Abstract
Genetic recombinants that resulted from lateral gene transfer (LGT) have been detected in sexually transmitted disease isolates of Chlamydia trachomatis, but a mechanism for LGT in C. trachomatis has not been described. We describe here a system that readily detects C. trachomatis LGT in vitro and that may facilitate discovery of its mechanisms. Host cells were simultaneously infected in the absence of antibiotics with an ofloxacin-resistant mutant and a second mutant that was resistant to lincomycin, trimethoprim, or rifampin. Selection for doubly resistant C. trachomatis isolates in the progeny detected apparent recombinant frequencies of 10−4 to 10−3, ∼104 times more frequent than doubly resistant spontaneous mutants in progeny from uniparental control infections. Polyclonal doubly resistant populations and clones isolated from them in the absence of antibiotics had the specific resistance-conferring mutations present in the parental mutants; absence of the corresponding normal nucleotides indicated that they had been replaced by homologous recombination. These results eliminate spontaneous mutation, between-strain complementation, and heterotypic resistance as general explanations of multiply resistant C. trachomatis that originated in mixed infections in our experiments and demonstrate genetic stability of the recombinants. The kind of LGT we observed might be useful for creating new strains for functional studies by creating new alleles or combinations of alleles of polymorphic loci and might also disseminate antibiotic resistance genes in vivo. The apparent absence of phages and conjugative plasmids in C. trachomatis suggests that the LGT may have occurred by means of natural DNA transformation. Therefore, the experimental system may have implications for genetically altering C. trachomatis by means of DNA transfer.
Chlamydia trachomatis, the biology of which has been reviewed comprehensively (40), is the leading worldwide cause of sexually transmitted infection and, mostly in the developing world, of preventable blindness. The organism lacks many of the genes needed for extracellular existence, and its intracellular development has impeded its genetic manipulation in the laboratory. This impediment has hampered progress in managing the huge public health problem that comprises millions of C. trachomatis infections, many of which result in serious pathologies, such as pelvic inflammatory disease, ectopic pregnancy, infertility, and blindness. While many C. trachomatis infections appear to be cleared by treatment with antibiotics, follow-up screening reveals a substantial fraction of previously treated subjects who are infected because of reinfection, treatment failure, or the relative difficulty of eliminating latent infections (47). Efforts to develop protective vaccines for humans have failed (reviewed in reference 5).
Gene mutation, intrachromosomal genetic exchange, and lateral gene transfer (LGT) are means by which pathogens can produce genetic variants that have improved reproductive fitness and ability to evade host defenses. A role for mutation in evasion of protective immunity by C. trachomatis is suggested by the high frequencies of nonsynonymous nucleotide substitutions in the polymorphic ompA gene that encodes the major outer membrane protein (MOMP), which is thought to be an important target of antibody and cell-mediated immunity (4, 8, 9, 20, 22, 28, 33). Some MOMP variants in urogenital tract isolates could have resulted from intragenic recombinations that joined segments of ompA genes derived from two or three MOMP alleles (4, 8, 9, 15, 19, 23, 29, 31, 49). These sequence variations disrupted or created new combinations of epitopes in ways that might affect immune recognition. Thirty-five such mosaic MOMP alleles were found among a total of 1,950 sexually transmitted disease (STD) isolates, with an overall frequency of 1.8 percent. In addition, intergenic recombinations between the polymorphic ompA and pmpC loci were found among urogenital tract isolates (16) and 18 C. trachomatis reference strains (17).
The observations summarized above suggest that LGT-mediated recombination has occurred repeatedly in the history of C. trachomatis, but the mechanism and frequency of LGT events that generated the current high frequency of in vivo recombinants are not known. Some currently observed recombinants appear to be relics of LGT that occurred in the distant past (17), but generation of recombinants may be ongoing by unknown mechanisms of gene transfer in C. trachomatis. Laboratory studies that yield basic information about LGT in vitro might contribute to our understanding of the origins of recombinants in vivo and of their possible roles in the pathology and epidemiology of C. trachomatis. This report describes the first such in vitro genetic study of which we are aware.
C. trachomatis has a 1.04-Mb chromosome (Fig. 1) and a 7.5-kb, low-copy-number plasmid, pCT (34). The DNA is highly condensed in the infectious form, the elementary body (EB), which is metabolically inactive. Attachment of the EB to a host cell triggers its internalization into a membrane-bound vesicle in which it is subsequently converted into an enlarged reticulate body (RB). The DNA decondenses, general metabolism commences, and replication ensues. The vesicle becomes a gradually enlarging inclusion, in which the RBs multiply by binary fission. Starting at 20 to 24 h postinfection (p.i.), some RBs are converted into EBs, while other RBs continue to multiply. After 48 to 72 h p.i., depending upon the strains and conditions, the greatly enlarged inclusion liberates several hundred to about 1,000 EBs that can infect other cells and that can be assayed as inclusion-forming units (IFU) on monolayers of host cells in vitro.
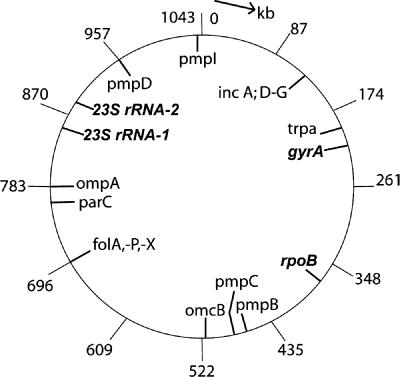
FIG. 1.
The 1.04-Mb C. trachomatis chromosome, showing the locations of mutant genes (32) (in bold and italics) that we used to isolate LGT recombinants. Also shown are loci that have relatively numerous between-strain polymorphic differences that could be used to analyze the sizes and possible directionality of chromosome segments that are transferred by LGT. The parC locus is shown because mutations in it can confer resistance to ofloxacin; the potential significance of this is mentioned in the Discussion.
The system described below was adapted to the constraints imposed by the C. trachomatis life cycle and provides a way to study LGT in vitro. Throughout, the complete nucleotide sequence of the C. trachomatis genome (41) (www.stdgen.lanl.gov/) was an invaluable resource in conducting our genetic experiments. The system has already revealed high-frequency LGT involving genes distributed over at least half of the C. trachomatis chromosome. Further studies may discover LGT mechanisms that generate recombinants and that may disseminate antibiotic resistance genes in vivo. The apparent absence of phages and conjugative plasmids in C. trachomatis combined with BLAST analysis of genes involved in DNA transformation suggests that LGT in C. trachomatis may occur by means of natural DNA transformation and may yield clues to making planned genetic modifications by means of DNA transfer.
MATERIALS AND METHODS
C. trachomatis strains and their cultivation.
C. trachomatis serovar L1 (a gift from G. Byrne) was propagated in HeLa 229 cells (American Type Culture Collection, Gaithersburg, MD) in antibiotic-free growth medium (GM) consisting of Dulbecco's modified Eagle's medium (89 vol) (Invitrogen catalog no. 11965-092 or equivalent), fetal bovine serum (10 vol) (HyClone catalog no. SH30070.03), Eagle's 100× nonessential amino acids (1 vol) (Invitrogen catalog no. 11140-050), and cycloheximide (1 μg per ml). Unless noted otherwise, infections were performed in 12.5-cm2 flasks (Falcon no. 353107) containing ∼1.5 × 106 cells.
Infections of HeLa flasks were performed by draining the medium from the flasks, rinsing them once with 2 ml of Ca-free phosphate-buffered saline (PBS) (Invitrogen catalog no. 70011-044), and inoculating them with 1.0 ml of infection medium (IM) containing the desired number of IFU. IM consisted of Dulbecco's modified Eagle's medium supplemented with 1× nonessential amino acids and cycloheximide (1.0 μg per ml). The flasks were placed on a Bellco rocker platform (Bellco Biotechnology, Vineland, NJ) at two excursions per minute for 60 min at room temperature and then rested for 60 min prior to addition of 4 ml of IM supplemented with 13 percent serum and 1.0 μg per ml of cycloheximide in order to constitute GM. C. trachomatis development occurred in a CO2 incubator at 37°C.
Harvesting EBs.
C. trachomatis stocks were prepared by inoculation of HeLa flasks with a multiplicity of infection (MOI) of 5 to 10 IFU per cell in GM as described above. At about 48 h p.i., the flasks were rinsed once with 2.0 ml of PBS and frozen at −80°C. EBs were liberated from the cells by thawing the flasks at room temperature, adding 1.0 ml of IM plus ∼1 ml sterile, 3-mm glass beads (Fisher no. 11-312A) to each flask, and shaking back and forth 100 times. The resultant bead harvests (BHs) were centrifuged at 500 × g for 5 min, and the clarified supernatant fluids were aliquoted and stored at −80°C.
Late in the course of this study, we adapted hypotonic harvesting as a convenient alternative to bead harvesting for the purpose of harvesting IFU from limiting dilution clones (see below). Clone-containing wells on 48-well plates were drained and rinsed once with 500 μl of PBS. The wells received 200 μl of water and were frozen after 20 min at room temperature. After thawing at room temperature, they received 200 μl of 2× IM and were clarified by centrifugation at 500 × g for 5 min. The clonal hypotonic harvests were used to infect cells in 24-well plates prior to further expansion in flasks.
IFU assays were performed in HeLa flasks that were infected as described above with inocula intended to contain 1 × 105 to 2 × 105 IFU. At 40 to 48 h p.i., inclusions in fields of measured area were counted with an inverted phase-contrast microscope. At least 10 evenly spaced fields along each of two tracks on opposite sides of each flask were examined in order to count at least 100 inclusions. These numbers allowed calculation of the IFU titers of undiluted samples being assayed, which were ≥90% of more reliable titers obtained by limiting dilution cloning (data not shown).
