an optical density at 600 nm (OD600) of 1.0, and a 1:50 inoculum was transferred anaerobically into 5 ml of prewarmed eTSB medium in individual Teflon screw-cap test tubes (13 × 100 mm). Tubes were capped tightly and incubated at 37°C in a water bath outside the anaerobic chamber. At predetermined time points, tubes were vortexed and the absorbances at 600 nm were recorded using a SpectraMax Plus spectrophotometer. Growth curves for each clone were examined on four replicate samples, and samples were collected at an OD600 of 0.8 (mid-log phase) and after 48 h (early stationary phase) of growth for protease activity assays and gene transcription assays. Cells were preserved in RNAprotect (QIAGEN) and stored at −80°C immediately after collection to preserve intact mRNA for real-time PCR studies.
Plasmid construction for rgpA deletion inactivation.
A 1-kb flanking region 5′ to the rgpA gene (The Institute for Genomic Research [TIGR] accession no. PG2024) was amplified by PCR with RgpAFrAKpnIF and RgpAFrASmaIR primers, using Accuprime Pfx DNA polymerase (Invitrogen Inc.) and incorporating KpnI and SmaI sites for cloning into pUC19 plasmid (New England Biolabs Inc.) to give a modified plasmid, pURA-B. All primers used in this paper are listed in Table S1 in the supplemental material. A promoterless chloramphenicol resistance (cat) gene was amplified (using catBluntF and catXbaIR primers) from the plasmid pLysS in the E. coli strain BL21(DE3)pLysS (Promega Inc.) with a blunt 5′ end and an XbaI restriction site at the 3′ end for incorporation into the plasmid pURA-B described above at SmaI and XbaI sites, respectively. The resultant plasmid, pURAc-G, was further modified by the incorporation of an amplified 1-kb flanking region 3′ to the rgpA gene (using RgpAFrBXbaI and RgpAFrBSalI primers) at XbaI and SalI sites to create the final construct pURgpA-A (see Fig. S1 in the supplemental material). The correct placement and orientation of the DNA segments were confirmed by sequencing.
Plasmid construction for rgpB mutagenesis studies.
Using a similar method to the pURgpA-A plasmid construction method described above, a section of the rgpB gene (TIGR accession no. PG0506) including its terminator stem-loop was amplified using RgpBFrAEcoRIF and RgpBFrASmaIR primers and ligated into pUC19 plasmid via EcoRI and SmaI sites to give plasmid pUr-A. An erythromycin resistance cassette (ermF-ermAM) active in both E. coli and P. gingivalis was amplified from plasmid pVA2198 (14) using ermFAMSmaIF and ermFAMSalR primers and ligated into SmaI and SalI sites in pUr-A to give rise to pUre-C. Finally, the region immediately 3′ to the amplified segment of the rgpB gene described above was amplified using RgpBFrBSalIF and RgpBFrBPstIR primers for ligation into pUre-C at SalI and PstI sites to complete the master pURgpB-E construct to be used for mutagenic studies (see Fig. S2 in the supplemental material). The correct placement and orientation of the DNA segments were confirmed by sequencing.
Deletion and site-directed mutagenesis of the master pURgpB-E construct.
By using a modified version of the site-directed, ligase-independent mutagenesis (SLIM) method of Chiu et al. (4), various truncated and site-directed mutagenic mutants were constructed using the master pURgpB-E plasmid as the template. Briefly, for each mutagenesis reaction, a set of four primers with an 18-base complementary overhang was employed to amplify the whole plasmid starting from the region to be mutated (Fig. 2; see Table S1 in the supplemental material). To 25 μl of this amplified PCR mixture was added 4 μl of buffer D (20 mM MgCl2, 20 mM Tris, pH 8.0, and 5 mM dithiothreitol) and 10 U of DpnI restriction enzyme. Template plasmid digestion was carried out at 37°C for 3 h before it was stopped by the addition of 30 μl of buffer H (300 mM NaCl, 50 mM Tris, pH 9.0, and 20 mM EDTA). The mixture was denatured at 95°C for 1 min, and the DNA fragments were rehybridized using two cycles of 68°C for 5 min, 60°C for 5 min, 55°C for 5 min, and 30°C for 10 min. An aliquot of 20 μl of the rehybridized mixture was used to heat shock transform RbCl2 competent E. coli cells using a modified version of the Hanahan method (19). Clones were selected on antibiotic selective LB agar and screened for the correct mutation by DNA sequencing of the pertinent region.
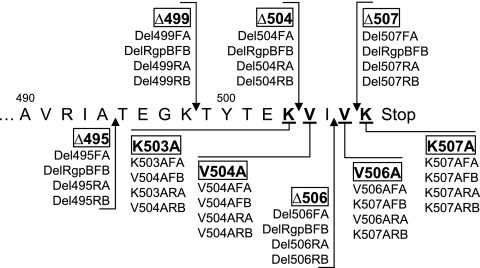
FIG. 2.
Truncated and site-directed mutagenic map of the carboxyl terminus of rgpB. Arrows point to the sites of truncation, and underlined residues are mutated to the amino acid alanine. rgpB mutant names are boxed, and the primer set used for the SLIM mutagenesis method employed is listed below each name. The sequences of these primers are detailed in Table S1 in the supplemental material.
Generation of isogenic mutants via homologous recombination.
Integration of modified plasmid constructs into the P. gingivalis genome by a double-crossover recombination event was achieved by the electrotransformation method of Smith et al. (48). Briefly, a 50-ml mid-log-phase culture of P. gingivalis was harvested by centrifugation at 6,000 × g for 15 min, washed three times in electroporation buffer (10% glycerol [vol/vol], 1 mM MgCl2), and resuspended in 0.5 ml of the same buffer. All steps were carried out at 4°C. Two hundred microliters of the cell suspension was transferred into a prechilled 0.2-cm-gap width electroporation cuvette (Bio-Rad Laboratories, CA) along with 1 μg of plasmid DNA. Electroporation was carried out at 2.5 kV and 10 μF capacitance using a Micropulser electroporation apparatus (Bio-Rad Laboratories), resulting in time constants of approximately 4.5 ms. After electroporation, the cells were allowed to recover on ice for 2 min before the addition of 1 ml of eTSB medium to the suspension. Cells were allowed to grow anaerobically at 37°C overnight before being plated onto antibiotic selective media. Resistant clones were further subcultured on selective plates, and genomic integration by a double crossover event was confirmed by PCR. Southern blotting with digoxigenin-labeled ermF-ermAM or cat gene probes was used to confirm the presence of only one crossover site in the genome in each mutant. Deletion and site-directed mutagenic mutants were further verified by DNA sequencing of the pertinent region of the genome.
Enzyme activity assay.
The amidolytic activities of Rgp and Kgp enzymes were assessed by the hydrolysis of the chromogenic substrate benzoyl-l-arginine-p-nitroanilide (BApNA) and carboxybenzoyl-l-lysine p-nitroanilide (zKpNA; Novabiochem, Germany), respectively. In a 96-well format, 20-μl samples were preincubated in assay buffer (200 mM Tris-HCl, 100 mM NaCl, 5 mM CaCl2 [pH 7.6], supplemented with fresh l-cysteine to 10 mM) for 2 min prior to the addition of 1 mM substrate in a total volume of 200 μl. For activity measurement of Sarkosyl-treated membrane fractionations (see Materials and Methods), a 0.125 mM concentration of a synthetic arginine substrate pyro-glutamyl-glycyl-l-arginine-p-nitroanilide (pyroEGRpNA; Pharmacia-Harper, Uppsala, Sweden) was used instead of BApNA due to precipitation of the BApNA substrate in the presence of the Sarkosyl detergent. The presence of 0.1% Sarkosyl detergent in the assay did not affect the rate of substrate hydrolysis as determined with purified RgpB (data not shown). The rate of formation of p-nitroanilide was measured at 405 nm using a SpectraMax Plus spectrophotometer (Molecular Devices Inc., CA). For ease of comparison between mutants and statistical analyses of independent repetitions, activity units were defined as the total activity present in the RgpB+ control mutant culture equaling 100 U for culture partitioning studies, the total activity in the RgpB+ control mutant cells equaling 100 U for cellular fractionation studies, or the total activity in the RgpB+ control mutant membranes equaling 100 U for membrane fractionation studies.
