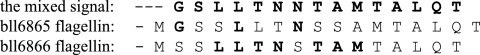Abstract
Bradyrhizobium japonicum is one of the soil bacteria that form nodules on soybean roots. The cell has two sets of flagellar systems, one thick flagellum and a few thin flagella, uniquely growing at subpolar positions. The thick flagellum appears to be semicoiled in morphology, and the thin flagella were in a tight-curly form as observed by dark-field microscopy. Flagellin genes were identified from the amino acid sequence of each flagellin. Flagellar genes for the thick flagellum are scattered into several clusters on the genome, while those genes for the thin flagellum are compactly organized in one cluster. Both types of flagella are powered by proton-driven motors. The swimming propulsion is supplied mainly by the thick flagellum. B. japonicum flagellar systems resemble the polar-lateral flagellar systems of Vibrio species but differ in several aspects.
Bradyrhizobium japonicum is a nitrogen-fixing bacterial species that forms root nodules specifically on soybean (Glycine max) roots. Because soybeans are a good dietary source of protein for vegetarians, soybeans and hence B. japonicum are agriculturally important. The complete genome of B. japonicum has been sequenced (8). It retains both flagellar and type III secretion systems, which play a crucial role in plant-microorganism interactions, especially for bacterial adhesion to the root hair surfaces.
Flagella of rhizobacterial species are different from those of enteric species. For example, Rhizobium lupini or Sinorhizobium meliloti has peritrichous flagella, but the flagellar filament shows a zigzag pattern on the surface, which is called the complex filament. The complex filament exhibits a prominent helical pattern of alternating ridges and grooves, thus appearing more complex than plain filaments of enteric bacteria (14, 16). Azospirillum brasilense has two sets of flagellar systems (6): a polar flagellum and lateral flagella, similar to those of Vibrio parahaemolyticus (4, 11). V. parahaemolyticus cells swim in aqueous (low-viscosity) conditions by using a single polar flagellum as a screw, while they swarm on the viscous surface by using lateral flagella (9).
In this study, we first observed B. japonicum cells by electron microscopy and were amazed about the unusual set of flagella, one thick flagellum and a few thin flagella, both growing from the side of the cell body. Thus, they are distinctive from the similar set of flagella of V. parahaemolyticus: polar flagellum and lateral flagella. We have purified the flagella separately from mutants, analyzed the component proteins by amino acid sequencing, and identified the genes encoding those proteins. We have also examined the role of each flagellum by microscopic observations of mutants that carry only one set of the flagella.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. japonicum strain 110spc4 is a mutant derivative of B. japonicum USDA3I1b110. BJDΔ283 is a mutant with the deletion of flagellin genes bll6865 and bll6866, which are part of the set 2 cluster of flagellar genes. They encode the thin filament. BJDΔ293 is a mutant with the deletion of flagellin genes bll5846 to bll5843 of the set 1 cluster of flagellar genes. The genes encode the thick filament. BJDΔ289-Δ293 is a double mutant with deletions of bll6865 and bll6866 and of bll5846 to bll5843, resulting in a strain that has no flagella.
B. japonicum was cultivated at 30°C in arabinose-gluconate medium (HM salts medium supplemented with 0.1% yeast extract, 0.1% l-arabinose, and 0.1% sodium gluconate) (13) supplemented with the appropriate antibiotics (100 μg/ml kanamycin, 50 μg/ml gentamicin, and 100 μg/ml streptomycin). Cells were typically grown by gentle shaking for 2 days for motility assays and 3 days for flagellum preparation.
Sinorhizobium meliloti was a kind gift from Rudy Schmitt, and V. parahaemolyticus was a kind gift from Linda McCarter. The cells were grown by conventional methods.
Construction of the flagellin deletion mutants B. japonicum BJDΔ283, BJDΔ293, and BJDΔ289-Δ293.
The deletion of the genes bll6865 and bll6866 was accomplished by site-directed mutagenesis. For this, flanking fragments were amplified by specific primers and cloned into pSUPPOL2SCA (10). A kanamycin resistance cassette was cloned between the fragments and used as a selection marker. The plasmid was introduced into B. japonicum 110spc4 by conjugation as described previously by Krause et al. (10). As a result, the genomic region ranging from nucleotide position 7561071 to nucleotide position 7563315, based on the chromosomal map described previously by Kaneko et al. (8), was replaced by the kanamycin resistance cassette, yielding strain BJDΔ283.