Crosses were performed to detect LGT. Among the seven crosses described below, crosses no. 1 to no. 4 have the same basic format (Fig. 2). In each cross, 10-flask sets of primary (P-0) flasks containing HeLa cells were infected with one parental mutant (set A) or another (set B); these sets were controls for enumerating C. trachomatis that became resistant to both antibiotics used in the cross by means of spontaneous mutation. Ten set C P-0 HeLa flasks were infected with a mixture of both parental mutant strains at an MOI (see Table 4) that ensured infection of virtually every cell with at least one IFU of each parental strain (mixed infection). Should LGT require cooccupancy of an inclusion by C. trachomatis between which DNA is being transferred, inclusions that are separately initiated in the same cell by different kinds of parental EBs normally fuse early enough in development to permit LGT (cross no. 6 [see below]). C. trachomatis development occurred in the absence of antibiotics in P-0 flasks and was terminated by draining and freezing (D/F) the flasks at about 48 h p.i. BHs of the IFU were prepared, and rare doubly resistant C. trachomatis strains produced in P-0 flasks were amplified by using 1/10 of each P-0 BH to initiate four serial passages (P-1 through P-4) in the presence of both antibiotics being used to select for recombinants. IFU assays of sample P-0 BHs (see Table 6) indicated that the use of 1/10 of a P-0 BH to infect a P-1 flask resulted in MOIs of 20 to 47. Each P-2, P-3, and P-4 flask was infected with one-half of the BH of the preceding passage. P-4 flasks were microscopically scanned for the presence of doubly resistant inclusions. Deviations from the basic protocol are explained in the descriptions of crosses no. 5 to no. 7.
FIG. 2.
Basic format of crosses no. 1 to no. 5, as illustrated with cross no. 1 (OFXr-1 × LINr-1). (P-0) Sets of 10 primary flasks (P-0) containing HeLa cells (pale outlines) were infected with OFXr-1 (○) (set A), LINr-1 (•) (set B), or both (set C). Separately initiated inclusions in a cell fused to form a single inclusion (darkly outlined ovoid objects) that, in the case of set C, allowed both parental types to multiply in the same inclusion in the absence of antibiotics. The thin arrow points to an OFXr LINr recombinant (⊙) that originated in a set C P-0 flask. (P-1) One-tenth of each BH prepared from a P-0 flask was used to infect a first-passage (P-1) HeLa flask containing medium supplemented with OFX and LIN. Excepting rare spontaneous mutants, only OFXr LINr recombinants could multiply in set C P-1 flasks, as shown. (P-2 through P-4) One-half of each bead harvest prepared from a P-1 flask was used to infect a P-2 flask containing OFX plus LIN medium. OFXr LINr IFU present in a P-1 inclusion were liberated by the EB harvesting procedure and initiated separate doubly resistant inclusions in P-2 flasks. P-3 and P-4 flasks were infected with one-half of their antecedent BHs and contained OFX plus LIN medium. (P-4) Phase-contrast microscope images of typical ×200 fields in P-4 flasks. Crosses no. 2 to no. 4 differed from cross no. 1 only with respect to the parental mutant strains and antibiotics that were used (see text describing individual crosses).
TABLE 4.
Summary of crosses no. 1 to no. 5, in which HeLa host cells were simultaneously infected with two kinds of antibiotic-resistant mutantsa
| Cross no. | Flask set | Parental MOI
|
No. of P-0 flasks with IFUb
|
Fisher's exact P value | ||
|---|---|---|---|---|---|---|
| First mutant | Second mutant | + | − | |||
| 1 (OFXr-1 × LINr-1) | A | 5.0 | 0 | 10 | ||
| B | 5.0 | 2 | 8 | |||
| C | 5.0 | 5.0 | 10 | 0 | <10−3 | |
| D | 5.0 | 0.5 | 10 | 0 | <10−3 | |
| 2 (OFXr-2 × LINr-1) | A | 5.0 | 0 | 10 | ||
| B | 5.0 | 2 | 8 | |||
| C | 5.0 | 5.0 | 10 | 0 | <10−3 | |
| 3 (OFXr-1 × RIFr-1) | A | 6.8 | 3 | 7 | ||
| B | 5.1 | 5 | 5 | |||
| C | 5.1 | 6.8 | 10 | 0 | <10−2 | |
| 4 (OFXr-1 × TMPr-1) | A | 5.0 | 0 | 10 | ||
| B | 5.0 | 1 | 9 | |||
| C | 5.0 | 5.0 | 10 | 0 | <10−3 | |
| 5 (OFXr-1 × LINr-1) | I | 5.0 | 10−1 | 10 | 0 | NA |
| II | 5.0 | 10−2 | 10 | 0 | NA | |
| III | 5.0 | 10−3 | 10 | 0 | NA | |
| IV | 5.0 | 10−4 | 10 | 0 | NA | |
| V | 5.0 | 10−5 | 10 | 0 | NA | |
The numbers of infected P-0 flasks that yielded IFU simultaneously resistant to two selection antibiotics are shown. In crosses no. 1 to no. 4, P-0 HeLa flasks were infected with one kind of mutant (set A), the other kind of mutant (set B), or both (set C and others as specified) and were then passaged with selection as described in the text. P-4 flasks either lacked doubly-resistant inclusions (−) or had numerous doubly-resistant inclusions (+) (Fig. 3). The presence of abundant inclusions in P-4 flasks identified antecedent P-0 flasks that contained spontaneous mutants (sets A and B) or LGT recombinants (sets C and, in cross no. 1, D). Uniparental infections were not necessary in cross no. 5 and were omitted. Instead, 10-flask sets (sets I to V) of HeLa flasks were simultaneously infected with both parental strains at the MOI shown (first and second mutants refer to the mutants in the order listed for the cross in the far-left column). Each P value is the probability that the difference between uniparental and biparental infections in the numbers of flasks marked + and flasks marked − resulted from chance, alone, according to Fisher's exact test. NA, not applicable.
For crosses no. 1, 2, and 5, OFXr LINr IFU; for cross no. 3, OFXr RIFr IFU; and for cross no. 4, OFXr TMPr IFU.
TABLE 6.
Direct enumeration of antibiotic-resistant C. trachomatis recombinants produced in primary mixed-infection cellsa
| Cross no. (conditions) | Total IFU
|
Recombinant IFU
|
|||
|---|---|---|---|---|---|
| Yield per P-0 cell | Plated per P1 flask (y) | Per ×200 field | Per P1 flask (x) | Frequency (x/y) | |
| 1 (OFXr-1 × LINr-1, D/F at 48 h p.i.) | 467 | 5.8 × 106 | 124/204 fields (0.61 per field) | 2.3 × 103 | 4.0 × 10−4 |
| 3 (OFXr-1 × RIFr-1, D/F at 48 h p.i.) | 342 | 2.4 × 107 | 316/102 fields (3.1 per field) | 1.2 × 104 | 5.0 × 10−4 |
| 4 (OFXr-1 × TMPr-1, D/F at 46 h p.i.) | 405 | 1.6 × 105 | 370/110 fields (3.4 per field) | 1.4 × 103 | 8.5 × 10−4 |
| 6 (OFXr-1 × LINr-1, D/F at 22 h p.i.) | 3.9 | 7.6 × 105 | 34/108 fields (0.32 per field) | 1.2 × 103 | 1.6 × 10−3 |
| 6 (OFXr-1 × LINr-1, D/F at 44 h p.i.) | 350 | 1.5 × 107 | 290/60 fields (4.8 per field) | 1.8 × 104 | 1.2 × 10−3 |
For each cross (see text for details of individual crosses), assays of total IFU in BHs from mixed-infection P-0 flasks grown in the absence of antibiotics were performed and used to calculate the total IFU per P-0 cell at the times the P-0 flasks were D/F. The numbers of doubly resistant recombinants in the P-0 BHs were determined by plating aliquots in P-1 HeLa flasks that contained OFX plus LIN (crosses no. 1 and no. 6), OFX plus RIF (cross no. 3), or OFX plus TMP (cross no. 4). Recombinant inclusions (Fig. 3) were counted by scanning 12 to 22 ×200 phase-contrast microscope fields per each of five F12s of each cross at 48 h p.i. (crosses no. 1, no. 4, and no. 6) or at 67 h p.i. (cross no. 3). The average number of recombinant IFU per ×200x field was used to calculate the total number of recombinant IFU per P-1 flask (x) (see Materials and Methods). The total numbers of IFU per P-1 flask (y) were calculated from the total IFU titers of the BHs, and the frequencies of recombinants were expressed as x/y. Doubly-resistant inclusions were not detected in P-1 F12s derived from P-0 flasks that had been infected with just one kind of mutant.
The method of passaging.
C. trachomatis-infected cultures rather than reliance on plaque formation were used to select for mutants and recombinants because our initial experiments were exploratory and were intended merely to detect LGT at frequencies as low as 10−8 to 10−7. In order to detect plaques formed by LGT recombinants at such low frequencies, it might have been necessary to make P-1 platings (see above) of IFU from P-0 mixed-infection cells at an MOI high enough to impair the ability of host cells to support the several courses of C. trachomatis development in situ that would be required for plaque formation. Furthermore, the concentrations of oflaxacin (OFX), lincomycin (LIN), and trimethoprim (TMP) we thought appropriate for selection were still partially inhibitory toward mutant C. trachomatis and might have substantially increased the time (∼11 days [3]) needed to form visible plaques, during which time cell monolayers could deteriorate. These potential problems were circumvented by passaging IFU from cultures of mixed-infection cells in the presence of antibiotics. Four to five passages allowed us to quickly expand even single recombinant IFU present in P-0 flasks into populations of EBs sufficient for DNA isolation. Doubly resistant strains isolated in our study may now be useful for devising conditions for isolating recombinants using either plaque formation or limiting dilution.
Enumeration of recombinants and mutants.
