Abstract
Quorum sensing is involved in the regulation of multicellular behavior through communication via small molecules. Given the high number and diversity of the gastrointestinal microbiota, it is postulated that members of this community communicate to coordinate a variety of adaptive processes. AI-2 is suggested to be a universal bacterial signaling molecule synthesized by the LuxS enzyme, which forms an integral part of the activated methyl cycle. We have previously reported that the well-documented probiotic strain Lactobacillus rhamnosus GG, a human isolate, produces AI-2-like molecules. In this study, we identified the luxS homologue of L. rhamnosus GG. luxS seems to be located in an operon with a yxjH gene encoding a putative cobalamin-independent methionine synthase. In silico analysis revealed a methionine-specific T box in the leader sequence of the putative yxjH-luxS operon. However, transcriptional analysis showed that luxS is expressed mainly as a monocistronic transcript. Construction of a luxS knockout mutant confirmed that the luxS gene is responsible for AI-2 production in L. rhamnosus GG. However, this mutation also resulted in pleiotropic effects on the growth of this fastidious strain. Cysteine, pantothenate, folic acid, and biotin could partially complement growth, suggesting a central metabolic role for luxS in L. rhamnosus GG. Interestingly, the luxS mutant also showed a defect in monospecies biofilm formation. Experiments with chemically synthesized (S)-4,5-dihydroxy-2,3-pentanedione, coculture with the wild type, and nutritional complementation suggested that the main cause of this defect has a metabolic nature. Moreover, our data indicate that suppressor mutations are likely to occur in luxS mutants of L. rhamnosus GG. Therefore, results of luxS-related studies should be carefully interpreted.
There is a growing consumer and scientific interest in probiotic bacteria, which are defined as “live microorganisms which, when consumed in adequate amounts, confer a health benefit on the host beyond basic nutrition” (19). Lactobacillus rhamnosus GG (ATCC 53103) is a well-documented and extensively studied probiotic organism (18, 30). Nevertheless, the basic molecular mechanisms of its probiotic action are mainly unknown. Moreover, the understanding of the physiology and genetics of this bacterium is still limited (12).
Many bacteria, including pathogens and commensals, communicate via diffusible signal molecules to coordinate multicellular behavior in a process referred to as “quorum sensing.” Quorum sensing is known to regulate important traits of bacteria such as virulence gene expression, adherence, competence, and stress response at the population level (57). Regarding the high density and diversity of the gastrointestinal microbiota, it is postulated that bacterial communication fulfills an important role in coordinating various processes in the gut (32). One class of intriguing bacterial signaling molecules is called AI-2 (41). AI-2 synthesis is catalyzed by the LuxS enzyme in many gram-positive and gram-negative bacteria and is proposed to be involved in interspecies bacterial communication (40).
The LuxS/AI-2 system was initially characterized and best studied in Vibrio harveyi, where it is one of the signals that regulate light production (46). Since then, many studies on gastrointestinal pathogens including Helicobacter pylori, Vibrio cholerae, Salmonella enterica serovar Typhimurium, and enterohemorrhagic Escherichia coli in which a role for LuxS or AI-2 has been demonstrated in multicellular behaviors like biofilm formation, virulence, and motility have been published (recently reviewed in reference 55). Production of AI-2 is, however, not limited to pathogenic bacteria. In fact, many commensal and potentially probiotic bacteria such as Bifidobacterium and Lactobacillus strains possess a luxS homologue (1, 33, 49) and can produce AI-2 (14). In a previous study, we have shown that the probiotic strain L. rhamnosus GG is able to produce AI-2-like molecules (14), but a role for the AI-2/LuxS system has not been assigned yet.
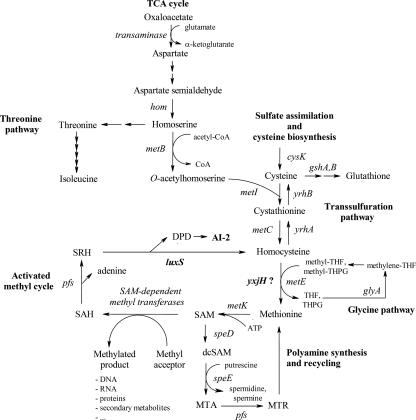
The biosynthetic pathway leading to AI-2 production is highly integrated in the central metabolism and physiology of bacteria since it forms an essential part of the activated methyl cycle (Fig. 1). AI-2 is produced from S-adenosylmethionine (SAM) in three enzymatic steps. During various methyltransferase reactions, SAM is converted to S-adenosylhomocysteine (SAH). Because of its structural similarity to SAM, SAH is a potent feedback inhibitor of the SAM-dependent methyltransferases (59). SAH is detoxified by the Pfs enzyme to yield adenine and S-ribosylhomocysteine (SRH). LuxS then catalyzes the conversion of SRH, yielding (S)-4,5-dihydroxy-2,3-pentanedione (DPD) and homocysteine. DPD undergoes spontaneous rearrangements to give rise to several furanone derivatives, collectively known as AI-2 (8, 41, 46). Alternatively, in most eukaryotes, archaea, α-proteobacteria, actinobacteria, and cyanobacteria, SAH is detoxified in a one-step conversion using SAH hydrolase. This reaction yields adenosine and homocysteine but no AI-2 (49, 59). Homocysteine is used to recycle methionine. In Escherichia coli, two enzymes can catalyze the transfer of the methyl group to homocysteine, namely, the cobalamin-dependent methionine synthase MetH and the cobalamin-independent methionine synthase MetE (47). Most lactobacilli contain a homologue of the MetE enzyme only (44). Additionally, an open reading frame (ORF) of unknown function, designated yxjH (22), can be found in these bacteria, which is suggested to be an alternative cobalamin-independent methionine synthase that is involved mainly in the SAM recycling pathway (44). Methionine is converted to SAM in a reaction catalyzed by SAM synthetase (MetK), which is an essential enzyme (58). Besides an activated methyl donor for the methylation of various cellular compounds (36), SAM is also decarboxylated to form cationic polyamines that can modulate the functions of RNA, DNA, and other acidic substances (27), implying an important physiological role for LuxS (60).
FIG. 1.
The activated methyl cycle, biosynthesis of AI-2, and directly coupled metabolic pathways in gram-positive bacteria. AI-2 is produced from SAM, which is essential for a large number of methylation processes and is used for polyamine synthesis, in three enzymatic steps. During various methyltransferase reactions, SAM is converted to SAH. SAH is detoxified by the Pfs enzyme to yield adenine and SRH. LuxS then catalyzes the conversion of SRH, yielding DPD and homocysteine. DPD undergoes spontaneous rearrangements to form AI-2. Homocysteine is used to recycle methionine. Methionine synthase is required by all organisms to ensure the regeneration of the methyl group of SAM. Most gram-positive bacteria contain a cobalamin-independent methionine synthase, MetE, and a YxjH enzyme, which is suggested to be an alternative cobalamin-independent methionine synthase that is mainly involved in the SAM recycling pathway. Methionine is then converted to SAM in a reaction catalyzed by SAM synthetase (MetK). Homocysteine can also be synthesized de novo from oxaloacetate in seven steps as part of the aspartate family. Notably, some organisms, such as L. plantarum and L. salivarius, can synthesize homocysteine from cysteine by direct sulfhydrylation involving the CysK enzyme. Homocysteine can also be converted to cysteine via the reverse transsulfuration pathway. The enzymes cystathionine β-lyase and cystathionine γ-synthase (encoded by metI and metC homologues) require pyridoxal-phosphate as cofactor. These complex branching pathways are coordinated by a variety of regulatory mechanisms, including multivalent feedback inhibition of enzyme activity by more than one end product, “metabolite-sensing” controllers of gene expression such as riboswitches and amino-acid-specific T boxes, and regulatory proteins. Moreover, the scheme presented here is simplified. For example, some gram-positive organisms can use alternative organic and inorganic sulfur sources, and some bacteria use O-succinylhomoserine instead of O-acetylhomoserine in the MetB-catalyzed reaction. Additionally, not all gram-positive bacteria contain the complete set of pathways. The common gene names for B. subtilis and E. coli are used. The pathway was constructed based on previously described data (13, 44), the KEGG PATHWAY website (http://www.genome.jp/kegg/pathway/map/map00271.html), and the LacplantCyc pathway genome database (53). TCA, tricarboxylic acid; MTA, methylthioadenosine; MTR, methylthioribose; THF, tetrahydrofolate; THPG, tetrahydropteroyltriglutamate; dcSAM, decarboxylated SAM.
