Abstract
Marine hydrocarbonoclastic bacteria, like Alcanivorax borkumensis, play a globally important role in bioremediation of petroleum oil contamination in marine ecosystems. Accumulation of storage lipids, serving as endogenous carbon and energy sources during starvation periods, might be a potential adaptation mechanism for coping with nutrient limitation, which is a frequent stress factor challenging those bacteria in their natural marine habitats. Here we report on the analysis of storage lipid biosynthesis in A. borkumensis strain SK2. Triacylglycerols (TAGs) and wax esters (WEs), but not poly(hydroxyalkanoic acids), are the principal storage lipids present in this and other hydrocarbonoclastic bacterial species. Although so far assumed to be a characteristic restricted to gram-positive actinomycetes, substantial accumulation of TAGs corresponding to a fatty acid content of more than 23% of the cellular dry weight is the first characteristic of large-scale de novo TAG biosynthesis in a gram-negative bacterium. The acyltransferase AtfA1 (ABO_2742) exhibiting wax ester synthase/acyl-coenzyme A:diacylglycerol acyltransferase (WS/DGAT) activity plays a key role in both TAG and WE biosynthesis, whereas AtfA2 (ABO_1804) was dispensable for storage lipid formation. However, reduced but still substantial residual TAG levels in atfA1 and atfA2 knockout mutants compellingly indicate the existence of a yet unknown WS/DGAT-independent alternative TAG biosynthesis route. Storage lipids of A. borkumensis were enriched in saturated fatty acids and accumulated as insoluble intracytoplasmic inclusions exhibiting great structural variety. Storage lipid accumulation provided only a slight growth advantage during short-term starvation periods but was not required for maintaining viability and long-term persistence during extended starvation phases.
Approximately 2 to 6 million tons of crude petroleum oil enters marine environments per annum, mainly from anthropogenic sources, but also pollution from natural marine oil seepages results in considerable input of petroleum oil into the sea. Petroleum oil is highly toxic to the majority of living organisms and can be tolerated only by relatively few species. Oil pollution events are therefore serious threats to sensitive marine ecosystems and result in severe ecological perturbations.
Fortunately, a substantial proportion of the petroleum oil entering marine habitats is degraded by indigenous microorganisms. Although many marine bacteria are capable of degrading petroleum hydrocarbons, only a few of them seem to be important for petroleum biodegradation in natural marine environments. They belong to a new taxonomic group of phylogenetically related oil-degrading γ-proteobacteria which have been isolated during the last decade from different sites all over the world. These so-called hydrocarbonoclastic bacteria so far comprise the genera Alcanivorax (42), Cycloclasticus (10), Marinobacter (15), Neptunomonas (21), Oleiphilus (16), Oleispira (41), and Thalassolituus (40). Among those bacteria, particularly members of the genus Alcanivorax seem to play a major role in the first steps of petroleum oil biodegradation. They are assumed to be of global importance for removal of crude oil contamination in marine environments due to their cosmopolitan distribution (18, 19, 25).
Alcanivorax sp. and other hydrocarbonoclastic bacteria exhibit a unique oligotrophic physiology. They are specialized for hydrocarbon degradation but have an otherwise highly restricted substrate spectrum, being capable of utilizing only a few organic acids like acetate and pyruvate, but not simple sugars, for growth (42). Alcanivorax spp. are present only in low abundance in pristine waters, but they multiply and grow rapidly in oil-polluted waters, where they can constitute 80 to 90% of the microbial community (19, 25, 36). After the initial bloom and rapid increase in abundance, the population size declines to much lower numbers within a few weeks, correlating with the biodegradation of the major portion of saturated hydrocarbons (20).
Despite the severe ecological problems caused by oil pollution, the vast majority of the world's ocean waters are still not polluted with petroleum oil. In fact, the marine habitat constitutes quite a nutrient-limited environment, and oil spills are rather rare events affecting relatively small areas. In view of their oligotrophic lifestyle and the sporadic availability of hydrocarbons as substrates for their growth, it seems obvious that marine hydrocarbonoclastic bacteria are frequently facing extended phases of starvation conditions. Therefore, these bacteria must have developed strategies and abilities for adapting to and surviving under such unfavorable environmental conditions which probably are important factors contributing to their successful ubiquitous distribution.
The accumulation of intracellular carbon storage compounds, like storage lipids, may be one strategy for the survival of starvation periods. Storage lipids occur very frequently among hydrocarbon-utilizing marine bacteria, with most of them accumulating specialized polymeric lipids like poly(hydroxybutyrate) or other poly(hydroxyalkanoic acids) (PHAs) (1). In addition to PHAs, which represent the most abundant class of lipophilic storage compounds produced by bacteria (33), prokaryotic storage lipids are less frequently found in the form of triacylglycerols (TAGs) and wax esters (WEs). The accumulation of TAGs in large quantities has so far been described only for certain gram-positive bacteria of the actinomycetes group, belonging to the genera Mycobacterium, Nocardia, Rhodococcus, and Streptomyces (2). In contrast, WEs (oxoesters of long-chain primary fatty alcohols and long-chain fatty acids), along with small amounts of TAGs, seem to be common storage lipids within the gram-negative genus Acinetobacter (12, 13, 14, 22). Recently, the key enzyme for storage lipid biosynthesis in Acinetobacter baylyi strain ADP1 (formerly Acinetobacter sp. strain ADP1) (38), the wax ester synthase/acyl coenzyme A (acyl-CoA):diacylglycerol acyltransferase (WS/DGAT), was discovered. This unspecific acyltransferase simultaneously synthesizes WEs and TAGs by utilizing fatty acid coenzyme A thioesters (acyl-CoA) in addition to long-chain fatty alcohols and diacylglycerols (DAGs), respectively, as substrates. Remarkably, this novel enzyme exhibits no homology to known acyltransferases involved in TAG or WE biosynthesis in eukaryotes, and it is widely distributed among TAG-accumulating actinomycetes (23).
The recently finished genome sequencing project for Alcanivorax borkumensis (17, 31) prompted us to search for the presence of genes with potential involvement in storage lipid biosynthesis. The identification of two gene products homologous to the WS/DGAT from A. baylyi strain ADP1 indicated the potential capability of A. borkumensis to synthesize and accumulate TAGs and/or WEs. This assumption was corroborated by the recent description of WE formation in the hydrocarbonoclastic strain Alcanivorax jadensis (formerly Fundibacter jadensis) (4). The present study describes the analysis of biosynthesis and accumulation of storage lipids in A. borkumensis and the use of biochemical characterization and directed mutagenesis to examine the role of the two WS/DGAT-homologous proteins in this pathway.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains and plasmids used in this study are listed in Table 1. A. borkumensis strains were cultivated aerobically at 30°C in ONR7a medium (10) containing 1% (wt/vol) sodium pyruvate or 0.5% (vol/vol) hexadecane for 72 h as described previously (42). Marinobacter hydrocarbonoclasticus (72 h at 30°C), Alcanivorax jadensis T9 (72 h at 30°C), and Thalassolituus oleivorans (96 h at room temperature) were grown aerobically in ONR7a medium containing 1% (wt/vol) sodium pyruvate. Cells of Escherichia coli were grown in LB medium at 37°C (30). Media were inoculated 1% (vol/vol) from saturated precultures. Solid medium contained 1.8% (wt/vol) agar. If appropriate, antibiotics were added at the following concentrations: ampicillin (Ap), 75 mg liter−1; kanamycin (Km), 50 mg liter−1; streptomycin (Sm), 100 mg liter−1; chloramphenicol (Cm), 34 mg liter−1; nalidixic acid, 10.0 mg liter−1; tetracycline, 12.5 mg liter−1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| Alcanivorax borkumensis strains | ||
| SK2 | Type strain, wild type | 42; DSM 11573 |
| atfA1ΩKm | atfA1 disruption mutant; Kmr; derivative of SK2 | This study |
| atfA2ΩSm | atfA2 disruption mutant; Smr; derivative of SK2 | This study |
| atfA1ΩKmatfA2ΩSm | atfA1 and atfA2 double disruption mutant; Kmr; Smr; derivative of atfA1ΩKm | This study |
| Alcanivorax jadensis T9 | Type strain, wild type | 5, 11; DSM 12178 |
| Marinobacter hydrocarbonoclasticus | Type strain, wild type | 15; DSM 8798 |
| Thalassolituus oleivorans | Type strain, wild type | 40; DSM 14913 |
| Escherichia coli strains | ||
| BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm (DE3) | Novagen, Madison, WI |
| S17-1 | recA1 thi1 hsdR17 (rK− mK+) proA tra-RP4 | 32 |
| Plasmids | ||
| pET23a(+) | Apr; T7 promoter-based expression vector | Novagen, Madison, WI |
| pET23a::atfA1 | Derivative of pET23a containing the atfA1 gene as a 1.4-kbp HindIII/XhoI fragment; Apr | This study |
| pET23a::atfA2 | Derivative of pET23a containing the atfA2 gene as a 1.4-kbp HindIII/XhoI fragment; Apr | This study |
| pET23a::atfA1ΩKm | ΩKmr cassette cloned into StuI site of pET23a::atfA1; Apr; Kmr | This study |
| pET23a::atfA2ΩSm | ΩSmr cassette cloned into EheI site of pET23a::atfA2; Apr; Smr | This study |
| pSKsymΩKm | Apr; Kmr; contains ΩKmr cassette | 27 |
| pCDFDuet-1 | Smr; source for ΩSmr cassette construction | Novagen, Madison, WI |
| pSUP202 | Apr; Cmr; Tcr; ColE1 origin; mob site; unable to replicate in A. borkumensis | 32 |
| pSUP202::pET23a::atfA1ΩKm | Fusion of pSUP202 and pET23a::atfA1ΩKm via HindIII sites; mob site; Apr; Cmr; Kmr | This study |
| pSUP202::pET23a::atfA2ΩSm | Fusion of pSUP202 and pET23a::atfA2ΩSm via HindIII sites; mob site; Apr; Cmr; Smr | This study |
Tc, tetracycline.
