Abstract
The obligate intracellular parasitic bacteria rickettsiae are more closely related to mitochondria than any other microbes investigated to date. A rickettsial putative peptidase (RPP) was found to resemble the α and β subunits of mitochondrial processing peptidase (MPP), which cleaves the transport signal sequences of mitochondrial preproteins. RPP showed completely conserved zinc-binding and catalytic residues compared with β-MPP but barely contained any of the glycine-rich loop region characteristic of α-MPP. When the biochemical activity of RPP purified from a recombinant source was analyzed, RPP specifically hydrolyzed basic peptides and presequence peptides with frequent cleavage at their MPP-processing sites. Moreover, RPP appeared to activate yeast β-MPP so that it processed preproteins with shorter presequences. Thus, RPP behaves as a bifunctional protein that could act as a basic peptide peptidase and a somewhat regulatory protein for other protein activities in rickettsiae. These are the first biological and enzymological studies to report that a protein from a parasitic microorganism can cleave the signal sequences of proteins targeted to mitochondria.
The endosymbiont hypothesis for the origin of mitochondria in eukaryotes is now widely accepted, accompanied by a growing interest in evolution as entire genomic sequences are revealed from various organisms. According to this hypothesis, a free-living bacterium as an organelle progenitor once entered an anaerobic organism, which is thought to be an archaebacterial ancestor, and established a constitutive endosymbiotic relationship with the host cells, mainly to supply ATP (7, 11, 17). Modern mitochondria originated when the parasitic invader lost most of its own genome and began to depend on nuclearly encoded proteins for its biogenesis, although it retained control of the eukaryotic cell viability via metabolic and apoptotic pathways.
Most mitochondrial proteins are encoded in the nucleus and synthesized by cytoplasmic ribosomes as preproteins with N-terminal presequences that are required for targeting to mitochondria (3, 12). These preproteins are unfolded and imported into the mitochondrial matrix across the double membrane through protein translocation machinery comprising a translocase on the outer mitochondrial membrane and a translocase on the inner mitochondrial membrane (20, 24, 26, 27). Finally, the presequences are cleaved by a matrix-located metalloendopeptidase, i.e., mitochondrial processing peptidase (MPP) (10, 13, 16). Overall, this proteolytic processing is involved in maturation of the mitochondrial proteins and is essential for eukaryotic cell viability from unicellular (30) to multicellular (22) organisms. Therefore, the α and β subunits of MPP (α- and β-MPP, respectively) tightly regulate the protease action and specifically cleave the preproteins.
The genome sequences of the obligate intracellular parasitic bacteria rickettsiae (the agents that cause typhus) reveal gene profiles strikingly similar to those of mitochondria (2, 18, 23). Among the bacteria examined to date, rickettsiae are more closely related to mitochondria than any other bacteria analyzed at the genome level. Interestingly, Rickettsia prowazekii gene 219 (RP219) encodes a putative peptidase (rickettsial putative protease [RPP]) that is highly similar to MPPs (2), and corresponding genes have also been found in other rickettsial species (18, 23). Similar to β-MPPs, which are the catalytic MPP subunits (14, 15), RPPs have a zinc-binding motif, HxxEHx75E, in which the histidine residues and final glutamate residue presumably participate in metal binding, while the first glutamate residue could be involved in water activation for hydrolysis of the peptide bond. These active-site motifs are found in the M16 protease family of metalloendopeptidases characterized by Escherichia coli pitrilysin and insulinases from mammals and insects, which appear to diverge widely from bacteria to higher eukaryotes (25).
In the present study, we analyzed the structural characteristics and biochemical activities of the RPP from R. prowazekii. The RPP primary structure resembled those of both of the MPP subunits, since the N- and C-terminal regions of RPP were similar to the N domains of β-MPPs and C domains of α-MPPs, respectively. The biphasic structure of RPP seemed to reflect dual functions, namely, catalytic and regulatory actions toward basic peptides and preprotein processing, respectively. Thus, the characteristics of the structures and functions of RPP and the MPP subunits could be inherited from a common ancestor protein in the parasite that led to the endosymbiotic evolution of mitochondria. Here, we discuss the evolutionary and functional relationships between proteins that resemble components of the mitochondrial transport-processing system on the basis of mitochondrial endosymbiotic evolution.
MATERIALS AND METHODS
Genetic analyses.
The amino acid sequences of MPP and RPP were assembled from the Swiss-Prot and Genome databases and aligned using the CLUSTAL W program (29). From this multiple sequence alignment, a consensus sequence for the active site of MPP was derived. The phylogeny was inferred by using the bootstrap method with 1,000 trials, and a bootstrap tree was drawn using TreeView 1.6.6. Genomic blastp searches to identify microbial MPP-like proteins were carried out using the BLAST website for microbial genomes at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi), using the whole protein sequence of yeast α- or β-MPP. The searches were performed with the EXPECT parameter set to 0.0001 and the FILTER set to default. In the resultant proteins, we collected MPP-like proteins with expected values under 1E−10 and confirmed whether each sequence had a glycine-rich loops (GRL) region.
Culture of rickettsiae and RPP expression analyses.