The aim of this study was to critically investigate the functional role of the luxS gene in the probiotic strain L. rhamnosus GG in relation to both AI-2-mediated quorum sensing and central metabolism. We first present a detailed analysis of the genomic organization of the luxS gene and its relation to the activated methyl cycle. Subsequently, the consequences of luxS inactivation resulting in reduced growth and biofilm formation are investigated in relation to the dual function of LuxS.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
L. rhamnosus GG and its derivatives (Table 1) were routinely grown at 37°C in de Man-Rogosa-Sharpe (MRS) medium (Difco) (16) under static conditions. For determinations of AI-2 activity, L. rhamnosus GG was grown in modified MRS medium in which glucose was replaced by galactose (14). Bacto Lactobacilli AOAC medium (Difco) containing 15 g/liter peptonized milk, 5 g/liter yeast extract, 10 g/liter glucose, 5 g/liter tomato juice, 2 g/liter monopotassium phosphate, and 1 g/liter polysorbate 80 was also used in this study. If required, antibiotics were used at the following concentrations: 10 μg/ml tetracycline, 100 μg/ml ampicillin, and 10 μg/ml (L. rhamnosus GG) or 100 μg/ml (E. coli) erythromycin. E. coli cells were grown in LB medium with aeration at 37°C (45). V. harveyi cells were grown overnight at 30°C in AB medium (21).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− φ80dlacZΔM15Δ (lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK−) supE44 λ−thi-1 girA96 relA1 | Gibco-BRL |
| L. rhamnosus GG | ||
| Wild type | Human isolate | ATCC 53103 (48) |
| CMPG5412 | luxS knockout mutant of L. rhamnosus GG; luxS::tet(M); reduced growth capacity | This study |
| CMPG5339 | CMPG5412 complemented by integration of pCMPG5339 | This study |
| CMPG5912 | CMPG5412 complemented by integration of pCMPG5912 | This study |
| CMPG5413 | luxS knockout mutant of L. rhamnosus GG; luxS::tet(M); putative suppressor mutant; wild-type growth capacity; no biofilm formation | This study |
| V. harveyi | ||
| BB170 | luxN::Tn5 | 3 |
| BB152 | luxM::Tn5 (AI-1− AI-2+) | 4 |
| Plasmids | ||
| pCRII-TOPO | Cloning vector; ampicillin and kanamycin resistance | Invitrogen |
| pFAJ5301 | Cloning vector; pUC18 derivative; erythromycin resistance | Unpublished results |
| pLAB1301 | E. coli-Lactobacillus shuttle vector; ampicillin and erythromycin resistance | 29 |
| pMD5057 | Tetracycline resistance plasmid from L. plantarum 5057 | 10 |
| pEM40 | pUC19E-derived integration vector (attB located at the 3′ end of the tRNALeu locus) containing a 1.6-kb int-attP cassette of phage A2; ampicillin and erythromycin resistance | 2 |
| pCMPG5332 | pFAJ5301-derived suicide vector to knock out the luxSLGG gene through insertion of the tet(M) gene from pMD5057 | This study |
| pCMPG5339 | pEM40-derived integration vector containing the luxSLGG gene and its putative promoter P2 | This study |
| pCMPG5912 | pEM40-derived integration vector containing the putative yxjH-luxS operon of L. rhamnosus GG, its putative promoter P1, and the putative methionine-specific T box in the 5′ UTR | This study |
DNA manipulations.
Routine molecular biology techniques were performed according to standard procedures (45). Restriction and modifying enzymes (from New England Biolabs) were used as recommended by the manufacturer. Plasmid DNA was prepared from E. coli by using QIAGEN Miniprep kits. Chromosomal DNA and plasmid DNA were isolated from L. rhamnosus GG as previously described (12).
Cloning and analysis of the L. rhamnosus GG luxS gene (luxSLGG) and flanking regions.
When we initiated this study, part of the genome sequence of Lactobacillus casei ATCC 334 was published online (http://spider.jgi-psf.org/JGI_microbial/html/). A tBLASTn search with the luxS gene of Lactobacillus plantarum WCFS1 (GenBank accession number NP_784522.1) (35) resulted in the identification of the luxS homologue of L. casei (recently published in the NCBI database under accession number ZP_00384741). Primers (Pro-198, Pro-199, and Pro-200) (Table 2) used for PCR amplification of the luxSLGG gene were derived from the conserved regions detected by the CLUSTAL W multiple alignment (54) of luxS sequences of Lactobacillus gasseri, L. plantarum, and L. casei and were 100% identical to the luxS sequence of L. casei. The resulting internal fragment of the luxS gene was cloned into the pCRII-TOPO vector (Invitrogen) and sequenced. A combination of inverse PCR and Southern hybridization was used to determine the complete DNA sequence of the luxSLGG gene and circa 1-kb flanking regions. “Inverse primers” (Pro-215 and Pro-216) were designed based on the known core region of the luxSLGG gene (Table 2). Genomic DNA of L. rhamnosus GG was digested with EclXI, followed by Southern hybridization with a digoxigenin (DIG)-labeled 382-bp luxS probe (amplified with primers Pro-198 and Pro-200) to determine the size of the fragments containing the luxSLGG gene. Subsequently, the digested genomic DNA was allowed to self-ligate, and an inverse PCR was performed with Pro-215 and Pro-216 to amplify the DNA sequences lying outside the boundaries of the known sequences. The PCR fragments were cloned in a pCRII-TOPO vector and sequenced via cycle sequencing using an ABI 3100-Avant DNA analyzer (Applied Biosystems) according to the manufacturer's instructions. A BLASTx search was performed to analyze the DNA sequence. A putative luxSLGG promoter region was determined based on the Lactobacillus consensus promoter sequence (39). A putative rho-independent terminator was determined by using the Mfold Web server (64).
TABLE 2.
Primer sequences used in this studya
| Primer | Sequence (5′-3′) |
|---|---|
| Pro-198 | CAGCTGTTAAAGCACCTTATGTTCG |
| Pro-199 | CCCCAAGTAATCAAGTGGAAACCGG |
| Pro-200 | CGCCCACTCCTTTGCTGAAAACAG |
| Pro-215 | CCAGCCTTGAAGCGATTGC |
| Pro-216 | CCATTCGGGCCTTTTTCG |
| Pro-221 (EcoRI) | GAATTCGAGATTCCTTTACAAATATGCTCTTAC |
| Pro-222 (EcoRI) | CGAATTCGTTCGGAATAGGTTATACTAGACAAAAG |
| Pro-238 | CGGTCAAATCTTCAGGTAATCGAA |
| Pro-239 | GCTGCCAGGTTCGCTATTCGAAC |
| Pro-240 | AGGTAAGCTATTGAGAAGGAGATCC |
| Pro-246 | CAATGCCGGTGACGGATAACAACTGG |
| Pro-247 | GATGCATGGAAGTATGAACGGGAC |
| Pro-340 | GATCACACTGCAGTTAAGGCGCC |
| Pro-341 | GTGTTCAATTGTATGCAACCCGGC |
| Pro-LuxF | CCTTCGCCGACTGAGTCGACGCTGCCAGGTTCGCTATTCGAAC |
| Pro-LuxR | CTCTCGAGGATTGAACGTATCCATGCTG |
| Pro-533 (BamHI) | ATGGATCCCAAGCCTCAGACAACCTGAC |
| Pro-538 (NdeI) | AACATATGACAGAAGAAGTTCAAA |
| Pro-569 (BamHI) | ATGGATCCGTTGTTCCATTGCTGCGTGT |
| Pro-578 (BamHI) | ATGGATCCGTTGTTCCCATGAGACACCTCC |
Note that restriction sites present in the primer sequence are underlined, and the respective restriction enzymes are indicated in parentheses.
In silico detection of methionine-specific T box upstream of yxjH and luxS.
Sequences of T boxes described previously by Rodionov et al. (44) were used to construct a T-box model. Specifically, seven sequences upstream of the yxjH gene of the species Enterococcus faecalis, Lactobacillus brevis, L. casei, L. gasseri, Leuconostoc mesenteroides, L. plantarum, and Pediococcus pentosaceus were used based on their phylogenetic closeness to L. rhamnosus GG. We used the algorithm MotifLocator (9, 38) to find predefined motifs for the methionine-specific T box (44) in the DNA sequences using an adapted position weight matrix scoring scheme. Since MotifLocator does not accommodate gaps, the T box was split into two halves, and each was used in an independent search for motifs. The results from both searches were checked manually for best scores and relative location to delineate a potential T box. The upstream sequences of all eight species were aligned with ClustalW (54) to further outline the specifier hairpin region described previously by Rodionov et al. (44).
Northern blot analysis.
L. rhamnosus GG was grown in different media, and total RNA was isolated from cultures in exponential and stationary phases by using the RNeasy kit (QIAGEN). Total RNA was denatured and transferred onto nylon membranes using standard procedures (45). A 170-bp luxS-specific probe was generated with primers Pro-340 and Pro-341 and DIG labeled (Roche). A 456-bp yxjH-specific probe was generated with primers Pro-238 and Pro-247. The presence of two luxS transcripts was further confirmed by reverse transcription-PCR. cDNA was made using the RevertAid H Minus first-strand cDNA synthesis kit (Fermentas) according to the manufacturer's instructions. Subsequently, a PCR was performed with primers Pro-340 and Pro-341 to amplify a 170-bp internal fragment of luxS. A PCR with primers Pro-247 and Pro-341 was used to amplify a 1.2-kb fragment of the yxjH-luxS cotranscript.
AI-2 assay.