Cell fractionation procedures.
A 2-day-old (stationary-phase) culture of each clone was adjusted to an OD600 of 1.5, and a 0.5-ml sample, designated the culture sample, was collected. All cellular fractionation steps were carried out at 4°C using a modified version of the method of Parker and Smith and in the absence of protease inhibitors to allow for enzymatic assaying of the fractions (36). Cells in 15 ml of the adjusted culture were collected by centrifugation at 6,000 × g for 15 min. The cell pellet was washed once with 15 ml phosphate-buffered saline (PBS), and the cell pellet was resuspended in 5 ml of 0.25 M sucrose and 30 mM Tris, pH 7.6, and left mixing gently for 10 min before being repelleted at 12,500 × g for 15 min. The outer membrane was disrupted by the rapid addition of 5 ml ice-cold distilled H2O, and the cells were left to mix gently for 10 min. The spheroplast suspension was pelleted by centrifugation at 12,500 × g for 15 min, and the supernatant was designated the periplasmic sample. The remaining spheroplast pellet was washed once with 15 ml PBS before being resuspended in 5 ml PBS and ultrasonicated in 10 5-s pulses (17 W per pulse) in an ice-water bath with 2 s of rest between each pulse. Cellular debris and membranes were pelleted by ultracentrifugation at 150,000 × g for 1 h, and the supernatant was designated the cytoplasmic sample. The remaining pellet was washed once with 30 ml PBS prior to resuspension in 5 ml cold PBS by sonication and is designated the membrane sample. Contamination of the periplasmic fraction with cytoplasmic content was checked by quantification of nucleic acids using ethidium bromide binding fluorescence. The assay was carried out using 20 μl of each fraction in 5 mM Tris-0.1 mM EDTA (TE buffer) with 0.5 μg/ml of ethidium bromide in a total volume of 100 μl. Fluorescence of the dye bound to nucleic acids was measured in a Spectramax Gemini EM (Molecular Devices Inc., CA) spectrophotometer with excitation at 525 nm and emission at 600 nm (27). By this method, there is approximately 25% contamination of cytoplasmic content in the periplasmic fraction.
Sarkosyl separation of cellular membranes.
Collected cells were washed once with PBS and lysed by ultrasonication as described above. The membranes were pelleted by ultracentrifugation at 150,000 × g for 1 h and washed once with 30 ml PBS to remove periplasmic and cytoplasmic proteins prior to being resuspended in 4.5 ml PBS by sonication. A 0.5-ml sample of a 10% (wt/vol) Sarkosyl (lauryl sarcosine) solution was added and left to mix gently for 20 min at 4°C to dissolve the inner membrane (IM) (13, 54). The residual Sarkosyl-resistant outer membranes were pelleted by ultracentrifugation at 150,000 × g for 1 h, and the supernatants were designated the IM samples. The pellets were washed once with 30 ml PBS before being resuspended in 5 ml PBS by sonication and designated the OM samples. Purity of the various fractions was checked by Western blotting with P. gingivalis anti-LPS 1B5 MAb. As LPS resides exclusively on the outer leaflet of the OM, the absence of detectable LPS in the IM fraction validated the fractionation technique (data not shown).
Immunoprecipitation of RgpB.
RgpB-specific monoclonal antibody 18E6 was cross-linked onto protein G-agarose beads using the Seize X protein G immunoprecipitation kit (Pierce Inc., IL) as per the manufacturer's instructions using the gentle wash protocol. RgpB was solubilized from 0.5-ml whole-culture samples of P. gingivalis by the addition of 2% Tween 20 (vol/vol) (final concentration) in the presence of a 5 mM concentration of the protease inhibitor tosyl-lysine chloromethyl ketone (TLCK) and gently mixed for 30 min at room temperature. Cellular debris was pelleted by centrifugation at 18,000 × g for 10 min, and the clarified supernatant was diluted 1:20 into PBS and incubated with the beads overnight at 4°C. Beads were washed extensively with PBS and boiled in nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer with 5 mM TLCK for 5 min prior to the addition of 10 mM β-mercaptoethanol and boiled for a further 5 min. Samples were subjected to the following Western blot procedures.
Western blot analysis.
Stationary-phase cultures or cell fractionation fractions were first boiled in nonreducing SDS-PAGE sample buffer containing 5 mM TLCK for 5 min to inactivate all proteases prior to the addition of 10 mM β-mercaptoethanol and boiled for a further 5 min for complete denaturation. Samples were electrophoresed using the discontinuous system by the method of Laemmli et al. (26). Proteins were subsequently electrotransferred onto 0.45-μm-pore-size nitrocellulose membranes and blocked in a PBS solution with 2% (wt/vol) bovine serum albumin overnight. The RgpB peptide backbone was detected using a 1:2,000 dilution of the primary MAb 18E6 (produced on-site; specific for the first half of the immunoglobulin domain of RgpB), and the RgpB carbohydrate epitope was detected using a 1:200 dilution of the primary MAb 1B5 (8) in TTBS (20 mM Tris, 500 mM NaCl, pH 7.5, supplemented with 0.1% Tween 20) for 3 h. Membranes were washed three times with TTBS before being probed for 2 h with a 1:2,000 dilution of an alkaline phosphatase-conjugated anti-mouse secondary antibody (Pierce Biotechnology Inc., IL). Development was carried out using the alkaline phosphatase conjugate substrate kit as per the manufacturer's instructions (Bio-Rad Laboratories, CA).
Real-time PCR procedure.
Total RNA from mid-log-growth cells at an OD600 of 0.8 was extracted using the RNeasy mini kit (QIAGEN) and quantified on a Nanodrop ND-1000 spectrophotometer (Nanodrop Technology Inc., DE). Reverse transcription of 20 ng total RNA was carried out in a volume of 10 μl using the TaqMan reverse transcription reagent kit (Applied Biosystems, CA) as per the manufacturer's instructions using random hexamer primers. Singleplex real-time PCR was performed on three replicate samples on an Applied Biosystems 7500 real-time PCR system using the Power SYBR green PCR master mix (Applied Biosystems, CA). In a total volume of 25 μl, 5 μl of a 1:200 dilution of the cDNA in the previous step was amplified using 300 nM primer sets against an endogenous control gene DNA gyrase (dnaA) and the target rgpB gene (see primer sequences in Table S1 in the supplemental material). The standard curve against dilutions of purified DNA gave a linear normalized cycle threshold (CT) relationship over 5 log units of dilution, and dissociation curves of the amplified product along with visual inspection on 2% agarose gel electrophoresis confirmed the presence of only one amplicon and the absence of primer dimer formation (data not shown). Fluorescence intensities were normalized against a passive fluorophore present in the Mastermix and converted to absolute quantities using the standard curves. Relative expression of rgpB transcripts in each mutant was expressed as the ratio between the average rgpB CT value divided by the average dnaA CT value for each sample, with the ratio in the reference RgpB+ strain being set at 1.00. Real-time PCR data were obtained from two separate experiments.
Protein motif search and sequence alignment.
Databases at TIGR, the Los Alamos Oral Pathogens Sequences, and TrEBML were searched for the protein motif K[LIV]x[LIV][KR]Stop initially and then for K[LIV]x[LIV][KRHEQ]Stop as the work progressed. The returned gene sequences were analyzed for the presence of a gram-negative signal peptide as predicted by the SignalP 3.0 program at ExPASy (www.expasy.org). The last 50 residues of proteins with a predicted signal peptide from P. gingivalis, Prevotella intermedia, and Tannerella forsythensis are aligned using the ClustalW program at the European Bioinformatics Institute (www.ebi.ac.uk/clustalw/) and displayed using the JalView program (6). Alignment of the remaining proteins without a predicted signal peptide is also available in Fig. S3 in the supplemental material.
