Abstract
The Burkholderia cenocepacia cepIR quorum-sensing system regulates expression of extracellular proteases, chitinase, and genes involved in ornibactin biosynthesis, biofilm formation, and motility. In a genome-wide screen we identified cepIR-regulated genes by screening a random promoter library of B. cenocepacia K56-2 constructed in a luminescence reporter detection plasmid for differential expression in response to N-octanoyl-l-homoserine lactone (OHL). Eighty-nine clones were identified; in 58 of these clones expression was positively regulated by cepIR, and in 31 expression was negatively regulated by cepIR. The expression profiles of the 89 promoter clones were compared in the cepI mutant K56-dI2 in medium supplemented with 30 pM OHL and K56-2 to confirm that the presence of OHL restored expression to wild-type levels. To validate the promoter library observations and to determine the effect of a cepR mutation on expression of selected genes, the mRNA levels of nine genes whose promoters were predicted to be regulated by cepR were quantitated by quantitative reverse transcription-PCR in the wild type and cepI and cepR mutants. The expression levels of all nine genes were similar in the cepI and cepR mutants and consistent with the promoter-lux reporter activity. The expression of four selected cepIR-regulated gene promoters was examined in a cciIR mutant, and two of these promoters were also regulated by cciIR. This study extends our understanding of genes whose expression is influenced by cepIR and indicates the global regulatory effect of the cepIR system in B. cenocepacia.
Strains of the Burkholderia cepacia complex (Bcc) have emerged as important opportunistic pathogens in patients with cystic fibrosis (CF) and chronic granulomatous disease, and infections often result in significant mortality rates (10, 27, 29, 41). The Bcc is comprised of nine species, all of which have been isolated from environmental and clinical sources (10). Burkholderia cenocepacia is the most prevalent Bcc species in CF respiratory infections (35, 41). There have been numerous cases of transmission of Bcc strains or clones between CF patients, resulting in rigorous infection control measures (6, 29, 30, 32, 41). The majority of transmissible clones belong to B. cenocepacia, which has also been reported to replace other Bcc species in CF respiratory infections (6, 30, 32).
The quorum-sensing (QS) system is a form of cell-cell communication that involves the production of small signaling molecules produced in a cell density-dependent manner. In gram-negative bacteria these signaling molecules are usually N-acyl-homoserine lactones (AHLs) (for reviews, see references 44 and 46). Two sets of QS genes, cepIR and cciIR, have been identified in B. cenocepacia (5, 23). CepI synthesizes two AHL molecules, N-octanoyl-l-homoserine lactone (OHL) and N-hexanoyl-l-homoserine lactone (23, 24). OHL is the predominant AHL produced by Bcc strains (15, 26). CepR is a transcriptional regulator that responds to the AHL signals and either positively or negatively regulates target genes (13, 24, 44). CciI produces predominantly N-hexanoyl-l-homoserine lactone and minor amounts of OHL (31). The cepIR quorum-sensing genes have been reported to be present in all Bcc species (15, 26), whereas the cciIR system is present only in epidemic B. cenocepacia strains containing the cenocepacia island (cci) (5). The cepIR system appears to be the ancestral system, and the cciIR system is incorporated into the cepIR regulatory network (31). CepR is required for expression of the cciIR genes, which are cotranscribed (12). Both QS systems have been implicated in virulence. Murine respiratory, nematode, and onion infection models have shown that cepIR contributes to the virulence of B. cenocepacia and B. cepacia (2, 21, 40). A B. cenocepacia cciI mutant was shown to have attenuated virulence in a rat chronic respiratory infection model (5).
QS systems regulate genes involved in the virulence of many bacterial species (11, 46, 48). The cepIR QS system was first identified in strains of B. cenocepacia because of its involvement in regulation of biosynthesis of the siderophore ornibactin (23) and its requirement for biofilm formation on abiotic surfaces (17). B. cenocepacia cepI or cepR mutants have been reported to be altered in extracellular protease production, chitinase activity, swarming motility, and mature biofilm development (17, 18, 20, 23, 24, 40, 43). Expression of the pvdA gene involved in ornibactin biosysthesis (24) and expression of the zinc metalloprotease genes, zmpA (40) and zmpB (20), have been shown to be negatively and positively regulated, respectively, in cepI and cepR mutants, suggesting that the expression of these genes is directly regulated by CepR. Expression of aidA, a protein having an unknown function that is important for virulence in nematodes, is also positively regulated by cepR in both B. cenocepacia and B. cepacia (3, 16, 36, 47). Little is currently known about cepR regulation of other genes that might influence the phenotypes described above or the virulence of B. cenocepacia.
Several approaches have been used to identify genes regulated by the cepIR QS system. Aguilar et al. identified positively regulated OHL-CepR-dependent promoters in B. cepacia ATCC 25416 using a library of ATCC 25416 fragments cloned into a vector containing cepR and a promoterless lacZ gene downstream from a multiple cloning site (3). The library was screened in Escherichia coli DH5α, and 28 putative gene promoters that exhibited CepR-dependent expression in the presence of OHL were identified (3). Proteomic analysis was used to compare B. cenocepacia H111 to its cepI mutant (36). Two-dimensional gel electrophoresis of the protein profiles revealed that there was differential expression of 55 of 985 detectable spots. Approximately 5% of the B. cenocepacia H111 proteome was downregulated and 1% was upregulated in the cepI mutant. Nineteen peptides from 11 proteins were identified by N-terminal amino acid sequencing, and these peptides included peptides from AidA, FimA, SodB, RpsD, a thermolysin metallopeptidase, and some hypothetical proteins (28). The thermolysin metallopeptidase was subsequently determined to be ZmpB and was shown to be cepIR regulated in B. cenocepacia K56-2 using zmpB::lux reporter fusions (20). Genes regulated by the cepIR QS system have also been identified using transposon mutagenesis. Seven genes, including cepI and aidA, were identified using a transposon that incorporates a promoterless lacZ reporter gene and screening for induction of β-galactosidase activity in the presence of OHL (47). Purified CepR was shown to directly bind to promoters upstream of cepI and aidA, confirming that the CepIR system directly regulates expression of these genes (47).
Although several cepIR-regulated genes have been identified using molecular and proteomic methods, our understanding of the cepIR regulon is far from complete. To identify additional cepIR-regulated genes in B. cenocepacia K56-2, we employed a random promoter library using a sensitive luminescent reporter system that provided a high-throughput method for measurement of real-time gene expression (7, 12). A random promoter library of B. cenocepacia K56-2 luxCDABE transcriptional fusions was generated and screened in a cepI deletion mutant grown in medium with or without OHL to identify OHL-responsive promoters. In addition, real-time reverse transcription (RT)-PCR was performed with representative genes to confirm that there was differential gene expression in K56-2 and cepI and cepR mutants of this strain. The results of this study increase our understanding of cepIR-mediated gene regulation in B. cenocepacia K56-2.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Strains and plasmids used in this study are listed in Table 1. E. coli and B. cenocepacia were cultured aerobically at 37°C in Luria-Bertani (LB) medium (Invitrogen Canada Inc., Burlington, ON, Canada). For analysis of the expression of pvdA and orbI promoters, cultures were grown in TSB-DC medium (33). When appropriate, antibiotics were used at the following concentrations: for B. cenocepacia, 100 μg/ml trimethroprim and 200 μg/ml tetracycline; and for E. coli, 25 μg/ml kanamycin and 1.5 mg/ml trimethroprim. Antibiotics were purchased from Sigma-Aldrich Canada Ltd. (Oakville, ON, Canada).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA gyrA96 thi-1 hsdR17 supE44 relA1 deoR | Invitrogen |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| B. cenocepacia strains | ||
| K56-2 | Cystic fibrosis respiratory isolate | 28 |
| K56-R2 | cepR::Tn5-OT182 derivative of K56-2, Tcr | 23 |
| K56-dI2 | ΔcepI derivative of K56-2 | 31 |
| Plasmids | ||
| pCR2.1TOPO | Cloning vector for PCR products, Apr Kmr | Invitrogen |
| pBS7 | pCR2.1TOPO containing 1.3-kb ndh fragment | 14 |
| pMS402 | lux-based promoter reporter plasmid, Kmr Tpr | 12 |
| pCC1 | pMS402 containing a zmpA::luxCDABE fusion, Kmr Tpr | 40 |
| pSLS225 | pUCP26 with 1.5-kb SphI-KpnI fragment containing cepI, Tcr | 25 |
| pRM445 | pMS402 containing a cepR::luxCDABE transcriptional fusion, Kmr Tpr | 31 |
| pCP300 | pMS402 containing a cepI::luxCDABE transcriptional fusion, Kmr Tpr | 31 |
| pMVbcscV | pMS402 containing a bcscV::luxCDABE transcriptional fusion, Kmr Tpr | This study |
| pBS18 | pMS402 containing a katB::luxCDBAE transcriptional fusion, Kmr Tpr | |
| pVDA01 | pMS402 containing a pvdA::luxCDBAE transcriptional fusion, Kmr Tpr | This study |
| pVDO301 | pMS402 containing an orbI::luxCDBAE transcriptional fusion, Kmr Tpr | This study |
Ap, ampicillin; Km, kanamycin; Tp, trimethoprim; Tc, tetracycline.
Construction of the B. cenocepacia K56-2 random promoter library.