The method of passaging sets of flasks initially provided a way to approximately enumerate antibiotic-resistant mutants and LGT recombinants that is related to a classical procedure for estimating mutation frequencies in bacteria (25). The presence of one or more resistant IFU in the sample of a P-0 bead harvest that was used to infect a P-1 flask resulted in the presence of abundant resistant inclusions in its descendant P-4 flask, but such P-4 flasks were not informative about the specific number of resistant IFU in their antecedent P-0 flasks. However, if the entire IFU yield of a P-0 flask was passaged, each P-4 flask that lacked inclusions would unambiguously identify an antecedent P-0 flask that lacked resistant IFU; the fraction of all P-4 flasks that lacked resistant IFU in a set would then equal the fraction of antecedent P-0 flasks in that set. When the average number of resistant IFU in a set of P-0 flasks was small, some flasks in the descendant set of P-4 flasks would lack resistant inclusions. The fraction of inclusion-free P-4 flasks in a set could be used as the P0 term in the Poisson distribution for approximately estimating the average number of resistant IFU in the set. This procedure was used to estimate the frequencies of spontaneous OFXr and rifampin-resistant (RIFr) mutants (see “Antibiotic-resistant mutants” in the supplemental material); its use for estimating the frequency of OFXr LINr recombinants in cross no. 5 set V flasks (see Results) required correction for the use of only 1/10 of each P-0 bead harvest to infect a P-1 flask. This correction was not required for the subsequently adopted method described in “Direct enumeration of recombinants” in Results.
Cloning by limiting dilution.
Aliquots (200 μl) of IM containing various dilutions of a preassayed EB preparation were used to infect wells containing 105 HeLa cells each on 48-well plates (Corning catalog no. 3548). After the infection process was completed, 800 μl of GM containing 13% serum and 1.3 μg of cycloheximide was added to each well and the plates were placed in a CO2 incubator. The EB dilutions were chosen to include at least one that was expected to yield an average of about 1/5 IFU per well, and two plates were used at each dilution. By 6 days postinfection, microscopic scanning (×100, non-phase contrast) detected unmistakable foci of inclusions resulting from multiple cycles of infection initiated by a single IFU in a small proportion of the wells.
DNA sequence analysis.
DNA was isolated as previously described (1), without the addition of cetyltrimethylammonium bromide-NaCl, from EBs that were harvested from one to three 12.5-cm2 flasks and collected by centrifugation at 12,000 × g for 10 min. EBs prepared from one flask usually yielded ∼10 μg of DNA.
The relevant parts of genes of interest were sequenced after PCR amplification in 50-μl reactions constituted as follows: 10 μl 5× Phusion HF buffer, 1 μl 10 mM deoxynucleoside triphosphate, 0.5 μl of Phusion polymerase (Finnzyme), 25 pmol of sense and antisense primers (Table 1), 50 ng of EB template DNA, and water. All PCRs had an initial denaturation step at 98°C for 30 s and a final extension step at 72°C for 7 min in addition to each cycle having a denaturation step at 98°C for 15 s. The following cycling conditions were used to amplify each region of interest: for gyrA, denaturation at 98°C for 10 s, annealing at 62°C for 15 s, and extension at 72°C for 20 s for 30 cycles; for the fol operon, denaturation at 98°C for 10 s, annealing at 61°C for 15 s, and extension at 72°C for 1 min 30 s for 35 cycles; for rpoB, denaturation at 98°C for 10 s, annealing at 60°C for 15 s, and extension at 72°C for 15 s for 35 cycles; and for 23S rRNA-1 and -2, denaturation at 98°C for 10 s, annealing at 60°C for 15 s, and extension at 72°C for 30 s for 35 cycles. PCR amplicons were purified using a QIAquick PCR purification kit (QIAGEN) as instructed by the manufacturer. Purified amplicons were eluted in 50 μl of EB buffer, and sequencing reactions were performed in mixtures containing the following: 2 μl of ET Terminator (GE Healthcare), 2 μl of sequencing buffer (20 mM Tris, pH 9.0, 5 mM MgCl2), 1 μl of 3.2 mM oligonucleotide (Table 2), approximately 50 to 100 ng of amplified product, and water to 20 μl. The following cycling conditions were used: denaturation at 96°C for 15 s, annealing at 50°C for 5 s, and extension at 60°C for 2 min, repeated for 30 cycles. Completed sequencing reactions were run on an ABI 3730 instrument in house.
TABLE 1.
Oligonucleotide pairs used for PCR amplification of relevant regions of genes that confer antibiotic resistance in mutant C. trachomatis
| Sense oligonucleotide (sequence) | Antisense oligonucleotide (sequence) | Region amplified |
|---|---|---|
| gyrAB F7 (TGGGATACAACAATGAATCCTG) | gyrAB RC6 (ACACGAATCCCGTCTTTGTC) | gyrB(2245) →gyrA(908) |
| CT616_929F (GGCAATTATTCGATGGGTTG) | CT611_170R (TCTGCTTTGCACATTCTTGC) | fol operon [CT616(929) → CT611(170)] |
| rpoB_F (ATGGGCGATGAGAAGACATC) | rpoB_R (TGCATGTTCGATCCCATAAG) | rpoB(1132) →rpoB(1892) |
| 23S rRNA5 (TAGGTGAAGTGCTTGCATGG) | 23S rRNA-13 (TGCTTTTGATCTTTCCAGCA) | CT610(2514) → CT611(6024) |
| 23S rRNA5 (TAGGTGAAGTGCTTGCATGG) | 23S rRNA-23 (CCGAAAATTCGATAATAAGGAGAA) | IGR617(2193) → IGR617(5623) |
TABLE 2.
Oligonucleotides used for sequencing relevant regions of genes that confer antibiotic resistance in mutant C. trachomatis
| Gene(s) sequenced | Sense oligonucleotide(s) (sequence[s]) | Antisense oligonucleotide(s) (sequence[s]) |
|---|---|---|
| gyrA | gyrAB F7 (TGGGATACAACAATGAATCCTG) | gyrAB R6 (ACACGAATCCCGTCTTTGTC) |
| folA | TMP F3 (GGGACGAAAGACTTGGGAGT) | CT611_170R (TCTGCTTTGCACATTCTTGC) |
| TMP F9 (ATTGAGCAAATGTTGCGTTG) | TMP R22 (CCGCCCATGCTTATACTTGT) | |
| folP_678_F (TTATCTACGCGTGCATCAGG) | ||
| rpoD promoter | CT616_929_F (GGCAATTATTCGATGGGTTG) | rpoD_112_R (ACGTGATGAACCCTTGATCC) |
| rpoB | rpoB_F (ATGGGCGATGAGAAGACATC) | rpoB_R (TGCATGTTCGATCCCATAAG) |
| rpoB_F1 (AACCCTGTCGCAGAATTGAC) | rpoB_R1 (TCGAACTTCAAACCCAGCTC) | |
| IGR610 and IGR617 | 23SrRNA1814F (CCCAATGCCAAAAGGTTAAA) | 23srRNA2428R (CCCCGGAGTACCTTTTATCC) |
RESULTS
Plan of the work.
In order to detect LGT, we performed crosses, in which host cells were infected with two different kinds of antibiotic-resistant mutants at once (mixed infection). We then selected for doubly resistant IFU among the progeny C. trachomatis produced in the mixed-infection cells. It was possible that growth in the presence of selection antibiotics could result from complementation between parental mutant strains or from heterotypic resistance (21, 39). Both phenomena would involve the presence of numerous antibiotic-sensitive organisms in isolates grown out in the presence of the two antibiotics used to select for recombinants in a cross. These possibilities were investigated by determining if C. trachomatis clones that were derived from doubly resistant isolates from mixed-infection cells were sensitive to either of the antibiotics used to select for recombinants. Doubly resistant C. trachomatis could also originate by means of spontaneous mutation in either of the two singly resistant parental mutants used in a cross. We distinguished between doubly resistant spontaneous mutants and LGT recombinants by isolating collections of mutants resistant to a single antibiotic and sequencing the genes in which mutations conferred antibiotic resistance on the parental mutant strains used in crosses. These surveys of spontaneous mutants showed that separately isolated mutants resistant to the same antibiotic often had diverse nucleotide substitutions, while recombinants would be expected to have only the specific resistance-conferring mutations initially present in the parental strains.
Mutants used to detect LGT (Table 3).
TABLE 3.
C. trachomatis serovar E/Bour mutant strains used to detect LGT
| Strain | Used in cross no.: | Response toa:
|
Gene and mutation | |||
|---|---|---|---|---|---|---|
| OFX | LIN | TMP | RIF | |||
| wt | s | s | s | s | wt | |
| OFXr-1 | 1, 3, 4, 5, 6, 7 | r | s | s | s | gyrA (CT189); T249→G |
| OFXr-2 | 2 | r | s | s | s | gyrA (CT189); A247→C |
| LINr-1 | 1, 2, 5, 6, 7 | s | r | s | s | 23S rRNA (IGR610, IGR617); A2039→C |
| RIFr-1 | 3, 7 | s | s | s | r | rpoB (CT315); T1383→G |
| TMPr-1 | 4 | s | s | r | s | Not known |
s, sensitive, i.e., does not form normal inclusions in the presence of the specified antibiotic; r, resistant, i.e., forms normal inclusions in the presence of the specified antibiotic. Antibiotics were used at the following concentrations: OFX at 1 μg per ml, LIN at 2 μg per ml, RIF at 0.015 μg per ml, and TMP at 15 μg per ml.