Preparation of crude extracts.
Cells of A. borkumensis strains were cultivated with pyruvate as described above. Cells of E. coli were grown in LB medium to an optical density at 600 nm of 0.5 at 37°C before isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and induced cultures were incubated at 37°C for 3 h. Cells were finally harvested by centrifugation at 4,000 × g for 20 min at 4°C, washed twice with 125 mM sodium phosphate buffer (pH 7.4), and resuspended in the same buffer. Cell disruption was done by ultrasonification. Protein concentrations were determined by the Bradford method (3).
Determination of enzyme activities.
WS/DGAT activity was determined in a total volume of 250 μl containing 12.5 μg ml−1 bovine serum albumin, 4.72 μM [1-14C]palmitoyl-CoA (specific activity, 1.961 Bq pmol−1; Hartmann Radiochemicals, Braunschweig, Germany), 125 mM sodium phosphate buffer (pH 7.4), and different acceptor molecules at a concentration of 3.75 mM. 1-Hexadecanol and 1,2-dipalmitin were used as standard substrates for assaying WS and DGAT activity, respectively. Water-insoluble substrates and bovine serum albumin were applied as double-concentrated stock solutions emulsified by ultrasonification. The assay mixtures were incubated for 20 min at 35°C, and the reactions were stopped by extraction with 500 μl chloroform/methanol (1:1, vol/vol). After centrifugation, the chloroform phase was withdrawn and evaporated to dryness, and 40 μg of unlabeled reference substances was added. The reaction products were separated by thin-layer chromatography (TLC) using different solvent systems for the separation of linear, cyclic, and aromatic WEs as well as monoacylglycerols (MAGs), DAGs, or TAGs, respectively, essentially as described previously (35). After separation of lipids by TLC and staining of TLC plates with iodine vapor, spots corresponding to the reaction products were scraped from the plates, and radioactivity was measured by scintillation counting. If reference substances were not available, the radioactive reaction products on the TLC plates were detected by autoradiography.
DNA manipulations.
DNA manipulations and other standard molecular biology techniques were performed according to the method described in reference 30. The oligonucleotide primers used for PCR amplifications and reverse transcription (RT)-PCR analysis are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study as primers for PCR, diagnostic PCR, and RT-PCR
| Primer | Sequencea |
|---|---|
| atfA1_5-end | 5′-TTTTTTAAGCTTAAGGAGAATATATGAAAGCGCTT-3′ |
| atfA1_3-end | 5′-TTTTTTCTCGAGCTATTTAATTCCTGCACCGATTT-3′ |
| atfA2_5-end | 5′-TTTTTTAAGCTTAAGGAGCAGCAAGTATGGCCCGT-3′ |
| atfA2_3-end | 5′-TTTTTTCTCGAGTCAAGGCTCCACCAGCG-3′ |
| atfA1_up | 5′-CAGCTGGCATGGAGAGTGCATAAC-3′ |
| atfA1_down | 5′-GATGCGGTTTAGTTCAGTTGCCAT-3′ |
| atfA2_up | 5′- GGCTGACTGTTGATAACCAAACGC-3′ |
| atfA2_down | 5′-CCCGTTAGCATGCCGGCAATGTG-3′ |
| atfA1_int1 | 5′-GACAGCAACCCATGCACGTAG-3′ |
| atfA1_int2 | 5′-GGTGAGGGCGGTGAAATTGAG-3′ |
| atfA2_int1 | 5′- AGCATCTGCCGCCCATTTAAC-3′ |
| atfA2_int2 | 5′-ACCGATCACGCCAAACTCAAG-3′ |
| aceA_int1 | 5′-GCGACGAAGCCAAGCAGAAAG-3′ |
| aceA_int2 | 5′-TCACAGTAGCGATGCCCTGAC 3′ |
| Abo_2743_int1 | 5′-GTGTTCAGCGGCAATATCAGC-3′ |
| Abo_2743_int2 | 5′-TGGAGCCAAAGTGAGCACATC-3′ |
| ΩSmr_5-end | 5′-TTTTTTCCCGGGCTCACGCCCGGAGCGTAGCGACC-3′ |
| ΩSmr_3-end | 5′-TTTTTTCCCGGGAACGACCCTGCCCTGAACCGACG-3′ |
Restriction sites used for cloning purposes are underlined. Start and stop codons of atfA1 and atfA2 are indicated in boldface.
Cloning of atfA1 and atfA2 from A. borkumensis SK2.
The atfA1 and atfA2 genes were amplified from total genomic DNA of A. borkumensis SK2 by tailored PCR using the oligonucleotides atfA1_5-end and atfA1_3-end or atfA2_5-end and atfA2_3-end (Table 2) as primers, respectively. The resulting PCR products were cloned as HindIII-XhoI fragments into the expression vector pET23a colinear to the T7 promoter, yielding pET23a::atfA1 or pET23a::atfA2, respectively, and were then transformed into E. coli BL21(DE3).
Gene disruption by biparental filter mating.
For gene inactivation of atfA1, an ΩKm cassette was isolated by SmaI digestion of plasmid pSKsymΩKm and cloned into the singular StuI site of atfA1, yielding pET23a::atfA1ΩKm. For gene inactivation of atfA2, an ΩSm cassette was amplified by PCR from the vector pCDFDuet-1, employing the primers ΩSmr_5-end and ΩSmr_3-end (Table 2), digested with SmaI, and cloned into the singular EheI site of atfA2, yielding pET23a::atfA2ΩSm. The plasmids were then linearized by HindIII digestion; fused to the HindIII restricted mobilizable suicide plasmid pSUP202, yielding plasmid pSUP202::pET23a::atfA1ΩKm or pSUP202::pET23a::atfA2ΩSm, respectively; and transformed into E. coli S17-1.
Subsequently, inactivation of the atfA1 and atfA2 genes in A. borkumensis was achieved by conjugational transfer of the suicide plasmids pSUP202::pET23a::atfA1ΩKm and pSUP202::pET23a::atfA2ΩSm from E. coli strain S17-1 (donor) to A. borkumensis strains SK2 and atfA1ΩKm (recipients), respectively, employing a biparental filter-mating technique. The donor strain was cultivated at 30°C in LB medium for 24 h, and the recipient strain was grown in ONR7a medium containing pyruvate for 48 h at 30°C. Cells were harvested, washed once with cLB mating medium (LB medium supplemented with 2% [wt/vol] pyruvate, 0.0445% [wt/vol] Na2HPO4 × 2H2O, 0.25% [wt/vol] NaNO3, 1.15% [wt/vol] NaCl, 0.375% [wt/vol] KCl, 0.0735% [wt/vol] CaCl2 × 2H2O), and concentrated 10-fold in cLB mating medium. The donor and recipient cells were mixed at a ratio of 1:4 (vol/vol), and 200 μl of this cell mixture was spotted on Millipore nitrocellulose membrane filters (diameter, 45 mm; pore size, 0.45 μm) and placed on cLB mating agar plates. After 24 h of incubation at 30°C, cells were washed from the filters with 10 mM MgSO4, and aliquots were plated onto ONR7a agar plates supplemented with pyruvate containing appropriate antibiotics for selection of transconjugants (Km or Sm) and nalidixic acid to select against E. coli. After incubation at 30°C for 7 days, the resulting transconjugants were picked and patched onto selective agar plates with or without Cm. Putative homozygous gene disruptant mutants resulting from homologous recombination with a double crossover event were identified by their Cm sensitivity.