Molecular biology procedures were generally performed as described by Sambrook et al. (38). K56-2 genomic DNA was isolated as described by Ausubel et al. (4) and was partially digested with Sau3AI (Invitrogen Canada Inc.). The digested DNA fragments were separated by sucrose density gradient centrifugation. Fragments in the range from 0.5 to 3 kb long were ligated into pMS402, which contains a promoterless luxCDABE operon (12), previously digested with BamHI (New England BioLabs, Mississauga, ON, Canada) and dephosphorylated with shrimp alkaline phosphatase (Roche, Mannheim, Germany). After enzyme inactivation, the dephosphorylated plasmid was purified using a QIAgen PCR purification kit (QIAGEN, Mississauga, ON, Canada). Ten independent ligation reactions were performed using various vector/insert ratios, and the products were electroporated into ElectroMAX DH10B cells (Invitrogen Canada Inc.) using a Gene Pulser (Bio-Rad Laboratories Inc., Hercules, CA). The efficiency of ligation was determined for 12 randomly selected transformants from each ligation using colony PCR performed with primers pZE05 (5′-CCAGCTGGCAATTCCGA-3′) and pZE06 (5′-AATCATCACTTTCGGGAA-3′) (12) corresponding to sequences that flank the BamHI site of pMS402. The pooled plasmid extracts from five high-efficiency ligation mixtures that contained approximately 60% inserts were subsequently electroporated into the cepI deletion mutant, K56-dI2.
Screening the random promoter library.
OHL was purified from B. cenocepacia K56-2(pSLS225) as previously described (9). The optimum concentration of OHL used for screening the library was determined by comparing the luminescence values for known quorum-sensing-regulated gene fusions, zmpA::lux, cepI::lux, and cepR::lux, in medium containing 0, 300 pM, 30 pM, 3 pM, 300 fM, and 30 fM OHL. The method used for screening the random promoter library was adapted from the method of Bjarnason et al. (7). Transformants containing the random promoter library clones were inoculated into clear 384-well plates (Costar; Corning Incorporated, Corning, NY) containing 70 μl of LB medium using a colony-picking robot (Norgren Systems, Palo Alto, CA) and were incubated overnight at 37°C. These cultures were subcultured into black 384-well plates (3710 Costar; Corning Incorporated, Corning, NY) containing LB medium and LB medium supplemented with 300 pM or 3 pM OHL using a 384-pin manual plate replicator (V&P Scientific Inc., San Diego, CA) and incubated at 37°C. Luminescence was measured with a Wallac Victor2 1420 multilabel counter (Perkin-Elmer Life Sciences, Boston, MA) after 16 h of incubation. Clones for which there were differences in promoter activity between the medium with OHL and the medium without OHL were rearrayed (using Norgren Systems software) and used in subsequent experiments.
A second screen was performed to reduce the incidence of false-responsive promoters. Clones with at least twofold differences in luminescence between the medium with OHL and the medium without OHL were rearrayed into clear 96-well plates (531172; Falcon, Franklin Lakes, NJ) containing 150 μl LB medium. The overnight cultures of clones exhibiting promoter activity were diluted to an optical density at 600 nm (OD600) of 0.05 in LB medium supplemented with 0, 3 pM, or 30 pM OHL in 96-well clear-bottom black plates (9520 Costar; Corning Incorporated, Corning, NY), and the luminescence and absorbance (OD600) were determined at 16 h. The expression levels were normalized to the OD600 to determine the relative ratio of number of cps to absorbance. Clones exhibiting at least a twofold difference in expression between the medium with 3 pM or 30 pM OHL and the medium without OHL were rearrayed and subsequently analyzed by DNA sequencing.
DNA sequencing and sequence analysis.
Plasmids containing OHL-responsive promoters were transformed into E. coli DH5α by heat shock at 42°C (38) to isolate plasmids that were pure enough for DNA sequencing. The size of the fragment inserted was determined by PCR using primers pZE05 and pZE06. Purified plasmids containing a <3-kb insert were sequenced at Macrogen Inc. (Seoul, Korea) using primers pZE05 and pZE06. The DNA sequences obtained were compared with the B. cenocepacia J2315 genome-sequencing project data (http://www.sanger.ac.uk/Projects/B_cenocepacia/) using BLASTN. The sequences obtained were compared to the unpublished annotation file for B. cenocepacia J2315 to identify identical sequence locations on the chromosomes. Artemis software (37) was used to view the annotation files and to identify open reading frames downstream of predicted promoters.
Potential cep box-like sequences were identified using the consensus sequence CTGTAAAAGTTACCAGTT based on the promoter sequences of six B. cenocepacia K56-2 genes positively regulated by CepR, including cepI, zmpA, aidA, and phuR (9a). Sequences 300 bp upstream and 50 bp downstream of the predicted start codon for the open reading frame identified in the promoter fusion were analyzed on both the sense and antisense strands, and sequences with at least 50% identity to the consensus were considered putative cep boxes.
Construction of luxCDBAE transcriptional fusions.
A 439-bp fragment containing the predicted promoter region of bcscV (BCAM 2057) was amplified using primers 5′-GGCCCTCGAGTCGGCGACGAGCAGCAGCAG-3′ and 5′-GGCCGGATCCGCCATCGTGCCGTCGTCCTG-3′ (the underlined sequences are XhoI and BamHI restriction sites, respectively). A 1.3-kb PCR fragment containing the katB upstream region was amplified with primers 5′-GGGCCTCGAGGCAGCGGTCGATGCCGATCT-3′ and 5′-CCGGGGATCCGGCCGTCTGCGAATTCTGGTTG-3′. These products were cloned into the XhoI-BamHI sites of pMS402.
A 329-bp fragment containing the orbI (BCAM1696) promoter region was amplified using primers 5′-GGAAGAAGGCTCGTGGC-3′ and 5′-GATGCAGCGCAGTCGGG-3′. A 471-bp fragment containing the region upstream of pvdA was amplified using primers 5′-CGCATTCGCGGACGGC-3′and 5′CAGCGTGAAGCGCACGC-3′. These PCR products were cloned into pCR2.1 TOPO (Invitrogen). An EcoRI fragment was excised from each of these constructs and cloned into pMS402. All pMS402 constructs were verified by sequencing with primers pZEO5 and pZEO6.
Real-time quantitative RT-PCR (qRT-PCR).
B. cenocepacia K56-2, K56-dI2, and K56-R2 were cultured, harvested after 8, 12, and 16 h, and used for extraction of total RNA. Total RNA was isolated from these strains using a Ribopure bacterial kit (Ambion, Austin TX), followed by DNase I treatment with 8 U of TURBO DNase and inactivation by DNase inactivation reagent (Ambion, Austin, TX). Oligonucleotide primers were designed with the Primer Express software (version 2; Applied Biosystems, Foster City, CA) and synthesized by University of Calgary Core DNA and Protein Services, Calgary, Alberta, Canada. They are listed in Table 2.
TABLE 2.
Oligonucleotide primer sequences used in real-time RT-PCRa
| Oligonucleotide | Open reading frame | Homolog | Forward primer | Reverse primer |
|---|---|---|---|---|
| F/RBS-B02 | BCAL2732 | Cold shock | GGGCGGTGAGGATCTGTTT | GCCTTCCTTCAGCGTCTTGA |
| F/RBS-B29 | BCAL3010 | spoT | CGCGAACATCGTCCATATTG | CGCTGACCTGGATCACGAA |
| F/RBS-B34 | BCAL3506 | fliM | GGAATGCGGCTACGGAATT | GCTCGCGCCGATCATCT |
| F/RBS-C53 | BCAL3524 | gspC | GGCCAGCTCGACAAGA | CGAGAATGCCGAACAG |
| F/RBS-E13 | BCAL1699 | pvdA | TGCAGATCTCGTTTCTGAAGGA | TTCGAACAGGTAGTTGATGAACGT |
| F/RBS-E62 | BCAS0409 | zmpA | CGCGGCAGACATCGACTAC | AGATGCCGTTGCGGTTGT |
| F/RBS-F105 | BCAM2041 | bcscR | CGTGCTGGTGCCAAGCTT | CGACGATGAACACGATGTAGATC |
| F/RBS-F135 | BCAM0931 | katB | CTGGAACATGATGACGAACGA | AGGTCCCACTTCGGGAAATC |
| F/RBS-E51 | pBCA054 | Regulator | GCCACCTCGCGATCCA | ATTCGGCGAACGCTGTCA |
| F/Rndh | BCAM0166 | ndh | GCGATCGGGCTGTACAAGTT | AGTGGCTCAGCGACTGGAA |
Oligonucleotide primers were designed based on the unpublished sequence of B. cenocepacia J2315 (http://www.sanger.ac.uk/Projects/B_cenocepacia/).
The NADH dehydrogenase gene ndh (BCAM0166) was determined not to be regulated by cepIR and was used as a reference standard. The ndh RNA was generated and purified from pBS7 as previously described (14). The purified ndh RNA (20 fg to 200 pg) was used to construct a standard curve for quantification using primers ndhF and ndhR to amplify the internal sequence.
Quantification was performed with a TaqMan 7500 instrument and SYBR green RT-PCR reagents (Applied Biosystems). The primer amplification efficiency for each primer pair was within 10% of the efficiency determined for ndh, which indicated the validity of using relative gene expression analysis. The cDNA from genes homologous to BCAS0409 (zmpA), BCAL3506 (fliM), pBCA054 (regulator), BCAL3010 (spoT), BCAM2041 (bcscR), BCAL1699 (pvdA), BCAL2732 (cold shock), BCAL3524 (gspC), and BCAM0931 (katB) were generated by using gene-specific primers listed in Table 2. Reverse transcription reactions were performed in 10-μl mixtures (1× TaqMan RT buffer, 2.5 μM forward primer, 2.5 μM reverse primer, 5.5 mM MgCl2, mixture containing each deoxynucleoside triphosphate at a concentration of 500 μM, 0.4 U/μl RNase inhibitor, 1.25 U/μl MultiScribe reverse transcriptase, 20 ng total RNA template), and 5 μl of cDNA diluted 1:20 was subjected to real-time PCR in 25-μl reaction mixtures (1× SYBR green PCR master mix, 50 nM forward primer, 50 nM reverse primer). The parameters for RT were 25°C for 10 min, 48°C for 30 min, and 95°C for 5 min, followed by 45 PCR cycles consisting of 95°C for 15 s and 60°C for 1 min. The concentrations of target mRNA in samples were normalized to concentrations of ndh mRNA. All quantitative real-time RT-PCR assays were repeated three times, and similar results were obtained.