Rickettsia typhi, Rickettsia conorii, and Rickettsia japonica were grown in Vero cells at 34°C and purified as described previously (31). Total RNA was extracted from purified R. typhi cells using a total RNA isolation system (Promega) according to the manufacturer's instructions. Next, cDNAs were synthesized from the total RNA using reverse transcriptase (Promega) and the primers 5′-TTGGATCCTTAAAATCCATTAAGATCATTC-3′ for rpp and 5′-TTAGAAGTCCATACCACC-3′ for groEL. Amplification of the DNA fragments was performed by PCR using the primer pairs 5′-ATGATAATCCTGATGACC-3′ and 5′-TTCTTCTGACTTATAGG-3′ for rpp and 5′-CTTCAAGAGCGTTTAGC-3′ and 5′-TATCAGAAGGTTCATCC-3′ for groEL. Proteins from purified rickettsiae were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting using a rabbit anti-RPP antibody and horseradish peroxidase-conjugated goat antirabbit immunoglobulin G.
Overproduction, purification, and characterization of recombinant proteins.
An R. prowazekii genomic clone including RP219 inserted into lambda-Zap (a kind gift from S. G. Andersson and C. G. Kurland [University of Uppsala]) was used as the source of the RPP gene. The DNA fragment for the open reading frame of RPP was amplified by PCR using an RPP-specific pair of 5′ and 3′ primers and ligated into the expression vector pET-23d (Novagen), which produces proteins with a C-terminal His6 tag, thereby constructing pET-RPP. E. coli BL21 cells were cotransformed with pET-RPP and pKY206, a plasmid encoding the E. coli GroE operon. After the cells were cultured in LB medium at 30°C for 24 h, the proteins were extracted and purified using a nickel-chelating Sepharose (Amersham Biosciences) column as described previously (15). The RPP-containing fractions eluted from the affinity column were pooled and incubated with 5 mM ATP at 0°C for 60 min to release the GroEL and ES associated with RPP. The protein solution was diluted by more than 10-fold with buffer A (20 mM Tris-HCl, pH 7.5, 30% glycerol, and 0.01% Tween 20) and loaded onto a DEAE Sepharose (Amersham Biosciences) column equilibrated with buffer A. After the column was washed, RPP was eluted with buffer A containing 100 mM NaCl. The last purification step was repeated once. Size exclusion chromatography was performed, using TSK-GEL SUPER SW 3000 (TOSOH) in 20 mM Tris-HCl (pH 7.5) containing 100 mM NaCl. The gene for PRR* (E52Q mutant) was engineered using a QuikChange site-directed mutagenesis system (Promega), and the protein was produced and purified as described above. Wild-type MPP, β-MPP* (E73Q mutant), and α-MPPΔGRL (Δ287-249 mutant) were purified as described previously (15, 19).
Assay for peptidase activity and identification of cleavage sites.
The peptides purchased for this study were dynorphin A (Peninsula Laboratories, Inc.), vasoactive intestinal peptide (Peninsula Laboratories, Inc.), mastoparan (Peninsula Laboratories, Inc.), α-melanocyte-stimulating hormone (αMSH) amide (C S Bio Co.), big endothelin 1 (Phoenix Pharmaceuticals, Inc.), peptide fragment of somatostatin receptor SSTR2A (Gramsch Laboratories), peptide fragment of μ opioid receptor MOR1A (Gramsch Laboratories), and gastrin I (Bachem). We also used various presequence peptides synthesized as described previously (21): mouse malate dehydrogenase, amino acids 2 to 28 (MDH2-28); yeast heat shock protein SSC1, amino acids 1 to 32; yeast MDH2-17; mouse aldehyde dehydrogenase, amino acids 1 to 29; human ornithine aminotransferase, amino acids 1 to 25; yeast ubiquinol cytochrome c reductase subunit 2, amino acids 1 to 32; bovine adrenodoxin, amino acids 18 to 57 (ADX18-57); and yeast cytochrome c oxidase subunit 4, amino acids 2 to 25 (COXIV2-25). Each peptide (1 μM) was incubated with RPP (25 nM) or yeast MPP (2.5 nM) at 30°C for 60 min before the reaction was stopped and analyzed by reverse-phase high-performance liquid chromatography (HPLC) as described previously (21). The cleavage efficiencies were determined from the ratios of the residual peptide amounts after incubation with or without peptidases. To define the cleavage sites, the eluates containing peptide fragments were independently collected, lyophilized, mixed with a matrix solution of sinapinic acid, and analyzed using a mass spectrometer (Voyager PR-HR Biospectrometry; ABI).
Assay for preprotein processing.
Processing assays of [35S]methionine-labeled preproteins were performed as described previously (15). RPP, MPP, or premixed combinations of RPP and MPPs were incubated with the preprotein at 30°C for 60 min in the processing buffer. The processing products were separated by SDS-PAGE and visualized using an imaging analyzer (Bas1000; Fujifilm). The processing efficiency was determined by quantifying the radioactivity of the cleaved protein relative to that of the total protein using Image Gauge 3.0 (Fuji Photo Film Co., Ltd., and Koshin Graphic Systems).
RESULTS
Evolutionary and structural characteristics of RPP.