Statistical analysis.
Prism v3.03 software (GraphPad Software Inc., CA) were used for all statistical analyses. Enzymatic activities and real-time PCR data were tested for normality distribution, and differences were analyzed using one-way analysis of variance (ANOVA) with Bonferroni's correction and 95% confidence intervals. P values below 0.05 were considered significant.
RESULTS
Generation of C-terminally truncated RgpB.
RgpA and RgpB together are responsible for all of the arginine-specific endopeptidase activity in P. gingivalis (38). Due to the high similarity between substrate specificity, biophysical behavior, and identical segments of gene sequences between RgpA and RgpB, it was essential to inactivate RgpA by deletion to eliminate background in the study of RgpB truncation. Although RgpA has been inactivated by insertion of an antibiotic cassette to disrupt the gene in other strains of P. gingivalis (7, 30), an isogenic mutant in the reference and genome sequenced strain of W83 is not available (31). Consequently, RgpA was inactivated in this study by the replacement of the gene by the introduction of a chloramphenicol resistance cassette (cat) in a pUC19-derived suicide plasmid, pURgpA-A, into P. gingivalis by electroporation (see Fig. S1 in the supplemental material). Isogenic mutants arising from homologous recombination were selected on antibiotic selective media, and the correct clones were confirmed by PCR and Southern blotting (data not shown). After the selection and confirmation of the correct rgpA deletion inactivation mutant, the absence of the rgpA gene made the subsequent culturing in chloramphenicol selective media to maintain the genotype unnecessary, as reversion to the wild-type state is not possible.
The strategy for truncating RgpB involved the initial construction of a master plasmid, pURgpB-E, harboring the insertion of an antibiotic cassette 3′ to the terminator stem-loop structure of the rgpB gene (see Fig. S2 in the supplemental material). Homologous recombination of this construct into the P. gingivalis genome resulted in a fully functional RgpB+ mutant to serve as a control against polar effects from genetic manipulations. From this master plasmid construct, various deletion mutagenic constructs were made, removing initially the last 13 residues from RgpB due to the high conservation of this region in the alignment with other P. gingivalis outer membrane proteins (53) and then leading progressively to smaller deletions from the C terminus (Fig. 2). Electroporation of these constructs into competent P. gingivalis cells resulted in genomic integration via double crossover of the homologous regions of the plasmid and giving rise to mutants with the desired truncation of RgpB (Table 1). All constructs were checked by DNA sequencing, and the resulting P. gingivalis mutants were confirmed by PCR and DNA sequencing of the pertinent region along with confirmation of the presence of only one antibiotic cassette integration into the genome by Southern blotting (data not shown).
TABLE 1.
P. gingivalis strains and mutants used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| W83 | Wild-type | Reference strain (31) |
| RgpA− | ΔrgpA (Cmr) | This study |
| E8 | ΔrgpA (Tcr) rgpB (Emr) | 1 |
| RgpB+ | ΔrgpA (Cmr) rgpB+(Emr) | This study |
| Δ495 | ΔrgpA (Cmr) rgpBΔ495 (Emr) | This study |
| Δ499 | ΔrgpA (Cmr) rgpBΔ499 (Emr) | This study |
| Δ504 | ΔrgpA (Cmr) rgpBΔ504 (Emr) | This study |
| Δ506 | ΔrgpA (Cmr) rgpBΔ506 (Emr) | This study |
| Δ507 | ΔrgpA (Cmr) rgpBΔ507 (Emr) | This study |
| K507A | ΔrgpA (Cmr) rgpB(K507A) (Emr) | This study |
| V506A | ΔrgpA (Cmr) rgpB(V506A) (Emr) | This study |
| V504A | ΔrgpA (Cmr) rgpB(V504A) (Emr) | This study |
| K503A | ΔrgpA (Cmr) rgpB(K503A) (Emr) | This study |
Arginine-specific endopeptidase activity of the RgpB truncated mutants.
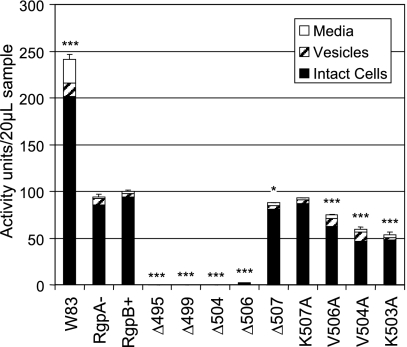
Deletion of the rgpA gene in the RgpA− mutant resulted in a significant 61% reduction in arginine-specific endopeptidase activity (or a decrease from 241 activity units in the wild type to 94 units in the RgpA− mutant; for ease of comparison between the mutants, activity units were defined as the total Arg-specific activity in the RgpB+ control mutant equaling 100 units) under the growth conditions employed (Fig. 3). This is in general agreement with the finding of others, reporting reductions of 63 to 66% in rgpA isogenic mutants in two different strains of P. gingivalis (30, 42). The introduction of the erythromycin resistance cassette after the rgpB gene in the RgpB+ mutant did not affect the level of Arg-specific activity compared to the level in the RgpA− mutant as expected. Using a well-established method of partitioning the culture into various soluble and cell-associated fractions (37), most of the enzyme activity across all strains was found to be associated with the cell fraction (Fig. 3). However, truncation of the C-terminal 13 residues (Δ495) resulted in a complete loss of all Arg-specific activity from the whole culture (Fig. 3). Progressive truncation of this region revealed that all truncated mutants except for the last residue truncation (Δ507) resulted in a similar loss of activity. The Δ507 mutant showed a minor reduction in extracellular production of RgpB (Fig. 3). The growth rates of all mutants were similar under the growth conditions employed (data not shown), and Kgp activity and culture distribution were generally unaffected by the truncation of RgpB (data not shown). The loss of Arg-specific activity was not at the transcriptional level, as the quantity of rgpB mRNA in the truncated mutants as determined by real-time PCR was generally unaffected compared to the quantity in the RgpB+ mutant (data not shown).
FIG. 3.
Arginine-specific activity partitioning in the bacterial culture. Early stationary-phase cultures of each mutant were separated into cellular, vesicular, and medium fractions, and 20-μl samples of each fraction were assayed against an Arg-specific synthetic substrate as per Materials and Methods. The total Rgp activity in the RgpB+ control mutant was set at 100 activity units. Data were obtained from duplicate enzymatic measurements from four independently grown cultures and were normally distributed. Differences in the total arginine-specific activity of each strain against the RgpB+ mutant were statistically analyzed using one-way ANOVA with Bonferroni's correction. Values that were significantly different from the value for the RgpB+ mutant are indicated by asterisks (*, P < 0.05; ***, P < 0.001).
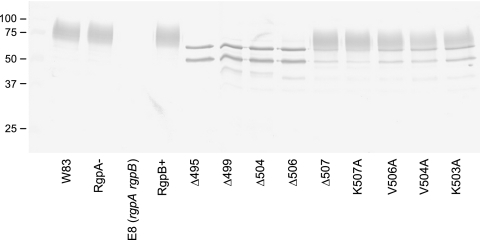
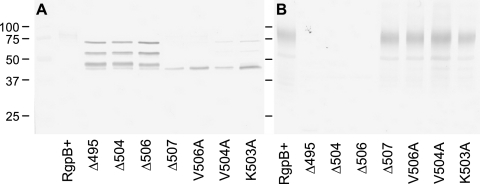
Western blotting of the cultures probed with an RgpB-specific monoclonal antibody, 18E6, revealed the presence of an equivalent quantity of RgpB in the inactive mutants, but in these mutants, there was a drastic shift in the molecular mass of the RgpB protein. The specificity of the MAb against RgpB was shown by the absence of any band in the RgpA and RgpB double-knockout E8 strain (Fig. 4). Normally, in the W83 wild-type strain, RgpB is glycosylated and migrates as a diffuse band in the region of 70 to 90 kDa (Fig. 4). This pattern is preserved in the RgpA− and RgpB+ mutants carrying the intact rgpB gene. However, in the inactive truncated mutants, RgpB migrates as two or three distinct bands of approximately 65 and 52 kDa and faintly at 48 kDa, all of which are larger than the predicted mature, unglycosylated RgpB of 45 kDa. The results suggested that RgpB enzymes in the truncated mutants are partially processed as was shown previously (28, 45) and that the presence of an intact C terminus, in particular, the penultimate valine residue, is critical in the proper processing and possibly, proper glycosylation of RgpB. There was minimal release of RgpB into the soluble medium fraction across all mutants as determined by Western blotting (data not shown).