Genes bll5846, bll5845, bll5844, and bll5843 were replaced by a gentamicin resistance gene using the same standard techniques as indicated above. In strain BJDΔ293, the deletion extends from nucleotide position 6408773 to nucleotide position 6418851. The gentamicin resistance was derived from pBSL114 (3).
To create a double mutant lacking both regions, B. japonicum strain BJDΔ293 was used. In this chromosomal background, genes bll6865 and bll6866 were deleted as described above, except that a spectinomycin-streptomycin resistance cassette derived from pHP45Ω (12) was used as selection marker. The double mutant strain was designated BJDΔ289-Δ293. Deletions were confirmed by Southern blot hybridization.
Purification of sheared flagella and intact flagella.
The thin flagella were recovered from cell-free culture media by polyethylene glycol (PEG) precipitation (adding 2% of PEG 6000 and 0.1 mM of NaCl into the culture medium and incubating it on ice for 1 h to form bundles of flagella, which were collected by low-speed centrifugation). To purify the thick flagellum, cell pastes were sheared in a blender at its maximum speed, and flagella that were freed from cells were collected by PEG precipitation.
For the preparation of the flagellar basal body, a conventional method was used, with minor modifications (2). The cells were harvested by low-speed centrifugation, washed once with STE buffer (100 mM NaCl, 10 mM Tris [pH 8.0], 1 mM EDTA) to remove sticky materials from the cell surface, and resuspended in sucrose solution (0.5 M sucrose, 0.15 M Trizma base) by gentle stirring. Lysozyme (final concentration, 0.1 mg/ml) and EDTA (final concentration, 3 mM) were gradually added to the suspension, and the mixture was incubated on ice with gentle stirring. After 30 min of incubation, most cells became spheroplasts as judged by dark-field microscopy. The cells were lysed with Triton X-100 (final concentration, 1%), and the cell debris and nonlysed cells were removed by low-speed centrifugation. Intact flagella were collected by PEG precipitation. The flagella were suspended in distilled water for further analysis.
Motility and swarm assays.
To measure the swimming speeds of cells, cultured cells, without washing, were directly observed with the dark-field microscope attached with a charge-coupled-device camera (WAT-902H2; Watec) and a video-recording system (Adobe Premiere Pro 1.5).
To examine the effects of antibiotics on the thick flagellum, BJDΔ283 cells were grown in arabinose-gluconate medium at 30°C, harvested by low-speed centrifugation, and suspended in 50 mM Tris-HCl buffer (pH 7.0 or pH 8.5). Carbonyl cyanide m-chlorophenyl hydrazone (CCCP) (Sigma Co.) was added at 0 to 20 μM, and phenamil (Sigma Co.) was added at 0 to 100 μM.
To observe the swarming ability of each strain, cells were inoculated on an agar plate (0.4% agar in nutrient broth) and incubated at 30°C for 3 weeks.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out using the Mini-Gel kit from Bio-Rad. The acrylamide concentration was 12.5%. Gels were stained by Coomassie brilliant blue R-250. A molecular mass standard protein marker was obtained from APRO.
Electron microscopy.
Samples were negatively stained with 2% phosphotungstic acid (pH 7.0) and observed with a JEM-1200EXII electron microscope (JEOL, Tokyo, Japan). Micrographs were taken at an accelerating voltage of 80 kV.
RESULTS
Electron microscopic observations of flagella.