Four kinds of antibiotic-resistant mutants were isolated from C. trachomatis serovar L1: ofloxacin resistant (OFXr), lincomycin resistant (LINr), rifampin resistant (RIFr), and trimethoprim resistant (TMPr). The isolation and characterization of the mutants by DNA sequencing are described in the supplemental material (see “Antibiotic-resistant mutants”). The LINr-1 and TMPr-1 mutants were used in our experiments because each of them was the only mutant of its kind obtained in selections against ≥2 × 109 wild-type (wt) IFU. Thus, the frequency of spontaneous LINr and TMPr mutants was ≤5 × 10−10. In contrast, the frequency of spontaneous OFXr mutants was 1.9 × 10−8 and that of spontaneous RIFr mutants was 4.1 × 10−8. These frequencies defined backgrounds above which we intended to detect LGT. Mutants named OFXr-1, OFXr-2, and RIFr-1 were chosen for use in our LGT experiments because they had the least common nucleotide substitutions among mutants of their kinds. The mutations and the concentrations of antibiotics used to select for resistance are listed in Table 3.
Cross no. 1 (OFXr-1 × LINr-1): the gyrA and 23S rRNA genes participate in LGT (Table 4) .
P-0 HeLa flasks were infected as follows: set A, OFXr-1; set B, LINr-1; sets C and D, OFXr-1 and LINr-1. Set D was included because the small total number of LINr-1 IFU (about 7.5 × 107) used to infect the flasks was expected to yield ≤1 spontaneous OFXr mutant of the LINr-1 parental strain. At 48 h p.i., in the absence of antibiotics, all P-0 flasks were D/F and 1/10 of each P-0 BH was used to initiate four passages in the presence of OFX plus LIN.
Table 4 shows that none of the P-4 flasks in control set A contained OFXr LINr inclusions, indicating the absence of spontaneous LINr mutants of the OFXr-1 parental strain. Two P-4 flasks in control set B contained spontaneous OFXr mutants of the LINr-1 parent. In contrast, all 20 P-4 flasks derived from mixed-infection set C and D P-0 flasks contained abundant OFXr LINr inclusions. The 2/20 versus 20/20 difference in the frequencies of flasks containing doubly resistant C. trachomatis is highly significant (P < 10−3 by Fisher's exact test). The presence of doubly resistant mutants in two set B flasks suggests that a similar number of set C flasks might have contained such mutants, but the small number of LINr-1 IFU used to infect set D flasks precluded the origin of OFXr LINr by means of spontaneous mutation. These numerical results suggest that OFXr LINr growth originated by means other than spontaneous mutation in mixed-infection flasks of sets C and D.
Could complementation or transient heterotypic resistance (21, 39) involving the two kinds of parental mutants explain the sustained, rapid multiplication of doubly resistant isolates from mixed-infection cells during at least four passages in the presence of OFX plus LIN? Both hypothetical explanations imply that numerous antibiotic-sensitive C. trachomatis isolates would be present in doubly resistant outgrowths and that individual C. trachomatis isolates would be incapable of initiating sustained rapid growth in the presence of OFX plus LIN. These possibilities were addressed by plating (MOI of <0.05) equal aliquots of OFXr LINr BHs derived from three set C P-4 flasks in the presence and absence of OFX plus LIN. Each inclusion that formed contained a clone that was initiated by a single IFU in >99 percent of the infected cells, thereby precluding complementation. Only ∼1 percent or less of the IFU in heterotypically resistant populations are phenotypically resistant after several passages in the presence of an antibiotic (39). The IFU titers of our doubly resistant isolates in the presence and in the absence of OFX plus LIN were not significantly different, indicating that individual IFU in all three OFXr LINr isolates were predominantly or exclusively OFXr LINr recombinants (data not shown).
Recombinants in this cross should always have the A2039→C mutations present in the 23S rRNA genes of the LINr-1 parent and the gyrA T249→G mutation of the OFXr-1 parent. Doubly resistant C. trachomatis resulting from spontaneous mutations might have diverse nucleotide changes, as shown by our survey of spontaneous OFXr mutants (see Table S1 and “Antibiotic-resistant mutants” in the supplemental material); in contrast, each of five set C and five set D OFXr LINr isolates that were sequenced had only the gyrA and 23S rRNA mutations of the parental strains (Table 5). While spontaneous OFXr mutants of the LINr-1 parent might have contributed other than gyrA T249→G mutant sequencing signals in a few set C flasks, those signals apparently were obscured by very high frequencies of recombinants. Such non-gyrA T249→G signals were not expected in set D flasks because of the improbability that set D flasks contained spontaneous OFXr mutants of the LINr-1 parent; all sequenced set D flasks contained only the parental gyrA T249→G and 23S rRNA A2039→C mutations expected in recombinants.
TABLE 5.
Relevant nucleotides in doubly resistant C. trachomatis strains isolated from the indicated flasks
| Construct | na | wt sequence and relevant ntb change
|
||
|---|---|---|---|---|
| 23S rRNA-1 and -2 (nt 2034-2044)c | gyrA (nt 243-253) | rpoB (nt 1378-1388) | ||
| wt | ACGAAAAGACC | AGAAAGTGTCA | ATGGATCAGAC | |
| Cross no. 1 | ||||
| OFXr-1 | ........... | ......G.... | NS | |
| LINr-1 | .....C..... | ........... | NS | |
| OFXr LINr flasks | ||||
| Set B | 2/10 | .....C..... | ......G.... | NS |
| Set C | 10/10 | .....C..... | ......G.... | NS |
| Set D | 10/10 | .....C..... | ......G.... | NS |
| Cross no. 2 | ||||
| OFXr-2 | ........... | ....C...... | NS | |
| LINr-1 | .....C..... | ........... | NS | |
| OFXr LINr flasks | ||||
| Set B | 1/10d | .....C..... | ........... | NS |
| 1/10 | .....C..... | ......A.... | NS | |
| Set C | 10/10 | .....C..... | ....C...... | NS |
| Cross no. 3 | ||||
| OFXr-1 | NSf | ......G.... | ........... | |
| RIFr-1 | NS | ........... | .....G..... | |
| OFXr RIFr flasks | ||||
| Set A | 5/10e | NS | ......G.... | ........... |
| Set B | 1/10d | NS | ........... | .....G..... |
| 2/10 | NS | .....T..... | .....G..... | |
| Set C | 10/10 | NS | ......G.... | .....G..... |
| Cross no. 5 | ||||
| OFXr-1 | ........... | ....C...... | NS | |
| LINr-1 | .....C..... | ........... | NS | |
| OFXr LINr flasks | ||||
| Set V | 10/10 | .....C..... | ....C...... | NS |
Number of flasks yielding isolates with the mutation/total number of flasks.
nt, nucleotide.
The two 23S rRNA genes were individually sequenced (Tables 1 to 3); both of the genes in an isolate always had identical sequences.
Spontaneous OFXr mutant that lacked a mutation in gyrA nt 1 to 438.
Spontaneous RIFr mutants in cross no. 3 that had a G1399→A, T1432→G, T1432→C, G1441→T, or C1525→T mutation in rpoB.
NS, not sequenced.
Definitive evidence indicating that individual OFXr LINr IFU were responsible for OFXr LINr growth in isolates from mixed-infection cells was obtained by isolating 15 clones (see Materials and Methods) from the three set C isolates used to test for complementation and heterotypic resistance (see above). The clones were isolated in the absence of OFX and LIN. DNA sequencing showed that each of the 15 clones had only the 23S rRNA mutations of the LINr-1 parent and the gyrA mutation of the OFXr-1 parent (data not shown). The apparently complete absence of the corresponding wt nucleotides indicates that they had been replaced by homologous recombination and confirms the genetic stability of the recombinants. Complementation and heterotypic resistance are conclusively ruled out as explanations of the outgrowth of OFXr LINr isolates from mixed-infection cells; the participation of either phenomenon would require the presence of parental mutant strain IFU in doubly resistant isolates and, therefore, detection of the wt nucleotides corresponding to the resistance-conferring parental mutations.
Interestingly, both 23S rRNA genes sequenced early in the isolation of uncloned recombinant populations, as well as in clones subsequently isolated from them, had the A2039→C mutation, as with the LINr-1 parental strain (see “Antibiotic-resistant mutants” in the supplemental material for a discussion of the presence of both mutant genes in recombinants). Most unexpectedly, both doubly resistant spontaneous OFXr mutants in set B control flasks had the gyrA T249→G nucleotide change present in the OFXr-1 parent. We attributed this to improbable coincidence resulting from randomness of the mutation process. Furthermore, while they were not expected to contain spontaneous OFXr mutants, all five sequenced set D flasks had the gyrA T249→G mutation of the OFXr-1 parental strain.
Cross no. 2 (OFXr-2 × LINr-1): LGT recombinants have the specific gyrA mutation present in their OFXr parental strain (Table 4).
Mutant OFXr-2 has an uncommon gyrA A247→C mutation and was used to demonstrate that OFXr LINr isolates from mixed-infection cells have the specific gyrA mutation of the OFXr parent used in a cross. P-0 HeLa flasks were infected in the absence of antibiotics with only OFXr-2 (set A), only LINr-1 (set B), or both (set C) and were passaged in the presence of OFX plus LIN. The difference between 2/20 control flasks and 10/10 mixed-infection P-4 flasks that had OFXr LINr C. trachomatis was highly significant (P < 10−3) (Table 4).
DNA sequencing around gyrA codon 83 (Table 5) showed that neither set B spontaneous OFXr mutant had a mutation that corresponded to the gyrA A247→C mutation present in the OFXr-2 parental strain. In contrast, all 10 mixed-infection set C P-0 flasks yielded OFXr LINr P-4 isolates that had the gyrA A247→C mutation present in the OFXr-2 parental strain and the A2039→C 23S rRNA mutation of the LINr-1 parental strain. These results reinforce our conclusion, based on cross no. 1, that OFXr LINr C. trachomatis isolates produced in cells that are simultaneously infected with LINr-1 and an OFXr mutant originate predominantly by means of LGT. The frequencies of LGT recombinants are estimated starting with cross no. 5 (see below).
