Abstract
Bordetellae are gram-negative bacteria that colonize the respiratory tracts of animals and humans. We and others have recently shown that these bacteria are capable of living as sessile communities known as biofilms on a number of abiotic surfaces. During the biofilm mode of existence, bacteria produce one or more extracellular polymeric substances that function, in part, to hold the cells together and to a surface. There is little information on either the constituents of the biofilm matrix or the genetic basis of biofilm development by Bordetella spp. By utilizing immunoblot assays and by enzymatic hydrolysis using dispersin B (DspB), a glycosyl hydrolase that specifically cleaves the polysaccharide poly-β-1,6-N-acetyl-d-glucosamine (poly-β-1,6-GlcNAc), we provide evidence for the production of poly-β-1,6-GlcNAc by various Bordetella species (Bordetella bronchiseptica, B. pertussis, and B. parapertussis) and its role in their biofilm development. We have investigated the role of a Bordetella locus, here designated bpsABCD, in biofilm formation. The bps (Bordetella polysaccharide) locus is homologous to several bacterial loci that are required for the production of poly-β-1,6-GlcNAc and have been implicated in bacterial biofilm formation. By utilizing multiple microscopic techniques to analyze biofilm formation under both static and hydrodynamic conditions, we demonstrate that the bps locus, although not essential at the initial stages of biofilm formation, contributes to the stability and the maintenance of the complex architecture of Bordetella biofilms.
In contrast to the planktonic or free swimming mode of existence in laboratory settings, bacteria predominantly form surface-attached communities known as biofilms in their natural habitats. Biofilms are commonly defined as highly structured communities of cells that are encased in a self-produced polymeric organic matrix (7). Biofilms are increasingly being recognized as important contributors to chronic bacterial diseases (8, 19, 48). Biofilms provide protection from both the innate and the adaptive components of the immune system, and bacteria in biofilms are extremely recalcitrant to antibiotic therapy and other antimicrobial agents (15, 20, 31, 40).
We are studying the members of the bacterial genus Bordetella as a model system in order to understand how bacteria adapt to cope with the selective pressures inside mammalian hosts. Given the demonstrated importance of biofilms in contributing to bacterial persistence, we are interested in characterizing the role that biofilms play in Bordetella physiology and the infectious cycle within hosts. Bordetellae are small, aerobic, gram-negative coccobacilli that colonize the respiratory tracts of humans and animals (42). Bordetella pertussis, the human pathogen, results in the disease known as whooping cough, while B. bronchiseptica mainly infects animals and causes a variety of respiratory diseases (42, 64). B. parapertussis strains can be divided into two genetically distinct types: those that infect humans, causing a pertussis-like illness, and those that cause respiratory infections in sheep (42). We and others have recently shown that Bordetella spp. are capable of living as biofilms on a number of abiotic surfaces (27, 44). We have also shown that B. bronchiseptica biofilms are highly resistant to a number of antimicrobial compounds, including compounds used clinically (44).