RESULTS
Development of an assay to identify CepR-regulated promoter fusions.
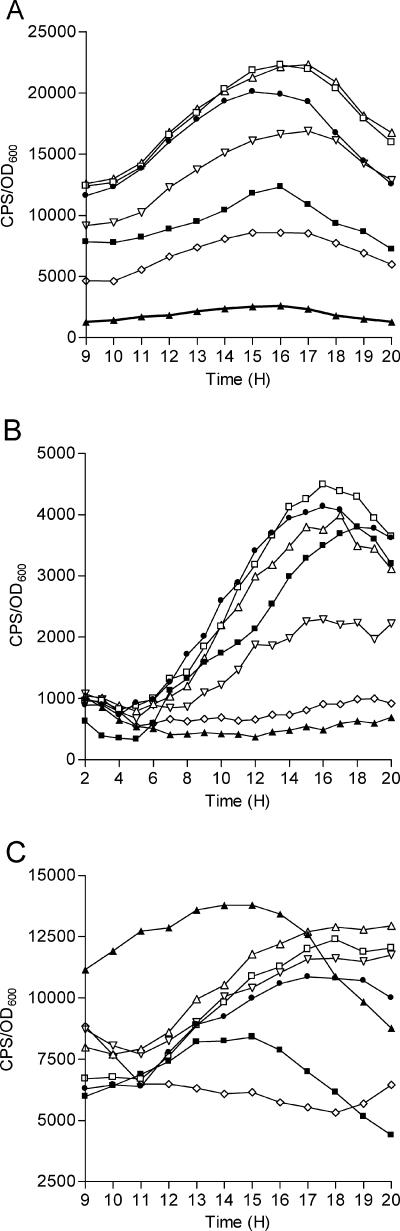
We developed an assay to identify both positively and negatively regulated promoters under control of the cepIR QS system by looking for differences in expression of genes between cultures in medium supplemented with OHL and cultures in medium not supplemented with OHL. Strain K56-dI2 has a deletion in cepI; therefore, the expression of positively regulated genes and the repression of negatively regulated genes by cepIR require addition of exogenous OHL. The appropriate concentrations of OHL to use in the assay were determined by comparing the levels of expression of cepI (pCP300), cepR (pRM432), and zmpA (pCC1) promoter-luxCDABE transcriptional fusions in K56-2 and K56-dI2. The culture medium for K56-dI2 was supplemented with 0, 300 pM, 30 pM, 3 pM, 300 fM, or 30 fM OHL, and the expression profiles of the promoter-lux fusions were compared for each condition. Luminescence and growth were measured hourly between 9 and 20 h (Fig. 1). The expression levels of the cepI::lux and zmpA::lux fusions in K56-dI2 were higher in medium supplemented with OHL than in medium without OHL. For example, at 16 h the levels of expression of cepI::lux and zmpA::lux were 8.6-fold and 18.6-fold higher, respectively, in medium with 30 pM OHL than in medium with no OHL added (Fig. 1A and B). The expression levels of cepR::lux in K56-2 and K56-dI2 in medium with 30 fM to 300 pM OHL were lower than the expression levels in K56-dI2 in medium without OHL at times between 9 and 16 h (Fig. 1C). At 16 h, the expression of the cepR::lux fusion in K56-dI2 was 2.4-fold lower in medium with 30 fM OHL than in medium with no OHL. These results are similar to previously reported observations that cepI and zmpA are positively regulated by cepIR and cepR is negatively regulated by cepIR (24, 40). Little difference in expression of the cepI or zmpA fusions was observed in media supplemented with between 3 and 300 pM OHL, and therefore OHL concentrations in this range were considered to be sufficient for identification of promoters positively regulated by CepR. OHL at these concentrations had no effect on the growth of these strains (data not shown).
FIG. 1.
Effects of various concentrations of OHL on expression of cepI::luxCDABE, zmpA::luxCDABE, and cepR::luxCDBAE fusions. Expression of promoter-luxCDABE fusions was measured throughout growth in LB medium containing 100 μg/ml trimethoprim supplemented with different concentrations of OHL. The values are the means for triplicate cultures. (A) cepI::luxCDABE. (B) zmpA::luxCDABE. (C) cepR::luxCDABE. ▪, K56-2; ▴, K56-dI2; •, K56-dI2 with 300 pM OHL; □, K56-dI2 with 30 pM OHL; ▵, K56-dI2 with 3 pM OHL; ▿, K56-dI2 with 300 fM OHL; ⋄, K56-dI2 with 30 fM OHL.
The levels of expression of the cepR::lux fusion decreased to wild-type levels in K56-dI2 grown in medium with 300 fM to 3 pM OHL during the late log and early stationary phases of growth; however, expression of this fusion increased over time, possibly because the culture either used up or broke down the OHL added to the medium (Fig. 1C). These data suggest that it would be possible to identify genes negatively regulated by cepR at 16 h but that it might not be possible to identify negatively regulated genes at later times. Although the optimum time for identification of negatively regulated genes in cultures was after 10 to 12 h of incubation, there is considerable variation in the growth rates of K56-dI2 in 384-well plates, making analysis of expression of the promoter fusions difficult at these times. Therefore, K56-dI2 containing the promoter clone library was screened at 16 h in medium with 0, 3, or 300 pM OHL. Two OHL concentrations were used since although there was little difference in the responses of the three test promoters to medium with 3 and medium with 300 pM OHL, it is possible that CepR might have different affinities for promoters depending on OHL availability.
Construction and screening of a random promoter library.
To identify genes controlled by the cepIR QS system, a random promoter library of B. cenocepacia K56-2 was generated using DNA fragments up to 3 kb long cloned upstream of luxCDABE in pMS402 in K56-dI2, a cepI mutant. The insertion efficiency of the random promoter library was estimated by averaging the efficiencies of the individual ligation reactions. Therefore, in a total of 36,096 clones, 21,952 clones (60.82%) were predicted to contain inserted fragments.
Of the 36,096 potential clones initially screened, 12,676 clones had luminescent activity either in medium with OHL or in medium without OHL. These data suggest that 57% of the potential clones contained promoters. The genome size of B. cenocepacia J2315 is 8,056 kb (http://www.sanger.ac.uk/Projects/B_cenocepacia/). If 2 kb of DNA were predicted to contain one promoter, this random promoter library should exhibit approximately threefold coverage of the predicted promoters in the genome. Of the 12,676 clones with luminescence activity in either medium, 2,282 clones produced luminescence with at least a twofold difference in expression between the medium with OHL and the medium without OHL after subtraction of the background luminescence activity obtained for K56-dI2 with the control vector pMS402. In order to rule out false-positive results potentially due to variations in growth or cross talk between wells in a 384-well plate, the 2,282 clones were rearrayed into black 96-well plates and rescreened in triplicate in medium with 0, 3 pM, or 30 pM OHL. Clones that showed at least a twofold difference between expression in media with the two OHL concentrations and expression in medium without OHL were selected for sequence analysis.
Identification and classification of genes.
A total of 440 plasmids containing promoter gene fragments were extracted, and the sizes of the insert fragments were determined by PCR using primers pZE05 and pZE06. Of the 440 plasmid clones, 200 clones had insert fragments smaller than 1.5 kb (45%), 140 clones had insert fragments between 1.5 and 3 kb long (32%), and 100 clones had insert fragments larger than 3 kb (23%). The nucleotide sequences of clones containing insert fragments that were <3 kb long (340 clones) were obtained using primers pZE05 and pZE06. The sequences were compared to the sequenced genome of B. cenocepacia J2315 (http://www.sanger.ac.uk/Project/B_cenocepacia/) using the BLASTN program to determine the chromosomal location and orientation of each clone. The location of each sequence was used to identify open reading frames downstream from predicted promoters using the unpublished annotation file of the B. cenocepacia J2315 genome. Many clones had identical or overlapping DNA sequences, suggesting that the screen was reliable since the same promoters were often identified more than once. A total of 134 unique promoter clones were differentially expressed in the presence or absence of OHL and therefore predicted to be regulated by cepIR; 91 were positively regulated, and 43 were negatively regulated.
Upon further analysis of the sequences of several promoter clones, we discovered that these clones contained sequences that were not adjacent on the J2315 genome and therefore were hybrid plasmids containing multiple inserts. To eliminate clones containing multiple fragments and possibly multiple promoters, the complete sequence of each clone was determined, which revealed that 45 (34%) of the 134 promoter clones contained inserts consisting of nonadjacent sequences in the genome or were hybrid clones.
Of the remaining 89 promoter clones, expression of 58 was positively influenced by OHL in the medium, whereas expression of 31 clones was negatively influenced by OHL (Table 3) . Five clones showed >5-fold differential luxCDABE expression in medium with OHL and medium without OHL, 57 clones showed between 2- and 5 fold differential expression, and 27 showed 2-fold differential expression in response to OHL (Table 4). These 89 clones were introduced into K56-2, and the levels of expression were compared to levels of expression in K56-dI2 (Table 3). For 47 clones the difference in expression of luminescence between K56-dI2 and K56-2 was 2- to 5-fold, for 12 clones the difference in expression was >5-fold, and for 10 clones the difference in expression was 2-fold; however, for 20 clones the difference in expression was <2-fold. The differences in the expression profiles of promoter clones between K56-dI2 in medium with 30 pM OHL and K56-dI2 in medium without OHL were generally similar to the differences between the promoter activities of the clones expressed in K56-dI2 and the promoter activities of the clones expressed in K56-2. For 14 clones with an approximately twofold difference between expression in K56-dI2 in medium with OHL and expression in K56-dI2 in medium without OHL the differences between expression in K56-2 and expression in K56-dI2 were between 1.4- and 1.9-fold.