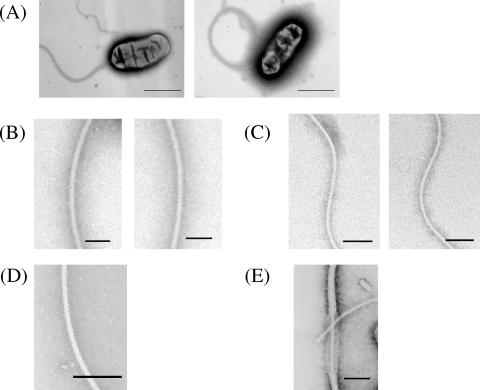
Bradyrhizobium japonicum wild-type cells were grown for 2 days, harvested at the early log phase of growth, and observed by electron microscopy. The cell is rod shaped and has two sets of flagella: one thick flagellum and a few thin flagella (Fig. 1A). These two sets of flagella resemble the two sets of flagella found in Vibrio species: one polar flagellum and several lateral flagella. The pitch of the thick flagellum (2.8 ± 0.3 μm; average of 10 samples) is four times larger than that of the thin one (0.7 ± 0.04 μm). Under the dark-field microscope, the thick flagellum appeared semicoiled, and the thin flagella had a tight-curly morphology, which are typical forms of polar and lateral flagella. The thickness of the thick flagellum (22 nm) is approximately two times larger than that of the thin one (12 nm) (Fig. 1B and C). The thick flagellum looks bushy but is neither sheathed nor complex compared to their microscopic appearance with authentic complex flagellum from Sinorhizobium meliloti (Fig. 1D) and sheathed flagellum from V. parahaemolyticus (Fig. 1E).
FIG. 1.
Electron micrographs of B. japonicum wild-type cells. (A) The wild-type cell typically has two types of flagella, a thick flagellum and a few thin flagella, both at the cell subpole. Enlarged images of (B) the thick flagellum, (C) the thin filament, (D) the complex flagellum from Sinorhizobium meliloti, and (E) the sheathed flagellum from V. parahaemolyticus are shown. A part of the sheath is broken, exposing the filament inside. Cells were negatively stained with 1% phosphotungstic acid (pH 7.0). Bars indicate 1 μm (A) and 100 nm (B to E).
The B. japonicum flagella grow near but not at the cell pole. To determine the precise positions of the flagellar bases, the distance of the flagellar base from one end of the cell was measured. The ratio of the base position relative to the cell length was then calculated. Bases of the thick flagellum distribute at one end of the cell from 10 to 26% of the cell length (the average of 35 cells). The average ratio is 18.7%. On the other hand, the bases of the thin flagella distribute widely from 9 to 44% with an average of 23.5%. Therefore, these flagella are literally neither polar nor lateral flagella but might be called subpolar flagella. We temporarily distinguish these two filaments as the thick flagellum and thin flagella and analyze them in comparison with V. parahaemolyticus flagella.
Multiple flagellin genes in the genome.
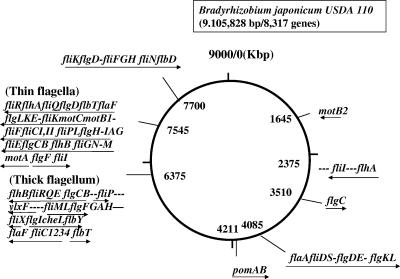
In the B. japonicum genome, there are two large clusters and several small, scattered spots of the flagellar genes (8). Since the two large clusters contain duplicate flagellar genes, they might represent two sets of flagellar systems encoding the component proteins of the thick and thin flagella observed.
In one cluster that locates at 6,375 kbp (set 1 cluster) (Fig. 2), there are four genes from bll5843 to bll5846 (tentative gene designations described by Kaneko et al. [8]) aligning in tandem on the chromosome. These four genes were similar to the flaA gene of the bacterial species that have a polar flagellar system (data not shown). Because they are essential for the formation of a subpolar flagellum (see below), we termed them fliC1234. The identity among these four gene products was 71.1%. The first 20 amino acid residues of Bll5844 and Bll5846 are identical. From this, Bll5843 (third residue) and Bll5845 (eighth residue) deviate at only one position. The deduced molecular masses of the four gene products are between 74.3 and 75.7 kDa.
FIG. 2.
Genome map of B. japonicum. The total size of the B. japonicum genome is 9,105,828 bp, containing 8,317 genes. The arrows under the genes show the directions of transcription, but the genes may not necessarily form an operon. The dashes among the flagellar genes indicate other genes or open reading frames that are not related to the flagella.
In the other cluster that locates at 7,545 kbp (set 2 cluster), there are two candidates for the flagellin gene: fliCI and fliCII (bll6865 and bll6866, respectively). The deduced molecular masses of these gene products are 32.5 and 32.2 kDa, respectively. The gene products of bll6865 and bll6866 are highly homologous (95.4% identity).