Transcription analysis of atfA1, atfA2, aceA, and Abo_2347 in A. borkumensis SK2.
RNA from cells cultivated with pyruvate was isolated using the RNeasy RNA purification kit (QIAGEN, Hilden, Germany) according to the manufacturer's protocol. RT-PCR was performed using the QIAGEN OneStep RT-PCR kit (QIAGEN, Hilden, Germany) according to the instructions provided, using the oligonucleotides atfA1_int1 and atfA1_int2 for analysis of atfA1, atfA2_int1 and atfA2_int2 for analysis of atfA2, aceA_int1 and aceA_int2 for analysis of aceA, or Abo_2743_int1 and Abo_2743_int2 for analysis of Abo_2743 (Table 2), and employing 0.5 ng RNA as template. Controls for DNA contamination experiments were set up by adding the RNA template after the reverse transcriptase step and before activating the Taq polymerase.
TLC.
TLC analysis of lipid extracts from whole cells was done as described previously (23) using the solvent system hexane:diethylether:acetic acid (80:20:1, vol/vol/vol) for WE and TAG analysis. Lipids were visualized by staining with iodine vapor. Oleyl oleate and triolein were used as reference substances for WEs and TAGs, respectively.
Fatty acid analysis.
Fatty acid analysis of whole cells was done by gas liquid chromatography (GC) according to reference (24) after derivatization to fatty acid methyl esters by sulfuric acid-catalyzed methanolysis. Fatty acid methyl esters were analyzed by GC using an Agilent 6850 GC (Agilent Technologies, Waldbronn, Germany) equipped with a BP21 capillary column (50 by 0.22 mm; film thickness, 250 nm; SGE, Darmstadt, Germany) and a flame ionization detector (Agilent Technologies, Waldbronn, Germany). A 2-μl portion of the organic phase was analyzed after split injection (1:5); hydrogen (constant flow, 0.6 ml min−1) was used as carrier gas. The temperatures of the injector and detector were 250°C and 275°C, respectively. The following temperature program was applied: 120°C for 5 min, increase of 3°C min−1 to 180°C, increase of 10°C min−1 to 220°C, 220°C for 31 min. Substances were identified by comparison of their retention times with those of authentic standard fatty acid methyl esters.
TEM.
After being washed three times in phosphate-buffered saline (PBS) (pH 7.3), the cells were fixed with 2.5% (wt/vol) glutaraldehyde in 0.1 M PBS (pH 7.3) for 16 h. After being washed three more times in PBS, each time for 20 min, the cells were postfixed with 1% (wt/vol) osmium tetroxide in 0.1 M PBS (pH 7.3) for 90 min and washed once with PBS for 15 min. Then the samples were dehydrated by a graded water-ethanol series (30%, 50%, 70%, 90%, 96%, and 100% ethanol and propylene oxide), with each step lasting for 15 min. For thin sectioning, the samples were embedded in SPURR resin with 50% (wt/vol) propylene oxide for 4 h and resin with 33% (wt/vol) propylene oxide for 16 h. The SPURR resin was changed every 24 h for a period of 3 days. The polymerization of the resin was performed at 70°C for 48 h. Sections 70 to 80 nm thick were made with an Ultracut Microtome (LEICA Mikroskopie und Systeme GmbH, Germany) using a diamond knife and were subsequently placed on a 200-mesh copper grid. Imaging was performed with an H-500 transmission electron microscope (TEM; Hitachi, Japan) in the bright-field mode at an acceleration voltage of 75 kV at room temperature.
Starvation experiments.
Cells of A. borkumensis strains were cultivated with pyruvate as described above to allow for accumulation of storage lipids. Cells were harvested, washed three times in ONR7a medium without any carbon source, resuspended in the same medium, and incubated at 30°C. At various time points aliquots of appropriate dilutions were plated onto ONR7a solid medium containing pyruvate for counting of viable cells.
RESULTS
Identification of two genes, atfA1 and atfA2, coding for WS/DGAT-homologous proteins in A. borkumensis.
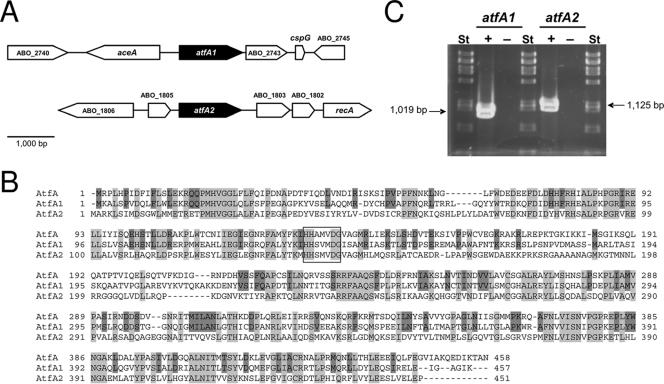
By using the WS/DGAT from A. baylyi strain ADP1, which is the first and best characterized member of this novel family of prokaryotic acyltransferases (23, 35), as the template, two open reading frames, termed atfA1 (ABO_2742) and atfA2 (ABO_1804), were identified by a BlastP search in the genome sequence of A. borkumensis SK2, whose translational products revealed significant homologies. AtfA1 and AtfA2 exhibited 49.1% and 40.2% amino acid identity to WS/DGAT, respectively, whereas AtfA1 and AtfA2 shared 46.0% amino acid identity to each other. Both acyltransferase candidates comprised a conserved putative active site motif (HHXXXDG [Fig. 1B ]), which has been proposed to be essential for catalytic activity (23).
FIG. 1.
Characterization of the WS/DGAT homologues AtfA1 and AtfA2 from A. borkumensis SK2. (A) Organization of the atfA1 and atfA2 gene loci. ABO_2740, hypothetical protein gene; aceA, isocitrate lyase gene; ABO_2743, hypothetical protein gene; cspG, cold-shock domain family protein gene; ABO_2745, hypothetical protein gene; ABO_1806, AMP-binding family protein gene; ABO_1805, hypothetical protein gene; ABO_1803, hypothetical protein gene; ABO_1802, CinA domain protein gene; recA, RecA protein gene. (B) Multiple amino acid sequence alignment of the WS/DGAT enzyme AtfA from A. baylyi strain ADP1 and AtfA1 and AtfA2 from A. borkumensis SK2. Analysis was done using the CLUSTAL W program (37). Residues identical in all sequences are shaded in light gray, and those conserved only in AtfA and AtfA1 are shaded in dark gray. A putative active site motif is boxed. (C) Transcription analysis of atfA1 and atfA2. Expression of atfA1 and atfA2 was analyzed by RT-PCR in samples derived from cells cultivated with 1% (wt/vol) sodium pyruvate as the carbon source. St, PstI-digested λ DNA. +, RT-PCR assay; −, control for DNA contamination experiments.
Neither atfA1 nor atfA2 was clustered with genes with any known relevance for storage lipid biosynthesis (Fig. 1A). Interestingly, atfA1 is situated in close proximity to the origin of replication of the chromosome, probably resulting in a high gene dosage.
Heterologous expression of atfA1 and atfA2 and biochemical characterization of the encoded acyltransferases.
The genes atfA1 and atfA2 were heterologously expressed in E. coli BL21(DE3) by use of the T7 promoter- and polymerase-based pET system. Although sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis revealed that most of the proteins were expressed as insoluble inclusion bodies (data not shown), functional expression of atfA1 as well as atfA2 in active form was achieved by using this system (Table 3). High levels of WS and lower DGAT activity were detected for AtfA1. In contrast, AtfA2 exhibited substantial WS activity, whereas DGAT activity was negligible (Table 3).
TABLE 3.