TABLE 3.
B. cenocepacia K56-2 genes with OHL-responsive promoters
| J2315 open reading framea | Clone (no. of times found)b | Gene homolog and/or predicted functionc | Fold induction with OHLd | K56-2 vs K56-dI2e | Insert size (kb)f | Possible cep boxg |
|---|---|---|---|---|---|---|
| BCAL0010 | PBS-B14 (1) | phhA, phenylalanine-4-hydroxylase | −3.4 | −3.3 | 0.46 | |
| BCAL0111 | PBS-B30 (1) | Putative TPR domain protein | 2.5 | 3.7 | 0.68 | |
| BCAL0153 | PBS-C39 (1) | Putative flavin mononucleotide flavoprotein, unknown | −2.1 | −2.1 | 0.3 | |
| BCAL0170 | PBS-C17 (1) | Hypothetical protein | −2.3 | −4.1 | 0.82 | |
| BCAL0305 | PBS-B24 (1) | Putative exported protein | 2.5 | 2.5 | 0.75 | GCGCGTAAGCTACCTGCT (+, −55) |
| BCAL0353 | PBS-F53 (1) | Putative membrane protein | −2 | −2 | 1.72 | CGGTGAATTAAACCAGGA (+, −35) |
| BCAL0380 | PBS-F106 (1) | Putative ABC transporter ATP-binding subunit | 6.3 | 2.7 | 2.01 | GTGTGAAAGTCAGAAGTG (+, −6) |
| BCAL0554 | PBS-B33 (1) | 5-Formyltetrahydrofolate cyclo-ligase family protein | 2.2 | 7.9 | 0.51 | |
| BCAL0685 | PBS-B37 (1) | IclR family regulatory protein | 2 | 2.5 | 0.89 | |
| BCAL0723 | PBS-C55 (1) | dctB, C4-dicarboxylate transport sensor protein | −2 | −1.8 | 0.83 | |
| BCAL0793 | PBS-C46 (1) | Putative cell division protein | 2 | 2.5 | 0.09 | |
| BCAL0812 | PBS-E21 (2) | Sigma 54 modulation protein | 2 | 2 | 0.21 | |
| BCAL0854 | PBS-F38 (1) | GntR family regulatory protein | 3.5 | 3.5 | 1.97 | |
| BCAL1040 | PBS-C01 (1) | Glycosyl transferase group 1 protein | 2.2 | 5.2 | 0.94 | |
| BCAL1182 | PBS-E27 (2) | Putative transcriptional regulator | 5.2 | 2.2 | 0.75 | GTGAAAAAGGTGTAAGCA (−, −17) |
| BCAL1374 | PBS-C44 (1) | Glyoxalase/bleomycin resistance protein | 2.3 | 1.6 | 1.1 | |
| BCAL1446 | PBS-B48 (2) | Putative lipoprotein | 2 | 1.8 | 0.23 | |
| BCAL1513 | PBS-C20 (1) | Putative transcriptional regulatory protein | 3.4 | 3.5 | 2.47 | |
| BCAL1550 | PBS-E46 (1) | Putative sugar ABC transport system | 2.8 | 1.4 | 0.1 | |
| BCAL1625 | PBS-C48 (1) | acoR, acetoin catabolism regulatory protein | −2.2 | −4.4 | 0.32 | |
| BCAL1651 | PBS-C47 (1) | lexA, LexA repressor | 2.7 | 1.8 | 0.82 | CTGTATAAAAATACAGCT (−, −42) |
| BCAL1814 | PBS-D11 (1) | Putative MerR family transcriptional regulator | −2.1 | −2.2 | 1 | ACGTTCGAGACTCCAGTG (+, −39) |
| BCAL1889 | PBS-F50 (2) | Putative 23S rRNA (uracil-5-)-methyltransferase/orthologue, ygcA | 2.5 | 2.5 | 1.27 | CTTCAACGTTTGTCAGGA (−, −25) |
| BCAL1914 | PBS-E02 (2) | Unknown | 2.6 | 2.6 | 0.7 | |
| BCAL1990 | PBS-F40 (1) | pgi, glucose-6-phosphate isomerase | 4.4 | 2.2 | 2.93 | |
| BCAL2037 | PBS-C30 (4) | Putative membrane protein | 8.4 | 2.4 | 1.17 | |
| BCAL2068 | PBS-E56 (1) | Putative membrane protein | 9.6 | 9.7 | 0.9 | |
| BCAL2170 | PBS-C51 (1) | Putative membrane protein | −2.1 | −1.6 | 0.48 | |
| BCAL2244 | PBS-C10 (1) | hutU, urocanate hydratase | 2 | 2.2 | 0.25 | |
| BCAL2301 | PBS-C50 (1) | Putative exported protein | 2.1 | 3.3 | 0.26 | |
| BCAL2302 | PBS-C09 (1) | uvrB, excinuclease ABC subunit B | −2.3 | −2 | 0.66 | |
| BCAL2321 | PBS-E09 (1) | Putative glutathione S-transferase-related protein | 2 | 2.6 | 0.8 | |
| BCAL2373 | PBS-C03 (1) | Putative globin | 2.1 | 2.5 | 0.74 | |
| BCAL2464 | PBS-B13 (1) | Short-chain dehydrogenase | 2 | 4.4 | 0.78 | CACAGAATGTGGCAAGTT (+, −34) |
| BCAL2630 | PBS-C21 (1) | hemC, putative porphobilinogen deaminase protein | −2.5 | −2.3 | 0.82 | |
| BCAL2644 | PBS-C04 (1) | Putative ATP-binding protein | −2 | −1.9 | 0.62 | |
| BCAL2652 | PBS-B27 (1) | panD, putative aspartate 1-decarboxylase | 2.2 | 1.8 | 0.15 | CTGGAAATCTGACGGGCG (+, −55) |
| BCAL2732 | PBS-B02 (1) | Cold shock protein, DNA binding | −2.5 | −2.5 | 0.9 | |
| BCAL2799 | PBS-C24 (1) | dalK, putative carbohydrate kinase | 2 | 2.1 | 0.51 | |
| BCAL2818 | PBS-E40 (1) | Putative sugar kinase protein | 2.5 | 7.4 | 0.56 | |
| BCAL2931 | PBS-E12 (1) | acpD, acyl carrier protein phosphodiesterase | −2.3 | −3.2 | 1.17 | CTGTATAAATGTACAGTA (−, −43) |
| BCAL3006 | PBS-F44 (1) | cspA, cold shock-like protein | 2.9 | 1.9 | 1.51 | CTGTCAATATTACCGGGA (+, −239) |
| BCAL3010 | PBS-B29 (1) | spoT, guanosine-3′,5′-bis(diphosphate) 3′-pyrophosphohydrolase | 2.2 | 6.3 | 1.21 | CTGAAGAAGGTGCCGGTG (−, −83) |
| BCAL3051 | PBS-E67 (1) | ribE, riboflavin synthase alpha chain | −2.2 | −1.4 | 0.58 | |
| BCAL3103 | PBS-E33 (1) | ureD, UreD family accessory protein | 2 | 1.6 | 0.26 | GCTTCCTCGTTTCCCGTT (+, −25) |
| BCAL3130 | PBS-F48 (1) | wtz, ABC transporter, ATP-binding component | 2 | 3 | 1.7 | |
| BCAL3443 | PBS-C35 (2) | ispB, octaprenyl-diphosphate synthase | 2.3 | 5.6 | 0.63 | CTGTCAGACGTACCGGCA (+, −73) |
| BCAL3506 | PBS-B34 (3) | fliM, flagellar motor switch protein FliM | 2.9 | 2.3 | 0.97 | |
| BCAL3523 | PBS-B40 (1) | gspG, general secretory pathway protein G | −2 | −2 | 0.2 | ATGTGAAATTTGCGTTGA (−, −155) |
| BCAL3524 | PBS-C53 (1) | gspC, putative general secretory pathway protein | −2 | −1.9 | 0.22 | |
| BCAM0003 | PBS-E08 (1) | Putative partition protein ParA | 2.8 | 1.4 | 0.54 | CTGTAGAATTCCAACGAC (+, −34) |
| BCAM0010 | PBS-E44 (1) | klb, 2-amino-3-ketobutyrate coenzyme A ligase | 2.5 | 6.9 | 1.15 | CTGTTATAGTCGTCGTTC (−, −67) |
| BCAM0038 | PBS-D03 (1) | feaR, AraC family regulatory protein | 3.3 | 6.1 | 2 | |
| BCAM0057 | PBS-C33 (1) | p-Hydroxybenzoate hydroxylase | −2 | −2.4 | 0.28 | GTTTTGAAGATACCGGCC (−, −94) |
| BCAM0130 | PBS-C37 (1) | Putative acyl-coenzyme A dehydrogenase | 2 | 2 | 0.58 | TGGTATGTATTACTAGAT (+, −80) |
| BCAM0300 | PBS-C41 (1) | Putative metallo-beta-lactamase family protein | 2.2 | 3.5 | 1 | |
| BCAM0341 | PBS-F130 (1) | Unknown | −2 | −9.3 | 3 | |
| BCAM0677 | PBS-E10 (1) | Putative transcriptional regulator, araC family | −2 | −1.7 | 0.6 | CGGTAGAATTCGCCATTC (−, −41) |
| BCAM0722 | PBS-B45 (1) | Putative O-methyltransferase-like protein | −2 | −4.2 | 0.25 | |
| BCAM0832 | PBS-F37 (1) | Putative dyd-type peroxidase conserved hypothetical protein | −2.3 | −2.1 | 2.52 | |
| BCAM0860 | PBS-D06 (1) | Putative glycosyltransferase | −2.9 | −11.5 | 1.2 | CTGAGCGCGTGACCGGGT (+, −73) |
| BCAM0881 | PBS-F108 (1) | Putative trehalose trehalohydrolase alpha-amylase | −3.9 | −3.6 | 1.61 | GCGTGACGGTCGCCCGTT (+, −190) |
| BCAM0901 | PBS-E28 (1) | amn, AMP nucleosidase | 2.7 | 1.7 | 0.23 | |
| BCAM0931 | PBS-F135 (1) | katB, catalase precursor | −2.2 | −2 | 2.18 | |
| BCAM1110 | PBS-B01 (1) | Unknown putative sugar transporter | −3.3 | −1.6 | 0.7 | |
| BCAM1142 | PBS-C32 (1) | Unknown | 2.1 | 2.2 | 0.9 | |
| BCAM1165 | PBS-D01 (1) | Putative membrane protein | 2.7 | 3.9 | 2 | GTGTTAGATTTCGCCGTC (+, −48) |
| BCAM1189 | PBS-F70 (1) | Putative transcriptional regulator, LysR family | 2.3 | 2.1 | 1.75 | |
| BCAM1227 | PBS-D09 (1) | pgaB, polysaccharide deacetylase | −2 | −2.3 | 2.7 | |
| BCAM1453 | PBS-E68 (1) | LysR family regulatory protein | 2.1 | 3.2 | 0.35 | |
| BCAM1501 | PBS-B06 (1) | Unknown | −2.5 | −2.3 | 0.19 | TGGTTCATGTGATCAGTA (+, −72), |
| BCAM1515 | PBS-F110 (1) | Conserved hypothetical protein | 2.1 | 2.7 | 2.19 | CTGTACAAATGTACAGTG (+, −27) |