The AI-2 luminescence reporter assay was performed essentially as described previously (21, 50), with modifications reported previously (14). Results are presented as a percentage of the induction level produced by the positive control (V. harveyi BB152). Each sample was tested threefold, and assays were performed at least in duplicate.
Construction and analysis of the L. rhamnosus GG luxS mutant.
A fragment of 2.5 kb containing the luxSLGG gene and circa 1-kb regions flanking the luxS gene upstream and downstream was amplified from an L. rhamnosus GG genomic DNA template using primers Pro-246 and Pro-247. This fragment was cloned in the EcoRI site of pFAJ5301, an erythromycin-resistant derivative of pUC18 (unpublished results). A tetracycline resistance cassette, tet(M), was amplified from plasmid pMD5057 of L. plantarum 5057 (10) with primers Pro-221 and Pro-222 containing EcoRI sites and inserted into the MfeI site of the luxSLGG gene. The resulting suicide vector, pCMPG5332, was electroporated into L. rhamnosus GG according to methods described previously by De Keersmaecker et al. (12). Three to four days after the electroporation, transformants were selected for resistance to 10 μg/ml tetracycline and sensitivity to 10 μg/ml erythromycin. Confirmation of DNA recombination was performed by PCR using primers Pro-239 and Pro-240 and Southern blotting. One of the luxS mutants was designated CMPG5412 and was further analyzed for its ability to produce AI-2 using the bioassay described above.
Constructs for genetic complementation analysis.
A functional luxS gene was introduced on an integrating plasmid into the L. rhamnosus GG luxS mutant for complementation and into the wild type for a control. Therefore, the luxS gene and its putative promoter P2 (Fig. 2A) were amplified by PCR with primers Pro-LuxF and Pro-LuxR and cloned in the EcoRI site of pEM40 (2), resulting in plasmid pCMPG5339. This vector allows site-specific genomic integration of the luxS gene with its putative promoter at the 3′ end of the tRNALeu locus. Introduction of genes at this locus has been previously shown to be successful in L. rhamnosus GG (12). Moreover, a second complementation construct was made to investigate the role of the coexpression of yxjH and luxS and cis-regulatory sequences in complementation for growth and biofilm formation. To this end, the putative operon yxjH-luxS with its promoter P1 was amplified with primers Pro-239 and Pro-578 and cloned into the BamHI site of pEM40, resulting in the integrating plasmid pCMPG5912. These integrating plasmids were then transformed into the luxS mutant and wild-type L. rhamnosus GG. Integration of the vectors was confirmed using PCR and Southern hybridization as previously described (12).
FIG. 2.
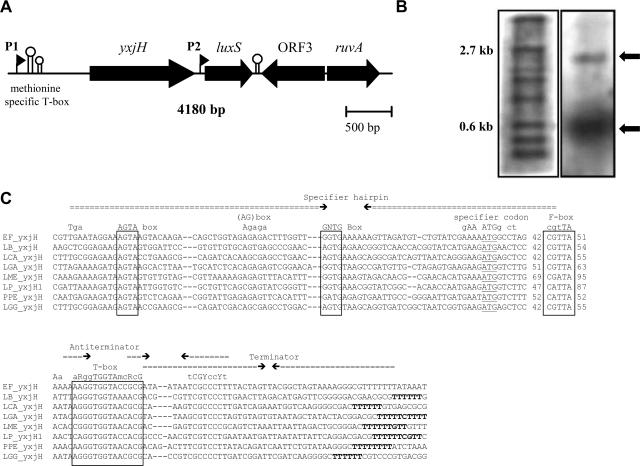
Genomic organization of the L. rhamnosus GG luxS gene. (A) Schematic representation of the relative orientations of the different ORFs within the luxS chromosomal region (4.2 kb) of L. rhamnosus GG as revealed by sequencing and BLAST analysis. The respective gene names are indicated in the figure. ORF3 encodes for a conserved hypothetical protein with unknown function in lactobacilli (see the text). The putative promoters (shown by flags) P1, corresponding to yxjH-luxS cotranscription, and P2, corresponding to luxS expression, were determined based on the Northern blot analysis shown in B. The methionine-specific T box is shown by two consecutive stem-loop structures representing the specifier hairpin and the mutually exclusive terminator-antiterminator hairpin as further explained below (C). The other stem-loop represents the putative rho-independent terminator of the luxS gene. The nucleotide sequence for this genomic region of L. rhamnosus GG has been deposited in the NCBI database (GenBank accession number DQ335218). (B) Transcriptional analysis of the luxSLGG gene. L. rhamnosus GG was grown in MRS medium, and total RNA was prepared from liquid cultures in exponential and stationary phases. Total RNA was then subjected to Northern blotting and hybridization using a DIG-labeled luxS-specific probe. The first lane of the nylon membrane (lane 1) contains a DIG-labeled RNA marker (Roche), and relevant sizes are indicated on the left side of this lane. The second lane (lane 2) contains total RNA of L. rhamnosus GG hybridized with the luxS-specific probe. The arrows mark the sizes of the luxSLGG mRNA transcripts under this condition. (C) Alignment of methionine-specific T boxes present in the leader sequence of yxjH genes of L. rhamnosus GG and related organisms. The complementary stems of the RNA secondary structure and positions of the hairpins and conserved boxes are shown in the upper lines. The names of the boxes and conserved structural elements crucial for the regulations are based upon data reported previously (24, 44). The first hairpin structure is the “specifier hairpin.” The specifier codon ATG for methionine is underlined. The other hairpin structures are the mutually exclusive antiterminator-terminator loops. The antiterminator contains the T-box sequence, which is the most highly conserved leader element. The effector molecule that signals limitation for the specific amino acid is the cognate uncharged tRNA, which can interact directly with the leader by making at least two contacts. The first is a codon-anticodon interaction with the specifier codon. The second occurs by base pairing between 5′-UGGN-3′ of the T box in the antiterminator side bulge and the 5′-NCCA-3′ acceptor end of uncharged tRNA, which stabilizes the antiterminator and prevents the formation of the competitive terminator helix (24). The binding of uncharged tRNA to the methionine-specific T-box sequence therefore promotes the formation of the antiterminator and results in the transcription of the yxjH-luxS operon of L. rhamnosus GG under methionine-limited conditions.
Nutritional complementation experiments.
Cultures grown in MRS medium overnight were brought to equal densities (5 × 108 CFU/ml) and 15,000-fold diluted in 300 μl fresh Lactobacilli AOAC medium (Difco). Bacteria were grown at 37°C, and the optical density at 600 nm (OD600) was measured automatically every 30 min during 3 days in a BioscreenC apparatus (Labsystems Oy, Zellik, Belgium). For each time point, the average optical density was calculated from three independent measurements. Generation time (g) was calculated as follows: g = [(t2 − t1) log2]/(log OD2 − log OD1), where t is time and 1 and 2 are successive time points in the exponential growth phase. The growth rate, μ, was calculated as follows: μ = ln2/g. The following components (Sigma or Merck) were added to the growth medium in different concentrations (ranging from 7 nM to 2 mM): methionine, SAM, homocysteine, cysteine-HCl, aspartic acid, isoleucine, threonine, lysine, folic acid, d-(+)-biotin, cyanocobalamin, pyridoxol-HCl, nicotinic acid, riboflavin, Ca-d-(+)-pantothenate, thiamine-HCl, spermine-4HCl, and spermidine (Fig. 1). Additionally, chemically synthesized DPD (15) was added to the growth medium in different concentrations ranging from 0.5 nM to 0.5 mM. Each experiment was repeated at least six times to prevent possible bias of the results by spontaneous reversion.
Calculation of reversion frequencies.
luxS mutant strain CMPG5412 was grown overnight in nutrient-rich MRS medium. Subsequently, a dilution series was plated out onto MRS plates and grown for 48 h. Two hundred colonies were then transferred into Lactobacilli AOAC medium and were grown for 5 days. After 5 days, these colonies were reinoculated in AOAC medium to select for colonies that showed a stable reversion to wild-type growth. All revertants were also checked for the absence of AI-2 production.
In vitro biofilm assays and extracellular complementation experiments.
A method for assaying biofilm formation of L. rhamnosus GG in vitro was optimized based on methods described previously by De Keersmaecker et al. (15), with minor modifications. Briefly, biofilm formation on polystyrene pegs hanging into microtiter plate wells was assayed. The pegs were placed into wells containing 200 μl medium and bacterial cells (ca. 3 × 107 CFU) and incubated in anaerobic jars for 72 h at 37°C. Wells containing sterile growth medium and wells containing the wild type in AOAC medium were included as negative and positive controls, respectively. Data were normalized to the control (L. rhamnosus GG in AOAC medium), which was taken to be 100%. Each strain and/or condition was tested eightfold. Each experiment was performed at least in triplicate. To investigate the role of AI-2 and other LuxS-derived signals in L. rhamnosus GG biofilm formation, different extracellular complementation experiments were performed. Conditioned supernatant of wild-type L. rhamnosus GG was added to AOAC medium as follows. L. rhamnosus GG cells were grown for 24 h and then centrifuged (10 min, 6,000 × g). The supernatant was collected, and 10% filter-sterilized conditioned medium was added to AOAC medium. Biofilm formation was assessed as described above. DPD (15) was added in different concentrations to AOAC medium (ranging from 1 nM to 100 μM). Alternatively, biofilms were grown in two coculture systems. A two-compartment system (63) in which each well of a 96-well polystyrene plate was separated into two compartments by 0.2-μm-pore-size Anopore membranes (Nunc tissue culture inserts, eight-well strip) was used. For nutritional complementation, SAM (1 mM), methionine (100 μM), cysteine-HCl (2 mM), Ca-d-(+)-pantothenate (50 μg/ml), folic acid (0.01 μg/ml), and d-(+)-biotin (10 μg/ml) were added.