Biofilm development is a multistep process that begins with bacterial attachment to surfaces and is sometimes followed by the formation of microcolonies and the establishment of an ordered 3-dimensional structure (1, 2, 61, 63). In certain species, this structure is composed of pillars of bacteria surrounded by water channels that are predicted to allow biofilm bacteria access to nutrients and to aid in the diffusion of toxic metabolites out of the biofilm (7). A defining characteristic of mature biofilms is that bacteria are generally embedded in a matrix composed of one or more macromolecules such as nucleic acids, proteins, and polysaccharides (4, 52, 53). A number of genetic loci that affect exopolysaccharide production have been shown to influence surface attachment, intercellular adhesion, and the development and maintenance of 3-dimensional structures of bacterial biofilms (reviewed in reference 4). We have investigated the role of one such Bordetella locus, which we have termed bpsABCD, in Bordetella biofilm formation. The bps (Bordetella polysaccharide) locus displays significant sequence similarity to several bacterial loci implicated in the synthesis of poly-β-1,6-N-acetyl-d-glucosamine (poly-β-1,6-GlcNAc), including the icaADBC locus of staphylococcal species (reviewed in references 22 and 57) and the pgaABCD locus of Escherichia coli and Actinobacillus species (33, 60). To investigate the contribution of the bps locus to biofilm formation, we constructed an isogenic derivative (Δbps) of the wild-type B. bronchiseptica strain RB50 that contains an in-frame deletion of the bps locus. Scanning electron microscopy (SEM) of biofilms formed by the wild-type and Δbps strains shows that while RB50 formed a thick, uniform, multilayered stack of cells, the Δbps strain was present as scattered cells across the coverslip. Confocal scanning laser microscopy (CSLM) of live, fully hydrated biofilms grown in flow cells demonstrates that while at earlier time points both RB50 and the Δbps strain adhered to the same degree, at later time points the Δbps strain failed to form differentiated 3-dimensional structures. Our results strongly suggest that the bpsABCD locus is required to maintain the complex architecture of Bordetella biofilms formed under in vitro conditions.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotides.
Strains, plasmids, and primers used in this study are described in Table 1.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description or sequence | Source or reference |
|---|---|---|
| Strains | ||
| RB50 | Wild-type strain of B. bronchiseptica | 9 |
| Bp536 | Wild-type strain of B. pertussis | 51 |
| Bpp12822 | Wild-type strain of B. parapertussis | 24 |
| ΔbpsABCD | RB50 derivative containing an in-frame deletion of the bpsABCD locus | This study |
| CC118λpir | pir, host for pir-dependent plasmids | 13 |
| DH5α | E. coli laboratory strain | Stratagene |
| Primers | ||
| GP1 | CTAGTCTAGAGGCGAAATTATACCGCGTT | This study |
| GP2 | CCCAAGCTTCCCCGCCACCAGCAGCCGAGT | This study |
| GP3 | CCCAAGCTTCAGCGGCAACCCGACGGACGCAT | This study |
| GP4 | CGGGGTACCGGGCGCGGCTGCTGCTGCAGG | This study |
| GP5 | CCGCTCGAGATCTGGCGTTTTCGTAGAA | This study |
| GP6 | CTAGTCTAGAGCGCGGCTATACGGACTA | This study |
| Plasmids | ||
| pRK2013 | Mobilization helper plasmid | 18 |
| pRE112 | Allelic exchange vector; Cmr | 16 |
| pGP8 | pRE112 derivative, bpsABCD deletion plasmid | This study |
| pMM11 | pBBR1MCS derivative containing the entire bpsABCD locus | |
| pKK223-3 | Expression vector containing the tac promoter | Pharmacia |
| pBBR1-GFP | Broad-host-range plasmid containing promoterless GFP | 46 |
| pTac-GFP | Constitutive GFP expression vector; tac promoter cloned upstream of the GFP cassette | This study |
| pBBR1MCS | Broad-host-range plasmid | 36 |
Molecular biology, genetics, and bioinformatics.
Standard procedures were used for plasmid isolation, restriction digestion, cloning, and transformation. Sequence analysis was performed, and homology was determined by conducting a BLASTP search at the NCBI website. Conserved protein domains and predicted membrane localization domains were obtained from the Entrez Protein Database at the NCBI website.
Growth conditions.
Bordetella strains were maintained on Bordet-Gengou agar (BG agar) supplemented with 7.5% defibrinated sheep blood. For planktonic and biofilm growth, the Bordetella strains were grown in Stainer-Scholte (SS) broth (54). For B. pertussis, the SS medium was supplemented with heptakis(2,6-di-O-methyl-β-cyclodextrin). E. coli strains were grown in Luria-Bertani medium. When needed, the various growth media were supplemented with the appropriate antibiotics, chloramphenicol (50 μg/ml) and streptomycin (50 μg/ml).