| BCAM1573 | PBS-B22 (1) | otsA, glycosyltransferase family | 2 | 1.9 | 0.99 | |
| BCAM1650 | PBS-E32 (1) | LysR transcriptional regulator | 3 | 3 | 0.45 | |
| BCAM2155 | PBS-E05 (1) | Putative cytochrome P450 oxidoreductase | 2 | 2 | 0.63 | CTGGCAGATCAGCCAGTC (+, −238) |
| BCAM2158 | PBS-B39 (1) | Putative DNA-binding protein | 2.6 | 2.7 | 1.46 | |
| BCAM2162 | PBS-E24 (2) | Putative MarR transcriptional regulator family | 2 | 2 | 0.75 | GTGGACAACCTTCCATTA (+, −276) |
| BCAM2176 | PBS-E37 (1) | glsA, thermolabile glutaminase | −2 | −2 | 0.45 | |
| BCAM2507 | PBS-E43 (1) | Putative sugar transport-related membrane protein | −2.1 | −1.5 | 1.19 | |
| BCAM2583 | PBS-F119 (1) | Putative amidohydrolase | −2.2 | −2.3 | 2.42 | |
| BCAM2615 | PBS-A13 (1) | Unknown | 2.4 | 2 | 0.5 | |
| BCAM2805 | PBS-F112 (1) | Periplasmic binding protein | 2.3 | 2.6 | 2 | |
| BCAM2835 | PBS-E20 (2) | estC, putative esterase | 2.2 | 2.6 | 0.45 | |
| BCAS0161 | PBS-C15 (1) | Putative transcriptional regulator, LysR family | −2.3 | −1.8 | 0.54 | ACATAATATTTACAACTT (−, −36) |
| BCAS0162 | PBS-C16 (1) | Putative periplasmic solute-binding protein | 3.2 | 6.5 | 0.82 | |
| BCAS0221 | PBS-C42 (1) | Putative ABC transporter (pseudogene) | 2 | 1.7 | 0.39 | |
| BCAS0409 | PBS-E62 (2) | zmpA, extracellular zinc metalloprotease | 2.3 | 2.6 | 1.3 | GTTTAAAAGTCATCACTT (+, −60) |
| BCAS0730 | PBS-E30 (2) | Putative Na+-dependent nucleoside transporter | 2 | 2.9 | 0.26 | |
| pBCA054 | PBS-E51 (20) | LuxR family regulatory protein | 8.6 | 15 | 1.68 | CAATACGAGTCGCCCGTC (+, −69) |
Open reading frame predicted with the unpublished annotation file of the B. cenocepacia J2315 genome (http://www.sanger.ac.uk/Projects/B_cenocepacia/).
Promoter clone designation. The number in parentheses indicates the number of times that the identical clone was found in the library.
Predicted gene or function based on the unpublished annotation of the B. cenocepacia J2315 genome (http://www.sanger.ac.uk/projects/B_cenocepacia/).
Fold difference in expression between K56-dI2 in LB medium with 30 pM OHL and K56-dI2 in LB medium without OHL.
Fold difference in expression between K56-2 and K56-dI2.
Size of the fragment inserted in pMS402.
Potential cep box. For the data in parentheses, a plus sign indicates that the box is on the coding strand, and a minus sign indicates that the box is on the antisense strand. The values indicate the direction and distance from the start codon (position 1).
TABLE 4.
Relative differential expression of promoter clones in response to OHL
| Regulation | No. of promoters fora:
|
|
|---|---|---|
| K56-dI2 with OHL vs K56-dI2 | K56-2 vs K56-dI2 | |
| Positive | ||
| 2- to 5-fold | 37 | 32 |
| >5-fold | 5 | 10 |
| 2-fold | 16 | 5 |
| <2-fold | 0 | 11 |
| Total | 58 | 58 |
| Negative | ||
| 2- to 5-fold | 20 | 15 |
| >5-fold | 0 | 2 |
| 2-fold | 11 | 5 |
| <2-fold | 0 | 9 |
| Total | 31 | 31 |
Numbers of promoters with fold differences indicated after subtraction of the background.
Genes were identified based on the unpublished annotation file for B. cenocepacia J2315 (Table 3). Genes expressed under the control of cepIR are involved in adaptation or resistance, type II and type III secretion systems, metabolism, membrane or surface structures, regulatory genes, and transport, and there are also genes with unknown functions. OHL-responsive promoters were identified on all three chromosomes and the plasmid.
Clone PBS-E62, which contained part of the zmpA gene and its promoter region promoter, had 4.6-fold-higher activity in K56-dI2 in medium with 30 pM OHL than in K56-dI2 in medium without OHL and 2.6-fold-higher activity in K56-2 than in K56-dI2. These results are similar to those previously reported for zmpA::lacZ fusions (40). Two clones containing the type II secretion system genes gspG (PBS-B40) and gspC (PBS-C53) were identified and showed twofold negative regulation. We obtained 20 copies of clone PBS-E51, which contains a promoter for the plasmid-encoded gene pBCA054, which exhibits similarity to genes encoding response regulator proteins of the LuxR family. This promoter was positively regulated; there was an 8.6-fold increase in expression in K56-dI2 in medium with OHL, and the level of expression was approximately 15-fold higher in K56-2 than in K56-dI2 (Table 3).
Clone PBS-F24 contained part of cepI and the cepI promoter region. This clone had 16-fold-higher lux activity in K56-dI2 grown in medium supplemented with 30 pM OHL than in medium without OHL and 3.6-fold-higher activity in K56-2 than in K56-dI2. These data are similar to data obtained in previous studies that demonstrated that cepI expression is positively regulated by cepR and is significantly decreased in a cepI mutant without OHL supplementation (24). This clone was determined to contain multiple fragments of the cepI promoter region, as well as a nonadjacent fragment, and therefore was not included in Table 3.
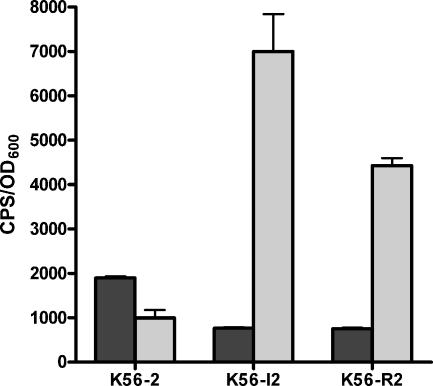
The sequence upstream of the pvdA gene was found in two identical clones, which unfortunately contained nonadjacent DNA fragments, and is also not listed in Table 3. The pvdA gene (BCAL1699) encodes l-ornithine 5-monooxygenase and has previously been shown to be negatively regulated by cepIR (24). Interestingly, the promoter clones that contained pvdA appeared to be positively regulated by cepIR. To investigate this discrepancy, a 300-bp fragment upstream of pvdA was cloned into pMS402 and designated pVDA01. The pvdA gene has recently been shown to be transcribed as part of a large operon from a promoter upstream of BCAL1696 or orbI (1). The promoter region upstream of BCAL1696 was cloned into pMS402 and designated pVDO301. The levels of expression of these two promoter-lux fusions in K56-2, K56-dI2, and K56-R2, the cepR mutant, were compared (Fig. 2). The expression of the lux fusion in pVDA01 was twofold higher in K56-2 than in the cepI or cepR mutant, indicating that this fragment did contain a promoter that was positively regulated by CepR. The levels of expression of the promoter in pVDO301, however, were approximately fivefold higher in the cepI and cepR mutants than in K56-2, indicating that this promoter was negatively regulated by CepR (Fig. 2). Expression of lux in pVDO301 was also repressed by addition of iron to the medium (data not shown). These data suggest that expression of pvdA may be influenced by two promoters that are regulated differently by CepR but the major iron-regulated promoter of this ornibactin biosynthesis operon is negatively regulated.
FIG. 2.
Comparison of the levels of expression of pvdA (pVDA01) and orbI::luxCDBAE (pVDO301) fusions in K56-2, K56-dI2, and K56-R2. Cultures were grown in TSB-DC medium containing 100 μg/ml trimethoprim. Dark gray bars, pVDA01; light gray bars, pVDO301. The values are means ± standard deviations for data obtained at 24 h for triplicate cultures and are representative of three experiments. Similar results were obtained at other times in the stationary phase.