Nucleotide sequence accession number.
A 4.2-kb nucleotide sequence of genomic DNA of L. rhamnosus GG has been deposited in the NCBI database under GenBank accession number DQ335218. This fragment contains the nucleotide sequence for the genes yxjH, luxS, and ruvA and an ORF encoding a conserved hypothetical protein with unknown function as described in Results.
RESULTS
Identification of the L. rhamnosus GG luxS gene and its flanking DNA sequences.
Since the genome sequence is unavailable, a homology-based strategy, described in Materials and Methods, was followed to isolate the L. rhamnosus GG luxS gene. Based on the luxS sequence of L. casei ATCC 334, a 4.2-kb sequence of L. rhamnosus GG genomic DNA was determined, which contains four ORFs, including luxSLGG, as presented in Fig. 2A. This sequence has been deposited in the NCBI database (GenBank accession number DQ335218). The in silico-translated sequence of luxS (474 bp) is 96% identical to LuxS of L. casei (accession number ZP_00384741) and 80% identical to LuxS of L. plantarum WCFS1 (accession number NP_784522) (33). A potential ribosome binding site, AGGAGA, is located 10 nucleotides upstream of the putative ATG start codon of luxS. The 3′ end of luxS is separated from the 3′ end of ORF3 by 86 nucleotides that contain features of a rho-independent transcriptional terminator with a ΔG value of −33 kcal/mol (64). The amino acid sequence of the gene (ORF3, 666 bp) downstream of luxS is 48% identical (E = 9 × 10−47) to that of a conserved hypothetical protein with unknown function in L. johnsonii NCC533 (accession number NP_965581.1). The amino acid sequence of ORF4 (597 bp) is 48% identical (E = 6 × 10−41) to RuvA, the Holliday junction DNA helicase of L. gasseri (ZP_00046063.1). Interestingly, ORF1 (yxjH, 1,137 bp) upstream of luxS encodes a protein that has strong similarity to the cobalamin-independent methionine synthase II of other lactic acid bacteria such as L. casei, L. plantarum, and L. gasseri, with E values ranging between 10−123 and 10−102. yxjH encodes a protein homologous to the C-terminal part of the cobalamin-independent methionine synthase MetE (44). Orthologues of this gene were identified in various gram-positive bacteria, several proteobacteria, and archaea (44).
Transcriptional organization of the putative yxjH-luxS operon.
We next examined the transcriptional organization of the luxSLGG gene. It has been previously suggested, based on in silico predictions (44), that yxjH and luxS are located in one operon in the closely related strain L. casei. In L. rhamnosus GG, the 136-bp intergenic region between yxjH and luxS contains no obvious rho-independent transcriptional terminator. However, Northern blot analysis revealed that luxS is transcribed mainly as a 0.5-kb monocistronic mRNA from promoter P2 in L. rhamnosus GG (Fig. 2B). Additionally, a second larger transcript (ca. 2.5 kb) also hybridized with the luxS-specific probe. Considering the genetic organization of the luxSLGG locus, it seems likely that the larger transcript corresponds to a bicistronic yxjH-luxS mRNA, transcribed from a second promoter, P1 (Fig. 2B). This was confirmed by reverse transcription-PCR (data not shown). Additionally, a yxjH-specific probe was used to ascertain that the ca. 2.5-kb signal did not originate from unspecific binding of the luxS probe to an unrelated transcript (data not shown). Under the growth conditions tested, i.e., growth in MRS and AOAC media, the bicistronic yxjH-luxS transcript seemed to be considerably less abundant than the monocistronic luxS transcript (data not shown).
In silico detection of a methionine-specific T-box regulatory sequence upstream of the putative yxjH-luxS operon.
Genes involved in methionine and SAM metabolism are known to be tightly regulated. In gram-positive bacteria, cis-regulatory elements and riboswitches that sense metabolites of the activated methyl cycle are implicated (44). To detect possible cis-regulatory sequences in the promoter and 5′ untranslated regions (5′ UTRs) of yxjH and luxS, we first made an in silico analysis of putative promoters (see Materials and Methods). A putative P2 luxS promoter, 5′-TTCACA-N17-TATGAT-3′, was determined, which matches the Lactobacillus consensus sequence (39). A DNA fragment containing the luxS gene with the P2 promoter was sufficient for complementation of the L. rhamnosus luxS mutants for AI-2 production as shown below (Fig. 3C). A second promoter region, 5′-TTGACA-N16-TATGAT-3′ (P1), was detected upstream of yxjH. Cotranscription of yxjH and luxS from P1 results in a long 5′ UTR leader sequence. Subsequently, we scanned the putative promoters and 5′ UTR of yxjH and luxS for known regulatory motifs (20, 44). Based on the sequences preceding yxjH in closely related species of L. rhamnosus GG (44), the presence of a methionine-dependent T box upstream of the yxjHLGG gene was detected. An alignment of the methionine-specific T-box sequences of different lactic acid bacteria is shown in Fig. 2C. Methionine-specific T boxes function through a mechanism known as transcriptional attenuation (24).
FIG. 3.
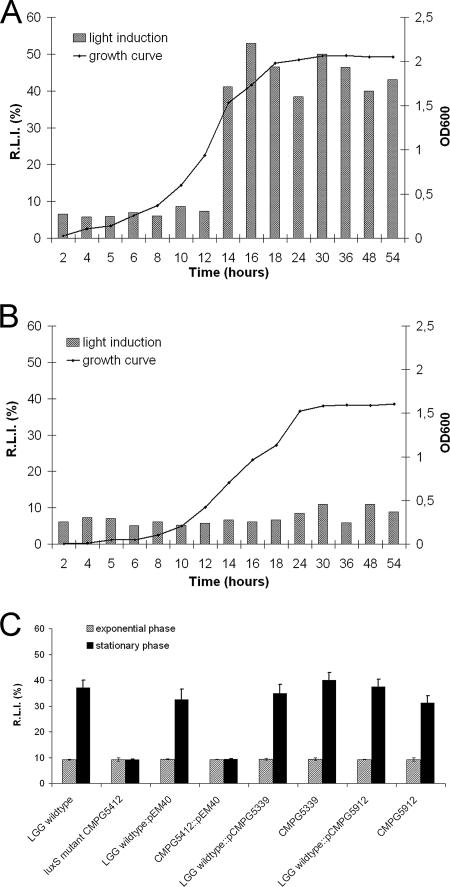
Detection of AI-2 activity in cell-free supernatant of L. rhamnosus GG strains by induction of V. harveyi BB170 luminescence. (A) Time course analysis of AI-2 activity in cell-free culture supernatant of wild-type L. rhamnosus GG (LGG) grown in modified MRS medium. For the detection of AI-2 activity, aliquots were removed for optical density measurements (line) and determinations of the capacity of conditioned medium to induce luminescence in V. harveyi BB170 at different time points (bars). The luminescence data are represented as relative light induction (R.L.I.) in percentages of induction by the positive control. The background level of luminescence was around 8% ± 2%. Time axes are not drawn to scale. (B) Lack of AI-2 activity in cell-free supernatant of luxS mutant CMPG5412. AI-2 activity was assessed as described above for the wild type. (C) Complementation of AI-2 production in the luxS mutant CMPG5412 with plasmid pCMPG5339 (P2-luxS) and pCMPG5912 (P1-yxjH-luxS). Strains with the empty cloning vector pEM40 served as a negative control. Conditioned medium was prepared from the exponential (OD600 of 0.3 to 0.4) and stationary (OD600 of 1.8 to 2) growth phases.
The luxS gene is required for AI-2 production by L. rhamnosus GG.