Enzymatic treatment of biofilms.
Biofilm formation was assessed using the microtiter dish assay as previously described (44, 45). Bordetella strains were cultured in a total volume of 100 μl in SS medium at 37°C in 96-well polyvinyl chloride plates. The individual wells were inoculated with the overnight cultures of the respective Bordetella strains, grown under shaking conditions, at an optical density at 600 nm (OD600) of 0.05. DspB was purified as previously described (28) and added separately, at a final concentration of 50 μg/ml, into the wells of the plates at the time of inoculation. At designated time points, the medium was removed and the plates were vigorously washed with water. The adhered cells were stained with crystal violet for 30 min, followed by washing with water. The crystal violet staining the cells was solubilized in 200 μl of 95% ethanol and quantified at an OD540 as described previously (44).
Immunoblot analysis.
Crude polysaccharide extracts were prepared using a method previously described for the purification of poly-N-acetylglucosamine (PNAG) from Staphylococcus aureus (37). Approximately 5 × 109 cells from an overnight broth culture of Bordetella were collected by centrifugation, resuspended in 100 μl of 0.5 M EDTA, and boiled for 5 min at 100°C. Cells were removed by centrifugation, and the supernatant was treated with proteinase K (final concentration, 1 mg/ml) for 60 min at 60°C. Proteinase K was heat inactivated by incubating for 30 min at 90°C. A 10-μl aliquot of the extract was spotted onto a nitrocellulose membrane and allowed to air dry overnight. The blot was blocked with 5% nonfat milk and then probed with a 1:5,000 dilution of a goat antibody raised against S. aureus PNAG conjugated to diphtheria toxoid (41). The secondary antibody used was a horseradish peroxidase-conjugated mouse anti-goat immunoglobulin G (IgG) antibody (Pierce) diluted 1:20,000 and detected with the Amersham ECL (enhanced chemiluminescence) Western blotting system.
SEM.
Bordetellae were cultured statically on glass coverslips vertically submerged in SS broth, resulting in the formation of biofilms at the air-liquid interface, as described previously (44). At the indicated times, the coverslips were removed, washed gently with sterile water, and fixed using 2.5% glutaraldehyde. Samples were then processed for SEM as previously described (44).
Deletion of the bpsABCD locus.
An in-frame nonpolar deletion of the bpsABCD locus was constructed utilizing allelic exchange as described previously (14, 16). A 575-bp XbaI-HindIII upstream fragment spanning regions 5′ to and including the first 20 codons of bpsA was amplified from the chromosome of RB50 using primers GP1 and GP2. A 563-bp HindIII-KpnI fragment containing regions 3′ to and including the last 20 codons of the bpsD open reading frame was similarly amplified utilizing primers GP3 and GP4. A three-way ligation containing the XbaI-HindIII- and HindIII-KpnI-digested fragments, along with the XbaI-KpnI-digested allelic exchange vector pRE112 (Cmr) (16), was carried out, resulting in plasmid pGP8. This plasmid was transformed in the E. coli strain CC118, which allows replication of plasmids containing the conditional replicon RK6ori. pGP8 was mobilized from CC118 into the wild-type B. bronchiseptica strain RB50 using plasmid pRK2013 by triparental mating. Exoconjugates were selected on BG agar containing chloramphenicol and streptomycin. Colonies that underwent second recombination events were selected on Luria-Bertani agar containing 7.5% sucrose as described elsewhere (14, 16). The genotype of the Δbps strain was confirmed by PCR and DNA sequencing of the PCR product.
Construction of pTac-GFP.