Flagellar mutants and their swimming behavior.
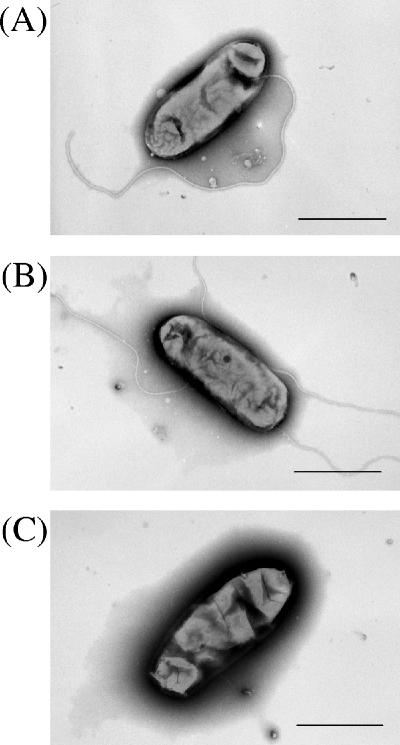
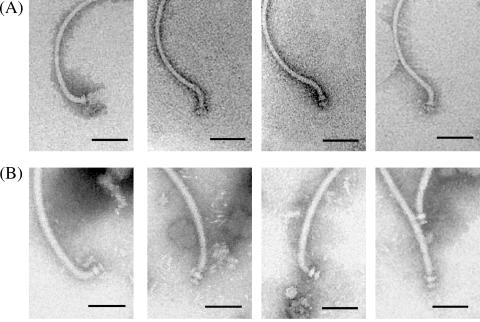
We constructed three deletion mutants of possible flagellin genes, including one mutant (strain BJDΔ283) with the deletion of two flagellin genes, fliCI (bll6865) and fliCII (bll6866), from set 2 flagellar cluster and a second mutant (strain BJDΔ293) with the deletion of the four flagellin genes, fliC1234 (bll5846 to bll5843), from the set 1 flagellar cluster. In the third mutant (BJDΔ289-Δ293), all six flagellin genes (fliCI, fliCII, and fliC1234) were deleted. As visualized by electron microscopy, BJDΔ283 cells had a thick flagellum but no thin flagella (Fig. 3A), while BJDΔ293 cells had the thin flagella but not the thick flagellum (Fig. 3B). The BJDΔ289-Δ293 cells consequently lacked both types of flagella (Fig. 3C). In conclusion, the flagellin genes of the set 1 cluster appear to encode the thick flagellum, and the flagellin genes of the set 2 cluster appear to encode the thin flagella.
FIG. 3.
Electron microscopic images of flagellar mutants. (A) Mutant BJDΔ283 has a thick flagellum without thin flagella. (B) Mutant BJDΔ293 has thin flagella without a thick flagellum. (C) Double mutant BJDΔ289-Δ293 has no filaments. Bars, 1 μm.
We then observed the behavior of those cells by light microscopy and measured the swimming speed of each mutant. The speeds were 30.4 ± 5.7 μm/s for the wild-type cells, 30.3 ± 2.9 μm/s for BJDΔ283 cells with thick filaments, and 16.8 ± 6.1 μm/s for BJDΔ293 cells with thin filaments. Swimming speeds of the wild-type cells and those with a thick flagellum are almost the same, but cells with only thin flagella are much slower. The swimming pattern of the cells with only thin flagella was aberrant and unstable. Therefore, we conclude that the swimming propulsive force of the wild-type cells is supplied mostly by the thick flagellum and not by the thin flagella.
Analysis of flagellins from purified flagella.
BJDΔ283 cells have only thick flagellum, while BJDΔ293 cells have only thin flagella. To identify flagellins of the two types of flagella, we have purified flagella from each mutant. Since the thick flagellum was firmly attached with the cells, flagella were sheared off from harvested cells by a blender or by passing them through a hypodermic needle. On the other hand, the thin flagella were often released during a 3-day incubation and were found floating in the culture medium. The thin flagella were purified directly from the media by PEG precipitation (see Materials and Methods).