To perform a functional analysis, the luxSLGG gene was disrupted by the insertion of a tetracycline resistance marker gene, tet(M), from L. plantarum MD5057 (10). The correct insertion of the tet(M) cassette in the luxSLGG gene was confirmed by PCR and Southern hybridization (data not shown). A luxS mutant strain of L. rhamnosus GG was selected for further functional analysis and designated CMPG5412 (see below). Wild-type AI-2 activity was shown to be maximal in late exponential phase and remained high in late stationary phase (Fig. 3A), as previously reported (14). No significant AI-2 activity was detected in conditioned medium of luxS mutant strain CMPG5412 (Fig. 3B). The AI-2 production defect of the luxS mutant could be restored by the introduction of the luxS gene under the control of its putative promoter P2 (CMPG5339) or by introducing the yxjH-luxS operon with its putative promoter P1 (CMPG5912) (Fig. 3C). These data indicate that the luxS gene is required for the production of AI-2-like molecules in L. rhamnosus GG that can be detected by the V. harveyi reporter. Additionally, increasing the luxS copy number in wild-type L. rhamnosus GG by the introduction of plasmid pCMPG5339 did not result in increased AI-2 production (Fig. 3C). This is in agreement with the idea that AI-2 production depends mainly on substrate availability, i.e., the metabolic flux through the activated methyl cycle, as previously reported for S. enterica serovar Typhimurium (5).
luxS is required for optimal growth of L. rhamnosus GG.
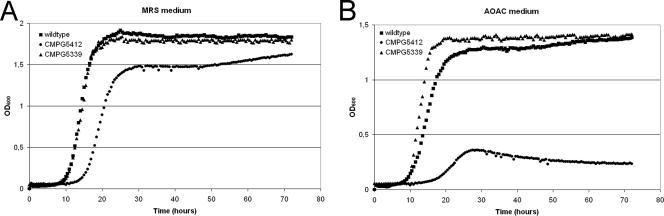
We detected a small growth defect of luxS mutant strain CMPG5412 of L. rhamnosus GG when grown in the complex MRS medium. An increased lag phase, a slightly reduced growth rate, and a lower density in stationary phase were observed (Fig. 4A). The luxS cells also showed a tendency to clump in MRS medium and resulted in slightly smaller colonies on MRS plates (data not shown). Additionally, growth in another complex medium, i.e., Lactobacilli AOAC medium, was assessed. Here, luxS mutant strain CMPG5412 showed a more dramatic growth defect (Fig. 4B). This growth phenotype in both media tested could be recovered by complementation with the functional luxS gene driven by its own promoter, P2 (CMPG5339) (Fig. 4), and with P1-yxjH-luxS (CMPG5912) (data not shown), indicating that the luxS gene is required for optimal growth of L. rhamnosus GG.
FIG. 4.
Role of luxS in optimal growth of L. rhamnosus GG. Growth of wild-type L. rhamnosus GG (square), its isogenic luxS mutant strain CMPG5412 (closed circle), and the complemented strains CMPG5339 (triangle) and CMPG5912 (data not shown) was assessed by measuring the OD600 every half hour for three consecutive days in MRS medium (A) and Lactobacilli AOAC medium (B). The complemented strain CMPG5912 showed the same growth characteristics as strain CMPG5339 (data not shown).
The growth defect of the L. rhamnosus GG luxS mutant can be nutritionally complemented.
To investigate the nature of the growth defect of luxS mutant strain CMPG5412 in Lactobacilli AOAC medium, different extracellular complementation experiments were performed (Table 3). Different components were added to the culture medium, and growth of the wild type and luxS mutant was assessed. The addition of these compounds did not significantly affect the growth of wild-type L. rhamnosus GG (Table 3). Since the luxS mutant does not produce AI-2, we first added chemically synthesized DPD in different concentrations, but no recovery of growth to the wild-type level was observed. To investigate the metabolic nature of the growth defect, we tried to complement growth in AOAC medium with components directly involved in the activated methyl cycle (Fig. 1), i.e., methionine, SAM, SAH, and cysteine. The exogenous addition of SAM could not complement growth, probably because L. rhamnosus GG cells lack a transport system for this molecule, as shown for E. coli (26). Significant increases in the growth rate and the final optical density in stationary phase were observed after the addition of cysteine, but still, the wild-type level was not reached (Table 3). Additionally, we investigated the toxic effect of SAH after exogenous addition (1 mg/ml) to the growth medium, but no influence on the growth of the wild type and the luxS mutant was observed (data not shown), probably because this intermediate, as hypothesized for SAM, cannot permeate L. rhamnosus GG cells. We therefore cannot exclude that the intracellular accumulation of SAH is an important cause of the observed growth defect.
TABLE 3.
Complex nutritional requirements of the L. rhamnosus GG luxS mutant CMPG5412 in Lactobacilli AOAC medium
| Component added | Concna | μmaxb
|
OD600 at 72 h
|
||
|---|---|---|---|---|---|
| Wild type (% of no addition)c | CMPG5413 (% of wild type)d | Wild type (% of no addition)c | CMPG5413e (% of wild type)d | ||
| None | 100f | 30.3 ± 5.9 | 100f | 33.8 ± 7.8 | |
| DPD | 0.5 mM | 96.8 ± 1.0 | 36.6 ± 9.8 | 95.6 ± 2.5 | 47.6 ± 9.7 |
| Methionine | 100 μM | 98.3 ± 1.2 | 28.5 ± 3.9 | 96.3 ± 0.1 | 27.3 ± 6.8 |
| SAM | 1 mM | 86.6 ± 4.6 | 37.7 ± 12.7 | 88.3 ± 5.2 | 42.9 ± 6.3 |
| Cysteine-HCl | 2 mM | 102.3 ± 2.6 | 72.9 ± 9.0 | 102.6 ± 0.1 | 83.5 ± 9.2 |
| Vitamins and cysteine | —g | 101.5 ± 2.9 | 83.2 ± 3.5 | 102.8 ± 2.8 | 90.5 ± 6.9 |
| Folic acid | 20 nM | 106.6 ± 1.9 | 55.9 ± 13.0 | 99.4 ± 0.1 | 60.4 ± 1.0 |
| Cyanocobalamin | 7 nM | 103.6 ± 1.6 | 33.1 ± 7.2 | 101.9 ± 0.9 | 35.5 ± 4.7 |
| d-(+)-biotin | 40 μM | 99.6 ± 0.6 | 61.1 ± 4.8 | 96.3 ± 0.8 | 61.8 ± 16.8 |
| Ca-d-(+)-pantothenate | 20 μM | 111.3 ± 3.0 | 74.2 ± 5.8 | 106.6 ± 0.2 | 77.6 ± 10.9 |
| Pyridoxol-HCl | 5 μM | 100.6 ± 0.6 | 37.4 ± 3.2 | 98.9 ± 0.1 | 43.9 ± 9.6 |
Concentration that resulted in the best complementation of growth in Lactobacilli AOAC medium.
Maximal growth rate, μmax, where μ equals ln2/g, g equals [(t2 − t1) log2]/(log OD2 − log OD1), and g equals the doubling time (hours).
Growth of wild-type L. rhamnosus GG in Lactobacilli AOAC medium.
Growth of the luxS mutant was compared to growth of wild-type L. rhamnosus GG under the same conditions.
A large lag phase was still observed when partial complementation could be achieved.
One hundred percent refers to the growth characteristics of wild-type L. rhamnosus GG in Lactobacilli AOAC medium without extra components added.
The vitamin solution contained folic acid (0.01 μg/ml), biotin (10 μg/ml), nicotinic acid (0.1 μg/ml), pantothenic acid (10 μg/ml), riboflavin (0.1 μg/ml), pyridoxol (1 μg/ml), and cyanocobalamin (0.01 μg/ml).
Since a number of metabolic pathways coupled to the activated methyl cycle could be affected by a luxS mutation (Fig. 1), we also tested the effects of adding the amino acids aspartic acid, threonine, and isoleucine and the polyamines spermine and spermidine. No significant increase in the growth rate was observed (data not shown). Finally, we also investigated the addition of a mixture of vitamins and cofactors in combination with cysteine and could observe growth to near-wild-type levels (Table 3). When we tested the vitamins individually, the addition of folate, biotin, and especially pantothenic acid (the precursor of acetyl coenzyme A) could partly complement the growth defect (Table 3). These nutritional complementation experiments suggest that a luxS mutation has pleiotropic effects and results in complex metabolic requirements for L. rhamnosus GG.
Occurrence of putative secondary site mutations in luxS mutants of L. rhamnosus GG.
The importance of the luxS gene for optimal growth and fitness of L. rhamnosus GG was further supported by the following observations. We sporadically noticed that in some of the many inoculation experiments performed, the growth capacity of luxS mutant strain CMPG5412 in AOAC medium was restored to near-wild-type levels after prolonged incubation (circa 5 days). When an inoculum of such 5-day-old cultures was taken for starting new cultures, wild-type growth characteristics were obtained. Moreover, among the luxS mutants initially isolated after electroporation (see Materials and Methods), a particular mutant, CMPG5413, showed the same growth capacity as wild-type L. rhamnosus GG in MRS and AOAC media (data not shown). This mutant, CMPG5413, showed the correct insertion of the tet(M) cassette in the luxSLGG gene (data not shown). Additionally, no AI-2 activity was detected in conditioned medium (data not shown). The AI-2 production defect of strain CMPG5413 could also be restored by the introduction of the luxS gene on the integrating plasmids pCMPG5339 and pCMPG5912 (data not shown). We speculate that in the above-described cases of prolonged incubation and in strain CMPG5413, suppressor secondary site mutations must have accumulated. Therefore, we calculated the percentage of putative suppressor mutants that could occur while growing strain CMPG5412 in AOAC medium during prolonged incubation. After 5 days, 22% showed a reversion to wild-type growth, while AI-2 production was not restored. In the more nutrient-rich MRS medium, none of the 200 inoculated colonies tested showed a reversion to wild-type growth.
luxS mutation influences monospecies biofilm formation of L. rhamnosus GG.