FIG. 4.
Forms of RgpB in truncated and site-directed mutants. Fifty-microliter samples of whole cultures of various mutants at early stationary growth phase were subjected to reducing SDS-PAGE, transferred onto nitrocellulose membranes, and probed with anti-RgpB MAb. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel.
Alanine scanning of the C-terminal residues of RgpB.
Multiple alignment of the last few residues at the C terminus of outer membrane proteins of P. gingivalis was reported to reveal a conserved terminal consensus sequence K[LIV]x[LIV]x(Stop) (53). With this information, alanine scanning of these residues to determine their relative importance in the correct processing of RgpB was carried out (Fig. 2). Although changing the terminal lysine 507 to alanine (K507A) did not significantly change the output of RgpB, changing valine 506 or 504 to alanine (V506A or V504A) significantly decreased the total extracellular production of RgpB to 75% and 59% of the RgpB+ control mutant, respectively (Fig. 3). Replacement of the conserved lysine 503 to alanine (K503A) led to a greater significant reduction of RgpB activity to 54% compared to RgpB+ (Fig. 3). Western blotting of these alanine mutants with anti-RgpB MAb showed an intermediate pattern of bands between the RgpB+ control mutant and the truncated mutants (Fig. 4). Taking into account that Western blotting is only semiquantitative, closer examination of the blot patterns of the K507A mutant compared to the other alanine mutants reveal a distinct difference (absence) in the relative quantity of the sharp bands of partially processed RgpB. The blot pattern of the K507A mutant is almost identical to that of the RgpB+ control mutant and is considerably different from those of the other alanine mutants. Although it cannot be accurately quantified in the blots, the approximate levels of the sharp bands in the mutants inversely correlate to the reduction of RgpB activity observed in the activity assays. These data together suggest a difficulty of the organism to correctly modify/process RgpB in these alanine-scanning mutants, hence, implicating the importance of the lysine-503 and the following bulky hydrophobic residues in mediating RgpB maturation.
Penultimately truncated RgpB is unglycosylated.
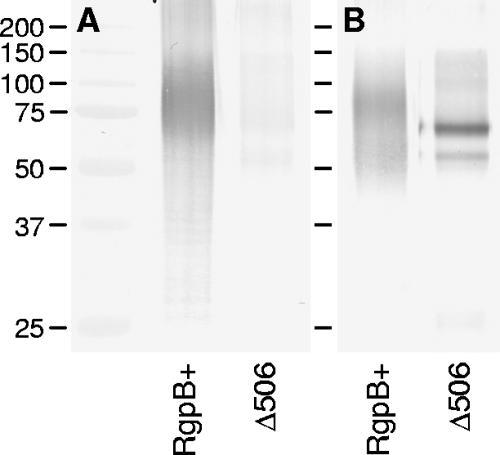
Loss of mass and the appearance of sharply distinct bands in the inactive truncated mutants suggested that the inactive RgpB has not been glycosylated or at least has not been fully glycosylated as in the wild type (Fig. 4). To investigate that possibility, immunoprecipitation of RgpB was carried out using anti-RgpB MAb to probe for the glycosylation status of the mutant RgpB. Using a monoclonal antibody that recognizes the glycosylated form of RgpB, 1B5 (8), the RgpB from Δ506 mutant was found to be indeed lacking the glycan addition of the wild-type RgpB (Fig. 5).
FIG. 5.
Failure of posttranslational glycosylation of RgpB in the truncated mutants. RgpB was immunoprecipitated from RgpB+ and Δ506 mutants using anti-RgpB MAb. Proteins were separated by reducing SDS-PAGE and blotted onto nitrocellulose membranes prior to being probed with an antibody against P. gingivalis LPS carbohydrate residues (1B5 MAb in panel A) and anti-RgpB antibody (18E6 MAb in panel B). The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel.
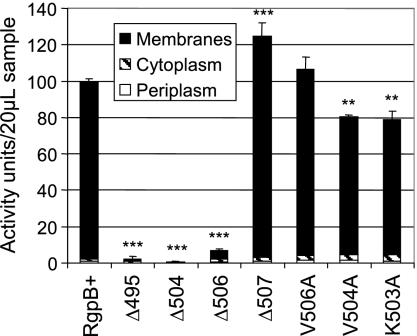
Cellular fractionation of RgpB in the mutants.
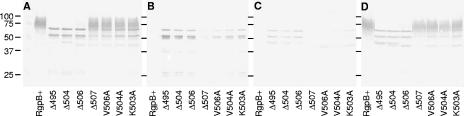
Due to the similarity between the K507A strain and the RgpB+ control mutant in both activity assays and in Western blots, we have chosen not to perform more detailed analysis on this mutant, since the others displayed more significant differences compared to the wild type. In order to quantify the relative amounts of enzyme and activity in each cellular compartment, all cultures were grown simultaneously and standardized to an equivalent number of cells (equivalent OD600 absorbance) at the collection stage prior to the extraction procedures and were fractionated at the same time. Using an established osmotic shock technique for extracting periplasmic proteins (36), we have found that there is approximately 25% contamination of cytoplasmic content in the periplasmic fraction as determined by ethidium bromide fluorescence binding to nucleic acids (see Materials and Methods). Nevertheless, almost all of the detectable enzyme activity was found to partition predominantly to the insoluble membrane fraction (Fig. 6). The Δ507 mutant showed a significantly higher total cellular Arg-specific activity than the control RgpB+ mutant, and the V506A mutant also showed the same trend though significance was not reached (Fig. 6). This is in contrast with the lower overall extracellular activity in these mutants compared to the activity in the control (Fig. 3), perhaps reflecting some difficulty of active RgpB in these mutants to traverse through the outer membrane, hence, the buildup of Arg-specific activity within the cells. Lower levels of intracellular Arg-specific activity were found in the V504A and K503A mutants and at minimal levels in the Δ495, Δ504, and Δ506 mutants (Fig. 6). This is in contrast with Western blots with anti-RgpB MAb where RgpB was present in significant quantities across all strains (Fig. 7). The level of active RgpB in the strains correlates with the presence of diffuse glycosylated bands in the 70- to 90-kDa range, whereas the presence of sharp, distinct bands appear to be more common in the inactive mutants (Fig. 7A). Moreover, glycosylated RgpB enzymes are partitioned to the membrane fraction and the unglycosylated, partially processed forms are partitioned predominantly to the periplasmic and membrane fractions in the inactive mutants along with trace amounts in the cytoplasm (Fig. 7B, C, and D). Taking into account that Western blotting is only semiquantitative, the quantity of unglycosylated RgpB in the intracellular fractions appears to be higher in the inactive mutants and is also slightly elevated in the periplasmic fraction of the alanine-scanning mutants. It is interesting to note the relative absence of partially processed forms of RgpB in the intracellular fractions of the control RgpB+ and the Δ507 mutant by Western blotting (Fig. 7B and C), suggesting that in the wild-type protein and in the Δ507 variety, RgpB is quickly and efficiently processed and exported out to the outer membrane after translation of the polypeptide.
FIG. 6.