The tac promoter was obtained by digesting plasmid pKK223-3 (Pharmacia) with BamHI. The resultant 268-bp BamHI fragment was cloned into the BglII site of plasmid pBBR1-GFP (46), resulting in plasmid pTac-GFP. pBBR1-GFP is a broad-host-range plasmid that contains a promoterless gfp cassette, gfpmut3, encoding green fluorescent protein (GFP) (46). pTac-GFP was mobilized in either the wild-type strain RB50 or the Δbps strain by triparental mating as described previously (14), and exconjugates were selected on BG agar containing chloramphenicol and streptomycin. Randomly picked colonies containing pTac-GFP were grown in SS broth with chloramphenicol and were analyzed for GFP expression utilizing a Nikon Eclipse TE300 inverted microscope. One of the GFP-expressing clones corresponding to each of the strains was chosen for experimental analysis. Comparison of the GFP-expressing strains with the respective parental strains not containing the pTac-GFP plasmid revealed no differences in growth in batch cultures or colony morphology on BG agar containing blood.
Genetic complementation of the bpsABCD locus.
A 6,057-bp fragment containing the entire bps locus plus 29 bp upstream of the putative translational start site of bpsA and 38 bp downstream of the termination codon of bpsD was amplified from RB50 chromosomal DNA with primers GP5 and GP6 by PCR utilizing Pfu DNA polymerase. The resulting PCR fragment containing flanking XhoI and XbaI restriction sites was cloned into the corresponding sites of plasmid pBBR1MCS (36), resulting in the complementation plasmid pMM11. This plasmid was transformed into DH5α and subsequently mobilized into the Δbps strain as described above.
Continuous flow confocal microscopy.
Three-chamber flow cells were obtained as sterile units from Stovall. Suspensions (200-μl) of exponentially growing bacterial cultures at an OD600 of 0.1 were aseptically inoculated directly into individual chambers of the flow cell by using a 25 5/8-gauge needle. Bacteria were allowed to adhere for 2 h, after which the flow cell was inverted and flow was resumed at a constant rate of ≈0.3 ml/min/chamber. Biofilms were visualized in situ every 24 h for 4 days using a Zeiss LSM 510 confocal scanning laser microscope. For displaying biofilm images, the LSM Image Browser software was utilized. Biofilm images were analyzed for biofilm parameters such as biomass, average thickness, and maximum thickness using the COMSTAT computer software package (26). Statistical analyses were carried out using an unpaired two-tailed Student t test.
Immunoelectron microscopy.
Biofilms were grown as described above for SEM analysis for 120 h and were fixed in 4% paraformaldehyde and 0.5% glutaraldehyde. Fixed samples were washed twice with phosphate-buffered saline (PBS), blocked with 5% normal rat serum for 30 min, and washed in PBS. The biofilm was probed with a 1:5,000 dilution of the goat anti-PNAG antibody for 1 h as described above, washed with PBS, and incubated with the secondary antibody (donkey anti-goat IgG) conjugated to 6-nm colloidal gold for 1 h. The samples were subsequently washed in PBS, fixed in 1% osmium tetroxide for 1 h, and washed three times in PBS. The samples were then dehydrated through a graded series of ethanol and flat embedded in Spurr's resin using a Chien embedding mold. The glass coverslips were removed from the cured epoxy-embedded samples by immersion in liquid nitrogen. Thin sections (thickness, 80 nm) were cut with a Reichert Ultracut E microtome, collected on Formvar-coated copper grids, and stained with uranyl acetate and lead citrate. The grids were observed using a Philips transmission electron microscope (TEM) 400 at 60 kV.
RESULTS
Bordetellae produce a polysaccharide similar to poly-β-1,6-N-acetylglucosamine.