Cross no. 3 (OFXr-1 × RIFr-1): rpoB is a third locus that participates in LGT (Table 4).
The potential value of using diverse selection markers to investigate LGT involving different segments of the C. trachomatis chromosome prompted us to investigate RIFr rpoB mutants. P-0 HeLa flasks were infected in the absence of antibiotics with RIFr-1 (control set A; MOI of 5.1), OFXr-1 (control set B), or both (set C). All P-0 flasks were D/F at 50 h p.i., and four passages were performed in the presence of OFX plus RIF. Three set A P-4 control flasks contained spontaneous OFXr mutants of the RIFr-1 parental strain, and five set B P-4 control flasks contained spontaneous RIFr mutants of the OFXr parental strain; in contrast, all 10 of the P-4 set C flasks derived from mixed-infection P-0 flasks had abundant OFXr RIFr inclusions. The difference between 8 of 20 control flasks that contained OFXr LINr growth and 10 of 10 mixed-infection set C flasks that contained such growth was significant (P < 10−2 by Fisher's exact test).
As with cross no. 1, we verified that neither complementation between parental strains nor heterotypic resistance was responsible for the outgrowth of C. trachomatis in the presence of OFX plus RIF in mixed-infection set C flasks. Platings of the IFU yields from three set C P-4 flasks such that >99% of inclusions were initiated by a single IFU yielded closely similar IFU titers in the presence and absence of OFX plus RIF (data not shown).
DNAs from OFXr RIFr isolates descended from 10 set C P-0 flasks were sequenced (Table 5); only the gyrA T249→G and rpoB T1383→G mutations present in the parental mutant strains were detected, as expected of recombinants. This result was confirmed by isolating 18 clones in the absence of antibiotics from 4 of the 10 uncloned set C isolates that had been sequenced; all of the 18 clones had only the resistance-conferring mutations of the two parental strains. In contrast, none of the eight OFXr RIFr mutants from control sets A and B had parental mutant nucleotides that were expected to be present in recombinants (data not shown). Diverse spontaneous mutants probably were present in some mixed-infection set C flasks, but their mutant sequencing signals were not detected because, as shown below (see “Cross no. 5” and “Direct enumeration of recombinants”), such mutants comprised less than 10−4 of the OFXr RIFr IFU present in such flasks. Recombination between OFXr-1 and RIFr-1 was confirmed by the isolation of OFXr RIFr recombinants in the context of cross no. 7 (see below).
Cross no. 4 (OFXr-1 × TMPr-1): a fourth locus that participates in LGT (Table 4).
While the mutation that renders TMPr-1 resistant to TMP has not been identified, we determined that the mutant locus could participate in LGT by performing mixed infections with OFXr-1 and TMPr-1. P-0 HeLa flasks were infected in the absence of antibiotics with OFXr-1 (set A), TMPr-1 (set B), or both (set C). All P-0 flasks were D/F at 46 h p.i. and were passaged in the presence of OFX plus TMP. The difference between set A plus set B control flasks versus set C mixed-infection flasks with regard to the numbers of flasks that contained doubly resistant inclusions is highly significant (P < 10−3 by Fisher's exact test). It was not necessary to sequence DNAs from isolates in this experiment: TMPr spontaneous mutants were absent, and sequencing of the sole OFXr spontaneous mutant would have been redundant. The results summarized above strongly indicate that the unidentified locus responsible for TMP-resistance in TMPr-1 can participate in LGT.
Cross no. 5: at least one per eight mixed-infection cells yielded LGT recombinants (Table 4).
Having established the occurrence of LGT in C. trachomatis with crosses no. 1 to no. 4, we attempted to estimate the frequencies of LGT recombinants. In a “range-finding” attempt, we held the MOI of the OFXr-1 parent constant at ∼5 in P-0 flasks and simultaneously infected sets of 10 of the flasks with varied MOIs of LINr-1 that ensured essentially all mixed-infection cells received just one LINr-1 IFU. Thus, the numbers of LINr-1 IFU used in each set of P-0 flasks determined the numbers of mixed-infection cells and resulted in 1.5 × 105 (set I) to 15 (set V) mixed-infection cells per P-0 flask. As in previous crosses, antibiotics were absent during development in these P-0 flasks but were present throughout the subsequent four passages.
P-4 flasks of all sets had abundant OFXr LINr inclusions (Table 4). The total of 150 LINr-1 IFU used to infect the 10 set V P-0 flasks was about 5 orders of magnitude too small to yield spontaneous OFXr mutants of the LINr-1 parental strain in those flasks. Similarly, LINr-1 is the only LINr mutant detected in all of our crosses involving selection for LIN resistance (mutant frequency of <5 × 10−10), making it virtually impossible for spontaneous LINr mutants of the OFXr-1 parent to contribute to the OFXr LINr growth observed with set V P-4 flasks. This strong evidence that doubly resistant growth originated by means of LGT-mediated recombination was verified by sequencing of DNAs of OFXr LINr isolates descended from five set V P-0 flasks; all of the flasks yielded only the mutant gyrA T249→G and 23S rRNA A2039→C sequencing signals characteristic of the parental mutants used in the cross (Table 5).
According to the Poisson distribution, if a set of 10 P-0 flasks contained an average of two recombinant-yielding cells per flask, two of the flasks might lack recombinants, as observed with set V. Set V flasks were initiated with an average of 15 mixed-infection cells, and the absence of recombinant-free set V flasks suggests that at least 2 per 15 mixed-infection cells yielded OFXr LINr recombinants. Assuming yields of 400 IFU per mixed-infection cell, this would correspond to a minimum LGT recombinant frequency of one per 3,000 IFU (3.3 × 10−4) if the entire IFU yield of each primary flask were used to infect a P-1 flask. Since only 1/10 of the IFU yield of each primary flask was passaged, the recombinant frequency was probably 3.3 × 10−4 to 3.3 × 10−3 and between approximately one per eight and all mixed-infection cells in set V flasks yielded recombinants (see Materials and Methods, “Enumeration of recombinants and mutants”). This result suggested that it should be practical to microscopically enumerate recombinants as antibiotic-resistant inclusions in P-1 flasks without additional passaging.
Direct enumeration of recombinants at the first passage from mixed-infection cells (Table 6).
Frozen remainders of five set C P-0 BHs from cross no. 1 were thawed, and IFU assays were performed in the absence of antibiotics (for total IFU) and in the presence of OFX plus LIN (for recombinant IFU). At 48 h p.i., many cells in antibiotic-containing flasks infected with set C P-0 BHs had a small, inhibited inclusion, while an OFXr LINr inclusion was observed in every few fields (Fig. 3C). Resistant inclusions were not observed during exhaustive scanning of P-1 flasks infected with BHs derived from control flasks that had been infected with either OFXr-1 (set A) or LINr-1 (set B) alone (not shown). By dividing the numbers of OFXr LINr inclusions in P-1 flasks by the total numbers of IFU plated in the flasks (Table 6), we derived a recombinant frequency of 4.0 × 10−4 that is fairly consistent with the minimum estimate of 3.3 × 10−4 obtained with the same parental mutant strains in cross no. 5.
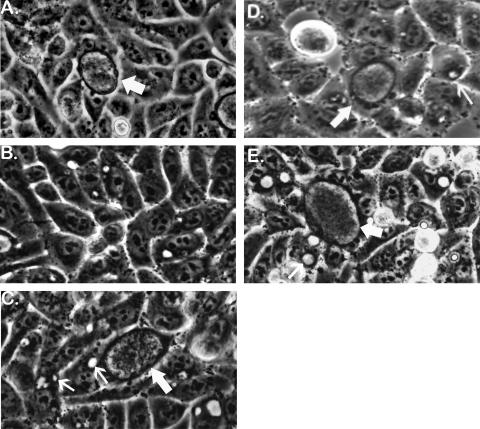
FIG. 3.
Phase-contrast microscopic detection of doubly resistant recombinant inclusions at the first passage (P-1) of IFU produced in cells that were simultaneously infected with two kinds of antibiotic-resistant mutants. (A) A normal 48-h p.i. inclusion (thick arrow) formed in the absence of antibiotics used to select for recombinants. Inclusions formed by each strain of C. trachomatis used in this study would have similar appearances in antibiotic-free culture medium or in a medium containing just that antibiotic to which the strain was resistant. (B) A typical 46-h p.i. field in a cross no. 1 (OFXr-1 × LINr-1) set A P-1 flask derived from a primary (P-0) flask in which all cells were infected with only parental strain OFXr-1. The P-1 flask contained OFX plus LIN. OFXr-1 is sensitive to LIN and formed tiny, inhibited inclusions in some cells. Similarly, LINr-1, the other parental strain in cross no. 1, is sensitive to OFX and would also be unable to form normal inclusions in OFX plus LIN medium (not shown). (C) A 48-h p.i. field in a cross no. 1 set C P-1 flask derived from a P-0 flask in which all cells were simultaneously infected with OFXr-1 and LINr-1. The P-1 flask contained OFX plus LIN medium, in which parental IFU formed inhibited inclusions (small arrows) and OFXr LINr recombinants formed normal inclusions (large arrow). (D) A typical 68-h p.i. field in a cross no. 3 set C P-1 flask derived from a P-0 flask in which all cells had been simultaneously infected with OFXr-1 and RIFr-1. The P-1 flask contained OFX plus RIF medium. An OFXr RIFr recombinant inclusion is shown (large arrow) along with numerous inhibited inclusions (small arrow) formed by nonrecombinant parental IFU. (E) A typical 46-h p.i. field in a cross no. 4 set C P-1 flask derived from a P-0 flask in which all cells were simultaneously infected with OFXr-1 and TMPr-1. The P-1 flask contained medium supplemented with OFX and TMP. An OFXr TMPr recombinant inclusion is shown (large arrow). Also shown are examples of relatively large, empty-appearing inclusions (small arrow) that are typically formed by TMP-sensitive strains of C. trachomatis (OFXr-1 in this experiment) in the presence of TMP. Spontaneous mutants of parental strains would also be able to form doubly resistant inclusions in P-1 flasks containing two antibiotics, but their rarity (<10−7) would prevent their detection by microscopic scanning in practice.