Rgp activity in cellular compartments. Cellular fractions of various mutants were assayed for Arg-specific activity using 20 μl of each sample and synthetic substrates as per Materials and Methods. Total Rgp activity in the RgpB+ control mutant was set at 100 activity units. Data were obtained from duplicate enzymatic measurements from two independent runs. Differences in the total arginine-specific activity of each strain against the RgpB+ mutant were statistically analyzed using one-way ANOVA with Bonferroni's correction. Values that were significantly different from the value for the RgpB+ mutant are indicated by asterisks (**, P < 0.01; ***, P < 0.001).
FIG. 7.
Cellular localization of RgpB. Cellular fractionation techniques were employed to separate cellular compartments into whole cells (A) and periplasm (B), cytoplasm (C), and membrane (D) fractions. Fifty-microliter samples from each fraction were separated by reducing SDS-PAGE and transferred onto nitrocellulose membranes prior to detection with anti-RgpB MAb. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel.
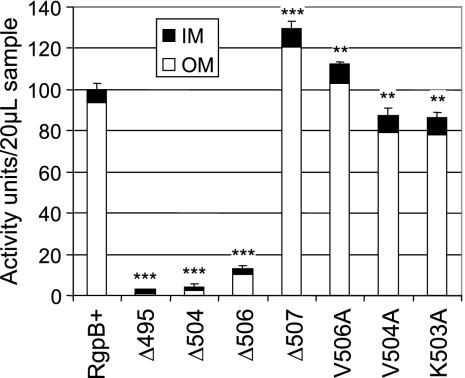
Membrane fractionation of RgpB in the mutants.
The ability of the detergent Sarkosyl to preferentially solubilize the inner membrane in gram-negative bacteria was utilized to separate it from the outer membrane in the total membrane fraction (13, 54). In the wild type, RgpB activity was found to partition predominantly to the outer membrane fractions, and a similar trend is evident in the Δ507 and alanine mutants (Fig. 8). Again, the Δ507 and V506A mutants showed higher overall membrane-bound RgpB activity, with most of this increase being associated with the outer membrane. Since this elevated activity was not detected from outside of the intact cells (Fig. 3), the buildup of active enzyme in these mutants must be occurring on the periplasmic side of the outer membrane. On the other hand, both V504A and K503A mutants showed smaller amounts of active enzyme than in the RgpB+ control (Fig. 8), suggesting a folding or processing difficulty in these mutants. Some low Arg-specific activity was detected associated with the outer membrane fraction in the Δ506 mutant, whereas minimal activity was detected in the longer truncated mutants (Fig. 8). On Western blots, there was obvious demarcation of the fully glycosylated forms of RgpB to the outer membrane (Fig. 9B). The inner membrane fraction of the inactive mutants revealed three or possibly four distinct bands migrating at a higher molecular mass than the predicted fully processed, unglycosylated form of RgpB at 45 kDa. These entities are presumably partially processed forms of RgpB, as recombinant forms of this protein in a yeast host have been previously shown to exhibit sequential autolytic processing at two locations within the propeptide domain and one location in the C-terminal domain of the protein (28). Furthermore, the detection of multiple, N-terminal partially processed forms of a recombinant RgpB truncated at the last 72 residues were consistent with our findings here (45). The Δ507 and alanine-scanning mutants showed a single band at approximately 45 kDa in the inner membrane fraction, which may be responsible for some Arg-specific activity observed in this fraction (Fig. 8). Moreover, on the basis of the lack of glycosylated RgpB in the cytoplasm and periplasm (Fig. 7B and C) and inner membrane fractions (Fig. 9A), the seemingly exclusive presence of glycosylated forms of RgpB in the outer membrane (Fig. 9B) suggested that the protein is glycosylated when it becomes associated with the outer membrane or the glycosylated protein has a high affinity for the outer membrane and partitions with it during the detergent treatment. Taken together, these results suggested that there are difficulties in the folding of the nascent RgpB polypeptide and its subsequent export across the outer membrane, particularly in mutants missing the C-terminal two or more residues, resulting in the buildup of inactive, partially processed, and unglycosylated forms in the periplasm, on the inner membrane and in the cytoplasm.
FIG. 8.
Partitioning of RgpB activity between the outer and inner membranes. Membrane fractionation by treatment of the membrane fraction with 1% Sarkosyl, and 20 μl of each sample was assayed for Arg-specific activity as per Materials and Methods. The total Rgp activity in the RgpB+ control mutant was set at 100 activity units. Data were obtained from duplicate enzymatic measurements from two independent runs. Differences in the total arginine-specific activity of each strain against the RgpB+ mutant were statistically analyzed using one-way ANOVA with Bonferroni's correction. Values that were significantly different from the value for the RgpB+ mutant are indicated by asterisks (**, P < 0.01; ***, P < 0.001).
FIG. 9.
Membrane localization of RgpB. Separation of the membranes into inner (A) and outer (B) membrane fractions were mediated by dissolution of the inner membrane with Sarkosyl detergent. Fifty-microliter samples from each fraction were separated by reducing SDS-PAGE and transferred onto nitrocellulose membranes prior to detection with anti-RgpB MAb. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel.
DISCUSSION
During the preparation of the manuscript, a new report appeared describing the characterization of an RgpB mutant missing the C-terminal 72 residues (45). The authors employed a recombinant form of RgpB, encoded on a shuttle plasmid, and expressed it in a P. gingivalis host strain that was derived from random mutagenesis of a different wild-type strain to create a restriction-deficient derivative that could tolerate the foreign plasmid (57). The behavior of this recombinant C-terminally 72-residue-truncated RgpB, though, seems similar to our C-terminal 13-residue-truncated mutant as reported here with the exception that we did not detect significant quantities of RgpB in the culture media by Western blotting. This difference could be real or could be a result of their concentration of the medium fraction by trichloroacetic acid precipitation. Consequently, the reported method cannot be used to quantify the relative amounts of RgpB in each culture partition. Furthermore, differences in the collection time of P. gingivalis cultures may have a significant influence on partitioning of the gingipains, as this organism has been known to undergo autolysis upon extended culturing, releasing the gingipains into the medium in significant quantities over time (25). In any case, we have extended their preliminary findings further by careful dissection of this aberrant RgpB export phenomenon and created a series of RgpB truncated mutants to determine the last two residues at the C terminus as the minimal motif necessary to elicit this effect.
The absence of glycosylation in the inactive mutants (Fig. 5) leads to the question whether glycosylation is necessary for RgpB activity or for correct folding of the protein. Indirect evidence against these theories could be found in two earlier reports. One paper described the inactivation of a glycan biosynthesis gene, porR, resulting in a mutant with nonglycosylated but fully functional gingipains that are found in abundance in the medium supernatant and are not cell associated as in the wild type (47). The second report detailed the autocatalytic processing of recombinant RgpB to an active form in a yeast host not in possession of the P. gingivalis glycosylation machinery (28). Moreover, due to the absence of a residue at the C terminus that is capable of being glycosylated, such as serine, threonine, tyrosine, or asparagine (51), truncation of the final two residues (VK) resulting in an unglycosylated form must mean that these few C-terminal residues are (i) essential in mediating the correct folding of the polypeptide directly or (ii) essential in mediating the correct folding through a chaperone(s), both of which result in an active enzyme and accessibility of glycosyltransferase(s) to the glycosylation sites or (iii) that the terminal residues are important recognition sites for glycosyltransferase(s) themselves to bind and catalyze the transfer of sugar residues to the peptide backbone.