Clone PBS-F105 contained the promoter region upstream of bcscR (BCAM2041), a putative type III secretion system gene. The expression of luxCDABE was positively regulated 2.4-fold (data not shown); however, this clone also contained multiple inserts. The positive regulation of bcscR was confirmed by qRT-PCR as described below. Two clusters of type III secretion genes have been identified in B. cenocepacia (34, 42). The bcscQ and bcscR genes are located upstream of virB1 and are predicted to be in the same operon. A second cluster of 12 genes is separated from bcscQR by approximately 10 kb and is transcribed in the opposite orientation. The first gene in this cluster is bcscV (BCAM2057). This cluster also includes bcscN, which has been shown to be important in virulence in an acute mouse respiratory infection model (42). To determine if genes in the bcscV cluster are also regulated by cepIR, the bcscV predicted promoter region was cloned into pMS402 to create a bcscV::luxCDABE reporter (pMVbcscV) and was introduced into K56, K56-dI2, and K56-R2. The luminescence activity was monitored over time. The level of expression of the bcscV::lux fusion was significantly higher in K56-2 than in either K56-dI2 or K56-R2 (P < 0.05, as determined by analysis of variance). For example, after growth for 16 h the cps/OD600 ratios were 9,345 ± 3,403 for K56-2, 2,465 ± 34 for K56-dI2, and 514 ± 92 for K56-R2 (means ± standard deviations). Therefore, bcscV is also positively regulated by the cepIR QS system.
Confirmation of the promoter gene expression analysis by quantitative RT-PCR.
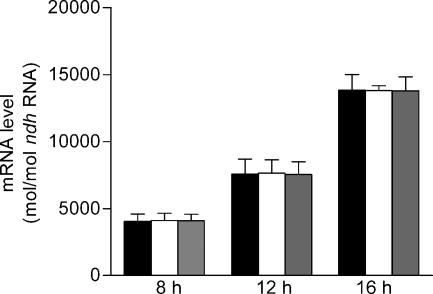
To confirm expression results obtained with the random promoter library and to determine the requirement for CepR for expression of genes responsive to OHL in the cepI mutant, nine genes downstream of the predicted promoters for BCAS0409 (zmpA), BCAL3506 (fliM), pBCA054 (regulator), BCAL3010 (spoT), BCAM2041 (bcscR), BCAL1699 (pvdA), BCAL2732 (cold shock), BCAL3524 (gspC), and BCAM0931(katB) that were differentially expressed in the random promoter library were selected for quantitative analysis of mRNA by real-time qRT-PCR. Since the two promoters cloned upstream of pvdA were regulated differently on the multicopy plasmid pMS402, pvdA (BCAL1699) was included in the analysis to determine the regulation of a single copy of this gene in the genome. BCAM2041 (bcscR) was included in the analysis even though the promoter clone contained multiple inserts to confirm that bcscR was regulated by cepIR, since the promoter for the second type III gene cluster upstream of bcscV was shown using the promoter lux fusions to be positively regulated by cepIR. Total RNA of K56-2, K56-dI2, and K56-R2 were extracted and used as templates in qRT-PCR. Quantitative expression levels of each gene were determined using sequence-specific primers for each target gene (Table 2). The ndh gene, encoding NADH dehydrogenase, was used as a reference gene for relative quantification in real-time RT-PCR. We first determined if expression of ndh was influenced by the cepIR QS system. Although the level of expression of ndh increased over time, the levels of expression were the same in all three strains, indicating that cepIR does not regulate ndh expression (Fig. 3). Therefore, in subsequent assays the ndh gene was used as a reference to compare the expression of the other genes predicted to be regulated by CepR.
FIG. 3.
Expression of the ndh gene in B. cenocepacia K56-2, K56-dI2, and K56-R2. Total RNA was extracted from 8, 12, and 16 cultures and subjected to real-time RT-PCR. The absolute expression levels were determined using an ndh standard curve. The values are means ± standard deviations for triplicate assays. Solid bars, K56-2; open bars, K56-dI2; gray bars, K56-R2.
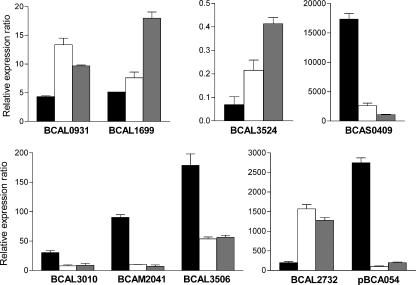
The results of the real-time RT-PCR demonstrated that the levels of expression of BCAS0409 (zmpA), BCAL3506 (fliM), pBCA054 (regulator), BCAL3010 (spoT), and BCAM2041 (bcscR) were 6.7-, 3.3-, 28-, 3.8-, 8.9-fold higher, respectively, and the levels of expression of BCAL1699 (pvdA), BCAL2732 (cold shock protein), BCAL3524 (gspC), and BCAM0931 (katB) were 1.5-, 8-, 3-, and 3-fold lower, respectively, in K56-2 than in K56-dI2 (Fig. 4). These results confirmed the expression data obtained in the random promoter luminescence assays, with the exception of the data for BCAL1699 (pvdA). The qRT-PCR data for pvdA are consistent with the lux expression of the promoter upstream of orbI, a previous report that expression of a chromosomally encoded pvdA::lacZ fusion was negatively regulated by cepIR (24), and the results of a transcriptional analysis indicating that pvdA is cotranscribed with orbI (1). Although the original clone (PBS-F105) which harbored bcscR contained additional fragments, the qRT-PCR data confirmed that the expression of this gene was positively influenced by cepI.
FIG. 4.
Quantitative RT-PCR of genes expressed from the random promoter library. The mean relative expression levels of the target gene were normalized to the levels of ndh and multiplied by 1,000. Solid bars, K56-2; open bars, K56-dI2; gray bars, K56-R2. The values are means ± standard deviations for six replicates.
We also used qRT-PCR to quantitate the expression of these genes in the cepR mutant. The relative mRNA levels of the nine genes in K56-R2 were similar to the relative mRNA levels in K56-dI2.The levels of expression of BCAS0409 (zmpA), BCAL3506 (fliM), pBCA054 (regulator), BCAL3010 (spoT), and BCAM2041 (bcscR) were 16.2-, 3.2-, 13.8-, 3.4-, 13-fold higher, respectively, and the levels of expression of BCAL1699 (pvdA), BCAL2732 (cold shock protein), BCAL3524 (gspC), and BCAM0931 (katB) were 3.5-, 6.5-, 6-, and 2.2-fold lower, respectively, in K56-2 than in K56-R2 (Fig. 4). These results indicate that CepR and the AHLs synthesized by CepI are required for regulation of these genes.
Influence of cciIR on expression of selected genes.
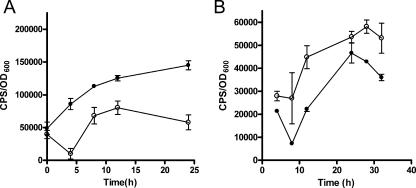
Since K56-2 also contains the CciIR quorum-sensing system, we decided to examine the expression of a few genes identified in the promoter library to see if they are also regulated by CciIR. Plasmids pBSC01 (BCAL1040::luxCDABE), pVDO301, pBS34 (fliM::luxCDABE), and pBS18 (katB::luxCDABE) were introduced into K56-2cciIR, which has a deletion of the cciIR promoter and therefore does not express either cciI or cciR (31). The expression of BCAL1040 and orbI was influenced by the cciIR mutation throughout growth in a manner similar to the expression in the cepI mutant. BCAL1040 expression was positively regulated by cciIR (Fig. 5A), and orbI expression was negatively regulated by cciIR (Fig. 5B). There was no difference in expression of either fliM or katB between K56-2 and K56-2cciIR. For example, the cps/OD600 ratios in these strains containing pBS-B34 (fliM) were 1.1 × 104 ± 206 and 1.4 × 104 ± 1,009, respectively. The cps/OD600 ratios for pBS18 (katB) were 670 ± 61and 620 ± 2 4, respectively. Therefore, some genes are regulated by both quorum-sensing systems, whereas others are likely regulated independently.
FIG. 5.
Comparison of the levels of expression of pBSCO1 (BCAL1040::luxCDABE) and pVDO301 (BCAL1696::luxCDABE) in K56-2 and K56cciIR. (A) pBSC01. Cultures were grown in LB medium containing 100 μg/ml trimethoprim. (B) pVDO301. Cultures were grown in TSB-DC medium containing 100 μg/ml trimethoprim. The values are means ± standard deviations for triplicate cultures.
Identification of potential cep boxes upstream of CepR-regulated genes.
To determine which promoters might be directly regulated by cepIR, a cep box consensus sequence was used to search the region upstream of the open reading frames identified in the promoter clones. Thirty genes had a potential cep box with at least 50% identity (Table 3). If more than one potential cep box was identified, the box with the highest levels of homology or closest to the start codon was selected. A cep box was also identified upstream of BCAL1696 (orbI) (CCGTTCGTCACACCAGTG) at position −42 bp relative to the start codon on the coding strand. These data suggest that most of the genes identified are likely to be indirectly regulated by CepR. Six potential regulatory genes that had putative cep boxes were identified. It is possible that these genes are intermediate regulators in the CepR regulatory cascade.
DISCUSSION
A random promoter library with a lux-based reporter was used to identify genes with promoters responsive to OHL. This approach provided a sensitive quantitative method to identify differentially expressed genes in high-throughput conditions. Eighty-nine promoter clones were differentially expressed at least twofold in the cepI mutant in the presence or absence of exogenous OHL. The majority of these clones were also differentially expressed at least twofold in the wild type and the cepI mutant. Both positively and negatively regulated promoters were identified, and the majority of these promoters have not previously been shown to be regulated by the cepIR QS system.