The frequencies of recombinants in crosses no. 3 and no. 4 were determined as for cross no. 1, with thawed remainders of their set C P-0 BHs derived from mixed-infection cells. For each cross, IFU assays of five P-0 BHs were performed in the absence of antibiotics to determine the total IFU titers and in the presence of either OFX plus RIF (cross no. 3) (Fig. 3D) or OFX plus TMP (cross no. 4) (Fig. 3E) in order to determine the titers of doubly resistant recombinants. The frequencies of OFXr RIFr recombinants (5.0 × 10−4) and of OFXr TMPr recombinants (8.5 × 10−4) were similar to those obtained in OFXr-1 × LINr-1 crosses no. 1 and no. 5 (Table 6).
Cross no. 6: LGT recombinants were present among EBs that appeared early during C. trachomatis development.
In order to characterize DNA segments that are transferred in individual LGT events, it would be desirable to analyze recombinants that have not participated in more than one LGT event because that could alter the makeup of an initially transferred segment. It is possible that EBs do not participate in LGT; if so, EBs that mature from an early pool of RBs have fewer chances to have participated in >1 LGT event than do EBs that mature later in development. Cross no. 6 was an initial attempt to determine when recombinant IFU first become detectable during C. trachomatis development. P-0 HeLa flasks were simultaneously infected in the absence of antibiotics with OFXr-1 and LINr-1, each at an MOI of ∼5. Groups of five flasks were D/F at 18 h p.i., 22 h p.i., and 44 h p.i., the last being included as a positive control for the production of LGT recombinants. Samples of BHs prepared from these mixed-infection P-0 flasks were used to infect P-1 flasks in the absence of antibiotics and in the presence of OFX plus LIN for determination of the frequencies of OFXr LINr recombinants as described in “Direct enumeration of recombinants” above. The numbers of OFXr LINr inclusions in each of the P-1 flasks at 44 to 46 h p.i. were determined.
Table 6 shows that flasks D/F at 44 h p.i. yielded 350 IFU per infected cell and that the frequency of OFXr LINr recombinants was 1.2 × 10−3, in fairly good agreement with the results of crosses no. 1 and no. 5, performed with the same parental strains. The frequency of recombinants was 1.6 × 10−3 in BHs prepared from mixed-infection cells that were D/F at 22 h p.i., when 3.9 IFU per infected cell were present. In contrast, recombinants were not detected in BHs from cells D/F at 18 h p.i., when <0.001 IFU per cell were present. These results indicate that LGT recombinants are already present among the early appearing new EBs in mixed-infection cells. The high recombinant frequency at 22 h p.i. would make it relatively easy to clone early recombinants.
Cross no. 7: some IFU can possess genes derived from three parental strains during one course of development.
We investigated the possibility that some IFU might have genes derived from more than one LGT episode in the course of development. Cells were simultaneously infected with three different kinds of antibiotic-resistant mutants followed by selection for triply resistant (i.e., triparental) recombinants among the progeny. HeLa cells (3 × 106) in a 25-cm2 flask were infected with OFXr-1 (MOI of 2.4), LINr-1 (MOI of 6.7), and RIFr-1 (MOI of 1.8). Development occurred in the absence of antibiotics, and the flask was D/F at 48 h p.i. The following determinations were made with aliquots of a 2.0-ml BH prepared from the infected cells.
(i) The IFU titer in the absence of antibiotics was determined to be 7.2 × 108 per ml, indicating a yield of 480 IFU per cell.
(ii) Positive controls for the production of biparental recombinants were five infected HeLa flasks that received OFX plus LIN (set I), five flasks that received OFX plus RIF (set II), and five flasks that received LIN plus RIF (set III). Biparental recombinants were directly enumerated as described above (“Direct enumeration of recombinants”) by scanning 40 ×200 phase-contrast microscope fields in each of the flasks for normal inclusions starting at 48 h p.i. The frequencies of biparental recombinants (data not shown) were as follows: for OFXr LINr (set I), 1.3 × 10−3, which was similar to that obtained with the same parental strains in crosses no. 1, no. 5, and no. 6; for OFXr RIFr (set II), 1.3 × 10−4, which was substantially lower than that obtained in cross no. 4 (Table 6) performed with the same parental strains (we cannot explain this discrepancy, but even the reduced number of recombinant inclusions we detected verifies that these particular biparental LGT events occurred); and for LINr RIFr (set III), 8.5 × 10−4, which was the first determination for a LINr × RIFr cross and suggests that the LGT events involving the rpoB-23S rRNA chromosome segments occur about as frequently as events involving the other chromosome segments studied in our crosses.
(iii) Selection for triparental recombinants was based on the expectation that the frequency of triparental recombinants would be too low to permit direct enumeration by microscopic scanning of P-1 flasks containing antibiotics. Therefore, triply resistant recombinants in the P-0 BH were amplified by passaging flask cultures in the presence of all three antibiotics. Sets of 10 P-1 flasks were infected with P-0 BH aliquots that contained either 7.2 × 106 IFU (set IV) or 7.2 × 105 IFU (set V) per flask in the presence of OFX plus LIN plus RIF. These P-1 flasks were passaged four times in the presence of all three antibiotics as in previous crosses, but at intervals of about 67 h p.i., and the P-4 flasks were scanned for OFXr LINr RIFr inclusions.
All 10 set IV and 8 set V P-4 flasks had abundant triply resistant inclusions. Since 2 of the 10 set V P-4 flasks lacked resistant inclusions, we used 0.20 as the P0 term of the Poisson distribution to estimate that set V P-1 flasks had been infected with an average of 1.6 triparental recombinants per 7.2 × 105 IFU (frequency of 2.2 × 10−6). The puzzling low frequency of OFXr RIFr recombinants noted above makes it difficult to estimate an expected frequency of triparental recombinants based on the assumption of random occurrences of LGT events among C. trachomatis in the course of development. However, the data do not suggest a large deviation from what might be expected if biparental recombinants participated in second LGT episodes with a frequency of about 10−3 during a single course of development. These results testify to a remarkable ability of C. trachomatis to transfer genes.
DISCUSSION
The central new observation in this report is that simultaneous infection of host cells with two different kinds of antibiotic-resistant mutants of C. trachomatis always resulted in the appearance of many progeny that were capable of sustained growth in the presence of both antibiotics to which the parental mutants were individually resistant. This observation was made with pairwise combinations of four mutant loci that were distributed over at least one half of the C. trachomatis chromosome (Fig. 1), indicating that the appearance of doubly resistant C. trachomatis was not idiosyncratic with regard to locus.
In principle, doubly resistant progeny could have resulted from spontaneous mutations in parental C. trachomatis strains that were initially resistant to just one antibiotic. Several kinds of quantitative and qualitative evidence disfavor spontaneous mutation as an explanation of double resistance, except in rare instances.
(i) Equal numbers of each parental mutant strain were used to infect mixed-infection cells and control cultures that were infected with just one strain or the other. Spontaneous mutations would have resulted in approximately equal numbers of doubly resistant isolates in uniparentally infected cultures and mixed-infection cultures. Contrary to this expectation, the numbers of doubly resistant isolates from mixed-infection cultures always significantly exceeded the numbers obtained from uniparental infections in crosses no. 1 to no. 4.
(ii) Enumeration of doubly resistant C. trachomatis isolates by two different methods (cross no. 5 and Table 6) indicated that frequencies of doubly resistant C. trachomatis isolates in IFU yields from mixed-infection cells were 3.3 × 10−4 to 1.6 × 10−3. In contrast, the frequencies of spontaneous mutants that were resistant to individual antibiotics were ≤5 × 10−10 to 4.1 × 10−8. Thus, the numbers of doubly resistant C. trachomatis isolates from mixed-infection cells exceeded those from uniparentally infected cells by at least ∼104-fold. This point is emphasized by the results of cross no. 7, in which cells were simultaneously infected with three different kinds of mutants. The frequency of progeny resistant to the three antibiotics used to select for triply resistant progeny was 2.2 × 10−6. The maximum frequency of triply resistant C. trachomatis isolates originating by means of two independent spontaneous mutations would have been attained in the LINr-1 parental strain and would have been about (1.9 × 10−8 for OFX resistance) × (4.1 × 10−8 for RIF resistant), i.e., 7.8 × 10−16; the total number of IFU subjected to triple antibiotic selection in cross no. 7 set V flasks was 7.2 × 106.
(iii) We showed that independent spontaneous mutants of the same kind had diverse nucleotide substitutions (see “Antibiotic resistant mutants” in the supplemental material, results of crosses no. 2 and no. 3). In contrast, sequencing of DNA from polyclonal doubly resistant isolates from mixed-infection cultures in crosses no. 1, no. 2, no. 3, and no. 5 revealed only the mutant nucleotides present in each of the parental strains used in the cross. The failure to detect corresponding wt nucleotides indicates that they had been replaced by homologous recombination.
Assuming a predominantly nonmutational origin of doubly resistant C. trachomatis strains in mixed infections, the alternatives of complementation between parental strains, heterotypic resistance, and LGT remain. Critical evidence excluding complementation and heterotypic resistance was obtained with 33 physically isolated clones derived from six polyclonal doubly resistant populations in crosses no. 1 and no. 3. Complementation and heterotypic resistance would have involved the presence of numerous antibiotic-sensitive C. trachomatis strains along with resistant organisms. Sequencing of DNA revealed that each clone had only the mutant nucleotide responsible for antibiotic resistance in the parental strains and not a trace of wt nucleotides that were associated with antibiotic sensitivity. These results demonstrate that LGT-mediated homologous recombination was responsible for double antibiotic resistance of individual IFU in the progeny C. trachomatis strains produced in cells simultaneously infected with two different kinds of antibiotic-resistant mutants.