WS and DGAT activities in A. borkumensis and E. coli strainsa
| Strain | Sp act (pmol mg−1 min−1) of enzyme:
|
|
|---|---|---|
| WS | DGAT | |
| E. coli BL21(DE3) | ||
| pET23a | 0.4 ± 0.1 | 0.2 ± 0.0 |
| pET23a::atfA1 | 308.2 ± 37.9 | 131.7 ± 4.3 |
| pET23a::atfA2 | 237.0 ± 30.5 | 4.6 ± 0.8 |
| pET23a::atfA1ΩKm | 1.0 ± 0.5 | 0.3 ± 0.1 |
| pET23a::atfA2ΩSm | 4.9 ± 0.3 | 0.3 ± 0.1 |
| A. borkumensis | ||
| SK2 | 64.0 ± 8.1 | 17.2 ± 0.3 |
| atfA1ΩKm | 11.3 ± 1.4 | 2.5 ± 0.2 |
| atfA2ΩSm | 38.5 ± 1.2 | 10.1 ± 1.6 |
| atfA1ΩKmatfA2ΩSm | 0.4 ± 0.2 | 1.6 ± 0.8 |
Assays were done employing crude extracts corresponding to 50 μg total protein for recombinant E. coli strains or 100 μg total protein for pyruvate-grown A. borkumensis strains. Data are mean values ± standard deviations from experiments done in triplicate. Control experiments without addition of acyl acceptor substrates revealed WS activities of 14.5 and 6.4 pmol mg−1 min−1 and DGAT activities of 10.3 and 0.3 pmol mg−1 min−1 for E. coli strains expressing atfA1 and atfA2, respectively. One picomole per milligram per minute corresponds to 117.66 and 235.32 cpm for E. coli and A. borkumensis strains, respectively.
Both enzymes were then further characterized biochemically by analyzing the specificity of these acyltransferases for various acyl acceptor molecules. Both enzymes could comparably utilize a broad range of short-, medium-, and long-chain linear alcohols with the highest specificity for medium-chain alcohols (1-decanol) (Table 4). Furthermore, both acyltransferases were also highly active with cyclic or phenolic alcohols (Table 4). In clear contrast to this, dramatic differences between AtfA1 and AtfA2, regarding their specificity for different acylglycerol substrates, were observed (Table 5). Whereas AtfA2 highly efficiently utilized all tested MAGs, AtfA1 showed a clear preference for 1-MAG. Most strikingly, only AtfA1 possessed significant DGAT activity, whereas AtfA2 was virtually completely inactive with both DAGs used as substrates (Table 5). In summary, these results unequivocally demonstrated that AtfA1 and AtfA2 are truly potent acyltransferases, both exhibiting very broad but substantially different substrate specificities.
TABLE 4.
Acyl acceptor specificities of AtfA1 and AtfA2 from A. borkumensis SK2 for different alcohol substratesa
| Acyl acceptor | Activity (% of hexadecanol control)
|
|
|---|---|---|
| AtfA1 | AtfA2 | |
| None | 4.4 ± 1.2 | 0.3 ± 0.0 |
| 1-Hexadecanol | 100.0 ± 3.2 | 100.0 ± 12.9 |
| 1-Butanol | 114.9 ± 16.7 | 72.7 ± 18.1 |
| 1-Decanol | 161.6 ± 26.4 | 115.6 ± 8.3 |
| 1-Tetracosanol | 18.2 ± 4.2 | 7.5 ± 2.2 |
| Cyclohexanol | 99.0 ± 2.2 | 129.9 ± 55.4 |
| Cyclohexylethanol | 131.6 ± 9.4 | 167.0 ± 13.4 |
| 2-Phenylethanol | 109.8 ± 10.7 | 154.3 ± 19.9 |
Assays were done employing crude extracts corresponding to 50 μg total protein from recombinant E. coli BL21(DE3) strains harboring plasmid pET23a::atfA1 or pET23a::atfA2 for heterologous expression of AtfA1 or AtfA2, respectively. Values are indicated as relative specific activities in comparison to hexadecanol as acyl acceptor. Data are mean values ± standard deviations from experiments done in triplicate. One hundred percent activity is equivalent to 324.9 and 237.0 pmol mg−1 min−1 for AtfA1 and AtfA2, respectively.
TABLE 5.
Acyl acceptor specificities of AtfA1 and AtfA2 from A. borkumensis SK2 for glycerol and acylglycerolsa
| Enzyme and acyl acceptor | Formation (pmol mg−1 min−1) of:
|
||||
|---|---|---|---|---|---|
| 1- + 3-MPG | 2-MPG | 1,2- + 2,3-DPG | 1,3-DPG | TPG | |
| AtfA1 | |||||
| Glycerol | ND | ND | NT | NT | NT |
| 1-Monopalmitoylglycerol | 32 ± 2 | 237 ± 16 | NT | ||
| 2-Monopalmitoylglycerol | 85 ± 27 | 41 ± 10 | NT | ||
| 3-Monopalmitoylglycerol | 31 ± 2 | 74 ± 3 | NT | ||
| 1,2-Dipalmitoylglycerol | 181 ± 9 | ||||
| 1,3-Dipalmitoylglycerol | 100 ± 36 | ||||
| AtfA2 | |||||
| Glycerol | ND | ND | NT | NT | NT |
| 1-Monopalmitoylglycerol | 22 ± 1 | 345 ± 27 | NT | ||
| 2-Monopalmitoylglycerol | 232 ± 15 | 134 ± 5 | NT | ||
| 3-Monopalmitoylglycerol | 19 ± 1 | 378 ± 33 | NT | ||
| 1,2-Dipalmitoylglycerol | 4 ± 1 | ||||
| 1,3-Dipalmitoylglycerol | 2 ± 0 | ||||
Assays were done employing crude extracts corresponding to 50 μg total protein from recombinant E. coli BL21(DE3) strains harboring plasmid pET23a::atfA1 or pET23a::atfA2 for heterologous expression of AtfA1 or AtfA2, respectively. 1,2- and 2,3-dipalmitoylglycerol as well as 1- and 3-monopalmitoylglycerol could not be separated under the applied TLC conditions and are therefore shown together. Data are averages ± standard deviations from experiments done in triplicate. Abbreviations: MPG, monopalmitoylglycerol; DPG, dipalmitoylglycerol; TPG, tripalmitoylglycerol; ND, not detectable; NT, not tested.
TAGs and WEs are the principal storage lipids in A. borkumensis.
The A. borkumensis genome comprises two differentially expressed putative PHA synthase genes (phaC1 and phaC2), which is a hint of the presence of PHAs in this bacterium (29). The identification of two WS/DGAT-homologous acyltransferases indicated that TAGs and/or WEs also might be synthesized in A. borkumensis. We therefore tested the presence of potential storage lipid species in alkane-grown and pyruvate-grown cells of A. borkumensis by means of GC and TLC analyses. Despite the existence of two PHA synthase genes, we were unable to detect significant amounts of any 3-hydroxy fatty acid, which are the monomer constituents of PHAs, by GC analysis regardless of the carbon source used for cultivation (in total, less than 1% of the cellular dry weight [CDW]). In conclusion, PHAs could be present only in irrelevant amounts in A. borkumensis under those conditions.
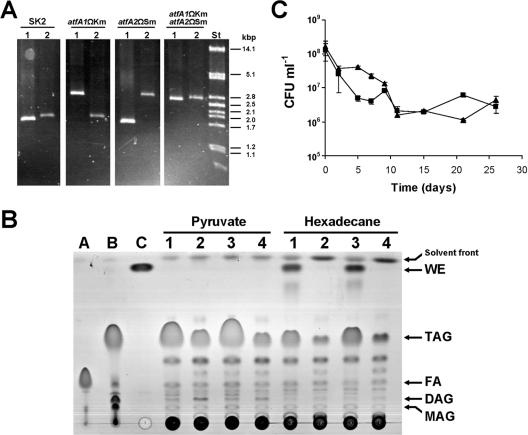
In contrast, large amounts of TAGs accumulated in cells cultivated with pyruvate, whereas WEs were not produced under such conditions (Fig. 2B). However, WEs were synthesized together with larger amounts of TAGs when hexadecane was used as the substrate. Besides TAGs and WEs, numerous other lipophilic substances of yet unknown chemical structure could be detected by TLC analysis. However, they were only of rather low abundance and were not further analyzed in the present study (Fig. 2B). The neutral lipids, mainly consisting of TAGs, amounted to a corresponding cellular fatty acid content of 23.2% of the CDW during growth on pyruvate, whereas the amounts of accumulated lipids were smaller when the cells were cultivated with hexadecane (total fatty acid content, 9.2% of CDW) (Table 6) . Interestingly, TAGs isolated from pyruvate-grown cells exhibited a saturated fatty acid (particularly palmitic acid) content that was significantly higher than the total lipid fatty acid composition. The storage lipids produced during cultivation with hexadecane contained almost exclusively saturated fatty acids, although monounsaturated fatty acids (oleic acid and palmitoleic acid) still constituted a significant portion (33.8 mol%) of the total cellular fatty acids under these culture conditions (Table 6). TAGs and WEs isolated from these cells contained, besides palmitic acid (main constituent) and myristic acid (minor constituent), only 2.1 mol% and traces of unsaturated fatty acids, respectively (Table 6). This strong bias towards saturated fatty acids indicates that the acyltransferases involved in TAG and WE biosynthesis in A. borkumensis strain SK2 probably have a high preference for saturated acyl-CoAs, although this hypothesis was not confirmed experimentally in this study. Hexadecanol, derived from oxidation of the growth substrate hexadecane, was the only fatty alcohol component of WEs isolated from alkane-grown cells detectable by GC analysis (data not shown).