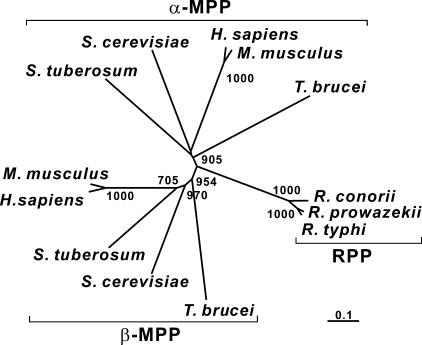
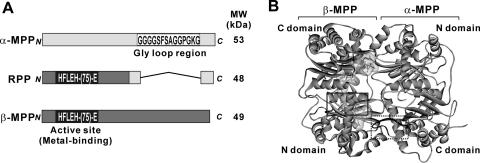
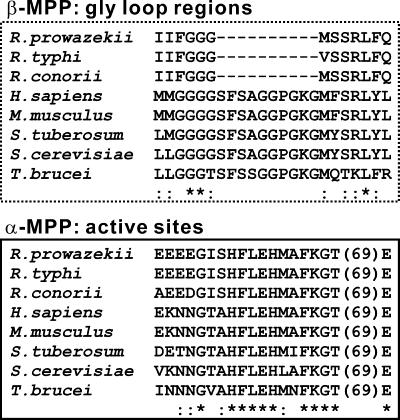
RPPs were identified in the genomes of several rickettsial species and were found to be highly conserved with each other. When three RPPs from R. prowazekii, R. typhi, and R. conorii were compared with MPP subunits (from protozoan to mammalian) using statistical genetics, the potential phylogenetic relationship between them revealed that RPPs were genetically close to either α-MPP or β-MPP and were more closely related to protozoan MPPs than to other eukaryotic MPPs (Fig. 1). Figure 2A shows structural schematic diagrams of R. prowazekii RPP and the yeast Saccharomyces cerevisiae MPP subunits. It was of interest to note that the N- and C-terminal regions of the RPP resembled the terminal domains of β-MPP and α-MPP (N and C domains), respectively. In particular, the N-terminal region of the RPP included an active-site motif, HxxEHx75E, that was previously characterized in a subfamily of M16 proteases. Moreover, the rickettsial amino acid sequences around the motif were highly conserved in the active-site sequences of β-MPPs (Fig. 3), indicating that RPPs potentially possess peptidase activity. On the other hand, the GRLs that are typically preserved in the C domains of α-MPPs were mostly lacking in the C domains of RPPs, whereas the rickettsial amino acid sequences corresponding to regions around the GRLs were comparatively similar to the primary structures of α-MPPs (Fig. 3), suggesting a lesser importance for the GRL function among RPPs. The three-dimensional structure of yeast MPP complexed with a synthetic presequence peptide is represented in Fig. 2B according to the coordination data reported by Taylor et al. (28). Each yeast MPP subunit contains N and C domains of ∼210 residues with nearly identical folding topologies, which are related by an approximately twofold rotation. Thus, the higher-order structure of RPPs appears to resemble those of the MPP subunits and is probably more similar to that of β-MPP, such that RPPs would be expected to show peptidase activity.
FIG. 1.
Phylogenetic analysis of MPPs from various organisms, namely, Homo sapiens, Mus musculus, Solanum tuberosum, Saccharomyces cerevisiae, and Trypanosoma brucei, and RPPs from three Rickettsia species. The bootstrap values (>50%) are shown beside each branch. The scale bar indicates the estimated sequence divergence per branch length.
FIG. 2.
Structure comparison of RPPs and MPPs. A, schematic diagrams of the primary structures of RPP and the yeast S. cerevisiae MPP subunits. Light and dark gray regions indicate α- and β-MPPs, respectively, and their homologous regions in RPP. B, crystal structure of yeast MPP complexed with a synthetic presequence peptide. The structure is shown as a ribbon diagram for the MPP subunits and a wire frame in a semitranslucent space-filling model for the yeast COXIV2-25 peptide. Quadrangles outlined with solid or dotted lines include the active site or GRL regions, respectively.
FIG. 3.
Alignments of the GRL regions (upper) and active site regions (lower) of RPPs and MPP subunits. Identical or similar amino acids are indicated by asterisks or colons under the alignments, respectively.
Gene and protein expression of RPP.
Since R. prowazekii is unavailable in Japan, we used another rickettsial species, R. typhi, whose RPP gene is closely related to the R. prowazekii one (Fig. 1). The RPP gene expression in R. typhi infecting simian cells was examined by reverse transcription-PCR (RT-PCR). Reverse transcripts from R. typhi total RNA extracts were amplified using specific primer pairs designed for the RPP-coding region, while RT-PCR for the GroEL-coding region served as a control. DNA fragments of 680 and 500 bp for the RPP and GroEL genes, respectively, were amplified from the transcripts in a reverse transcriptase-dependent manner (see Fig. S1A in the supplemental material). The DNA sequences of these fragments revealed that the RT-PCR products were the same as the corresponding part of each gene, indicating the existence of RPP mRNA in parasitic rickettsial cells. Three species of rickettsiae appeared to produce RPP proteins (see Fig. S1B in the supplemental material). Immunoreactive proteins of around 50 kDa were detected, although the rickettsial proteins were slightly smaller than the recombinant RPP, probably due to the additional C-terminal His6 tag. To characterize the RPP, we tried to purify the recombinant protein from an extract of E. coli cells transformed with a protein expression vector carrying the RPP gene. After centrifugation of the cell lysate, the RPP was barely recovered in the soluble protein fraction (see Fig. S1C in the supplemental material). However, we succeeded in producing the soluble protein by coexpression of the E. coli molecular chaperonins GroEL and GroES (see Fig. S1C in the supplemental material). The recombinant RPP was purified using a combination of nickel affinity and ion exchange chromatographies, and the protein preparation was estimated to show >99% purity by SDS-PAGE and protein staining (see Fig. S1C in the supplemental material). Size exclusion chromatography revealed that the nondenatured molecular size was approximately 50 kDa (data not shown), suggesting that the recombinant RPP was in a monomeric state.