Possible mechanisms of in vitro LGT in C. trachomatis.
Our study does not allow one to distinguish between the participation of reciprocal recombination versus nonreciprocal gene conversion in the origin of LGT recombinants in C. trachomatis. It has required special plasmid-based systems that are not available with C. trachomatis to make this distinction in other microbes (reviewed in reference 38), and this issue will not be discussed further here.
The three main modes of LGT in microbes are reviewed in reference 43. Phage-mediated transduction in C. trachomatis appears to be unlikely because a C. trachomatis phage has not been reported, although phages are present in other species of Chlamydia (12). Conjugation-mediated DNA transfer occurs in two main ways: (i) an extrachromosomal plasmid is transferred from a donor to a recipient bacterium, and (ii) a chromosome-mobilizing plasmid interacts with the DNA donor chromosome in a way that initiates transfer of chromosomal DNA into a recipient bacterium that lacks the plasmid. C. trachomatis does possess a low-copy-number, 7.5-kb plasmid, pCT (34), but our BLAST analysis did not detect C. trachomatis orthologues of conserved genes that are associated with conjugative plasmids. Nevertheless, mixed infections with strains lacking pCT (13, 42) might be informative about the involvement of the plasmid in LGT. Remnant insertion sequences that are associated with transposable elements were observed in the vicinity of recombined DNA segments in some LGT clinical isolates, but means for transferring the insertion sequences between C. trachomatis strains were not identified (16).
Natural DNA transformation after uptake and transfer of environmental DNA without facilitation by agencies, such as electroporation, has been described for at least 44 species of bacteria (24). The high frequency of recombinants we observed could result from uptake by RBs of DNA released from other C. trachomatis strains in inclusions occupied by genetically different strains. The possible necessity for cooccupancy of inclusions by C. trachomatis exchanging DNA could be investigated with mixed infections using fusion-defective variants of the same serovar (37, 44) or of different C. trachomatis serovars that have nonfusogenic inclusions. The possibility that LGT in C. trachomatis is accomplished by uptake of exogenous DNA is especially intriguing because of long-standing frustration with attempts to create planned genetic modifications by means of DNA transfer. Perhaps an understanding of how LGT works could lead to ways of transforming C. trachomatis. Uptake of exogenous DNA by C. trachomatis may have evolved to provide a source of nucleic acid building blocks for an obligate intracellular parasite (14, 36). In well-studied highly transformable bacteria, uptake and transfer of DNA are followed by degradation of single strands of trans-DNA and recombination of the intact DNA strand with recipient chromosomes (reviewed in reference 7). The degradation products of trans-DNA could be recycled for nucleic acid synthesis by recipients; C. trachomatis has lost the capacities for de novo synthesis and salvaging of purine and pyrimidine nucleotides but has the enzymes needed to convert deoxynucleoside monophosphates released from degraded DNA to the triphosphates used for DNA synthesis (27).
The possibility that natural DNA transformation is a mechanism of LGT in C. trachomatis is emphasized by BLAST analysis, which has detected in four C. trachomatis genes (identified by designations in parentheses below) sequences that are orthologous to five Neisseria gonorrhea genes that are listed in reference 7 as being involved in DNA uptake and transfer: pilF (CT571), pilG (CT570), pilT (also in CT571), pilQ (CT674), and comE (CT545). We did not detect clear evidence for the presence in C. trachomatis of the type IV secretion system genes that N. gonorrhea uses to secrete DNA (18). This suggests that DNA used for natural LGT by C. trachomatis may be derived by autolysis of some C. trachomatis within the inclusion (26) or by some other means. It is also noteworthy that the frequencies of in vitro LGT we report are in the range of in vitro natural transformation frequencies in other microbes (24).
Could LGT of the kind we report here generate the kinds of recombinants that have been observed in surveys of fresh clinical isolates and established strains? The opportunity for LGT between strains in vivo clearly exists. Simultaneous infection with at least two serovars has been detected in 2 to 8 percent of the isolates in various clinical surveys (2, 9, 10, 23, 35, 45); a recent survey that used very sensitive techniques detected a mixed-infection frequency of 13 percent (48). There are an estimated 92 million new C. trachomatis STD infections worldwide annually (46), with 3 million of them in the United States alone (6). If it is assumed that 5 percent of those infections were with more than one serovar, hundreds of thousands of those subjects might have multistrain infections. The high frequency of in vitro LGT we observed might then have the potential to generate many between-strain recombinants in vivo.
Furthermore, LGT might disseminate rare nucleotide substitutions within as well as between serovars. For instance, we presented here evidence suggesting that the rate of appearance of macrolide-resistant mutants that have 23S rRNA mutations in C. trachomatis is very low because of the possible need for multiple genetic changes (see “Antibiotic-resistant mutants” in the supplemental material). Yet, three STD subjects in the same clinic and short time interval yielded azithromycin-resistant isolates that had different ompA alleles but had identical nucleotide substitutions (A2039→C and T2611→C) in both of their 23S rRNA genes (30). We are unaware of other published reports of clustered occurrences of identical rare 23S rRNA mutations in STD subjects treated with azithromycin, which is one of the antibiotics most often used to treat C. trachomatis genital tract infections. Similarly, it is possible that multiple examples of tetracycline resistance associated with the presence of a plasmid-related tet(C) island in Chlamydia suis (11) resulted from a rare initial insertion of the resistance gene by an unspecified mechanism that was followed by relatively frequent LGT of insert-containing chromosomal DNA to several serovars.
The production of between-serovar recombinants in vitro would allow analysis of chromosome segments that are recombined and, possibly, of polarity that may be associated with the LGT process. With regard to such analysis, we emphasize that many loci that are not noted for being polymorphic have small numbers of polymorphic nucleotides; for instance, we found such polymorphisms in the 23S rRNA and gyrA genes (see “Antibiotic-resistant mutants” in the supplemental material) and suggest an alleged “triple mutation” in the L22 gene (30) may be a polymorphism. Such ubiquitous polymorphic nucleotides could serve as unselected genetic markers that could be used to delineate DNA segments that accompany selection markers located in different parts of the C. trachomatis chromosome into LGT recombinants. It may also become possible to create in vitro LGT recombinant strains for functional studies. Our three attempts to isolate an OFXr mutant from a total of >5 × 109 serovar D IFU for this purpose failed. Available time and funding will not permit us to continue attempts to isolate mutants from non-serovar L1 strains of C. trachomatis. We are publishing our results with serovar L1 with the hope that interested investigators will be able to isolate such mutants and cross them with our L1 mutants.
ADDENDUM
After submission of the manuscript and its final acceptance, we succeeded in isolating from serovar D a rifampin-resistant mutant, D:RIFr-1, that has a single nucleotide change in the rpoB gene. Cells infected with both D:RIFr-1 and the L1:OFXr-1 mutant described in this paper yielded OFXr RIFr recombinants that had the following properties: (i) only the mutant rpoB and gyrA nucleotides present in the parental mutant strains were detected at the relevant nucleotide positions, and (ii) single alleles of D versus E naturally polymorphic marker nucleotides that were linked to the selection nucleotides in rpoB of the serovar D parent and gyrA of the serovar E parent were detected. These results demonstrate that between-serovar LGT followed by homologous recombination had occurred. The recombinants have proven to be readily clonable and will be sequenced extensively in order to define their LGT segments. Some recombinants (“neomorphs”) formed inclusions that are morphologically distinct from both parents, suggesting that some D-plus-E genetic interactions altered C. trachomatis development.
Supplementary Material
Acknowledgments
This investigation was supported by grants RO1 AI 1056124 and R21 AI 05872801 from the National Institutes of Health.
David Watkins generously provided laboratory facilities in which this study could be continued. We appreciate helpful criticism of the manuscript by Joseph Dillard (University of Wisconsin) and Paula Kavathas (Yale University) and thank Bernard Weisblum (University of Wisconsin) for much useful advice concerning antibiotic resistance.
This study was performed in a facility and with protocols that were approved by the University of Wisconsin (Madison) Biological Safety Committee. We ascertained that all parental mutant strains used in this study were as sensitive as wt C. trachomatis to doxycycline. Doubly resistant recombinants isolated from crosses no. 1, no. 3, and no. 4 were also doxycycline sensitive.
Footnotes
Published ahead of print on 22 November 2006.
Supplemental material may be found at http://jb.asm.org/.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology, vol. 1. John Wiley and Sons, Inc., Hoboken, NJ.