The role of luxS in the adhesion of L. rhamnosus GG to surfaces was investigated as an example of multicellular behavior possibly regulated by quorum sensing. We observed that L. rhamnosus GG is able to form biofilms on polystyrene surfaces after 3 days under different conditions (S. Lebeer et al., unpublished data). luxS mutant strain CMPG5412 had a diminished capacity to form biofilms in AOAC medium. This phenotype could be complemented with a functional luxS gene, indicating that LuxS is required for monospecies in vitro biofilm formation of L. rhamnosus GG under these conditions (Fig. 5A).
FIG. 5.
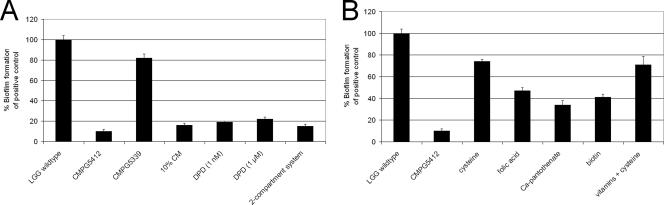
Role of AI-2 and luxS in monospecies biofilm formation by L. rhamnosus GG. (A) Biofilm formation capacities of wild-type L. rhamnosus GG (LGG) and luxS mutant strain CMPG5412 were compared. Biofilm formation of wild-type L. rhamnosus GG in AOAC medium served as a positive control in all the experiments and was set to 100%. The error bars represent standard deviations. Intracellular genetic complementation of biofilm formation of the luxS mutant could be achieved with the constructs pCMPG5339 (P2-luxS) (shown) and pCMPG5912 (P1-yxjH-luxS). Additionally, the role of AI-2 in the monospecies biofilm of L. rhamnosus GG was investigated using different extracellular complementation experiments to recover biofilm growth of luxS mutant strain CMPG5412, namely, 10% conditioned medium (CM), chemically synthesized DPD in different concentrations, and coculture with the wild type in a two-compartment system (0.22 μm). As a control, these conditions were also tested for wild-type L. rhamnosus GG, but no influence on biofilm formation was observed (data not shown). (B) Nutritional complementation experiments to investigate the metabolic cause of the defective biofilm formation of L. rhamnosus GG luxS mutant strain CMPG5412. SAM (1 mM) (data not shown), methionine (100 μM) (data not shown), cysteine (2 mM), folic acid (0.01 μg/ml), biotin (5 μg/ml), and pantothenic acid (10 μg/ml) were added to AOAC medium, and biofilm formation of wild-type L. rhamnosus GG (control) (not shown) and luxS mutant strain CMPG5412 (shown) was assessed. Additionally, the addition of the vitamin mix combined with cysteine (as described in Table 3) was investigated.
To investigate whether the biofilm formation defect of L. rhamnosus GG luxS mutant strain CMPG5412 is due to the absence of the AI-2 signaling molecule, different extracellular complementation experiments were conducted. To uncouple the roles of LuxS in the synthesis of AI-2 on one side and the recycling of homocysteine on the other side, complementation assays were performed with AI-2 using different sources and concentrations. First, AI-2 molecules present in the supernatant of a wild-type culture were added to the growth medium as 10% conditioned medium (Fig. 5A) but could not complement the biofilm formation of luxS mutant strain CMPG5412 (Fig. 5A). Still, conditioned medium contains many unknown compounds that could both positively and negatively affect biofilm formation. Therefore, we subsequently tried to complement biofilm formation with chemically synthesized DPD in different concentrations (ranging from 1 nM to 100 μM). We were unable to rescue biofilm formation (Fig. 5A). In an attempt to mimic the dynamics of AI-2 accumulation that are possibly necessary for the different phases of biofilm development, extracellular complementation experiments were also conducted in two-compartment systems in which the wild type and mutant were separated by a 0.22-μm membrane. Restoration of biofilm formation by diffusion of wild-type factors to the luxS mutant could not be observed in this coculture system (Fig. 5A).
Therefore, the biofilm defect was next examined in relation to the central metabolic role of LuxS. First, components directly involved in the activated methyl cycle were investigated. A significant increase in biofilm formation was observed after the addition of cysteine, but wild-type levels were not reached (Fig. 5B). The vitamins folic acid, pantothenic acid, and biotin could also induce biofilm formation to some extent (Fig. 5B), further confirming the complex metabolic defect of the luxS mutant. None of the above-mentioned compounds had a significant effect on biofilm formation by wild-type L. rhamnosus GG (data not shown).
Interestingly, luxS mutant strain CMPG5413, introduced above, was also impaired in biofilm formation. However, in contrast to strain CMPG5412, biofilm formation of CMPG5413 could not be complemented, either nutritionally or genetically, with a functional copy of the luxS gene. These findings further support the hypothesis that an as-yet-unidentified suppressor mutation(s) must have occurred in strain CMPG5413.
DISCUSSION
In this study, we identified the gene encoding the AI-2-producing LuxS enzyme of the probiotic bacterium L. rhamnosus GG and aimed for its functional characterization by construction and analysis of a luxS knockout mutant. The LuxS enzyme is of interest in relation to its role in AI-2-mediated quorum sensing and the regulation of multicellular behavior. However, the metabolic role of LuxS in the recycling of homocysteine after SAM-dependent methylation reactions should not be overlooked (55, 60). This dual role of LuxS indicates that the AI-2 quorum-sensing system is highly integrated in the central metabolism and physiology of bacterial cells, making analyses of phenotypes of luxS mutants more complex than initially anticipated. Here, we describe the results of different experiments that especially point to a central physiological role for the luxS gene in L. rhamnosus GG.
In L. rhamnosus GG, the luxSLGG gene is preceded by yxjH, a gene encoding a putative cobalamin-independent methionine synthase. YxjH is suggested to be an alternative methionine synthase involved mainly in the SAM recycling pathway (44), since mutation analysis using Bacillus subtilis has shown that yxjH is not required for de novo methionine synthesis (23). The gene arrangement of luxS in L. rhamnosus GG supports the view of LuxS as a primarily metabolic enzyme involved in sulfur amino acid metabolism. The same genomic organization of the luxS gene has been found in L. acidophilus (1), L. casei, and L. gasseri (44). In a considerable number of bacteria, particularly Firmicutes, luxS is often located adjacent to genes involved in either methionine recycling or, more often, de novo synthesis (44, 60). For example, Lactobacillus delbrueckii contains two luxS orthologues that are located in the operons metB-luxS1-metQ2 and luxS2-metE-metF (44).
In contrast to what has been proposed for L. casei (44), Northern blot analysis revealed that luxSLGG is transcribed mainly as a 0.5-kb monocistronic mRNA in L. rhamnosus GG. A second larger but less abundant yxjH-luxS transcript also hybridized with the luxS-specific probe, and a cis-regulatory methionine-dependent T box was found in the corresponding 5′ UTR. However, the fact that luxS is also expressed from its own, apparently constitutive, promoter P2 suggests that this gene needs to be expressed under all conditions. We did not yet search for other cis-regulatory sequences in the long 5′ UTR or upstream of the promoter of yxjH, but their presence is highly likely, since many genes of methionine metabolism are regulated by different mechanisms that act synergistically (44, 47). Clearly, the regulatory mechanisms of luxS, yxjH, and other genes of the activated methyl cycle are of great interest for future investigations.
We attempted to investigate the function of LuxS in relation to its dual role in metabolism and quorum sensing by constructing an L. rhamnosus GG luxS mutant. In this way, we could confirm that LuxS is responsible for the observed AI-2 production in L. rhamnosus GG, which we reported previously (14). This is an important finding since some lactobacilli are known to produce furanone-like molecules by other pathways that do not directly involve luxS (42). Additionally, we found that the luxS mutation causes a medium-dependent growth defect in L. rhamnosus GG, especially in AOAC medium. Genetic complementation experiments confirmed that the growth defect was indeed caused by the luxS mutation. Nutritional complementation experiments point to a complex metabolic defect caused by the luxS mutation in L. rhamnosus GG, since cysteine and the cofactors pantothenic acid, biotin, and folic acid could partly complement the growth defect. It is not yet clear why the addition of these components stimulated the growth of the L. rhamnosus GG luxS mutant. A detailed analysis of the fluxes through the activated methyl cycle is beyond the scope of this paper. However, it is important to note that the ability of lactobacilli to produce amino acids and vitamins is very scarce, and most species are auxotrophic for these compounds (11, 31). Clearly, a luxS mutation results in even more complex nutritional requirements for L. rhamnosus GG. For example, the rather high requirement of luxS mutant strain CMPG5412 for cysteine could be partly explained by an increased requirement for glutathione (Fig. 1A). The biosynthesis of glutathione, important for maintaining the cytoplasm in a reduced state and for protection against various stresses, is known to use considerable amounts of cysteine (60).