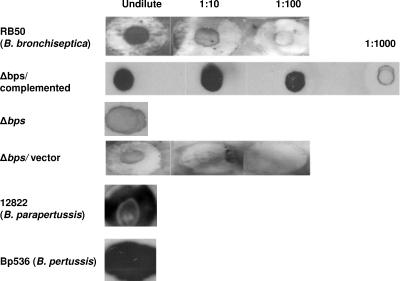
Given the demonstrated importance of polysaccharides in contributing to bacterial biofilm formation (4), we explored the role of carbohydrate moieties in Bordetella biofilm development. Specifically, we asked whether bordetellae produce poly-β-1,6-GlcNAc-like material. Our reasoning for this specific line of investigation was the previous identification of a locus in the three Bordetella species (B. bronchiseptica, B. pertussis, and B. parapertussis) that is homologous to the pgaABCD loci of E. coli and Actinobacillus species (33, 60). The pgaABCD locus of E. coli is necessary for the production of cell surface-associated poly-β-1,6-GlcNAc (60). Previous studies have also implicated poly-β-1,6-GlcNAc (known as PGA in E. coli and Actinobacillus species and as PIA, SAE, and PNAG in staphylococcal species) as an essential factor for biofilm formation (23, 33, 39, 43, 60). To determine whether bordetellae produce a similar poly-β-1,6-GlcNAc, we probed cell surface extracts (Materials and Methods) with an affinity-purified goat antibody raised to a conjugate of deacetylated PNAG from S. aureus (37, 41). Immunoblot assays showed that the antibody recognized a PNAG-like material in the remaining polysaccharides present in the extracts prepared from all three Bordetella species tested (Fig. 1). Thus, these assays indicate that bordetellae produce a polysaccharide that is antigenically similar to S. aureus PNAG.
FIG. 1.
Detection of crude exopolysaccharides by immunoblot analysis. Exopolysaccharides were extracted from overnight cultures of different Bordetella strains (designated on the left). Ten microliters of boiled EDTA extracts treated with proteinase K was spotted onto a nitrocellulose membrane and probed with a 1:5,000 dilution of an affinity-purified goat antiserum raised against a conjugate of S. aureus deacetylated PNAG and diphtheria toxoid (41), followed by mouse anti-goat IgG conjugated to horseradish peroxidase.
Disruption of Bordetella biofilms by an enzyme that specifically cleaves poly-β-1,6-GlcNAc.
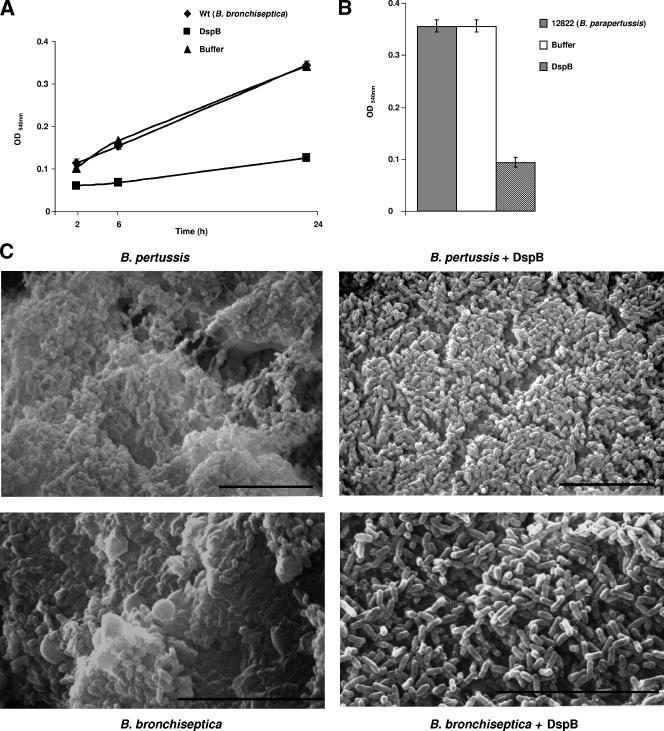
We next determined whether poly-β-1,6-GlcNAc contributes to the structural integrity of Bordetella biofilms. We reasoned that if poly-β-1,6-GlcNAc is an essential part of the Bordetella biofilm matrix, then enzymatic depolymerization of this compound will disrupt biofilms. Previously, it was demonstrated that dispersin B (DspB), an enzyme from Actinobacillus actinomycetemcomitans, specifically cleaves the glycosidic linkage of poly-β-1,6-GlcNAc (28, 32, 33, 50). The wild-type strains of B. bronchiseptica and B. parapertussis were cultivated separately on microtiter dishes either with a highly purified preparation of recombinant DspB or with a buffer control. As shown in Fig. 2A and B, growth of both B. bronchiseptica and B. parapertussis in the presence of DspB resulted in inhibition of biofilm formation. The inhibitory effect on biofilm formation was specific to DspB, since incubation with the DspB buffer (10 mM sodium phosphate) did not result in a significant reduction in the biofilm-forming ability of the respective wild-type strains (Fig. 2A and B). Additionally, there were no discernible differences in the planktonic growth of either B. bronchiseptica or B. parapertussis in the presence or absence of DspB (data not shown).