FIG. 2.
Characterization of A. borkumensis atfA1 and atfA2 gene disruption mutants. (A) Genotypic characterization of A. borkumensis mutants. Mutants were analyzed by diagnostic PCR using oligonucleotide primer pairs binding up- and downstream of the atfA1 (lanes 1) or atfA2 (lanes 2) gene, respectively. Expected sizes: native atfA1, 1.5 kbp; native atfA2, 1.6 kbp; atfA1ΩKm, 2.5 kbp; atfA2ΩSm, 2.6 kbp. St, PstI-digested λ DNA. (B) TLC analyses of storage lipid accumulation in A. borkumensis mutants. Cells were cultivated in ONR7a medium containing 1% (wt/vol) sodium pyruvate or 0.5% (vol/vol) hexadecane, respectively, for 72 h and analyzed by TLC. Lipid extracts obtained from 1.5 mg lyophilized cells were applied per lane. Lanes: A, oleic acid; B, triolein; C, oleyl oleate; 1, SK2; 2, atfA1ΩKm; 3, atfA2ΩSm; 4, atfA1ΩKmatfA2ΩSm. FA, free fatty acids. (C) Survival of A. borkumensis strains SK2 (triangles) and atfA1ΩKmatfA2ΩSm (squares) during carbon starvation. After 72 h of preincubation in ONR7a medium with pyruvate to allow for the accumulation of storage lipids, cells were transferred into ONR7a medium containing no carbon source, and survival was monitored by determining viable cell counts. Values are means ± errors of results from duplicate experiments.
TABLE 6.
Fatty acid content and fatty acid composition of A. borkumensis strains and fatty acid composition of purified lipids isolated from strain SK2a
| Carbon source | Strain or lipid | Total fatty acids (% of CDW) | Composition (mol%) of fatty acid:
|
||||
|---|---|---|---|---|---|---|---|
| C14:0 | C16:0 | C16:1 | C18:0 | C18:1 | |||
| Pyruvate | SK2 | 23.2 | 5.1 | 46.8 | 9.7 | 3.3 | 35.1 |
| atfA1ΩKm | 10.1 | 3.4 | 39.7 | 12.1 | 3.7 | 41.1 | |
| atfA2ΩSm | 26.9 | 5.1 | 47.1 | 9.4 | 3.4 | 35.0 | |
| atfA1ΩKmatfA2ΩSm | 9.6 | 2.9 | 36.8 | 13.9 | 5.2 | 41.2 | |
| TAGs purified from SK2 | ND | 7.0 | 60.8 | 5.0 | 4.4 | 22.7 | |
| Hexadecane | SK2 | 9.2 | 8.5 | 55.3 | 11.5 | 2.4 | 22.3 |
| atfA1ΩKm | 4.0 | 4.9 | 42.1 | 19.7 | 2.9 | 30.4 | |
| atfA2ΩSm | 11.7 | 9.5 | 54.5 | 10.8 | 2.6 | 22.7 | |
| atfA1ΩKmatfA2ΩSm | 5.3 | 5.3 | 42.5 | 19.4 | 2.8 | 30.1 | |
| TAGs purified from SK2 | ND | 15.8 | 76.0 | 1.4 | 5.9 | 0.7 | |
| WEs purified from SK2 | ND | 6.4 | 85.3 | tr | 8.3 | tr | |
Cells were cultivated for 72 h in ONR7a medium with 1% (wt/vol) sodium pyruvate or 0.5% (vol/vol) hexadecane. Lyophilized cells were analyzed by GC. TAGs and WEs were purified from cells of the wild type (SK2) by preparative TLC prior to subjection to GC analysis. ND, not determined; tr, trace amount (<0.5mol%).
Functional analysis of atfA1 and atfA2 in A. borkumensis by directed gene insertion mutagenesis.
For evaluating the functional role of the acyltransferases AtfA1 and AtfA2 and their relative contribution to storage lipid biosynthesis in A. borkumensis, we generated isogenic atfA1 and atfA2 single knockout mutants as well as a double knockout mutant defective in both genes. This was achieved by disrupting the coding gene regions via insertion of a kanamycin (in the case of atfA1) or streptomycin (in the case of atfA2) gene cassette. The loss of acyltransferase activities in recombinant E. coli isolates proved that by this method, both enzymes were efficiently inactivated (Table 3). Homozygous gene disruption mutants generated by homologous recombination via a double crossover event in A. borkumensis were obtained by employing a conjugative biparental filter-mating technique. Correct gene replacement in the mutant strains was confirmed by diagnostic PCR employing oligonucleotide primers binding to DNA regions up- and downstream of atfA1 and atfA2, respectively, which were not present in the constructs used for gene inactivation (Fig. 2A).
Wild-type A. borkumensis SK2 cultivated with pyruvate exhibited a moderate level of WS activity (64.0 pmol mg−1 min−1) and an approximately fourfold-lower level of DGAT activity (Table 3). Inactivation of atfA1 led to a drastic reduction (approximately 80%) of both WS and DGAT activity in vitro, leaving only a very low residual level of DGAT activity (2.5 pmol mg−1 min−1). In contrast, inactivation of atfA2 caused a relatively low reduction of both acyltransferase activities. In the double knockout mutant WS activity was completely abolished, whereas a very low but significant basic level of DGAT activity was maintained (Table 3).
In parallel to the highly diminished acyltransferase rates observed in vitro, inactivation of atfA1 resulted in a strong decrease of TAG accumulation during cultivation on pyruvate or hexadecane as revealed by TLC analysis (Fig. 2B). Furthermore, during growth on hexadecane, biosynthesis of WEs was totally abrogated in the atfA1 knockout mutant. In utter contrast, disruption of atfA2 did not seem to have any effect on TAG or WE accumulation under either culture condition as revealed by TLC. In addition, no significant alterations in the profiles of other neutral lipids were observed (Fig. 2B). Remarkably, even in the atfA1ΩKmatfA2ΩSm double knockout mutant, reduced but still substantial amounts of TAGs were present in cells cultivated on either substrate (Fig. 2B), accounting for a fatty acid content of ca. 5 to 10% of the CDW (Table 6). Strains with inactivated atfA1 (atfA1ΩKm and atfA1ΩKmatfA2ΩSm) exhibited an altered fatty acid composition, comprising lower palmitic acid content and higher unsaturated fatty acid content, whereas disruption of atfA2 had no effect on the fatty acid profile (Table 6).
In order to rule out the possibility that the phenotypes observed in atfA1 knockout strains were influenced by polar effects, we performed RT-PCR analyses of the adjacent genes aceA and Abo_2743 (Fig. 1A). Both genes were similarly expressed in the wild type and in the atfA1ΩKm mutant during cultivation with pyruvate, confirming that polar effects on neighboring genes caused by atfA1 disruption can be excluded (data not shown).
Although showing distinct acyltransferase activity in vitro, loss of AtfA2 function had surprisingly no detectable influence on storage lipid accumulation in vivo, suggesting that atfA2 might be silent in A. borkumensis. However, RT-PCR analysis demonstrated definitively that atfA2 was expressed in the cells (Fig. 1C). Functional expression of atfA2 was also documented by the fact that somehow reduced WS activity levels were observed in vitro in atfA2 knockout mutants (Table 3).