The lack of significant Arg-specific activity in the medium fraction of the inactive mutants (Fig. 3) or detection of the same by Western blotting (data not shown) suggested that the C-terminal motif is important for direct recognition of the outer membrane translocator or in interaction with a periplasmic chaperone, holding them in an glycosylation/translocation-compatible structure (17). Without these interactions, RgpB may remain trapped inside the cell in the form of inactive, nonglycosylated, aberrantly processed proteins (Fig. 7). Previously published data seemed to discount the glycosylation-requirement-for-export hypothesis by reporting that the construction of mutants with defective glycosylation machinery still resulted in the release of gingipains into the culture medium despite contradictory reports on whether they are in an active state (47, 52). However, these observations must be taken in the context that defects in glycosylation pathways may result in a defective outer membrane, giving a “leaky” phenotype with the concomitant release of periplasmic contents (43). Despite this uncertainty, our truncation of the last two C-terminal residues of RgpB in a fully functioning glycosylation background strain is sufficient to result in an inactive RgpB with glycosylation and exportation defects suggested that this terminal motif is most likely to be involved in mediating the correct folding of the nascent RgpB to enable accessibility to the potential glycosylation sites and for subsequent export through the outer membrane of the organism.
Recently, Sato et al. reported the construction of a Kgp′-′RgpB chimeric protein consisting of the Kgp catalytic domain fused to the C-terminal 98 residues of RgpB (44). This chimera was able to successfully export and attach itself to the outer membrane and displayed full Kgp activity as for the wild type. This is in contrast to the expression of the Kgp catalytic domain on its own where no activity could be detected either on the cells or in the culture medium (50). As a consequence, the 98 residues at the C terminus of RgpB are essential for the correct processing and export of the gingipains but have no role in the catalysis of the enzyme. In this study, we extend this finding further by careful dissection of this phenomenon, showing that the last two C-terminal residues of RgpB are essential for the correct processing and maturation of the protein. Although alanine scanning of the terminal five residues revealed some importance of the bulky hydrophobic valine residues at positions 504 and 506 along with the lysine 503 in facilitating the efficient export of RgpB, the absence of these few residues at the C terminus resulted in a drastic impairment of maturation and export of this enzyme. A similar phenomenon has been reported in a number of outer membrane porins in gram-negative bacteria involved in the movement of proteins and solutes across the outer membrane (20). These integral outer membrane proteins form a channel through the outer membrane by an arrangement of 8 to 16 antiparallel, amphipathic β-sheets spanning the lipid bilayer with either all sheets being located within one monomeric porin or divided evenly between multimers (20, 40). Although these porins differ considerably in the primary structure sequence, their extreme C termini share a stretch of alternating polar-hydrophobic residues and invariably terminate with a nonpolar aromatic residue, such as phenylalanine or tryptophan, which has been suggested to insert and anchor the protein into the lipid bilayer (49). The truncation of the extreme C termini of these porins resulted in the failure of the proteins to assemble in the outer membrane with a concomitant accumulation in the periplasm (2, 21). In particular, a member of the type V protein secretion machinery, the Hap autotransporter in Haemophilus influenzae has been reported to be unable to assemble into the outer membrane with the truncation of the final three C-terminal residues. Furthermore, the mutation of these C-terminal residues did not wholly prevent their incorporation into the outer membrane, but the efficiency of the process was affected (21, 49). This is reminiscent of our findings, but the prime difference is that the C terminus of RgpB has no resemblance to the C termini of these porins. The RgpB C terminus does not contain stretches of amphipathic β-sheets, and it lacks the terminal aromatic residue. Of further interest, a new report detailing the existence of a C-terminal signal sequence for a specialized secretion system of a virulence factor from Mycobacterium tuberculosis revealed that the recognition signal resides within the final seven C-terminal residues of the protein to be excreted (3). The RgpB C terminus, however, has no resemblance to the reported motif, and the reported secretion system in that gram-positive organism is unlikely to operate in two-membrane gram-negative organisms such as P. gingivalis.
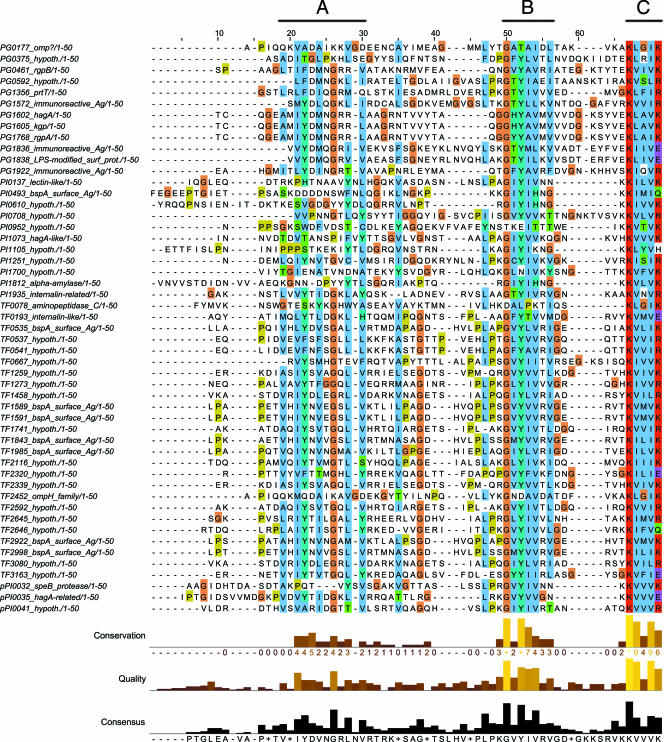
A protein motif search at The Institute of Genomic Research-Comprehensive Microbial Resource (TIGR-CMR; http://cmr.tigr.org) database and the Los Alamos Oral Pathogen Sequence (http://www.oralgen.lanl.gov) database using the search string K[LIV]x[LIV][KR]Stop against 325 microbial genomes revealed a significant presence of this terminal motif in a number of organisms belonging to the Cytophaga-Flavobacterium-Bacteroidetes phylum, such as P. gingivalis, Prevotella intermedia, Tannerella forsythensis, Cytophaga hutchinsonii, Psychroflexus torquis, Flavobacterium johnsoniae, and Flavobacterium bacterium. Interestingly, the first three organisms are human oral pathogens that have been implicated as possible etiologic agents in periodontal disease. Further analysis using 63 proteins with the C-terminal motif from the three dental pathogens, P. gingivalis, P. intermedia, and T. forsythensis, revealed that 42 (67%) proteins possess a predicted gram-negative signal peptide, suggesting that this C-terminal motif may play a significant role in the posttranslational modification/proteolytic processing and exporting of proteins to be inserted into the outer membrane or for translocation out into the extracellular milieu of these organisms. While it is possible that the remaining proteins without a predicted signal peptide may still be exported out to the periplasm by the conventional pathways, manual scanning of their N terminus did not reveal the presence of stretches of hydrophobic residues associated with membrane targeting of the protein (9). Searches using a random five C-terminal residues in these genomes returned very few hits and the presence of a predicted signal peptide of less than 10% (data not shown). Given that our data showing a tolerance to the presence or absence of the last residue, lysine 507, along with tolerance to its mutation to alanine, a wider search was performed within the genomes of this group for redundancy of this terminal residue. It was found that when a large terminal polar residue, such as a lysine, arginine, histidine, glutamate, or glutamine, is present at the end of the motif, the abundance of proteins with a signal peptide holds true (51 out of 77 [66%]). Alignment of the C-terminal 50 residues of these proteins with signal peptides revealed two stretches of well-conserved sequences further upstream of the terminal motif (Fig. 10; alignment of proteins without signal peptides is available in Fig. S3 in the supplemental material). The first of these regions (region A) is comprised of an alternating hydrophobic-polar residue motif surrounding a conserved glycine. Region B has a highly conserved GhYhhph motif where h denotes a hydrophobic residue and p denotes a polar residue. The terminal region C contains the terminal motif being studied in this paper. The functions of regions A and B are currently unknown, but previous speculation that they may act as a membrane-anchoring motif, a glycosylation site, or a recognition site for the effector glycosyltransferase cannot be ruled out at this stage (53). However, we speculate that region C may act as (i) a binding site for a periplasmic chaperone(s) that helps the nascent protein that has passed through the inner membrane to refold and maintain a structure compatible for glycosylation and/or export through the outer membrane, (ii) a novel outer membrane targeting signal, or (iii) a recognition site for glycosylation of the protein, at which point, it is rapidly partitioned to the outer membrane. Due to the abundance of inactive forms of RgpB in the truncated mutants, the first scenario seemed more likely. The presence of this terminal motif is important in mediating the correct folding of the nascent protein, which is then transported across the periplasm to be fully glycosylated during its translocation across the outer membrane for anchorage to the outer leaflet of the membrane.