- 2.Barnes, R. C., R. J. Suchland, S. P. Wang, C. C. Kuo, and W. E. Stamm. 1985. Detection of multiple serovars of Chlamydia trachomatis in genital infections. J. Infect. Dis. 152:985-989. [DOI] [PubMed] [Google Scholar]
- 3.Binet, R., and A. T. Maurelli. 2005. Frequency of spontaneous mutations that confer antibiotic resistance in Chlamydia spp. Antimicrob. Agents Chemother. 49:2865-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunham, R., C. Yang, I. Maclean, J. Kimani, G. Maitha, and F. Plummer. 1994. Chlamydia trachomatis from individuals in a sexually transmitted disease core group exhibit frequent sequence variation in the major outer membrane protein (omp1) gene. J. Clin. Investig. 94:458-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunham, R. C., and J. Rey-Ladino. 2005. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat. Rev. Immunol. 5:149-161. [DOI] [PubMed] [Google Scholar]
- 6.Cates, W., Jr., et al. 1999. Estimates of the incidence and prevalence of sexually transmitted diseases in the United States. Sex. Transm. Dis. 26:S2-S7. [DOI] [PubMed] [Google Scholar]
- 7.Chen, I., and D. Dubnau. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2:241-249. [DOI] [PubMed] [Google Scholar]
- 8.Dean, D., and K. Millman. 1997. Molecular and mutation trends analyses of omp1 alleles for serovar E of Chlamydia trachomatis. Implications for the immunopathogenesis of disease. J. Clin. Investig. 99:475-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean, D., E. Oudens, G. Bolan, N. Padian, and J. Schachter. 1995. Major outer membrane protein variants of Chlamydia trachomatis are associated with severe upper genital tract infections and histopathology in San Francisco. J. Infect. Dis. 172:1013-1022. [DOI] [PubMed] [Google Scholar]
- 10.Dean, D., and R. S. Stephens. 1994. Identification of individual genotypes of Chlamydia trachomatis from experimentally mixed serovars and mixed infections among trachoma patients. J. Clin. Microbiol. 32:1506-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dugan, J., D. D. Rockey, L. Jones, and A. A. Andersen. 2004. Tetracycline resistance in Chlamydia suis mediated by genomic islands inserted into the chlamydial inv-like gene. Antimicrob. Agents Chemother. 48:3989-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everson, J. S., S. A. Garner, P. R. Lambden, B. A. Fane, and I. N. Clarke. 2003. Host range of chlamydiaphages phiCPAR39 and Chp3. J. Bacteriol. 185:6490-6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farencena, A., M. Comanducci, M. Donati, G. Ratti, and R. Cevenini. 1997. Characterization of a new isolate of Chlamydia trachomatis which lacks the common plasmid and has properties of biovar trachoma. Infect. Immun. 65:2965-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finkel, S. E., and R. Kolter. 2001. DNA as a nutrient: novel role for bacterial competence gene homologs. J. Bacteriol. 183:6288-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitch, W. M., E. M. Peterson, and L. M. de la Maza. 1993. Phylogenetic analysis of the outer-membrane-protein genes of chlamydiae, and its implication for vaccine development. Mol. Biol. Evol. 10:892-913. [DOI] [PubMed] [Google Scholar]
- 16.Gomes, J. P., W. J. Bruno, M. J. Borrego, and D. Dean. 2004. Recombination in the genome of Chlamydia trachomatis involving the polymorphic membrane protein C gene relative to ompA and evidence for horizontal gene transfer. J. Bacteriol. 186:4295-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomes, J. P., A. Nunes, W. J. Bruno, M. J. Borrego, C. Florindo, and D. Dean. 2006. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. J. Bacteriol. 188:275-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton, H. L., N. M. Dominguez, K. J. Schwartz, K. T. Hackett, and J. P. Dillard. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol. Microbiol. 55:1704-1721. [DOI] [PubMed] [Google Scholar]
- 19.Hayes, L. J., R. L. Bailey, D. C. Mabey, I. N. Clarke, M. A. Pickett, P. J. Watt, and M. E. Ward. 1992. Genotyping of Chlamydia trachomatis from a trachoma-endemic village in the Gambia by a nested polymerase chain reaction: identification of strain variants. J. Infect. Dis. 166:1173-1177. [DOI] [PubMed] [Google Scholar]
- 20.Hayes, L. J., S. Pecharatana, R. L. Bailey, T. J. Hampton, M. A. Pickett, D. C. Mabey, P. J. Watt, and M. E. Ward. 1995. Extent and kinetics of genetic change in the omp1 gene of Chlamydia trachomatis in two villages with endemic trachoma. J. Infect. Dis. 172:268-272. [DOI] [PubMed] [Google Scholar]
- 21.Jones, R. B., B. Van der Pol, D. H. Martin, and M. K. Shepard. 1990. Partial characterization of Chlamydia trachomatis isolates resistant to multiple antibiotics. J. Infect. Dis. 162:1309-1315. [DOI] [PubMed] [Google Scholar]
- 22.Kim, S. K., and R. DeMars. 2001. Epitope clusters in the major outer membrane protein of Chlamydia trachomatis. Curr. Opin. Immunol. 13:429-436. [DOI] [PubMed] [Google Scholar]
- 23.Lampe, M. F., R. J. Suchland, and W. E. Stamm. 1993. Nucleotide sequence of the variable domains within the major outer membrane protein gene from serovariants of Chlamydia trachomatis. Infect. Immun. 61:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenz, M. G., and W. Wackernagel. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58:563-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luria, S. E., and M. Delbrück. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto, A. 1988. Structural characteristics of chlamydial bodies. CRC Press, Boca Raton, FL.
- 27.McClarty, G. 1999. Chlamydial metabolism as inferred from the complete genome sequence. ASM Press, Washington, DC.
- 28.Millman, K., C. M. Black, R. E. Johnson, W. E. Stamm, R. B. Jones, E. W. Hook, D. H. Martin, G. Bolan, S. Tavare, and D. Dean. 2004. Population-based genetic and evolutionary analysis of Chlamydia trachomatis urogenital strain variation in the United States. J. Bacteriol. 186:2457-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Millman, K. L., S. Tavare, and D. Dean. 2001. Recombination in the ompA gene but not the omcB gene of Chlamydia contributes to serovar-specific differences in tissue tropism, immune surveillance, and persistence of the organism. J. Bacteriol. 183:5997-6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Misyurina, O. Y., E. V. Chipitsyna, Y. P. Finashutina, V. N. Lazarev, T. A. Akopian, A. M. Savicheva, and V. M. Govorun. 2004. Mutations in a 23S rRNA gene of Chlamydia trachomatis associated with resistance to macrolides. Antimicrob. Agents Chemother. 48:1347-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morre, S. A., J. M. Ossewaarde, J. Lan, G. J. van Doornum, J. M. Walboomers, D. M. MacLaren, C. J. Meijer, and A. J. van den Brule. 1998. Serotyping and genotyping of genital Chlamydia trachomatis isolates reveal variants of serovars Ba, G, and J as confirmed by omp1 nucleotide sequence analysis. J. Clin. Microbiol. 36:345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myers, G., T. Brettin, R. Leach, E. Sutton, and N. Thayer. 2003. STD sequence databases. Los Alamos National Laboratory Bioscience Division. http://www.stdgen.lanl.gov.
- 33.Ortiz, L., K. P. Demick, J. W. Petersen, M. Polka, R. A. Rudersdorf, B. Van der Pol, R. Jones, M. Angevine, and R. DeMars. 1996. Chlamydia trachomatis major outer membrane protein (MOMP) epitopes that activate HLA class II-restricted T cells from infected humans. J. Immunol. 157:4554-4567. [PubMed] [Google Scholar]
- 34.Palmer, L., and S. Falkow. 1986. A common plasmid of Chlamydia trachomatis. Plasmid 16:52-62. [DOI] [PubMed] [Google Scholar]
- 35.Poole, E., and I. Lamont. 1992. Chlamydia trachomatis serovar differentiation by direct sequence analysis of the variable segment 4 region of the major outer membrane protein gene. Infect. Immun. 60:1089-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redfield, R. J. 2001. Do bacteria have sex? Nat. Rev. Genet. 2:634-639. [DOI] [PubMed] [Google Scholar]
- 37.Rockey, D. D., W. Viratyosin, J. P. Bannantine, R. J. Suchland, and W. E. Stamm. 2002. Diversity within inc genes of clinical Chlamydia trachomatis variant isolates that occupy non-fusogenic inclusions. Microbiology 148:2497-2505. [DOI] [PubMed] [Google Scholar]
- 38.Santoyo, G., and D. Romero. 2005. Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol. Rev. 29:169-183. [DOI] [PubMed] [Google Scholar]
- 39.Somani, J., V. B. Bhullar, K. A. Workowski, C. E. Farshy, and C. M. Black. 2000. Multiple drug-resistant Chlamydia trachomatis associated with clinical treatment failure. J. Infect. Dis. 181:1421-1427. [DOI] [PubMed] [Google Scholar]
- 40.Stephens, R. S. 1999. Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, DC.
- 41.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 42.Stothard, D. R., J. A. Williams, B. Van Der Pol, and R. B. Jones. 1998. Identification of a Chlamydia trachomatis serovar E urogenital isolate which lacks the cryptic plasmid. Infect. Immun. 66:6010-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Streips, U. N., and R. E. Yasbin. 2002. Modern microbial genetics, 2nd ed. Wiley-Liss Publishers, New York, NY.
- 44.Suchland, R. J., D. D. Rockey, J. P. Bannantine, and W. E. Stamm. 2000. Isolates of Chlamydia trachomatis that occupy nonfusogenic inclusions lack IncA, a protein localized to the inclusion membrane. Infect. Immun. 68:360-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suchland, R. J., and W. E. Stamm. 1991. Simplified microtiter cell culture method for rapid immunotyping of Chlamydia trachomatis. J. Clin. Microbiol. 29:1333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor-Robinson, D. 2000. Genitourinary chlamydial infections. Int. J. STD AIDS 11:272. [DOI] [PubMed] [Google Scholar]
- 47.Whittington, W. L., C. Kent, P. Kissinger, M. K. Oh, J. D. Fortenberry, S. E. Hillis, B. Litchfield, G. A. Bolan, M. E. St. Louis, T. A. Farley, and H. H. Handsfield. 2001. Determinants of persistent and recurrent Chlamydia trachomatis infection in young women: results of a multicenter cohort study. Sex. Transm. Dis. 28:117-123. [DOI] [PubMed] [Google Scholar]
- 48.Xiong, L., F. Kong, H. Zhou, and G. L. Gilbert. 2006. Use of PCR and reverse line blot hybridization assay for rapid simultaneous detection and serovar identification of Chlamydia trachomatis. J. Clin. Microbiol. 44:1413-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang, C. L., I. Maclean, and R. C. Brunham. 1993. DNA sequence polymorphism of the Chlamydia trachomatis omp1 gene. J. Infect. Dis. 168:1225-1230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.