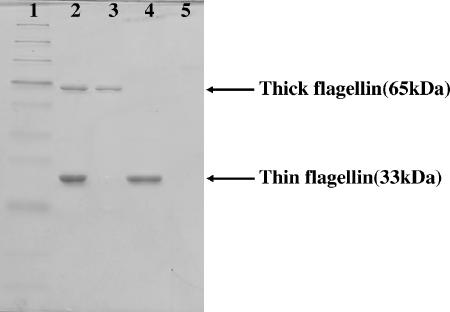
By SDS-PAGE, flagella isolated from wild-type cells give two bands at apparent molecular masses of 65 and 33 kDa (Fig. 4, lane 2). The thick flagellum from BJDΔ283 yields the 65-kDa band, and the thin flagella from BJDΔ293 give the 33-kDa band (Fig. 4, lanes 3 and 4).
FIG. 4.
SDS-PAGE of flagellins of B. japonicum. The molecular mass of the thick flagellin was 65 kDa, and that of the thin flagellin was 33 kDa. Lane 1, protein marker; lane 2, wild-type strain 110spc4 (both types of flagella exist); lane 3, BJDΔ283 (thin flagella are missing); lane 4, BJDΔ293 (thick flagellum is missing); lane 5, BJDΔ289-Δ293 (both types of flagella are missing).
There are multiple flagellin genes in the flagellar gene clusters as mentioned above. To identify the proteins incorporated into these flagella, we determined the N-terminal amino acid sequences. The sequence obtained for the 65-kDa protein was SGIVLSASVRQNLLSLQSTA, which is identical to the sequences deduced from the bll5844 and bll5846 genes. Microarray data indicated that genes bll5843 and bll5845 are expressed as well (unpublished data). Therefore, we analyzed the 65-kDa protein band by mass spectrometry and obtained fragments corresponding to all four proteins. This suggests that the thick flagellum might consist of all four proteins, albeit at an unknown stoichiometry.
The sequence of the first 15 amino acids of the 33-kDa protein was GSLLTNNTAMTALQT, which is predicted from neither bll6865 nor bll6866. But when we read the second peaks by taking the signal strength characteristic of each amino acid into account, the sequence could be read as a mixture of the two flagellins (Fig. 5). We conclude that the thin flagella are composed of two flagellins: FliCI and FliCII. Microarray data confirm that both genes are expressed (unpublished data).
FIG. 5.
Amino acid analysis of the N-terminal region of the thick flagellin. The first row is the determined sequence from the 33-kDa-band protein. The lower two columns are the deduced sequences of bll6865 flagellin (FliCI) and bll6866 flagellin (FliCII). The amino acids in boldface type give a strong signal in the analysis.
Gene organization in the genome and structural analysis of the flagellar basal body.
From analyses of flagellins, we assume that the set 1 genes are for the thick flagellum and that the set 2 genes are for the thin flagellum. Flagellar gene organization on the genome is summarized in Fig. 2. By comparison with a standard set of flagellar genes from Salmonella (1), two sets of flagellar gene clusters are incomplete to form the whole flagellar structure. The master genes flhDC, which activate the entire flagellar genes in Salmonella, are missing in B. japonicum. In the set 1 cluster, flagellar genes are not well organized but are fragmental. There are many genes essential for flagellar assembly that are missing. However, there are several islands of flagellar genes that would supplement the missing genes: the fliI operon at 2,375 kbp, the flaA operon at 4,085 kbp, the pomAB genes at 4,211 kbp, and the fliK operon at 7,700 kbp (Fig. 2). Even when all these genes are combined, there are still some genes missing from a standard set of flagellar genes: flgJMN, flhFG, and fliAJST. The set 2 cluster is more compactly organized and contains most essential genes but lacks the following genes: flgJMN and fliADHJST (Table 1) (see Discussion).
TABLE 1.