Several approaches have been used to identify cepIR-regulated genes in B. cenocepacia or B. cepacia. A promoter library approach employing a lacZ reporter plasmid that also contained cepR was used to identify B. cepacia cepIR-regulated genes expressed in E. coli (3). The vector used in our study enabled screening in the natural host rather than E. coli, which should make it possible to identify clones that require additional genes for expression that are present in B. cenocepacia but not present in E. coli. A promoterless lacZ-based transposon approach was used to identify OHL-induced genes in a cepI mutant of B. cenocepacia (47). In the studies using the lacZ reporter systems (3, 47), the initial screens were performed using blue-white selection, which is less sensitive for determining differences in expression than the luxCDBAE reporter, and this resulted in identification of genes whose expression was detectable only in the presence of OHL. The pMS402 lux reporter system has an advantage over the two lacZ reporter systems, since it is possible to detect genes that are expressed in the absence of cepI and OHL but exhibit increased expression in the presence of cepI and OHL. It was also possible to use this system to identify negatively regulated genes as well as positively regulated genes. Although we screened only for promoters expressed in stationary phase, the library could be used for detection of promoters expressed at other stages of growth. A proteomics approach was used to identify 11 differentially expressed proteins in B. cenocepacia H111 (36). Interestingly, each of the approaches used to identify genes influenced by cepI has led to identification of a different set of genes (2, 36, 47). This is not surprising since three different strains and two different species were used in these studies. One gene consistently identified in the other three studies but not in our study was the aidA gene (2, 36, 47), although we have determined that this gene is regulated by cepIR in B. cenocepacia K56-2 (E. Lutter and P. A. Sokol, unpublished observations). The cepI gene was identified in both our study and the study of Weingart et al. (47). The lack of an annotated genome sequence makes it difficult to compare the CepR-regulated genes identified in different studies; however, the transcriptional regulator pBCA54, shown to be positively regulated in our study, appears to be the gene designated bqiD that was identified by Weingart et al. (47).
The genome of B. cenocepacia J2315 contains 8.056 Mb in three chromosomes (3.870, 3.217, and 0.876 Mb) and a 92.7-kb plasmid containing approximately 7,200 coding sequences (http://www.sanger.ac.uk/Projects/B_cenocepacia/). The 89 promoter clones characterized were randomly distributed throughout the genome; 50 clones contained genes from the large chromosome, 33 clones contained genes from the medium chromosome, 5 clones contained genes from the small chromosome, and 1 clone contained genes from the plasmid. This indicates that there was a lack of bias in cloning of the promoters and suggests that good coverage of the genome was obtained. Although we estimated that our random promoter library should have threefold coverage of the genome, it clearly did not contain promoters for many of the previously identified genes regulated by cepIR, including zmpB (20), cepR (24), cciIR (31), and aidA (2, 36, 47). The conditions used to construct a library with a high insertion efficiency in pMS402 resulted in a very significant number of clones with multiple inserts, and 34% of our unique sequenced clones contained nonadjacent DNA sequences. The frequency of hybrid clones likely masked identification of some potential promoters. Careful sequencing of all promoter clones is clearly necessary when random promoter libraries are used due to the frequent occurrence of hybrid clones. Environmental factors such as medium composition and oxygen availability, as well as the growth phase, can also affect the transcription of many genes that are QS regulated. Thus, a number of cepIR-regulated genes may not have been detected under the conditions employed. All of the strategies used to date have led to identification of a portion of the cepIR QS regulon, but additional assay conditions and methods must be employed before this global regulatory network is fully described.
The random promoter library has led to identification of a number of novel genes in B. cenocepacia that are influenced by OHL concentrations and a functional cepI gene; these genes include katB, gspC, gspG, bcscR, spoT, and cspA. Expression of the katB (PBS-F135) gene was negatively regulated by the cepIR QS system. B. cenocepacia strain C5452 contains two catalase/peroxidase genes, katA and katB (22). KatB is the major catalase/peroxidase that plays a role in cellular protection against oxidative stress. A katB mutant exhibited reduced growth and hypersensitivity to hydrogen peroxide (22).
Cold shock proteins are small proteins (∼7 kDa) that are involved in mRNA folding, protein synthesis, and/or freeze protection. It is believed that they can function as RNA chaperones to reduce the secondary structure of RNA at low temperatures (49). Expression of the cspA gene (PBS-F44), which encodes a putative cold shock protein, was induced 2.9-fold in medium with OHL, indicating that it is positively regulated by the cepIR system. A cold shock protein transcriptional regulator, cspC, was identified using signature-tagged mutagenesis and was determined to be important for survival of B. cenocepacia in a chronic respiratory infection model (19).
Two clones containing different promoters for general secretion pathway genes were identified. Expression of both gspG (PBS-B40) and gspC (PBS-C53) was negatively influenced by the presence of OHL, and there was twofold negative induction in medium with OHL or when the parent and the cepI mutant were compared. It is interesting that expression of genes encoding the type II secretion machinery proteins are negatively regulated by cepIR; however, expression of at least two genes encoding proteins predicted to be secreted using this pathway, zmpA and zmpB, is positively influenced by cepI and cepR. Expression of bcscR and bcscV, the first genes in the two major type III secretion gene clusters, was shown to be positively influenced by OHL and cepI, indicating that genes in both type II and type III secretion systems are regulated by the cepIR QS system but there is an opposite effect on the expression of these secretion genes. Genes involved in type II and type III secretion have previously been shown to be regulated by the lasRI or rhlRI QS systems in Pseudomonas aeruginosa (8, 39, 45). Control of type II and III secretion system gene expression differs in P. aeruginosa and B. cenocepacia, however, since type II secretion genes are positively regulated and type III genes are negatively regulated in P. aeruginosa. Although quorum sensing may be a common mechanism of gene regulation in bacterial pathogens, clearly the targets of these regulatory genes are different in different pathogens.
To validate the results obtained for the promoter library, qRT-PCR was used to confirm expression of genes downstream of nine cepIR-regulated promoters. The expression levels of eight genes were consistent with the promoter library observations and similar for the cepI and cepR mutants. BCAS0409 (zmpA), BCAL3506 (fliM), pBCA054 (regulator), BCAL3010 (spoT), and BCAM2041 (bcsCR) were positively regulated, and BCAL2732 (cold shock), BCAL3524 (gspC), and BCAM0931 (katB) were negatively regulated. BCAL1699 (pvdA) was negatively regulated, which was consistent with the promoter upstream of the operon but not the putative promoter directly upstream of the pvdA. It is not clear why the sequence upstream of pvdA functions as a promoter in pMS402. There are no predicted promoter elements in the 300 bp upstream of pvdA, such as −35 or −10 sequences; however, at least when this sequence is present on a multicopy plasmid, it appears to have promoter activity.
Although we showed that the cepIR QS system influences expression of the genes identified using the random promoter library, we did not determine whether CepR directly regulates transcription of these genes. It is possible that CepR influences expression indirectly by regulating expression of an intermediate regulatory gene that subsequently alters the expression of other promoters. Several regulatory genes were identified in the promoter library and may be candidates for intermediate regulators in the quorum-sensing network. Inactivation of cepI might also influence expression of some promoters regulated by other regulators. We identified possible cep boxes upstream of 30 of 89 genes that we identified. These data should be interpreted with caution since direct binding of CepR has not been demonstrated and in most cases the putative cep box is not highly conserved. The cep boxes upstream of zmpA, spoT, cspA, and BCAL0380 were among the cep boxes with the greatest similarity. The 59 genes for which cep boxes were not identified are likely to be indirectly regulated by the cepIR system. It should also be noted that we measured only gene expression, not protein levels or any effects of posttranslational modification.
Recently, our laboratory has demonstrated that the cciIR QS system interacts with the cepIR QS system in B. cenocepacia K56-2 (31). Expression of some genes is influenced by both systems, whereas other genes appear to be regulated by only one QS system. For example, expression of zmpA (31, 40), BCAL1040, and BCAL1696 (orbI) is regulated in similar ways by cepIR and cciIR. Expression of zmpB is markedly reduced in cepI, cepR, and cciR mutants but is significantly enhanced in a cciI mutant (20). The cepIR genes, but not the cciIR genes, influence the expression of katB and fliM. It is not known whether CciR binds to the same binding site as CepR or whether it has a different specificity for target genes. Further studies are under way to investigate the influence of cciIR on gene expression and to further elucidate the QS gene regulatory network in B. cenocepacia.
Acknowledgments
This study was supported by grants from the Canadian Cystic Fibrosis Foundation and the CCFF Special Initiative in memory of Michael O'Reilly.
We thank S. Guillemette for technical assistance; M. G. Surette for providing pMS402 and advice on promoter library construction; A. Beeston, R. Malott, and J. Bjarnason for helpful discussions; J. Parkhill and M. Holden at the Welcome Trust Institute for access to the annotation data for the B. cenocepacia J2315 genome sequence prior to publication; and J. Fox and S. Wong at the Provincial Laboratory for Public Health, Calgary, AB, Canada, for advice on the qRT-PCR experiments.
Footnotes
Published ahead of print on 22 November 2006.
REFERENCES
- 1.Agnoli, K., C. A. Lowe, K. L. Farmer, S. I. Husnain, and M. S. Thomas. 2006. The ornibactin biosynthesis and transport genes of Burkholderia cenocepacia are regulated by an extracytoplasmic function σ factor which is a part of the Fur regulon. J. Bacteriol. 188:3631-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar, C., I. Bertani, and V. Venturi. 2003. Quorum-sensing system and stationary-phase sigma factor (rpoS) of the onion pathogen Burkholderia cepacia genomovar I type strain, ATCC 25416. Appl. Environ. Microbiol. 69:1739-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilar, C., A. Friscina, G. Devescovi, M. Kojic, and V. Venturi. 2003. Identification of quorum-sensing-regulated genes of Burkholderia cepacia. J. Bacteriol. 185:6456-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, NY.