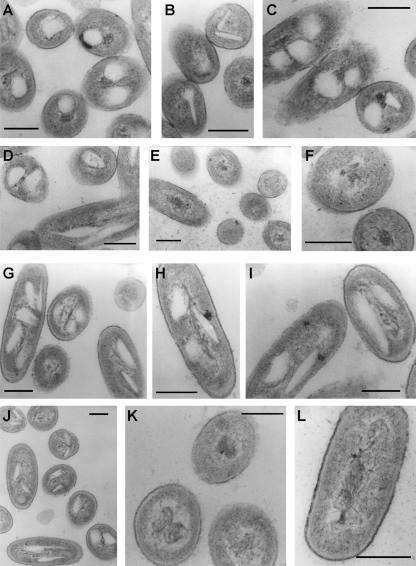
Ultrastructure of intracellular storage lipid inclusions in A. borkumensis.
Storage lipids predominantly consisting of TAGs synthesized during cultivation of A. borkumensis on pyruvate accumulated in the form of electron transparent intracytoplasmic inclusions as shown by TEM (Fig. 3A to D). Remarkably, these lipid inclusions were highly heterogeneous in size and shape, comprising mainly spherical inclusions but also rectangular, disc-shaped, needle-like, and irregularly shaped inclusions (Fig. 3A to D). During growth on hexadecane, intracellular storage lipids consisted of TAGs and WEs (Fig. 2B). Under these conditions spherical inclusions were only very rarely present (Fig. 3G to J). The structures of the inclusions were generally highly variable, many of them being quite long and sprawling, occasionally reaching a length of 500 nm. Although the double knockout mutant was still able to accumulate significant amounts of TAGs (Fig. 2B), TEMs of the A. borkumensis strain atfA1ΩKmatfA2ΩSm cells did not show obvious lipid inclusions in pyruvate-grown (Fig. 3E and F) or hexadecane-grown cells (Fig. 3K and L).
FIG. 3.
Structure of neutral lipid inclusions in A. borkumensis. Cells were cultivated for 72 h in ONR7a medium containing 1% (wt/vol) sodium pyruvate (A to F) or 0.5% (vol/vol) hexadecane (G to L). Ultrathin sections of cells were analyzed by transmission electron microscopy. The scale bars correspond to 200 nm. A. borkumensis SK2 (A to D and G to J) and A. borkumensis atfA1ΩKmatfA2ΩSm (E, F, K, and L) are shown.
Role of storage lipid accumulation for survival of carbon starvation.
Cells of wild-type A. borkumensis and of the double knockout mutant were preincubated under conditions promoting storage lipid accumulation and then transferred to carbon source-free medium. Survival of the cells under these conditions was then monitored by measuring remaining viable cell counts (CFU) over the time course of carbon starvation (Fig. 2C). Wild-type cells showed a two-log-fold decrease in viability within the first 10 days of starvation, but after that viability remained relatively constant even over an extended 4-week period of starvation. Storage lipid-reduced cells of the double knockout mutant seemed to lose viability slightly more rapidly than wild-type cells between days 2 and 8 of starvation, but finally only a two-log-fold decrease was observed, also reaching a plateau after 10 days, after which no further decrease in viability occurred even after a prolonged starvation period (Fig. 2C).
Occurrence of storage lipids in other hydrocarbonoclastic bacteria.
As described above, A. borkumensis accumulates only TAGs during cultivation with pyruvate, but no WEs are synthesized de novo (Fig. 2B). We then investigated other hydrocarbonoclastic bacteria for their ability to accumulate neutral storage lipids under these conditions (Fig. 4). In contrast to A. borkumensis, all three investigated hydrocarbonoclastic strains were able to synthesize substantial amounts of WEs de novo. In addition to this, only Alcanivorax jadensis produced substantial amounts of TAGs simultaneously, whereas Marinobacter hydrocarbonoclasticus accumulated only low levels and Thalassolituus oleivorans accumulated just trace amounts of TAGs (Fig. 4). In all tested strains, no significant amounts of 3-hydroxy fatty acid monomers were detectable by GC analysis, indicating that PHAs were absent or present only in very small amounts (data not shown).
FIG. 4.
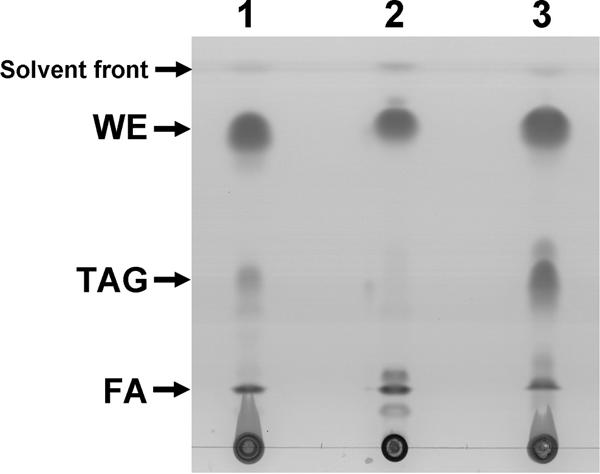
Storage lipid accumulation in different hydrocarbonoclastic marine bacteria. Cells were cultivated in ONR7a medium containing 1% (wt/vol) sodium pyruvate for 72 h (Marinobacter hydrocarbonoclasticus and Alcanivorax jadensis T9) or 96 h (Thalassolituus oleivorans) and analyzed by TLC. Lipid extracts obtained from 1.5 mg lyophilized cells were applied per lane. Lanes: 1, M. hydrocarbonoclasticus; 2, T. oleivorans; 3, A. jadensis T9. FA, free fatty acids.
DISCUSSION
In this study we have demonstrated the ability of the hydrocarbonoclastic marine bacterium A. borkumensis to synthesize and intracellularly accumulate large amounts of TAGs de novo from the unrelated substrate pyruvate, up to a corresponding fatty acid content of more than 23% of the CDW. In gram-negative bacteria, TAGs have so far been reported only as a minor constituent in Acinetobacter species (23, 28). Thus, to the best of our knowledge the present study represents the first description of substantial TAG accumulation in a gram-negative prokaryote. Furthermore, we refute hereby the so far largely accepted assumption that large-scale TAG accumulation in bacteria is restricted to the gram-positive group of actinomycetes (2, 39).
Despite the presence of two differentially expressed PHA synthase genes in A. borkumensis SK2 (29), we have shown that TAGs and WEs are the only predominating hydrophobic storage compounds occurring in this strain. Although very small amounts of PHAs might be synthesized by A. borkumensis, our data prove that PHAs play no role as storage compounds under the tested cultivation conditions. This is corroborated by the fact that no obvious intracellular lipophilic inclusions remain in mutant cells impaired in TAG and WE biosynthesis. PHA accumulation has been occasionally described for some other hydrocarbonoclastic bacteria (5, 11), but this was solely based on positive results from staining with lipophilic dyes and was not supported by chemical analyses. Such lipophilic dyes are not specific and cannot differentiate between PHAs, TAGs, WEs, and other neutral lipids, such as hydrocarbon inclusions. Based on our results using A. borkumensis and some other selected species it can be assumed that most probably TAGs and WEs, but not PHAs, are the principal storage lipids present in hydrocarbonoclastic marine bacteria. Since genes coding for key enzymes such as glycogen synthase, ADP-glucose pyrophosphorylase, or cyanophycin synthetase, which are required for biosynthesis of other bacterial carbon storage compounds like glycogen or cyanophycin, are lacking in the A. borkumensis genome (31), TAGs and WEs are most likely the only relevant carbon storage compounds produced in this bacterium.
The presence of two WS/DGAT homologues in A. borkumensis SK2 actually suggests a redundant function. However, although both enzymes (AtfA1 and AtfA2) exhibited robust acyltransferase activity in in vitro tests, only AtfA1 seems to be involved in vivo in TAG and WE biosynthesis and in fact undoubtedly plays a pivotal role in storage lipid accumulation in this organism. Surprisingly, AtfA2 is fully dispensable for this process despite contributing to the WS activity observed in vitro in A. borkumensis. Thus, due to the lack of an obvious phenotype, the function of this acyltransferase remains unclear, but it might be involved in production of a yet unknown fatty acid ester substance which could not be detected by the applied methods. Both acyltransferases exhibited little and partially overlapping substrate specificity; however, there were drastic differences regarding their specificity towards acylglycerols as substrates. Specifically, AtfA2 was almost totally devoid of DGAT activity. Thus, sequence comparison of AtfA1 and AtfA2 from A. borkumensis and WS/DGAT from A. baylyi shows that amino acid residues conserved in AtfA1 and WS/DGAT, but not AtfA2, might specify those domains that are important for DGAT activity, whereas amino acids conserved among all three enzymes indicate regions that are probably necessary for general acyltransferase catalytic activity (Fig. 1B).