FIG. 2.
Dispersin B disrupts biofilm formation by Bordetella species. (A) Time course of biofilm development by B. bronchiseptica strain RB50 cultured in SS medium either alone (diamonds) or supplemented with the enzyme buffer control (triangles) or with 50 μg/ml of DspB (squares). Biofilm formation was assessed by microtiter plate assays as described in Materials and Methods. Each data point is an average for three wells. Error bars, standard errors. Each experiment was repeated at least three times with similar results. (B) Wild-type B. parapertussis strain 12822 was cultured in SS medium either alone (shaded bar) or supplemented with the enzyme buffer control (open bar) or with 50 μg/ml of DspB (striped bar). Biofilm formation was assessed at 96 h as described for Fig. 2A. (C) SEM of Bordetella biofilms. B. pertussis strain Bp 536 (upper panels) and B. bronchiseptica strain RB50 (bottom panels) were grown statically on vertically submerged coverslips in SS medium alone (left panels) or containing 50 μg/ml of DspB (right panels). After 120 h (Bp536) or 96 h (RB50) of growth, the biofilms formed at the air-liquid interface were visualized by SEM. Bars, 10 μm.
We previously found that, unlike B. bronchiseptica and B. parapertussis, B. pertussis does not grow in microtiter dishes under static conditions (44). However, as shown by scanning electron microscopy, these bacteria formed biofilms when grown on glass coverslips (44). Therefore, we grew B. pertussis on vertically submerged coverslips in the presence of a medium containing DspB or with the buffer control. In agreement with our previously published results (44), SEM analyses of the biofilm formed by the wild-type bacteria demonstrated that it was composed of clusters of bacterial cells with a characteristic 3-dimensional structure (Fig. 2C). It was also apparent from these analyses that the cells were encased in an amorphous opaque extracellular material resembling the matrix frequently observed for other bacterial biofilms. Growth in the presence of DspB led to an almost complete loss of this amorphous encasing material (Fig. 2C). In addition, a similar loss of the opaque encasing material was also observed when B. bronchiseptica was grown on glass coverslips in the presence of DspB (Fig. 2C). For both B. bronchiseptica and B. pertussis, the presence of DspB did not appear to have any significant effect on either the attachment of the bacteria to the glass coverslip or the clustering of the cells (Fig. 2C). This suggests that DspB does not inhibit the initial surface attachment of the bacterial cells or the formation of microcolonies but leads to the dissolution of the extracellular polymeric substance that accumulates within the biofilm matrix, thereby inhibiting biofilm formation (Fig. 2A to C).
The Bordetella bpsABCD locus.