Peptidase activity of RPP.
Since RPPs showed conservation of the active-site sequence of metalloendopeptidases of the M16 protease family, we expected that the recombinant RPP would show proteolytic activity. To investigate whether RPP inherently hydrolyzes bound peptides, several synthetic peptides were incubated with the purified RPP in vitro and analyzed by reverse-phase HPLC. Basic peptides from various sources, namely dynorphin A, vasoactive intestinal peptide, mastoparan, and αMSH amide, produced detectable amounts of fragments (Table 1; also see Fig. S2 in the supplemental material) following incubation with RPP, whereas cleavage of neutral and acidic peptides (peptides E to H in Table 1) by RPP was undetectable. Thus, RPP showed preferential cleavage of the more basic peptides among the peptides investigated. Mass spectrometry was carried out to determine the molecular masses of the peptide fragments and reveal the cleavage sites. RPP frequently hydrolyzed peptide bonds before hydrophobic residues and sometimes attacked sites beside basic residues (Table 1). These experiments demonstrated that RPP has intrinsic peptidase activity for basic peptides. Next, we further investigated the proteolytic activity of RPP. Due to its preferential cleavage of basic sequences, the typical basic proteins lysozyme and cytochrome c were tested under native and denaturing conditions. RPP showed no proteolytic activity toward the proteins either in nondenaturing buffer or under intensive denaturing conditions with urea when the proteins were analyzed by SDS-PAGE and stained (see Fig. S3 in the supplemental material). After the RPP reactions, the proteins were analyzed by reverse-phase HPLC, and the elution time of the lysozyme solution that had been subjected to the denaturing conditions was delayed compared to that for the nondenatured lysozyme (data not shown), suggesting that the substrate protein was unfolded during the RPP action. The remaining urea in the RPP reaction solution did not affect the peptidase activity (data not shown). These results imply that RPP alone behaves as a basic peptide peptidase in organisms.
TABLE 1.
RPP cleaves basic peptides
| Peptidea | Peptide sequence and sites of cleavage by RPPb | Degradationc (%) | Net charged |
|---|---|---|---|
| A | YGGF↓LRR↓IRP↓K↓L↓KWDNQ | 76.4 | +4 |
| B | HSDAVFTDN↓YT↓R↓L↓RKQ↓M↓ AKKYLNSILN | 36.2 | +3 |
| C | INLKA↓LAA↓LAK↓KIL | 7.2 | +3 |
| D | Ac-SYSMEH↓F↓RWGKPV-NH2 | 15.7 | +1 |
| E | VNTPEHVVPYGLGSPRS | <1.0 | 0 |
| F | CETQRTLLNGDLQTSI | <1.0 | −1 |
| G | LENLEAETAPLP | <1.0 | −3 |
| H | Pyr-GPWLEEEEEAYGWMDF-NH2 | <1.0 | −6 |
Peptide A, dymorphin A; B, vasoactive intestinal peptide; C, mastoparan; D, αMSH amide; E, big endotheline; F, peptide fragment of somatostatin receptor SSTR2A; G, peptide fragment of μ opioid receptor MOR1A; H, gastrin I.
Arrows denote RPP cleavage sites for peptides, which were identifiable. Basic residues are indicated in italics.
Cleavage efficiencies are determined as described in Materials and Methods.
Net charges are caluculated from acidic and basic amino acid residues in each peptide in the reaction condition.
Cleavage of mitochondrial presequences by RPP.
RPP resembles the catalytic subunit of MPPs, attacks basic sequences, and particularly cleaves bonds before hydrophobic residues and around basic residues, which MPPs also prefer. These insights into the relationship between RPP and MPPs regarding their structures and substrate specificities led us to investigate whether RPP cleaves mitochondrial presequences. When a 16-residue presequence peptide, which included the normal presequence cleavage site between the residues Ala-8 and Phe-9, derived from the preprotein of yeast malate dehydrogenase (MDH2-17), were incubated with RPP and analyzed by HPLC and mass spectrometry, it was found that RPP cleaved one site between the alanine and phenylalanine residues that was also processed by yeast MPP (Table 2; also see Fig. S4 in the supplemental material). Since an RPP mutant in which the glutamate residue in the HxxEH motif was substituted with glutamine (RPP*) was inactive (see Fig. S5 in the supplemental material), the peptidase activity was specific and depended on the glutamate residue. RPP also cleaved other synthetic presequence peptides of various lengths from different organisms at one or more sites, with the exception of the yeast COXIV2-25 peptide (Table 2). Among the eight peptides tested, RPP attacked five of them at peptide bonds corresponding to MPP processing sites (Table 2), although its activities were lower than those of MPP (see Fig. S4 in the supplemental material). Excess amounts of recombinant yeast α-MPP or β-MPP did not affect the substrate specificity of RPP or enhance its activity (data not shown). It is likely that interaction between RPP and MPP subunits is not necessary to cleave the peptides.