- 5.Baldwin, A., P. A. Sokol, J. Parkhill, and E. Mahenthiralingam. 2004. The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia. Infect. Immun. 72:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernhardt, S. A., T. Spilker, T. Coffey, and J. J. LiPuma. 2003. Burkholderia cepacia complex in cystic fibrosis: frequency of strain replacement during chronic infection. Clin. Infect. Dis. 37:780-785. [DOI] [PubMed] [Google Scholar]
- 7.Bjarnason, J., C. M. Southward, and M. G. Surette. 2003. Genomic profiling of iron-responsive genes in Salmonella enterica serovar Typhimurium by high-throughput screening of a random promoter library. J. Bacteriol. 185:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bleves, S., C. Soscia, P. Nogueira-Orlandi, A. Lazdunski, and A. Filloux. 2005. Quorum sensing negatively controls type III secretion regulon expression in Pseudomonas aeruginosa PAO1. J. Bacteriol. 187:3898-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers, C. E., M. B. Visser, U. Schwab, and P. A. Sokol. 2005. Identification of N-acylhomoserine lactones in mucopurulent respiratory secretions from cystic fibrosis patients. FEMS Microbiol. Lett. 244:297-304. [DOI] [PubMed] [Google Scholar]
- 9a.Chambers, C. E., P. Law, M. B. Visser, E. Lutter, and P. A. Sokol. 2004. Abstr. 119B, p. 73. In Abstr. 2nd ASM Conf. Cell-Cell Signaling.
- 10.Coenye, T., and J. J. LiPuma. 2003. Molecular epidemiology of Burkholderia species. Front. Biosci. 8:55-67. [DOI] [PubMed] [Google Scholar]
- 11.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan, K., C. Dammel, J. Stein, H. Rabin, and M. G. Surette. 2003. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 50:1477-1491. [DOI] [PubMed] [Google Scholar]
- 13.Eberl, L. 2006. Quorum sensing in the genus Burkholderia. Int. J. Med. Microbiol. 296:103-110. [DOI] [PubMed] [Google Scholar]
- 14.Gingues, S., C. Kooi, M. B. Visser, B. Subsin, and P. A. Sokol. 2005. Distribution and expression of the ZmpA metalloprotease in the Burkholderia cepacia complex. J. Bacteriol. 187:8247-8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gotschlich, A., B. Huber, O. Geisenberger, A. Togl, A. Steidle, K. Riedel, P. Hill, B. Tummler, P. Vandamme, B. Middleton, M. Camara, P. Williams, A. Hardman, and L. Eberl. 2001. Synthesis of multiple N-acylhomoserine lactones is wide-spread among the members of the Burkholderia cepacia complex. Syst. Appl. Microbiol. 24:1-14. [DOI] [PubMed] [Google Scholar]
- 16.Huber, B., F. Feldmann, M. Kothe, P. Vandamme, J. Wopperer, K. Riedel, and L. Eberl. 2004. Identification of a novel virulence factor in Burkholderia cenocepacia H111 required for efficient slow killing of Caenorhabditis elegans. Infect. Immun. 72:7220-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517-2528. [DOI] [PubMed] [Google Scholar]
- 18.Huber, B., K. Riedel, M. Kothe, M. Givskov, S. Molin, and L. Eberl. 2002. Genetic analysis of functions involved in the late stages of biofilm development in Burkholderia cepacia H111. Mol. Microbiol. 46:411-426. [DOI] [PubMed] [Google Scholar]
- 19.Hunt, T. A., C. Kooi, P. A. Sokol, and M. A. Valvano. 2004. Identification of Burkholderia cenocepacia genes required for bacterial survival in vivo. Infect. Immun. 72:4010-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kooi, C., B. Subsin, R. Chen, B. Pohorelic, and P. A. Sokol. 2006. Burkholderia cenocepacia ZmpB is a broad specificity zinc metalloprotease involved in virulence. Infect. Immun. 74:4083-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kothe, M., M. Antl, B. Huber, K. Stoecker, D. Ebrecht, I. Steinmetz, and L. Eberl. 2003. Killing of Caenorhabditis elegans by Burkholderia cepacia is controlled by the cep quorum-sensing system. Cell. Microbiol. 5:343-351. [DOI] [PubMed] [Google Scholar]
- 22.Lefebre, M. D., R. S. Flannagan, and M. A. Valvano. 2005. A minor catalase/peroxidase from Burkholderia cenocepacia is required for normal aconitase activity. Microbiology 151:1975-1985. [DOI] [PubMed] [Google Scholar]
- 23.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewenza, S., and P. A. Sokol. 2001. Regulation of ornibactin synthesis and N-acyl-l-homoserine lactone production by CepR in Burkholderia cepacia. J. Bacteriol. 183:2212-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewenza, S., M. B. Visser, and P. A. Sokol. 2002. Interspecies communication between Burkholderia cepacia and Pseudomonas aeruginosa. Can. J. Microbiol. 48:707-716. [DOI] [PubMed] [Google Scholar]
- 26.Lutter, E., S. Lewenza, J. J. Dennis, M. B. Visser, and P. A. Sokol. 2001. Distribution of quorum-sensing genes in the Burkholderia cepacia complex. Infect. Immun. 69:4661-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahenthiralingam, E., A. Baldwin, and P. Vandamme. 2002. Burkholderia cepacia complex infection in patients with cystic fibrosis. J. Med. Microbiol. 51:533-538. [DOI] [PubMed] [Google Scholar]
- 28.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. W. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahenthiralingam, E., T. A. Urban, and J. B. Goldberg. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144-156. [DOI] [PubMed] [Google Scholar]
- 30.Mahenthiralingam, E., P. Vandamme, M. E. Campbell, D. A. Henry, A. M. Gravelle, L. T. Wong, A. G. Davidson, P. G. Wilcox, B. Nakielna, and D. P. Speert. 2001. Infection with Burkholderia cepacia complex genomovars in patients with cystic fibrosis: virulent transmissible strains of genomovar III can replace Burkholderia multivorans. Clin. Infect. Dis. 33:1469-1475. [DOI] [PubMed] [Google Scholar]
- 31.Malott, R. J., A. Baldwin, E. Mahenthiralingam, and P. A. Sokol. 2005. Characterization of the cciIR quorum-sensing system in Burkholderia cenocepacia. Infect. Immun. 73:4982-4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDowell, A., E. Mahenthiralingam, K. E. Dunbar, J. E. Moore, M. Crowe, and J. S. Elborn. 2004. Epidemiology of Burkholderia cepacia complex species recovered from cystic fibrosis patients: issues related to patient segregation. J. Med. Microbiol. 53:663-668. [DOI] [PubMed] [Google Scholar]
- 33.Ohman, D. E., J. C. Sadoff, and B. H. Iglewski. 1980. Toxin A-deficient mutants of Pseudomonas aeruginosa PA103: isolation and characterization. Infect. Immun. 28:899-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parsons, Y. N., K. J. Glendinning, V. Thornton, B. A. Hales, C. A. Hart, and C. Winstanley. 2001. A putative type III secretion gene cluster is widely distributed in the Burkholderia cepacia complex but absent from genomovar I. FEMS Microbiol. Lett. 203:103-108. [DOI] [PubMed] [Google Scholar]
- 35.Reik, R., T. Spilker, and J. J. Lipuma. 2005. Distribution of Burkholderia cepacia complex species among isolates recovered from persons with or without cystic fibrosis. J. Clin. Microbiol. 43:2926-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riedel, K., C. Arevalo-Ferro, G. Reil, A. Gorg, F. Lottspeich, and L. Eberl. 2003. Analysis of the quorum-sensing regulon of the opportunistic pathogen Burkholderia cepacia H111 by proteomics. Electrophoresis 24:740-750. [DOI] [PubMed] [Google Scholar]
- 37.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 39.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sokol, P. A., U. Sajjan, M. B. Visser, S. Gingues, J. Forstner, and C. Kooi. 2003. The CepIR quorum sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology 149:3649-3658. [DOI] [PubMed] [Google Scholar]
- 41.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomich, M., A. Griffith, C. A. Herfst, J. L. Burns, and C. D. Mohr. 2003. Attenuated virulence of a Burkholderia cepacia type III secretion mutant in a murine model of infection. Infect. Immun. 71:1405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomlin, K. L., R. J. Malott, G. Ramage, D. G. Storey, P. A. Sokol, and H. Ceri. 2005. Quorum-sensing mutations affect attachment and stability of Burkholderia cenocepacia biofilms. Appl. Environ. Microbiol. 71:5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venturi, V., A. Friscina, I. Bertani, G. Devescovi, and C. Aguilar. 2004. Quorum sensing in the Burkholderia cepacia complex. Res. Microbiol. 155:238-244. [DOI] [PubMed] [Google Scholar]
- 45.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319-346. [DOI] [PubMed] [Google Scholar]
- 47.Weingart, C. L., C. E. White, S. Liu, Y. Chai, H. Cho, C. S. Tsai, Y. Wei, N. R. Delay, M. R. Gronquist, A. Eberhard, and S. C. Winans. 2005. Direct binding of the quorum sensing regulator CepR of Burkholderia cenocepacia to two target promoters in vitro. Mol. Microbiol. 57:452-467. [DOI] [PubMed] [Google Scholar]
- 48.Williams, P., M. Camara, A. Hardman, S. Swift, D. Milton, V. J. Hope, K. Winzer, B. Middleton, D. I. Pritchard, and B. W. Bycroft. 2000. Quorum sensing and the population-dependent control of virulence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:667-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamanaka, K. 1999. Cold shock response in Escherichia coli. J. Mol. Microbiol. Biotechnol. 1:193-202. [PubMed] [Google Scholar]