FIG. 10.
Alignment of the C-terminal 50 residues of proteins from P. gingivalis, P. intermedia, and T. forsythensis carrying the K[LIV]x[LIV][KRHEQ] C-terminal motif that also has a signal peptide as predicted by the SignalP program. Gene accession numbers and annotation are from the Los Alamos Oral Pathogens Sequences Database (www.oralgen.lanl.gov) (hypoth., hypothetical). Regions of similarity are labeled above the alignment.
A possible candidate for a chaperone protein(s) could be the porT locus in P. gingivalis. A recent report describing an isogenic porT mutant having problems with posttranslational processing and export of the gingipains gave us the idea that this inner membrane-anchored periplasmic protein may act as a chaperone for the folding of nascent outer membrane proteins in general (44). The authors reported that a BLAST search of the porT gene returned the presence of its homologues in only two other genomes: C. hutchinsonii and P. intermedia. A more recent BLAST search by our group has revealed the presence of porT homologues in the recently released genomes of T. forsythensis, P. torquis, F. johnsoniae, and F. bacterium as well as in a number of incomplete genomes belonging to the Cytophaga-Flavobacterium-Bacteroidetes phylum. A C-terminal motif (K[LIV]x[LIV][KRHEQ]Stop) search for the whole phylum in the TrEBML protein database returned a significant number of proteins originating from the organisms with a porT homologue, and a large number of these proteins (113 out of 171 sampled [66%]) harbor a predicted signal peptide. It would either be a lucky coincidence that these organisms have the same C-terminal motif in their exported proteins and the presence of the porT gene in their genome or that the presence of both in the same genome and in the same cellular compartment may be related to a novel system of protein secretion in these organisms. These exciting possibilities are currently under investigation at our laboratory.
Supplementary Material
Acknowledgments
We thank Mike A. Curtis for the 1B5 MAb and the E8 Rgp-deficient P. gingivalis mutant and Lindsey N. Shaw for advice on some aspects of the molecular work.
This work was supported by an NIH grant (DE 09761) to J.T. and a grant from the MNiSW (Warsaw, Poland) to J.P. J.P. is also a Subsydium Profesorskie award recipient from the Foundation for Polish Science (FNP, Warszawa, Poland).
Footnotes
Published ahead of print on 1 December 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aduse-Opoku, J., N. N. Davies, A. Gallagher, A. Hashim, H. E. Evans, M. Rangarajan, J. M. Slaney, and M. A. Curtis. 2000. Generation of lys-gingipain protease activity in Porphyromonas gingivalis W50 is independent of Arg-gingipain protease activities. Microbiology 146:1933-1940. [DOI] [PubMed] [Google Scholar]
- 2.Bosch, D., M. Scholten, C. Verhagen, and J. Tommassen. 1989. The role of the carboxy-terminal membrane-spanning fragment in the biogenesis of Escherichia coli K12 outer membrane protein PhoE. Mol. Gen. Genet. 216:144-148. [DOI] [PubMed] [Google Scholar]
- 3.Champion, P. A., S. A. Stanley, M. M. Champion, E. J. Brown, and J. S. Cox. 2006. C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science 313:1632-1636. [DOI] [PubMed] [Google Scholar]
- 4.Chiu, J., P. E. March, R. Lee, and D. Tillett. 2004. Site-directed, ligase-independent mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 32:e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cianciotto, N. P. 2005. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13:581-588. [DOI] [PubMed] [Google Scholar]
- 6.Clamp, M., J. Cuff, S. M. Searle, and G. J. Barton. 2004. The Jalview Java alignment editor. Bioinformatics 20:426-427. [DOI] [PubMed] [Google Scholar]
- 7.Curtis, M. A. 1997. Analysis of the protease and adhesin domains of the PrpRI of Porphyromonas gingivalis. J. Periodontal Res. 32:133-139. [DOI] [PubMed] [Google Scholar]
- 8.Curtis, M. A., A. Thickett, J. M. Slaney, M. Rangarajan, J. Aduse-Opoku, P. Shepherd, N. Paramonov, and E. F. Hounsell. 1999. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect. Immun. 67:3816-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danese, P. N., and T. J. Silhavy. 1998. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu. Rev. Genet. 32:59-94. [DOI] [PubMed] [Google Scholar]
- 10.Desvarieux, M., R. T. Demmer, T. Rundek, B. Boden-Albala, D. R. Jacobs, Jr., R. L. Sacco, and P. N. Papapanou. 2005. Periodontal microbiota and carotid intima-media thickness: the Oral Infections and Vascular Disease Epidemiology Study (INVEST). Circulation 111:576-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichinger, A., H. G. Beisel, U. Jacob, R. Huber, F. J. Medrano, A. Banbula, J. Potempa, J. Travis, and W. Bode. 1999. Crystal structure of gingipain R: an Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J. 18:5453-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ezzo, P. J., and C. W. Cutler. 2003. Microorganisms as risk indicators for periodontal disease. Periodontol. 2000 32:24-35. [DOI] [PubMed] [Google Scholar]
- 13.Filip, C., G. Fletcher, J. L. Wulff, and C. F. Earhart. 1973. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J. Bacteriol. 115:717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fletcher, H. M., H. A. Schenkein, R. M. Morgan, K. A. Bailey, C. R. Berry, and F. L. Macrina. 1995. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect. Immun. 63:1521-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher, A., J. Aduse-Opoku, M. Rangarajan, J. M. Slaney, and M. A. Curtis. 2003. Glycosylation of the Arg-gingipains of Porphyromonas gingivalis and comparison with glycoconjugate structure and synthesis in other bacteria. Curr. Protein Pept. Sci. 4:427-441. [DOI] [PubMed] [Google Scholar]
- 16.Garcia, R. I., M. M. Henshaw, and E. A. Krall. 2001. Relationship between periodontal disease and systemic health. Periodontol. 2000 25:21-36. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh, P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68:771-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson, F. C., III, C. Hong, H. H. Chou, H. Yumoto, J. Chen, E. Lien, J. Wong, and C. A. Genco. 2004. Innate immune recognition of invasive bacteria accelerates atherosclerosis in apolipoprotein E-deficient mice. Circulation 109:2801-2806. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 20.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendrixson, D. R., M. L. de la Morena, C. Stathopoulos, and J. W. St. Geme III. 1997. Structural determinants of processing and secretion of the Haemophilus influenzae hap protein. Mol. Microbiol. 26:505-518. [DOI] [PubMed] [Google Scholar]
- 22.Holt, S. C., J. Ebersole, J. Felton, M. Brunsvold, and K. S. Kornman. 1988. Implantation of Bacteroides gingivalis in nonhuman primates initiates progression of periodontitis. Science 239:55-57. [DOI] [PubMed] [Google Scholar]
- 23.Imamura, T. 2003. The role of gingipains in the pathogenesis of periodontal disease. J. Periodontol. 74:111-118. [DOI] [PubMed] [Google Scholar]
- 24.Imamura, T., J. Travis, and J. Potempa. 2003. The biphasic virulence activities of gingipains: activation and inactivation of host proteins. Curr. Protein Pept. Sci. 4:443-450. [DOI] [PubMed] [Google Scholar]