TABLE 2.
RPP cleaves mitochondrial presequence peptides
| Peptidea | Peptide sequence and sites of cleavage by RPP and MPPb | Degradation by RPPc (%) |
|---|---|---|
| 1 | LSALARPVGAA↓LRRS↓↑FSTSAQNNAKVA | 59 |
| 2 | MLAAKNILNRSSLSSS↓↑FRIATRL↓QSTKVQGSA | 39 |
| 3 | LSRVAKRA↓↑FSSTVANP | 20 |
| 4 | MLRAA↓LTTVRRGPRLSR↓L↓↑LSAAATSAVPA | 17 |
| 5 | MFS↓KLAHL↓Q↓RPA↓VLSRG↑VHSSVASA | 17 |
| 6 | MLSAARLQFAQGSVRR↓↑LTVSARDAPTKISTLA | 9.5 |
| 7 | GRWRLLVRPRAGAGG↓LRGSRGPGLGGGAVATRT↑LSV | 7.3 |
| 8 | LSLRQSIRFFKPATRT↑LCSSTYLL | <1 |
Peptide 1, mouse MDH2-28; 2, yeast heat shock protein SSC1 (amino acids 1 to 32); 3, yeast MDH2-17; 4, mouse aldehyde dehydrogenase (amino acids 1 to 29); 5, human ornithine aminotransferase (amino acids 1 to 25); 6, yeast ubiquinol cytochrome c reductase subunit 2 (amino acids 1 to 32); 7, bovine ADX18-57; 8, yeast COXIV2-25.
Arrows (↓) denote RPP cleavage sites for peptides that can be detected or MPP cleavage sites (↑).
Cleavage efficiencies are determined as described in Materials and Methods.
Regulation of preprotein processing by RPP.
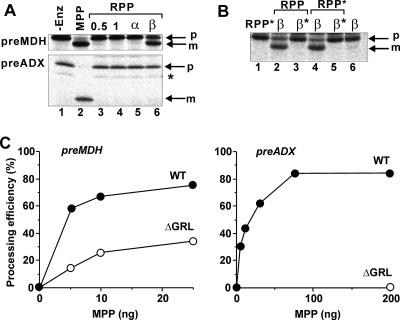
Next, we examined whether RPP could cleave mitochondrial preproteins using mouse precursor of MDH (preMDH) and bovine precursor of ADX (preADX), which contain different presequence lengths of 16 and 58 residues, respectively. RPP alone could not cleave either preMDH or preADX (Fig. 4A). To our surprise, however, a stoichiometric mixture of RPP and yeast β-MPP (RPP-β-MPP) processed preMDH, whereas a similar stoichiometric mixture of RPP and yeast α-MPP did not. On the other hand, preADX was not cleaved by either RPP-β-MPP or RPP-α-MPP. A functional association between RPP and β-MPP was established, since β-MPP alone did not show any preprotein processing activities. However, the stable RPP-β-MPP interaction was not demonstrated by β-MPP pull-down assays using the His6 tag of RPP and affinity beads (data not shown), probably due to their weak association with each other. To address which active sites were involved in the preMDH processing, RPP and β-MPP carrying a mutation of the catalytic glutamate residue (RPP* and β-MPP*, respectively) were added to the processing assay either separately or together. As shown in Fig. 4B, the processing of preMDH depended on the catalytic glutamate residue in β-MPP but not on that in RPP, since RPP*-β-MPP cleaved the preprotein, whereas RPP-β-MPP* and RPP*-β-MPP* did not, indicating that the active site of β-MPP is involved in the processing activity. Thus, RPP cooperates with β-MPP to recognize the preprotein and activates the catalytic site of β-MPP. A single β-MPP never cleaves preproteins, and RPP is also inert for the cleavage. The three-dimensional structure of MPP indicates that the substrate around the cleavage point is accommodated in β-MPP. Taken together, these findings indicate it is (highly) likely that the β-MPP-RPP complex processes preMDH at the correct site, though the actual peptide bond cleaved within preMDH has not been identified heretofore.
FIG. 4.