Remarkably, even in the double knockout mutant a reduced but still substantial level of TAG accumulation remained, although both WS/DGAT homologues were inactivated. This is compelling evidence for the existence of an alternative, non-WS/DGAT-dependent TAG biosynthesis pathway in this bacterium. Trace amounts of residual TAGs in a WS/DGAT knockout mutant of A. baylyi have previously provided hints that such an alternative pathway may exist (23). Due to the much higher TAG content in A. borkumensis this became more obvious and unambiguous in the present study. In eukaryotes, three different routes for TAG formation, all using DAG as the substrate, are known. Members of the eukaryotic DGAT1 and -2 protein families, which exhibit no sequence homology to each other, catalyze the acylation of DAG using acyl-CoA as the substrate in a manner similar to that of the bacterial WS/DGAT (6, 7). Besides this most widespread pathway, an acyl-CoA-independent reaction for TAG synthesis is catalyzed by a phospholipid:DAG acyltransferase. This enzyme uses phospholipids as acyl donors for DAG esterification and has been found in plants and bakers' yeast (9). The third mechanism of TAG synthesis is described for DAG:DAG transacylases, which were identified in animals and plants utilizing DAG as the acyl donor as well as the acyl acceptor (26, 34). However, no genes coding for such DAG:DAG transacylases are known yet. The nature of the alternative TAG biosynthesis route in A. borkumensis is not known so far. The genome comprises numerous putative acyltransferase genes of yet unknown function (31; see Table S1 in the supplemental material), but none of these exhibits reasonable homology to any of the above-mentioned eukaryotic TAG-synthesizing enzymes. Residual but fairly low DGAT activities still observable in atfA1 knockout strains of A. borkumensis make it questionable that an alternative, so far unknown DGAT isoenzyme is responsible for TAG biosynthesis in these mutants. However, it has to be emphasized that such a potential DGAT isoenzyme might nevertheless be present but only poorly active under the applied in vitro assay conditions due to differing requirements (pH, buffer, salts). Whether phospholipid:DAG acyltransferase or DAG:DAG transacylase activities are measurable in A. borkumensis has not been examined in the present study.
In contrast to the other three investigated hydrocarbonoclastic bacteria, A. borkumensis was only able to synthesize WEs during cultivation on hexadecane, not on the unrelated substrate pyruvate, indicating that this strain is probably impaired in de novo formation of fatty alcohols. In A. baylyi, fatty alcohols are formed starting from long-chain acyl-CoAs by two consecutive NADPH-dependent reduction reactions catalyzed by a fatty acyl-CoA reductase and a yet unidentified fatty aldehyde reductase (28). The A. borkumensis genome does not comprise a fatty acyl-CoA reductase homologue (data not shown), which might be the reason for the lack of de novo fatty alcohol biosynthesis capability in this bacterium. During oxidative degradation of alkanes, fatty alcohols are formed as first intermediate and are thus also available as substrates for AtfA1, enabling WE formation despite the absence of de novo fatty alcohol biosynthesis.
TAG and WE inclusions in lipid accumulating bacteria are usually organized in the form of spherical intracytoplasmic lipophilic inclusions (overview in reference 39). In contrast to this, storage lipid inclusions in A. borkumensis showed an extremely high structural diversity comprising spherical, rectangular, disc-shaped, needle-like, and irregularly shaped forms. A peculiarity of storage lipids from A. borkumensis, compared to those from other bacteria, is their relatively low content of unsaturated fatty acids. In particular, WEs and TAGs produced during cultivation on hexadecane almost exclusively contained saturated fatty acids. Such unsaturated lipids are supposed to be largely solid at physiological temperatures, thus resulting in a relatively high degree of crystallinity, which might in turn lead to the formation of such diverse and heterogeneous inclusion structures. Furthermore, this biased fatty acid composition also strongly indicates that AtfA1, the key enzyme for neutral lipid biosynthesis in strain SK2, probably has a strong preference for saturated acyl-CoAs. Although experimental proof for this is pending, the relative changes in the fatty acid profile caused by atfA1 disruption corroborate that assumption.
Nutrient starvation is assumed to be an environmental stress factor frequently challenging marine hydrocarbonoclastic bacteria in their natural habitat. Accumulation of storage lipids serving as endogenous carbon sources during starvation periods might thus be one strategy to cope with this and to prolong viability under such unfavorable conditions. Although we could not definitively address this hypothesis due to the lack of a mutant completely lacking storage lipids, storage lipid accumulation seems to provide a growth advantage only during short periods of starvation but is not required for maintaining viability during long-term starvation phases. Interestingly, after the majority of the population (ca. 99%) rapidly lost viability after only a few days during starvation, a subpopulation of A. borkumensis went into a stable, nonreplicating but viable state which allowed survival even during extended starvation periods. This long-term persistence capability, putatively involving the stringent response (8), might be a key factor for the successful global distribution of A. borkumensis and other hydrocarbonoclastic bacteria and for their adaptability to an ever-changing environment.
Supplementary Material
Acknowledgments
This study was supported by a grant of the Deutsche Forschungsgemeinschaft to A.S. (Ste 386/7-3). P.N.G. and K.N.T. acknowledge support from the German Ministry for Education and Research (BMBF) within the framework of the GenoMik network “Genome Research on Bacteria Relevant for Agriculture, Environment and Biotechnology.”
We thank Biggy-Nadine Wendt and Chlud Kaddor for skillful technical assistance.
Footnotes
Published ahead of print on 22 November 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alvarez, H. M., O. H. Pucci, and A. Steinbüchel. 1997. Lipid storage compounds in marine bacteria. Appl. Microbiol. Biotechnol. 47:132-139. [Google Scholar]
- 2.Alvarez, H. M., and A. Steinbüchel. 2002. Triacylglycerols in prokaryotic microorganisms. Appl. Microbiol. Biotechnol. 60:367-376. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Bredemeier, R., R. Hulsch, J. O. Metzger, and L. Berthe-Corti. 2003. Submersed culture production of extracellular wax esters by the marine bacterium Fundibacter jadensis. Mar. Biotechnol. 5:579-583. [DOI] [PubMed] [Google Scholar]
- 5.Bruns, A., and L. Berthe-Corti. 1999. Fundibacter jadensis gen. nov., sp. nov., a new slightly halophilic bacterium, isolated from intertidal sediment. Int. J. Syst. Bacteriol. 49:441-448. [DOI] [PubMed] [Google Scholar]
- 6.Cases, S., S. J. Smith, Y. W. Zheng, H. M. Myers, S. R. Lear, E. Sande, S. Novak, C. Collins, C. B. Welch, A. J. Lusis, S. K. Ericson, and R. V. Farese, Jr. 1998. Identification of a gene encoding an acyl-CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA 95:13018-13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cases, S., S. J. Stone, P. Zhou, E. Yen, B. Tow, K. D. Lardizabal, T. Voelker, and R. V. Farese, Jr. 2001. Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J. Biol. Chem. 276:38870-38876. [DOI] [PubMed] [Google Scholar]
- 8.Chatterji, D., and A. K. Ojha. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4:160-165. [DOI] [PubMed] [Google Scholar]
- 9.Dahlqvist, A., U. Ståhl, M. Lenman, A. Banas, M. Lee, L. Sandager, H. Ronne, and S. Stymne. 2000. Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 97:6487-6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyksterhouse, S. E., J. P. Gray, R. P. Herwig, J. C. Lara, and J. T. Staley. 1995. Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int. J. Syst. Bacteriol. 45:116-123. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez-Martinez, J., M. J. Pujalte, J. Garcia-Martinez, M. Mata, E. Garay, and F. Rodriguez-Valeral. 2003. Description of Alcanivorax venustensis sp. nov. and reclassification of Fundibacter jadensis DSM 12178T (Bruns and Berthe-Corti 1999) as Alcanivorax jadensis comb. nov., members of the emended genus Alcanivorax. Int. J. Syst. Evol. Microbiol. 53:331-338. [DOI] [PubMed] [Google Scholar]
- 12.Fixter, L. M., M. N. Nagi, J. G. McCormack, and C. A. Fewson. 1986. Structure, distribution and function of wax esters in Acinetobacter calcoaceticus. J. Gen. Microbiol. 132:3147-3157. [Google Scholar]
- 13.Fixter, L. M., and M. K. Sherwani. 1991. Energy reserves in Acinetobacter. In: Towner, K. J., E. Bergogne-Bérézin, and C. A. Fewson, (ed.) The Biology of Acinetobacter: Taxonomy, clinical importance, molecular biology, physiology, industrial relevance. Plenum press, New York, USA, pp. 273-294.