Strikingly, a luxS mutation did not affect growth in most bacterial strains published to date (55). However, some growth defects were reported. A luxS mutant of Streptococcus pyogenes showed a medium-dependent growth defect (37). Additionally, a luxS mutant of Bacillus anthracis was characterized by a longer lag phase and a significantly lower cell density in stationary phase (28). luxS mutants of Staphylococcus aureus exhibited a growth defect in a sulfur-limited defined medium (17). A luxS mutant of enterohemorrhagic Escherichia coli showed a severe growth defect in minimal medium (56). Interestingly, a pfs mutant of E. coli was reported to show a growth defect in complex medium that could be fully complemented with the vitamin biotin (7). This observation together with the results presented in this study indicate that the effects of mutations in the activated methyl cycle are not limited to sulfur amino acid metabolism but are integrated in a complex network of metabolic pathways, as comprehensively reviewed by Winzer et al. (60).
To investigate the dual role of luxS in more detail, biofilm formation was also studied, since this multicellular behavior is possibly regulated by quorum sensing (52). L. rhamnosus GG was shown to form biofilms under certain conditions (Lebeer et al., unpublished). Interestingly, luxS mutant strains CMPG5412 and CMPG5413 of L. rhamnosus GG showed an in vitro biofilm formation defect. In contrast to CMPG5413, the biofilm defect of CMPG5412 could be restored after the introduction of a functional luxS gene or by the addition of certain nutritional compounds (cysteine, biotin, folic acid, and pantothenic acid). Biofilm formation was not observed after the addition of various exogenous sources of AI-2. These findings suggest that the monospecies biofilm formation deficiency of the L. rhamnosus GG luxS mutant has an important metabolic cause and is not due merely to the growth defect. In fact, it is frequently observed that biofilm growth occurs under conditions of starvation, since a low growth rate often induces the production of an extracellular matrix (6, 34). However, this process seems to be determined by the limiting nutrients present. In the case of L. rhamnosus GG, biofilm formation is triggered by a high N-to-C ratio of the medium (Lebeer et al., unpublished). In such a nitrogen-rich environment, luxS mutant strain CMPG5412 of L. rhamnosus GG is not impaired in biofilm formation, in contrast to the situation with AOAC medium (Lebeer et al., unpublished).
Both negative and positive correlations between LuxS and biofilm formation have been reported for a considerable number of bacteria (55), and they are explained mostly in relation to quorum sensing. However, extracellular complementation experiments are crucial in supporting a role for AI-2 as a signal for biofilm formation. For example, the biofilm defect of the luxS mutant of S. enterica serovar Typhimurium could not be chemically complemented by DPD, SAM, or methionine (15). In Lactobacillus reuteri strain 100-23, the increased biofilm thickness of a luxS mutant could not be complemented with AI-2-containing conditioned medium (52). In contrast, Yoshida et al. (63) previously observed extracellular complementation of a S. mutans luxS mutant by wild-type compounds diffused through a 0.22-μm membrane. In Staphylococcus epidermis, the addition of AI-2-containing culture filtrate restored the biofilm formation of the luxS mutant to wild-type levels (62). In some of these reports, however, it was not unambiguously shown that AI-2 was the active compound in complementation, and the role of interfering medium compounds was not excluded. Only recently was a role for chemically synthesized DPD in the complementation of biofilm formation clearly shown (43). Interestingly, here, it concerned a mixed-species phenotype, i.e., the mutualistic biofilm growth of two oral commensal bacteria, Actinomyces naeslundii and Streptococcus oralis (43).
The observed discrepancy in growth and biofilm phenotypes of luxS mutants of different bacterial species highlights the need to study the role of luxS in relation to the genomic context and the metabolic pathways that are present in the different bacterial strains. The question to be answered is whether there are back-up pathways present in some bacteria to overcome the growth defect caused by the luxS mutation, aiding the detoxification of SAH and the recycling of homocysteine. For example, it would be interesting to investigate the supposed discrepancy in the metabolic capacities of two strains of H. pylori where luxS inactivation resulted in different outcomes (35). Additionally, the lost fitness of luxS mutations could be compensated for by secondary site suppressor mutations, as shown in this study. L. rhamnosus GG luxS mutant strain CMPG5413 displays wild-type growth but a defect in biofilm formation, which cannot be complemented nutritionally or genetically. These findings stress the importance of genetic complementation experiments for all phenotypes. Interestingly, it was also recently reported that second-site mutations affecting biofilm formation occurred in some luxS mutants of S. aureus (17). Therefore, experiments with luxS mutants should be carefully interpreted. Additionally, the metabolism of sulfur amino acids and SAM needs to be studied more into detail in different bacterial strains to better understand the role of the LuxS/AI-2 system.
The question remains, however, whether AI-2 serves a particular function in L. rhamnosus GG. It is important to note that we do not yet have indications that L. rhamnosus GG is able to import extracellular AI-2. As indicated in Fig. 3A, L. rhamnosus GG does not seem to take up AI-2 in stationary phase, in contrast to other species (51, 61). Additionally, Lactobacillus species do not have a LuxP homologue to import AI-2 (49), although alternative AI-2 receptors could exist (25). Mutants of the as-yet-unknown receptor of AI-2 and its downstream signaling pathway are interesting for investigating whether L. rhamnosus GG can detect AI-2 to regulate certain phenotypes and/or whether L. rhamnosus GG only produces AI-2 to thwart AI-2-regulated processes in other bacteria. Additionally, it is still plausible that AI-2 serves an important intracellular function.
Conclusively, in this study, we described functional analyses of the luxS gene in the probiotic bacterium L. rhamnosus GG. luxS is a gene that is widely studied in pathogenic bacteria in relation to quorum sensing and biofilm formation. Our results point especially to a central metabolic role for LuxS in L. rhamnosus GG. Regarding the importance of the activated methyl cycle, disruption of the luxS gene results in pleiotropic physiological effects in this fastidious strain. Many interdependent metabolic fluxes through this important cycle seem to be disturbed, as indicated by the complex nutritional requirements of the luxS mutant. Additionally, we have demonstrated that a luxS mutation can affect monospecies biofilm formation of L. rhamnosus GG indirectly, but not directly due to AI-2-mediated quorum sensing. In the future, different genes that define probiotic functions need to be determined for a better understanding of the probiotic-host relationship, including other putative quorum-sensing-related and signaling receptor genes. To this end, genome information and functional analyses such as the construction of knockout mutants are needed.
Acknowledgments
S.L. and S.C.J.D.K. are research assistants of the Belgian Fund for Scientific Research (FWO-Vlaanderen), aspirant and postdoctoral fellow, respectively. Additionally, this work was supported by the IWT through research projects STWW-00162 and GBOU-20160.
We gratefully acknowledge B. Bassler, M. Alvarez, and M. Danielsen for kindly providing strains and plasmids used in this study. We thank C. Varszegi for the chemical synthesis of DPD and for carefully reading the manuscript.
Footnotes
Published ahead of print on 10 November 2006.
REFERENCES
- 1.Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 102:3906-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez, M. A., M. Herrero, and J. E. Suarez. 1998. The site-specific recombination system of the Lactobacillus species bacteriophage A2 integrates in gram-positive and gram-negative bacteria. Virology 250:185-193. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 5.Beeston, A. L., and M. G. Surette. 2002. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3450-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branda, S. S., S. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 13:20-26. [DOI] [PubMed] [Google Scholar]
- 7.Cadieux, N., C. Bradbeer, E. Reeger-Schneider, W. Koster, A. K. Mohanty, M. C. Wiener, and R. J. Kadner. 2002. Identification of the periplasmic cobalamin-binding protein BtuF of Escherichia coli. J. Bacteriol. 184:706-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 9.Coessens, B., G. Thijs, S. Aerts, K. Marchal, F. De Smet, K. Engelen, P. Glenisson, Y. Moreau, J. Mathys, and B. De Moor. 2003. INCLUSive: a web portal and service registry for microarray and regulatory sequence analysis. Nucleic Acids Res. 31:3468-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danielsen, M. 2002. Characterization of the tetracycline resistance plasmid pMD5057 from Lactobacillus plantarum 5057 reveals a composite structure. Plasmid 48:98-103. [DOI] [PubMed] [Google Scholar]
- 11.Deguchi, Y., and T. Morishita. 1992. Nutritional requirements in multiple auxotrophic lactic acid bacteria: genetic lesions affecting amino acid biosynthetic pathways in Lactococcus lactis, Enterococcus faecium, and Pediococcus acidilactici. Biosci. Biotechnol. Biochem. 56:913-918. [DOI] [PubMed] [Google Scholar]
- 12.De Keersmaecker, S. C., K. Braeken, T. L. Verhoeven, V. M. Perea, S. Lebeer, J. Vanderleyden, and P. Hols. 2006. Flow cytometric testing of green fluorescent protein-tagged Lactobacillus rhamnosus GG for response to defensins. Appl. Environ. Microbiol. 72:4923-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Keersmaecker, S. C., K. Sonck, and J. Vanderleyden. 2006. Let LuxS speak up in AI-2 signaling. Trends Microbiol. 14:114-119. [DOI] [PubMed] [Google Scholar]
- 14.De Keersmaecker, S. C., and J. Vanderleyden. 2003. Constraints on detection of autoinducer-2 (AI-2) signalling molecules using Vibrio harveyi as a reporter. Microbiology 149:1953-1956. [DOI] [PubMed] [Google Scholar]
- 15.De Keersmaecker, S. C., C. Varszegi, N. van Boxel, L. W. Habel, K. Metzger, R. Daniels, K. Marchal, D. De Vos, and J. Vanderleyden. 2005. Chemical synthesis of (S)-4,5-dihydroxy-2,3-pentanedione, a bacterial signal molecule precursor, and validation of its activity in Salmonella typhimurium. J. Biol. Chem. 280:19563-19568. [DOI] [PubMed] [Google Scholar]
- 16.de Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 17.Doherty, N., M. T. Holden, S. N. Qazi, P. Williams, and K. Winzer. 2006. Functional analysis of luxS in Staphylococcus aureus reveals a role in metabolism but not quorum sensing. J. Bacteriol. 188:2885-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doron, S., D. R. Snydman, and S. L. Gorbach. 2005. Lactobacillus GG: bacteriology and clinical applications. Gastroenterol. Clin. N. Am. 34:483-498. [DOI] [PubMed] [Google Scholar]
- 19.FAO/WHO. 2001. Evaluation of health and nutritional properties of powder milk and live lactic acid bacteria, In Food and Agriculture Organization of the United Nations and World Health Organization expert consultation report. FAO, Rome, Italy.