Preprotein processing by RPP and MPP. A, processing of preproteins by a combination of RPP and β-MPP. Preprotein (preMDH or preADX) processing was analyzed as described in the Materials and Methods. Lane 1, no enzyme; lane 2, yeast MPP (0.1 μg); lane 3, RPP (0.5 μg); lane 4, RPP (1 μg); lane 5, RPP-α-MPP (1 μg each); lane 6, RPP-β-MPP (1 μg each). p, preprotein; m, mature protein. An asterisk indicates a faint band under the band of preADX which may be mistranslated or degraded by proteases during synthesis of the preprotein in an in vitro translation system. B, activation of β-MPP by RPP. Each lane contains 0.2 μg of each protein. Lane 1, RPP*; lane 2, RPP-β-MPP; lane 3, RPP-β-MPP*; lane 4, RPP*-β-MPP; lane 5, RPP*-β-MPP*; lane 6, β-MPP. RPP*, inactive mutant of RPP; MPP*, inactive mutant of β-MPP; p, preprotein; m, mature protein. C, processing of preproteins by a complex of α-MPPΔGRL-β-MPP. Preprotein processing was analyzed as described in Materials and Methods. WT, wild-type MPP complex (filled circles); ΔGRL, α-MPPΔGRL-β-MPP (open circles).
GRL is required for cleavage of long presequences.
Since RPP behaved as a regulatory subunit toward preMDH processing, similar to α-MPP, we next investigated why RPP-β-MPP was unable to cleave preADX. As mentioned above, there is a marked difference in the presequence lengths between the two preproteins, and RPP lacks a GRL region. GRL in yeast α-MPP was previously shown to play an essential role in cleaving a synthetic preMDH peptide (19). Here we analyzed whether an enzyme comprised of GRL deletion variants of yeast α-MPP (α-MPPΔGRL) and β-MPP (MPPΔGRL) could process the preproteins. MPPΔGRL was able to cleave preMDH, albeit with a lower processing activity than that of wild-type MPP, but showed inefficient preADX processing (Fig. 4C). An excess amount of α-MPPΔGRL in relation to β-MPP was null for the processing efficiency (data not shown). A pull-down assay of α-MPPΔGRL by β-MPP-His6 previously revealed a stoichiometric association of α-MPPΔGRL with β-MPP (19). These results therefore suggest that the GRL region is required for the cleavage of long presequences but does not influence the association between the subunits. The RPP-β-MPP complex behaved as MPPΔGRL at least for processing of preMDH and preADX. The reason for the inability of the RPP-β-MPP complex to process preADX could be lack of GRL in RPP. To confirm whether the GRL is sufficient to cleave long presequences, we tried to construct some variants of the GRL-inserting RPP and to examine whether these variants could process preADX. However, the variant proteins were misfolded due to the protein inclusion in E. coli, and they seemed to be unstable during the refolding in vitro.
DISCUSSION
In the present study, we addressed the peptidase activity of a eubacterial homologue of MPP and found a functional and evolutionary relationship between RPP and MPP. The RPP cleaved basic peptides, including mitochondrial targeting presequence peptides, with partial specificity for MPP cleavage sites. The most notable finding was that RPP was able to activate eukaryotic β-MPP and subsequently render it able to process preproteins. Thus, RPP retains not only the structural characteristics of the processing peptidases but also the bifunctional hallmarks of the catalytic and regulatory subunits of MPP. Considering mitochondrial evolution on the basis of the endosymbiont theory, RPP appears to have the closest remaining structural and functional hallmarks of a processing peptidase in primitive mitochondria.
Although RPP is expressed in rickettsial cells, its functions in vivo remain unknown. For instance, what is its function in rickettsiae and what is its physiological substrate? One of the reasons for this lack of knowledge is the difficulty associated with elucidating the exact actions of RPP in vivo, since rickettsiae can only live within other cells. However, its actions in these cells may be speculated on based on the functions of the ymxG gene, which encodes a type of M16 protease in the eubacterium Bacillus subtilis. The YmxG protein is highly homologous to MPPs (5) and notably shares a high degrees of similarity (∼60% identity and similarity) with RPPs and MPP-like proteins from the parasitic eubacteria Mycobacterium leprae and Mycobacterium tuberculosis (4). Analyses of a ΔymxG mutant strain revealed specific stimulation of the production of subtilisin (AprG), the major serine protease secreted by B. subtilis (4). This phenomenon appears to arise through negative regulation of aprE gene expression by YmxG rather than through the lack of YmxG proteolytic activity. As a consequence, Bolhuis et al. proposed two possible models for the actions of YmxG (4). First, the protein could act as a repressor via binding to the upstream sequence of the aprE gene, or second, it could indirectly modulate the activity of a transcriptional regulator of aprE, possibly through proteolysis. Considering the homology of RPP to YmxG and the RPP peptidase/regulatory activities revealed in the present study, RPP may play a key role in regulating protein expressions through its protease activity. Even though the RPP functions in vivo remain unknown, the fact that the eubacterial MPP-like protein YmxG is not essential for viability or cell growth is interesting (4) when we consider the origin of these preprotein processing enzymes according to mitochondrial endosymbiont evolution, as discussed below.