List of flagellar genes in Salmonella and B. japonicum
| Salmonella flagellar genesa |
Bradyrhizobium japonicum flagellar gene(s)b
|
||
|---|---|---|---|
| Set 1 genes (for the thick flagellum) | Set 2 genes (for the thin flagellum) | Other sites | |
| flgA, flgB, flgC, flgD, flgE, flgF, flgG, flgH, flgI, flgJ, flgK, flgL, flgM, flgN | flgA, flgB, flgC, flgF, flgG, flgH, flgI | flgA, flgB, flgC, flgD, flgE, flgF, flgH, flgI, flgK, flgL | flgC, flgD, flgE flgK, flgL |
| flhA, flhB flhC, flhD,cflhEd | flhB | flhA, flhB | flhA flhE |
| fliA, fliB, fliC, fliD, fliE, fliF, fliG, fliH, fliI, fliJ, fliK, fliL, fliM, fliN, fliO, fliP, fliQ, fliR fliS, fliT | fliC1234, fliE, fliL, fliM, fliO, fliP fliQ, fliR, | fliCI, fliCII, fliE, fliF, fliG, fliI, fliK, fliL, fliM, fliN, fliP, fliQ, fliR | fliD, fliF, fliG, fliH, fliI (fliJ),efliK, fliN, fliS |
| motA, motB | motA, motB1, motB2, motCf | pomA, pomBg | |
| flaF, flbT cheL, flbY, fliXh | flaF, flbTh | flaA, flbDh | |
The operon unit and the order of the genes are ignored in this table. The genes were put in the order of flg, flh, fli, mot, and others according to Salmonella flagellar gene clusters.
Set 1 and set 2 are the two largest clusters in the B. japonicum genome, locating at 6,375 kbp and 7,545 kbp, respectively. Flagellar genes locating at sites other than these two clusters are put in the column of “other sites.”
The master genes flhDC in Salmonella are not found in B. japonicum.
The flhE gene is a flagellar gene required for swarming motility (15).
The gene of interest resembles both fliI and fliJ, and thus, both genes were considered to be candidates.
The motC gene has been found in species that retain the polar flagellum. The function is unknown.
The pom genes are motility genes in Vibrio alginolyticus, corresponding to the mot genes of Salmonella.
These genes are not found in Salmonella. Many of them are regulatory genes in other organisms.
In order to confirm the assignments of the flagellar genes to each flagellar system, we have isolated the flagellar basal bodies and sequenced the hook protein. The basal structures from both types of flagella look alike (Fig. 6). The hook of the thin flagellum (the thin hook) (12 nm in diameter) was also thinner than that of the thick flagellum (the thick hook) (23 nm). The molecular size and sequences of both candidates for hook proteins are not so different (410 amino acids versus 420 amino acids; 33% identity). The amino acid sequencing of the thick hook protein was not successful because of N-terminal blockage, suggesting a chemical modification on the thick hook. The amino acid sequence of the thin hook protein was SLYGVMRTGVSGM, which was identical to that of FlgE deduced from bll6858.
FIG. 6.
Electron microscopic images of flagellar basal structures. The intact flagella attached to the hook-basal body were isolated according to a conventional method (2). The appearances of the hook-basal body of thin flagellum (A) and thick flagellum (B) are similar in terms of the number of rings and their sizes. The “thick” hook looks larger than the “thin” hook. Bars, 100 nm.
Both types of flagella are powered by proton-driven motors.
The Vibrio polar flagellum is powered by sodium-motive force (4). In order to determine whether B. japonicum thick flagellum (corresponding to the polar flagellum) is dependent on proton- or sodium-motive force, we have examined the effects of the protonophore CCCP and the sodium ionophore phenamil on BJDΔ283 cells. The cell motility was inhibited strongly with the increase in the CCCP concentration and stopped almost completely with 20 μM CCCP at both pHs tested. However, we have not seen any decrease in the motility rate by phenamil treatment (Table 2). Under the same conditions, the Vibrio polar flagellum was completely stopped by phenamil (data not shown).
TABLE 2.
Effects of ionophores on motility rates of BJDΔ283 cells
| Strain | Motility ratea
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CCCP (μM)
|
Phenamil (μM)
|
|||||||||||
| pH 7.0
|
pH 8.5
|
pH 7.0
|
pH 8.5
|
|||||||||
| 0 | 5 | 20 | 0 | 5 | 20 | 0 | 50 | 100 | 0 | 50 | 100 | |
| BJDΔ283 | ++ | − | − | ++ | + | − | ++ | ++ | ++ | ++ | ++ | ++ |
| BJDΔ293 | + | − | − | + | + | − | + | + | + | + | + | + |
+ and − indicate motility rates: more than 50% (++), 50% to 10% (+), and less than 10% (−) of the cells were swimming. The cells were observed for swimming ability under a dark-field microscope within a few minutes after the addition of ionophores. Numbers indicate the concentration of CCCP or phenamil in μM.