A cluster of three genes in B. bronchiseptica, BB1769, BB1768, and BB1767, displays a high degree of sequence similarity to the corresponding E. coli genes pgaA, pgaB, and pgaC, respectively (33, 60), and to the gene products of the icaADBC loci of staphylococcal species (22, 57) and the hmsHFRS locus of Yersinia pestis (17). All these bacterial loci either have been demonstrated or are predicted to be involved in the synthesis of components of the biofilm matrix, which in some cases is structurally related to poly-β-1,6-GlcNAc. Due to their similarity to the genes of the hmsHFRS locus, BB1767, BB1768, and BB1769 have been designated hmsR, hmsF, and hmsH, respectively, in the annotated Bordetella genome database. Based on the results of our immunoblot analyses (see below and Fig. 1), which demonstrate that this Bordetella locus is involved in the synthesis of a PNAG-like material, and to distinguish this locus from the Yersinia hmsHFRS locus, we propose to rename this locus as the bps locus, for Bordetella polysaccharide locus, and the respective genes as bpsA (BB1769), bpsB (BB1768), and bpsC (BB1767).
bpsA is predicted to encode a 661-amino-acid protein with four transmembrane domains and has 23% amino acid identity to both Y. pestis hmsH and E. coli pgaA. Specifically, Y. pestis HmsH has been demonstrated to be an outer membrane protein (49).
bpsB is predicted to encode a 701-amino-acid protein with a 26-amino-acid signal sequence. Staphylococcus epidermidis IcaB has been shown to be a surface-bound protein that deacetylates PIA and has 24% amino acid sequence identity to BpsB (56). Like IcaB, BpsB contains a putative polysaccharide N-deacetylase domain.
A search for conserved protein domains predicted that, like PgaC and IcaA, BpsC is a member of the glycosyltransferase 2 family (6). S. epidermidis IcaA shares 35% amino acid sequence identity with BpsC and has been shown to function as a processive glycosyltransferase (21). In addition, the five amino-acids that have been demonstrated to be essential for the glycosyltransferase activity of IcaA are all conserved in BpsC (D171, D251, Q292, R295, and W296) (21).
The fourth contiguous open reading frame (BB1766) that is in the same orientation as bpsA, bpsB, and bpsC does not show significant similarity to either icaD or pgaD. The only similarity among these three genes is that they all encode small proteins. IcaD has been demonstrated to enhance the activity of IcaA (21), and we have found that pgaD is needed for PGA synthesis in E. coli (Y. Itoh and T. Romeo, unpublished data). Despite the lack of apparent similarity, we propose to name BB1766 bpsD, since it might function in a manner similar to that of IcaD and PgaD.
The bpsABCD locus is needed for the production of a PNAG-like material.
Given the demonstrated role of the pgaABCD, icaADBC, and hmsHFRS loci in biofilm formation, we hypothesized that the bpsABCD locus might be involved in Bordetella biofilm formation. We utilized allelic exchange to construct the Δbps strain, which contains a nonpolar in-frame deletion of the bpsABCD locus. First, we used immunoblot assays to compare the Δbps strain to the wild-type B. bronchiseptica strain RB50 for the production of a PNAG/PIA-like material. As shown in Fig. 1, the deletion of the bpsABCD locus led to an almost undetectable level of the immunoreactive material. To further determine whether the lack of production of the PNAG-like material was due to the absence of the bps locus and not due to any pleiotropic effect, we cloned this locus into a broad-host-range plasmid (see Materials and Methods). As shown in Fig. 1, complementation of this locus in the Δbps strain resulted in the restoration of the polysaccharide expression. Thus, these results suggest that the bps locus is involved in the synthesis of a PNAG-like material in B. bronchiseptica.
The bpsABCD locus promotes biofilm development.