Genes for proteins containing zinc-binding HxxEH motifs characteristic of Μ16 proteases and sharing high degrees of similarity with MPPs were identified by sequence searches of many bacterial genomes. BLAST (1) searches of prokaryotic genomes revealed homologues of yeast MPP subunits (see Table S1 in the supplemental material). Notably, there were nearly 500 putative proteins resembling β-MPP in the 511 genomes of eubacteria examined, whereas proteins showing slight homology to MPP with genetic significance were encoded in 29 genomes of archaebacteria. In particular, BLAST searches of rickettsial genomes from a total of 20 species identified 33 genes encoding β-MPP-like proteins, indicating that at least one RPP gene could be carried on each rickettsial genome. Notably, no putative MPP-like proteins carrying GRLs were found in the bacterial genomes. Considering the genomic distributions of MPP-like genes described above, and since it is widely accepted that mitochondria originated from the α-proteobacterial order Rickettsiales based on phylogenetic analyses comparing the sequences of bacterial and mitochondrial genes (2, 9, 18, 23), MPP is unlikely to have originated from proteins in the host cells of an archaebacterial ancestor and is most likely to have arisen from an RPP-like progenitor in a parasitic bacterium. During the endosymbiotic evolution of mitochondria, the gene encoding the progenitor of MPP, which may be nonessential, similar to ymxG in B. subtilis, could be transferred from the endosymbiont to the host cell. At this stage, the nonessential gene conversion presumably succeeded during mitochondrial evolution due to the maintained viability of the imperfect symbiotic organelle. Alternatively, the genes may exist in both the endosymbiont and host genomes under the predominant circumstance of the immature eukaryotic cells. In any case, during or after the gene transfer, gain of the signal sequences and procurement of the protein transport-processing system must be one of the critical stages toward the success of endosymbiotic evolution in the primitive eukaryote cells.
Precise recognition of signal sequences by the protein translocation and processing system must be one of the critical stages for the biogenesis of mitochondria, plastids, and hydrogenosomes, which are thought to have arisen from endosymbiotic bacteria. Although it is unclear how the system was acquired during endosymbiotic evolution, the central components of the mitochondrial translocases may be genetically converted from eubacterial pore/channel-forming proteins and chaperones (8). The relationship between MPP and RPP suggests the same situation, as mentioned above. Since MPP primarily bears the traits of a parasitic bacterial peptidase, it appears that MPP and RPP have a common progenitor in ancient parasitic bacteria. During the evolution stage from endosymbiont to mitochondria, the progenitor gene was duplicated and the proteins were converted into two distinct components of the processing enzyme, which are now α- and β-MPP (see Fig. S6 in the supplemental material). It appears that this protein dimerization was required for the peptidase regulation and that gain of the GRL region was particularly involved in the efficient processing of longer transport signal sequences, since these lengths have tended to become greater. Interestingly, the stromal processing peptidase that cleaves the signal peptide of the plastidial protein precursor has an M16 protease active site and shares homology to MPPs, although it is active as a single polypeptide and carries no GRL region. To decipher the complex histories of preprotein transport and processing systems in endosymbiotic organelles, broad genetic investigations and biochemical analyses are required for parasitic organisms and some lower eukaryotes, since it was recently reported that Giardia mitosomes and Trichomonad hydrogenosomes share a common mode of protein targeting (6).
Supplementary Material
Acknowledgments
We thank S. G. Andersson and C. G. Kurland (University of Uppsala) for the genomic clones, including RP219, and Y. Akiyama (Kyoto University) for E. coli strain AD293 transformed with pKY206.
This work was supported in part by Grants-in-Aid for Scientific Research (to S.K.; no. 14658233) from the Ministry of Education, Science, Sports and Culture of the Japanese Government.
Footnotes
Published ahead of print on 8 December 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 3.Baker, K. P., and G. Shatz. 1991. Mitochondrial proteins essential for viability mediate protein import into yeast mitochondria. Nature 349:205-208. [DOI] [PubMed] [Google Scholar]
- 4.Bolhuis, A., E. Koetje, J.-Y. Dubois, J. Vehmaanper, G. Venema, S. Bron, and J. M. van Dijl. 2000. Did the mitochondrial processing peptidase evolve from a eubacterial regulator of gene expression? Mol. Biol. Evol. 17:198-201. [DOI] [PubMed] [Google Scholar]
- 5.Chen, N. Y., S. Q. Jiang, D. A. Klein, and H. Paulus. 1993. Organization and nucleotide sequence of the Bacillus subtilis diaminopimelate operon, a cluster of genes encoding the first three enzymes of diaminopimelate synthesis and dipicolinate synthase. J. Biol. Chem. 268:9448-9465. [PubMed] [Google Scholar]