- 20.Fuchs, R. T., F. J. Grundy, and T. M. Henkin. 2006. The SMK box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat. Struct. Mol. Biol. 13:226-233. [DOI] [PubMed] [Google Scholar]
- 21.Greenberg, E. P., J. W. Hastings, and S. Ulitzer. 1979. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch. Microbiol. 120:87-89. [Google Scholar]
- 22.Grundy, F. J., and T. M. Henkin. 1998. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 30:737-749. [DOI] [PubMed] [Google Scholar]
- 23.Grundy, F. J., and T. M. Henkin. 2002. Synthesis of serine, glycine, cysteine, and methionine, p. 245-254. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its relatives: from genes to cells. American Society for Microbiology Washington, DC.
- 24.Grundy, F. J., and T. M. Henkin. 2003. The T box and S box transcription termination control systems. Front. Biosci. 8:20-31. [DOI] [PubMed] [Google Scholar]
- 25.Herzberg, M., I. K. Kaye, W. Peti, and T. K. Wood. 2006. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J. Bacteriol. 188:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holloway, C. T., R. C. Greene, and C. H. Su. 1970. Regulation of S-adenosylmethionine synthetase in Escherichia coli. J. Bacteriol. 104:734-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Igarashi, K., and K. Kashiwagi. 2000. Polyamines: mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun. 271:559-564. [DOI] [PubMed] [Google Scholar]
- 28.Jones, M. B., and M. J. Blaser. 2003. Detection of a luxS-signaling molecule in Bacillus anthracis. Infect. Immun. 71:3914-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Josson, K., T. Scheirlinck, F. Michiels, C. Platteeuw, P. Stanssens, H. Joos, P. Dhaese, M. Zabeau, and J. Mahillon. 1989. Characterization of a gram-positive broad-host-range plasmid isolated from Lactobacillus hilgardii. Plasmid 21:9-20. [DOI] [PubMed] [Google Scholar]
- 30.Kalliomaki, M., S. Salminen, H. Arvilommi, P. Kero, P. Koskinen, and E. Isolauri. 2001. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet 357:1076-1079. [DOI] [PubMed] [Google Scholar]
- 31.Kandler, O., and N. Weiss. 1986. Genus Lactobacillus, p. 1063-1065. In P. H. A. Sneath, N. S. Mair, M. S. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. Williams & Wilkins Co., Baltimore, MD.
- 32.Kaper, J. B., and V. Sperandio. 2005. Bacterial cell-to-cell signaling in the gastrointestinal tract. Infect. Immun. 73:3197-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolter, R., and E. P. Greenberg. 2006. Microbial sciences: the superficial life of microbes. Nature 441:300-302. [DOI] [PubMed] [Google Scholar]
- 35.Lee, W. K., K. Ogura, J. T. Loh, T. L. Cover, and D. E. Berg. 2006. Quantitative effect of luxS gene inactivation on the fitness of Helicobacter pylori. Appl. Environ. Microbiol. 72:6615-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loenen, W. A. 2006. S-Adenosylmethionine: jack of all trades and master of everything? Biochem. Soc. Trans. 34:330-333. [DOI] [PubMed] [Google Scholar]
- 37.Lyon, W. R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparon. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 38.Marchal, K., S. De Keersmaecker, P. Monsieurs, N. van Boxel, K. Lemmens, G. Thijs, J. Vanderleyden, and B. De Moor. 2004. In silico identification and experimental validation of PmrAB targets in Salmonella typhimurium by regulatory motif detection. Genome Biol. 5:R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCracken, A., M. S. Turner, P. Giffard, L. M. Hafner, and P. Timms. 2000. Analysis of promoter sequences from Lactobacillus and Lactococcus and their activity in several Lactobacillus species. Arch. Microbiol. 173:383-389. [DOI] [PubMed] [Google Scholar]
- 40.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 41.Miller, S. T., K. B. Xavier, S. R. Campagna, M. E. Taga, M. F. Semmelhack, B. L. Bassler, and F. M. Hughson. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15:677-687. [DOI] [PubMed] [Google Scholar]
- 42.Ndagijimana, M., M. Vallicelli, P. S. Cocconcelli, F. Cappa, F. Patrignani, R. Lanciotti, and M. E. Guerzoni. 2006. Two 2[5H]-furanones as possible signaling molecules in Lactobacillus helveticus. Appl. Environ. Microbiol. 72:6053-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rickard, A. H., R. J. Palmer, Jr., D. S. Blehert, S. R. Campagna, M. F. Semmelhack, P. G. Egland, B. L. Bassler, and P. E. Kolenbrander. 2006. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 60:1446-1456. [DOI] [PubMed] [Google Scholar]
- 44.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2004. Comparative genomics of the methionine metabolism in gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res. 32:3340-3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 47.Sekowska, A., H. F. Kung, and A. Danchin. 2000. Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J. Mol. Microbiol. Biotechnol. 2:145-177. [PubMed] [Google Scholar]
- 48.Sherwood, L., and M. D. Gorbach. 1996. The discovery of Lactobacillus GG. Nutr. Today 31:2S-4S. [Google Scholar]
- 49.Sun, J., R. Daniel, I. Wagner-Dobler, and A. P. Zeng. 2004. Is autoinducer-2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways. BMC Evol. Biol. 4:36-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 52.Tannock, G. W., S. Ghazally, J. Walter, D. Loach, H. Brooks, G. Cook, M. Surette, C. Simmers, P. Bremer, F. Dal Bello, and C. Hertel. 2005. Ecological behavior of Lactobacillus reuteri 100-23 is affected by mutation of the luxS gene. Appl. Environ. Microbiol. 71:8419-8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teusink, B., F. H. van Enckevort, C. Francke, A. Wiersma, A. Wegkamp, E. J. Smid, and R. J. Siezen. 2005. In silico reconstruction of the metabolic pathways of Lactobacillus plantarum: comparing predictions of nutrient requirements with those from growth experiments. Appl. Environ. Microbiol. 71:7253-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vendeville, A., K. Winzer, K. Heurlier, C. M. Tang, and K. R. Hardie. 2005. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3:383-396. [DOI] [PubMed] [Google Scholar]
- 56.Walters, M., M. P. Sircili, and V. Sperandio. 2006. AI-3 synthesis is not dependent on luxS in Escherichia coli. J. Bacteriol. 188:5668-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319-346. [DOI] [PubMed] [Google Scholar]
- 58.Wei, Y., and E. B. Newman. 2002. Studies on the role of the metK gene product of Escherichia coli K-12. Mol. Microbiol. 43:1651-1656. [DOI] [PubMed] [Google Scholar]
- 59.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. Kirke, M. T. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]
- 60.Winzer, K., K. R. Hardie, and P. Williams. 2003. LuxS and autoinducer-2: their contribution to quorum sensing and metabolism in bacteria. Adv. Appl. Microbiol. 53:291-396. [DOI] [PubMed] [Google Scholar]
- 61.Xavier, K. B., and B. L. Bassler. 2005. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187:238-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu, L., H. Li, C. Vuong, V. Vadyvaloo, J. Wang, Y. Yao, M. Otto, and Q. Gao. 2006. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect. Immun. 74:488-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshida, A., T. Ansai, T. Takehara, and H. K. Kuramitsu. 2005. LuxS-based signaling affects Streptococcus mutans biofilm formation. Appl. Environ. Microbiol. 71:2372-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]