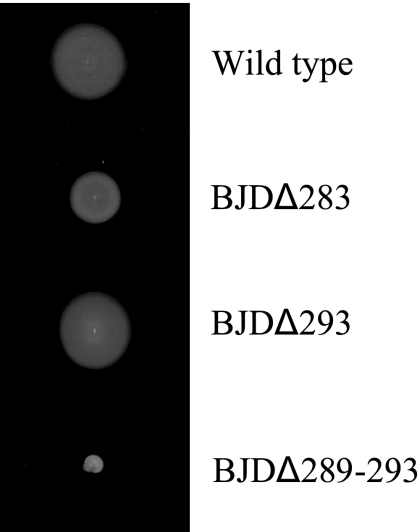
BJDΔ293 cells that have only thin flagella make a swarm ring larger than that made by BJDΔ283 cells on an agar plate (Fig. 7). However, BJDΔ293 cells do not swim smoothly in liquid but jiggle at one place, which is difficult for observing the effects of ionophores. Thus, we have prepared tethered cells of BJDΔ293. Although many cells were stuck on a glass slide for technical reasons, we could observe a few rotating tethered cells on the slide. None of those cells were rotating after CCCP treatment, while some were still rotating after phenamil treatment. In conclusion, both types of flagella of B. japonicum have proton-driven motors.
FIG. 7.
Colony types of B. japonicum mutants on an agar plate. Cells were inoculated on an agar plate (0.4% agar in nutrient broth) and incubated at 30°C for 3 weeks. BJDΔ293 cells made a swarm ring that was larger than that made by BJDΔ283 cells on the plate.
It should be noted that there are no mot genes in the set 1 cluster, but there are pomA and pomB (pomAB) genes at 4,211 kbp. pomAB of B. japonicum were annotated this way because the sequences resemble pomAB of V. parahaemolyticus more than the conventional mot genes. Since the pomAB genes were originally designated for the sodium-driven motor components, it is interesting to see how the PomAB complex, if formed and used, works in the proton-driven motor of B. japonicum.
Altogether, the B. japonicum flagellar system resembles that of V. parahaemolyticus in many aspects but is distinguishable in flagellar position on the cell surface and the energy source to power the flagellar motor.
DISCUSSION
Why are two sets of flagella necessary?
Why does B. japonicum have two sets of flagellar systems? Since they are close, albeit not exactly the same, to the polar-lateral flagellar system of V. parahaemolyticus, we may be able to employ the same logic of the latter; the thick flagellum is used for swimming, and the thin flagella are used for swarming. We observed that the thick flagellum is constitutively expressed, while the thin flagella are inducible by high viscosity in B. japonicum wild-type cells (data not shown; 9). The thick flagellum may be used for swimming to reach plant roots, while the thin flagella may be expressed for swarming on the root hair surface before nodule formation.
The mechanism that the lateral flagella use in swarming is not well understood. The jiggling motion of BJDΔ293 cells is not caused by frequent reverses of rotation of thin filaments, because tethered cells through thin filaments rotated in only one direction and never showed reverse rotation for as long as we observed them. It is believed that the polar flagellum in V. parahaemolyticus rotates in a way similar to that of Salmonella peritrichous flagella, although the energy source is not proton- but sodium-motive force (4). In B. japonicum, the thick flagellum corresponding to the polar flagellum in V. parahaemolyticus is driven by proton-motive force. This is probably because hydrogen ions are more available than sodium ions in soil environments.
Missing flagellar genes.