- 14.Gallagher, I. H. C. 1971. Occurrence of waxes in Acinetobacter. J. Gen. Microbiol. 68:245-247. [DOI] [PubMed] [Google Scholar]
- 15.Gauthier, M. J., B. Lafay, R. Christen, L. Fernandez, M. Acquaviva, P. Bonin, and J. C. Bertrand. 1992. Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. Int. J. Syst. Bacteriol. 42:568-576. [DOI] [PubMed] [Google Scholar]
- 16.Golyshin, P. N., T. N. Chernikova, W.-R. Abraham, H. Lünsdorf, K. N. Timmis, and M. M. Yakimov. 2002. Oleiphilaceae fam. nov., to include Oleiphilus messinensis gen. nov., sp. nov., a novel marine bacterium that obligately utilizes hydrocarbons. Int. J. Syst. Evol. Microbiol. 52:901-911. [DOI] [PubMed] [Google Scholar]
- 17.Golyshin, P. N., V. A. Martins Dos Santos, O. Kaiser, M. Ferrer, Y. S. Sabirova, H. Lunsdorf, T. N. Chernikova, O. V. Golyshina, M. M. Yakimov, A. Pühler, and K. N. Timmis. 2003. Genome sequence completed of Alcanivorax borkumensis, a hydrocarbon-degrading bacterium that plays a global role in oil removal from marine systems. J. Biotechnol. 106:215-220. [DOI] [PubMed] [Google Scholar]
- 18.Harayama, S., Y. Kasai, and A. Hara. 2004. Microbial communities in oil-contaminated seawater. Curr. Opin. Biotechnol. 15:205-214. [DOI] [PubMed] [Google Scholar]
- 19.Harayama, S., H. Kishira, Y. Kasai, and K. Shutsubo. 1999. Petroleum biodegradation in marine environments. J. Mol. Microbiol. Biotechnol. 1:63-70. [PubMed] [Google Scholar]
- 20.Head, I. M., D. M. Jones, and W. F. Roling. 2006. Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 4:173-182. [DOI] [PubMed] [Google Scholar]
- 21.Hedlund, B. P., A. D. Geiselbrecht, T. J. Bair, and J. T. Staley. 1999. Polycyclic aromatic hydrocarbon degradation by a new marine bacterium, Neptunomonas naphthovorans gen. nov., sp. nov. Appl. Environ. Microbiol. 65:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishige, T., A. Tani, Y. Sakai, and N. Kato. 2003. Wax ester production by bacteria. Curr. Opin. Microbiol. 6:244-250. [DOI] [PubMed] [Google Scholar]
- 23.Kalscheuer, R., and A. Steinbüchel. 2003. A novel bifunctional wax ester synthase/acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J. Biol. Chem. 278:8075-8082. [DOI] [PubMed] [Google Scholar]
- 24.Kalscheuer, R., H. Luftmann, and A. Steinbüchel. 2004. Synthesis of novel lipids in Saccharomyces cerevisiae by heterologous expression of an unspecific bacterial acyltransferase. Appl. Environ. Microbiol. 70:7119-7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasai, Y., H. Kishira, T. Sasaki, K. Syutsubo, K. Watanabe, and S. Harayama. 2002. Predominant growth of Alcanivorax strains in oil-contaminated and nutrient-supplemented sea water. Environ. Microbiol. 4:141-147. [DOI] [PubMed] [Google Scholar]
- 26.Lehner, R., and A. Kuksis. 1993. Triacylglycerol synthesis by an sn-1,2(2,3)-diacylglycerol transacylase from rat intestinal microsomes. J. Biol. Chem. 268:8781-8786. [PubMed] [Google Scholar]
- 27.Overhage, J., H. Priefert, J. Rabenhorst, and A. Steinbüchel. 1999. Biotransformation of eugenol to vanillin by a mutant of Pseudomonas sp. strain HR199 constructed by disruption of the vanillin dehydrogenase (vdh) gene. Appl. Microbiol. Biotechnol. 52:820-828. [DOI] [PubMed] [Google Scholar]
- 28.Reiser, S., and C. Somerville. 1997. Isolation of mutants of Acinetobacter calcoaceticus deficient in wax ester synthesis and complementation of one mutation with a gene encoding a fatty acyl coenzyme A reductase. J. Bacteriol. 179:2969-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabirova, J. S., M. Ferrer, D. Regenhardt, K. N. Timmis, and P. N. Golyshin. 2006. Proteomic insights into metabolic adaptations in Alcanivorax borkumensis induced by alkane utilization. J. Bacteriol. 188:3763-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual, p. A. 1, 2nd ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Schneiker, S., V. A. P. Martins dos Santos, D. Bartels, T. Bekel, M. Brecht, J. Buhrmester, T. N. Chernikova, R. Denaro, M. Ferrer, C. Gertler, A. Goesmann, O. V. Golyshina, F. Kaminski, A. N. Khachane, S. Lang, B. Linke, A. C. McHardy, F. Meyer, T. Nechitaylo, A. Pühler, D. Regenhardt, O. Rupp, J. S. Sabirova, W. Selbitschka, M. M. Yakimov, K. N. Timmis, F. J. Vorholter, S. Weidner, O. Kaiser, and P. N. Golyshin. 2006. Genome sequence of the ubiquitous hydrocarbon-degrading marine bacterium Alcanivorax borkumensis. Nat. Biotechnol. 24:997-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host-range mobilization system for in vivo genetic engineering transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 33.Steinbüchel, A. 1991. Polyhydroxyalkanoic acids. In: Biomaterials, Novel Materials. Byrom, D. (ed.). Basingstoke, MacMillan, pp. 123-213.
- 34.Stobart, K., M. Mancha, M. Lenman, A. Dahlqvist, and S. Stymne. 1997. Triacylglycerols are synthesised and utilized by transacylation reactions in microsomal preparations of developing safflower (Carthamus tinctorius L.) seeds. Planta 203:58-66. [Google Scholar]
- 35.Stöveken, T., R. Kalscheuer, U. Malkus, R. Reichelt, and A. Steinbüchel. 2005. The wax ester synthase/acyl coenzyme A:diacylglycerol acyltransferase from Acinetobacter sp. strain ADP1: characterization of a novel type of acyltransferase. J. Bacteriol. 187:1369-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Syutsubo, K., H. Kishira, and S. Harayama. 2001. Development of specific oligonucleotide probes for the identification and in situ detection of hydrocarbon-degrading Alcanivorax strains. Environ. Microbiol. 3:371-379. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaneechoutte, M., D. M. Young, L. N. Ornston, T. De Baere, A. Nemec, T. van der Reijden, E. Carr, I. Tjernberg, and L. Dijkshoorn. 2006. Naturally transformable Acinetobacter sp. strain ADP1 belongs to the newly described species Acinetobacter baylyi. Appl. Environ. Microbiol. 72:932-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wältermann, M., and A. Steinbüchel. 2005. Neutral lipid bodies in prokaryotes: recent insights into structure, formation, and relationship to eukaryotic lipid depots. J. Bacteriol. 187:3607-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yakimov, M. M., L. Giuliano, R. Denaro, E. Crisafi, T. N. Chernikova, W. R. Abraham, H. Luensdorf, K. N. Timmis, and P. N. Golyshin. 2004. Thalassolituus oleivorans gen. nov., sp. nov., a novel marine bacterium that obligately utilizes hydrocarbons. Int. J. Syst. Evol. Microbiol. 54:141-148. [DOI] [PubMed] [Google Scholar]
- 41.Yakimov, M. M., L. Giuliano, G. Gentile, E. Crisafi, T. N. Chernikova, W.-R. Abraham, H. Lünsdorf, K. N. Timmis, and P. N. Golyshin. 2003. Oleispira antarctica gen. nov., sp. nov., a novel hydrocarbonoclastic marine bacterium isolated from Antarctic coastal sea water. Int. J. Syst. Evol. Microbiol. 53:779-785. [DOI] [PubMed] [Google Scholar]
- 42.Yakimov, M. M., P. N. Golyshin, S. Lang, E. R. Moore, W. R. Abraham, H. Lunsdorf, and K. N. Timmis. 1998. Alcanivorax borkumensis gen. nov., sp. nov., a new, hydrocarbon-degrading and surfactant-producing marine bacterium. Int. J. Syst. Bacteriol. 48:339-348. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.