- 25.Kamaguchi, A., M. Nakano, M. Shoji, R. Nakamura, Y. Sagane, M. Okamoto, T. Watanabe, T. Ohyama, M. Ohta, and K. Nakayama. 2004. Autolysis of Porphyromonas gingivalis is accompanied by an increase in several periodontal pathogenic factors in the supernatant. Microbiol. Immunol. 48:541-545. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Latimer, L. J., and J. S. Lee. 1991. Ethidium bromide does not fluoresce when intercalated adjacent to 7-deazaguanine in duplex DNA. J. Biol. Chem. 266:13849-13851. [PubMed] [Google Scholar]
- 28.Mikolajczyk, J., K. M. Boatright, H. R. Stennicke, T. Nazif, J. Potempa, M. Bogyo, and G. S. Salvesen. 2003. Sequential autolytic processing activates the zymogen of Arg-gingipain. J. Biol. Chem. 278:10458-10464. [DOI] [PubMed] [Google Scholar]
- 29.Mougous, J. D., M. E. Cuff, S. Raunser, A. Shen, M. Zhou, C. A. Gifford, A. L. Goodman, G. Joachimiak, C. L. Ordonez, S. Lory, T. Walz, A. Joachimiak, and J. J. Mekalanos. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakayama, K., T. Kadowaki, K. Okamoto, and K. Yamamoto. 1995. Construction and characterization of arginine-specific cysteine proteinase (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis. Evidence for significant contribution of Arg-gingipain to virulence. J. Biol. Chem. 270:23619-23626. [DOI] [PubMed] [Google Scholar]
- 31.Nelson, K. E., R. D. Fleischmann, R. T. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. F. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Njoroge, T., R. J. Genco, H. T. Sojar, N. Hamada, and C. A. Genco. 1997. A role for fimbriae in Porphyromonas gingivalis invasion of oral epithelial cells. Infect. Immun. 65:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Brien-Simpson, N. M., P. D. Veith, S. G. Dashper, and E. C. Reynolds. 2003. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr. Protein Pept. Sci. 4:409-426. [DOI] [PubMed] [Google Scholar]
- 34.Offenbacher, S. 1996. Periodontal diseases: pathogenesis. Ann. Periodontol. 1:821-878. [DOI] [PubMed] [Google Scholar]
- 35.Paramaesvaran, M., K.-A. Nguyen, E. Caldon, J. A. McDonald, S. Najdi, G. Gonzaga, D. B. Langley, A. DeCarlo, M. J. Crossley, N. Hunter, and C. A. Collyer. 2003. Porphyrin-mediated cell surface heme capture from hemoglobin by Porphyromonas gingivalis. J. Bacteriol. 185:2528-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parker, A. C., and C. J. Smith. 1993. Genetic and biochemical analysis of a novel Ambler class A beta-lactamase responsible for cefoxitin resistance in Bacteroides species. Antimicrob. Agents Chemother. 37:1028-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potempa, J., R. Pike, and J. Travis. 1995. The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain. Infect. Immun. 63:1176-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Potempa, J., R. Pike, and J. Travis. 1997. Titration and mapping of the active site of cysteine proteinases from Porphyromonas gingivalis (gingipains) using peptidyl chloromethanes. Biol. Chem. 378:223-230. [DOI] [PubMed] [Google Scholar]
- 39.Potempa, J., A. Sroka, T. Imamura, and J. Travis. 2003. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr. Protein Pept. Sci. 4:397-407. [DOI] [PubMed] [Google Scholar]
- 40.Pugsley, A. P., O. Francetic, A. J. Driessen, and V. de Lorenzo. 2004. Getting out: protein traffic in prokaryotes. Mol. Microbiol. 52:3-11. [DOI] [PubMed] [Google Scholar]
- 41.Pukatzki, S., A. T. Ma, D. Sturtevant, B. Krastins, D. Sarracino, W. C. Nelson, J. F. Heidelberg, and J. J. Mekalanos. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA. 103:1528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rangarajan, M., J. Aduse-Opoku, J. M. Slaney, K. A. Young, and M. A. Curtis. 1997. The prpR1 and prR2 arginine-specific protease genes of Porphyromonas gingivalis W50 produce five biochemically distinct enzymes. Mol. Microbiol. 23:955-965. [DOI] [PubMed] [Google Scholar]
- 43.Ruiz, N., D. Kahne, and T. J. Silhavy. 2006. Advances in understanding bacterial outer-membrane biogenesis. Nat. Rev. Microbiol. 4:57-66. [DOI] [PubMed] [Google Scholar]
- 44.Sato, K., E. Sakai, P. D. Veith, M. Shoji, Y. Kikuchi, H. Yukitake, N. Ohara, M. Naito, K. Okamoto, E. C. Reynolds, and K. Nakayama. 2005. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J. Biol. Chem. 280:8668-8677. [DOI] [PubMed] [Google Scholar]
- 45.Seers, C. A., N. Slakeski, P. D. Veith, T. Nikolof, Y. Y. Chen, S. G. Dashper, and E. C. Reynolds. 2006. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J. Bacteriol. 188:6376-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi, Y., D. B. Ratnayake, K. Okamoto, N. Abe, K. Yamamoto, and K. Nakayama. 1999. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 274:17955-17960. [DOI] [PubMed] [Google Scholar]
- 47.Shoji, M., D. B. Ratnayake, Y. Shi, T. Kadowaki, K. Yamamoto, F. Yoshimura, A. Akamine, M. A. Curtis, and K. Nakayama. 2002. Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: cell surface polysaccharide as an anchorage for gingipains. Microbiology 148:1183-1191. [DOI] [PubMed] [Google Scholar]
- 48.Smith, C. J., A. Parker, and M. B. Rogers. 1990. Plasmid transformation of Bacteroides spp. by electroporation. Plasmid 24:100-109. [DOI] [PubMed] [Google Scholar]
- 49.Struyve, M., M. Moons, and J. Tommassen. 1991. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218:141-148. [DOI] [PubMed] [Google Scholar]
- 50.Sztukowska, M., A. Sroka, M. Bugno, A. Banbula, Y. Takahashi, R. N. Pike, C. A. Genco, J. Travis, and J. Potempa. 2004. The C-terminal domains of the gingipain K polyprotein are necessary for assembly of the active enzyme and expression of associated activities. Mol. Microbiol. 54:1393-1408. [DOI] [PubMed] [Google Scholar]
- 51.Upreti, R. K., M. Kumar, and V. Shankar. 2003. Bacterial glycoproteins: functions, biosynthesis and applications. Proteomics 3:363-379. [DOI] [PubMed] [Google Scholar]
- 52.Vanterpool, E., F. Roy, and H. M. Fletcher. 2005. Inactivation of vimF, a putative glycosyltransferase gene downstream of vimE, alters glycosylation and activation of the gingipains in Porphyromonas gingivalis W83. Infect. Immun. 73:3971-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veith, P. D., G. H. Talbo, N. Slakeski, S. G. Dashper, C. Moore, R. A. Paolini, and E. C. Reynolds. 2002. Major outer membrane proteins and proteolytic processing of RgpA and Kgp of Porphyromonas gingivalis W50. Biochem. J. 363:105-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Veith, P. D., G. H. Talbo, N. Slakeski, and E. C. Reynolds. 2001. Identification of a novel heterodimeric outer membrane protein of Porphyromonas gingivalis by two-dimensional gel electrophoresis and peptide mass fingerprinting. Eur. J. Biochem. 268:4748-4757. [DOI] [PubMed] [Google Scholar]
- 55.Weinberg, A., C. M. Belton, Y. Park, and R. J. Lamont. 1997. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 65:313-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoneda, M., T. Hirofuji, H. Anan, A. Matsumoto, T. Hamachi, K. Nakayama, and K. Maeda. 2001. Mixed infection of Porphyromonas gingivalis and Bacteroides forsythus in a murine abscess model: involvement of gingipains in a synergistic effect. J. Periodontal Res. 36:237-243. [DOI] [PubMed] [Google Scholar]
- 57.Yoshimoto, H., Y. Takahasi, D. Kato, and T. Umemoto. 1997. Construction of a plasmid vector for transformation of Porphyromonas gingivalis. FEMS Microbiol. Lett. 152:175-181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.