In the B. japonicum genome, there are two sets of flagellar gene clusters and several small operons containing one or a few flagellar genes. There are some missing genes as mentioned in Results: flgJMN, flhFG, and fliAJST in the set 1 cluster and flgJMN and fliADHJST in the set 2 cluster (Table 1). Are they essential or not?
flgM and fliA are regulatory genes for the peritrichous flagellar system and can be replaced by other genes in this system. The flgN, fliS, and fliT genes encode chaperones and are necessary for the efficient growth of flagella. The fliJ gene encodes a chaperone for FlgN. These small chaperones, although not necessarily essential, may be difficult to identify on the basis of protein homology. The flgJ gene is required for the penetration of the rod through the outer membrane and is thus essential for flagellar assembly (7). The corresponding protein of B. japonicum may have an amino acid sequence quite different from that of the FlgJ capping protein of other organisms and might be encoded by an unidentified gene. FliD is a capping protein necessary for filament growth. The flhFG genes are necessary for the localization and control of the number of polar flagella in Pseudomonas aeruginosa (5).
Acknowledgments
We thank Takanori Hirano, Kiwamu Minamisawa for discussion, and Tatsuaki Ebisawa and Kazuya Momma for their technical help.
We also thank the Japan Science and Technology Agency and the German BMBF for financial support.
Footnotes
Published ahead of print on 10 November 2006.
REFERENCES
- 1.Aizawa, S.-I. 2004. Flagella, p. 470-479. In M. Schaechter (ed.), The desk encyclopedia of microbiology. Academic Press, Washington, DC.
- 2.Aizawa, S.-I., E. D. Gay, J. J. Christopher, M. M. Robert, and S. Yamaguchi. 1985. Purification and characterization of the flagellar hook-basal body complex of Salmonella typhimurium. J. Bacteriol. 161:839-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexeyev, M. F., I. N. Shokolenko, and T. P. Croughan. 1995. Improved antibiotic-resistance gene cassettes and omega elements for Escherichia coli vector construction and in vitro deletion/insertion mutagenesis. Gene 160:63-67. [DOI] [PubMed] [Google Scholar]
- 4.Atsumi, T., L. McCarter, and Y. Imae. 1992. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature 355:182-184. [DOI] [PubMed] [Google Scholar]
- 5.Dasgupta, N., S. K. Arora, and R. Ramphal. 2000. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 182:357-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall, P. G., and N. R. Krieg. 1984. Application of the indirect immunoperoxidase stain technique to the flagella of Azospirillum brasilense. Appl. Environ. Microbiol. 47:433-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirano, T., T. Minamino, and R. M. Macnab. 2001. The role in flagellar rod assembly of the N-terminal domain of Salmonella FlgJ, a flagellum-specific muramidase. J. Mol. Biol. 312:359-369. [DOI] [PubMed] [Google Scholar]
- 8.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and M. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 9.Kawagishi, I., M. Imagawa, Y. Imae, L. McCarter, and M. Homma. 1996. The sodium-driven polar flagellar motor of marine Vibrio as the mechanosensor that regulates lateral flagellar expression. Mol. Microbiol. 20:693-699. [DOI] [PubMed] [Google Scholar]
- 10.Krause, A., A. Doerfel, and M. Göttfert. 2002. Mutational and transcriptional analysis of the type III secretion system of Bradyrhizobium japonicum. Mol. Plant-Microbe Interact. 12:1228-1235. [DOI] [PubMed] [Google Scholar]
- 11.McCarter, L. L. 2004. Dual flagellar systems enable motility under different circumstances. J. Mol. Microbiol. Biotechnol. 7:18-29. [DOI] [PubMed] [Google Scholar]
- 12.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 13.Sadowsky, M. J., R. E. Tully, P. B. Cregan, and H. H. Keyser. 1987. Genetic diversity in Bradyrhizobium japonicum serogroup 123 and its relation to genotype-specific nodulation of soybean. Appl. Environ. Microbiol. 53:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scharf, B., H. Schuster-Wolff-Bühring, R. Rachel, and R. Schmitt. 2001. Mutational analysis of Rhizobium lupini H13-3 and Sinorhizobium meliloti flagellin genes: importance of flagellin A for flagellar filament structure and transcriptional regulation. J. Bacteriol. 183:5334-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stafford, G. P., and C. Hughes. The flhE gene was identified as a flagellar gene required for swarming motility. Microbiology, in press. [DOI] [PMC free article] [PubMed]
- 16.Trachtenberg, S., and D. J. DeRosier. 1987. Three-dimensional structure of the complex flagellar filament of Rhizobium lupini and its relation to the structure of the plain filament. J. Mol. Biol. 195:603-620. [DOI] [PubMed] [Google Scholar]