- 6.Dolezal, P., O. Smid, P. Rada, Z. Zubacova, D. Bursac, R. Sutak, J. Nebesarova, T. Lithgow, and J. Tachezy. 2006. Giardia mitosomes and trichomonad hydrogenosomes share a common mode of protein targeting. Proc. Natl. Acad. Sci. USA 102:10924-10929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doolittle, W. F. 1998. A paradigm gets shifty. Nature 392:15-16. [DOI] [PubMed] [Google Scholar]
- 8.Dyall, S. D., M. T. Brown, and P. J. Johnson. 2004. Ancient invasions: from endosymbionts to organelles. Science 304:253-257. [DOI] [PubMed] [Google Scholar]
- 9.Emelyanov, V. V. 2001. Evolutionary relationship of Rickettsiae and mitochondria. FEBS Lett. 501:11-18. [DOI] [PubMed] [Google Scholar]
- 10.Gakh, O., P. Cavadini, and G. Isaya. 2002. Mitochondrial processing peptidases. Biochim. Biophys. Acta 1592:63-77. [DOI] [PubMed] [Google Scholar]
- 11.Gray, M. W., G. Burger, and B. F. Lang. 1999. Mitochondrial evolution. Science 283:1476-1481. [DOI] [PubMed] [Google Scholar]
- 12.Hartl, F.-U., N. Pfanner, D. W. Nicholson, and W. Neupert. 1989. Mitochondrial protein import. Biochim. Biophys. Acta 988:1-45. [DOI] [PubMed] [Google Scholar]
- 13.Ito, A. 1999. Mitochondrial processing peptidase: multiple-site recognition of precursor proteins. Biochem. Biophys. Res. Commun. 265:611-616. [DOI] [PubMed] [Google Scholar]
- 14.Kitada, S., K. Shimokata, T. Niidome, T. Ogishima, and A. Ito. 1995. A putative metal-binding site in the β subunit of rat mitochondrial processing peptidase is essential for its catalytic activity. J. Biochem. 117:1148-1150. [DOI] [PubMed] [Google Scholar]
- 15.Kitada, S., K. Kojima, K. Shimokata, T. Ogishima, and A. Ito. 1998. Glutamate residues required for substrate binding and cleavage activity in mitochondrial processing peptidase. J. Biol. Chem. 273:32547-32553. [DOI] [PubMed] [Google Scholar]
- 16.Luciano, P., and V. Géli. 1996. The mitochondrial processing peptidase: function and specificity. Experientia 52:1077-1082. [DOI] [PubMed] [Google Scholar]
- 17.Margulis, L. 1996. Archaeal-eubacterial mergers in the origin of Eukarya: phylogenetic classification of life. Proc. Natl. Acad. Sci. USA 93:1071-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLeod, M. P., X. Qin, S. E. Karpathy, J. Gioia, S. K. Highlander, G. E. Fox, T. Z. McNeill, H. Jiang, D. Muzny, L. S. Jacob, A. C. Hawes, E. Sodergren, R. Gill, J. Hume, M. Morgan, G. Fan, A. G. Amin, R. A. Gibbs, C. Hong, X. J. Yu, D. H. Walker, and G. M. Weinstock. 2004. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J. Bacteriol. 186:5842-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagao, Y., S. Kitada, K. Kojima, H. Toh, S. Kuhara, T. Ogishima, and A. Ito. 1998. Glycine-rich region of mitochondrial processing peptidase α-subunit is essential for binding and cleavage of the precursor proteins. J. Biol. Chem. 275:34552-34556. [DOI] [PubMed] [Google Scholar]
- 20.Neupert, W. 1997. Protein import into mitochondria. Annu. Rev. Biochem. 66:863-917. [DOI] [PubMed] [Google Scholar]
- 21.Niidome, T., S. Kitada, K. Shimokata, T. Ogishima, and A. Ito. 1994. Arginine residues in the extension peptide are required for cleavage of a precursor by mitochondrial processing peptidase. J. Biol. Chem. 269:24719-24722. [PubMed] [Google Scholar]
- 22.Nomura, H., S. Athauda, H. Wada, Y. Maruyama, K. Takahashi, and H. Inoue. 2006. Identification and reverse genetic analysis of mitochondrial processing peptidase and the core protein of cytochrome bc1 complex of Caenorhabditis elegans, a model organism of parasitic nematodes. J. Biochem. 139:967-979. [DOI] [PubMed] [Google Scholar]
- 23.Ogata, H., S. Audic, P. Renesto-Audiffren, P. E. Fournier, V. Barbe, S. Samson, V. Roux, P. Cossart, J. Weissenbach, J. M. Claverie, and D. Raoult. 2001. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 293:2093-2098. [DOI] [PubMed] [Google Scholar]
- 24.Pfanner, N., E. A. Craig, and A. Hönlinger. 1997. Mitochondrial preprotein translocase. Annu. Rev. Cell Dev. Biol. 13:25-51. [DOI] [PubMed] [Google Scholar]
- 25.Rawlings, N., and A. J. Barrett. 1995. Evolutionary families of metallopeptidases. Methods Enzymol. 248:183-228. [DOI] [PubMed] [Google Scholar]
- 26.Ryan, K. R., and R. E. Jensen. 1995. Protein translocation across mitochondrial membranes: what a long, strange trip it is. Cell 83:517-519. [DOI] [PubMed] [Google Scholar]
- 27.Schatz, G. 1996. The protein import system of mitochondria. J. Biol. Chem. 271:31763-31766. [DOI] [PubMed] [Google Scholar]
- 28.Taylor, A. B., B. S. Smith, S. Kitada, K. Kojima, H. Miyaura, Z. Otwinowski, A. Ito, and J. Deisenhofer. 2001. Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure 9:615-625. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yaffe, M. P., S. Ohta, and G. Schatz. 1985. A yeast mutant temperature-sensitive for mitochondrial assembly is deficient in a mitochondrial protease activity that cleaves imported precursor polypeptides. EMBO J. 4:2069-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan, Y., T. Uchiyama, and T. Uchida. 1993. Differentiation of Rickettsia japonica by restriction endonuclease fragment length polymorphism using products of polymerase chain reaction amplification with Rickettsia rickettsii 190-kilodalton surface antigen gene primers. Microbiol. Immunol. 37:441-445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.