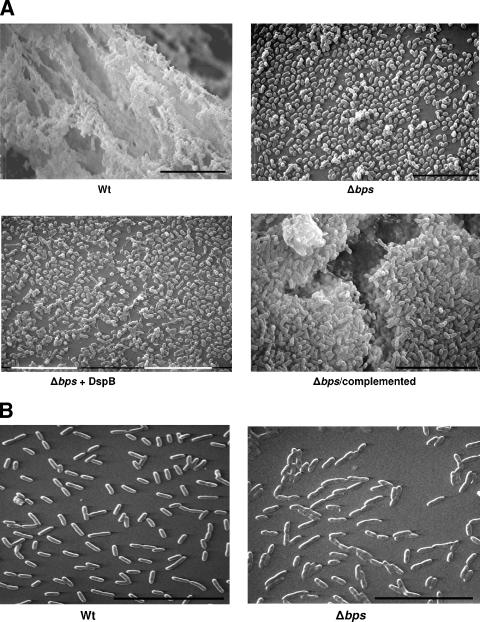
To investigate a role for the bps locus in contributing to biofilm formation, we cultured the wild-type and Δbps strains on glass coverslips, and the biofilms formed at the air-liquid interface were visualized by SEM. In contrast to the wild-type strain, which displayed the characteristic biofilm architecture of multicellular communities encased by an opaque matrix-like material, the Δbps strain existed as scattered cells across the coverslip (Fig. 3A). Addition of DspB during the growth of the Δbps mutant did not have any significant effect on attachment to the glass surface (Fig. 3A), suggesting that the inhibitory effect of DspB on B. bronchiseptica biofilms is specific to the product of the bps locus. Although the bps mutant by itself or in the presence of DspB appeared to exist mainly as a monolayer of cells, it was capable of forming some minute clusters. Note that there was no apparent defect in the growth of the wild-type and Δbps strains when they were grown in broth cultures (data not shown). SEM of biofilms formed by the complemented strain revealed the restoration of the multicellular appearance of the bacterial cells observed with the wild-type strain.
FIG. 3.
Scanning electron microscopy. Representative SEM micrographs of the bacterial strains grown for 120 h (A) or 2 h (B) at the liquid-air interface of glass coverslips are shown. Bar, 10 μm.
As revealed by SEM analyses, deletion of the bps locus resulted in a defect in biofilm formation that was much more severe than that observed in the biofilm formation of the wild-type B. bronchiseptica strain RB50 in the presence of DspB (compare Fig. 2C with Fig. 3A). This suggests that either DspB is not able to completely hydrolyze the poly-β-1,6-GlcNAc-like material or other factors are involved in stabilizing these biofilms. The inability of DspB to disperse the preformed biofilms of B. bronchiseptica is similar to its effect on Y. pestis biofilms. We have previously shown that biofilms formed by Y. pestis are not dispersed by DspB (28).
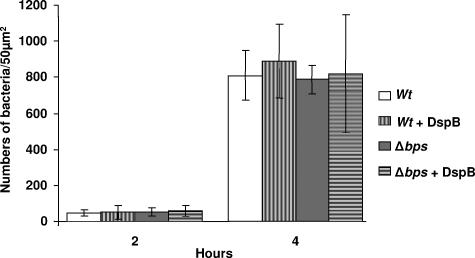
In contrast to the defect in biofilm formation at the late time point (120 h), both the wild-type and Δbps strains were spread around the disks and existed as a monolayer of cells at 2 h (Fig. 3B). To further investigate the role of Bps in initial adherence, we enumerated bacteria attached to the glass coverslips at 2 and 4 h after inoculation. These experiments were repeated multiple times with similar results. It is clear from Fig. 4 that there is no significant difference between the numbers of attached bacteria for the wild type and the Δbps strain at any of the early time points tested. Additionally, pretreatment of either the wild type or the Δbps strain with DspB did not result in a significant decrease in initial attachment (Fig. 4). Thus, these results, taken together, strongly suggest that the bps-encoded polysaccharide is not required for initial surface attachment but contributes to the formation and/or maintenance of multicellular communities.
FIG. 4.
Quantification of the initial bacterial attachment. Bacteria attached to the liquid-air interface of the glass coverslips at 2 and 4 h after inoculation of the wild-type (wt) and Δbps strains were counted under a light microscope. For DspB-treated samples, the strains were cultured overnight in presence of DspB, followed by inoculation of the fresh medium with the respective pretreated strains. Data are averages of at least four random 50-μm2 regions of the coverslip. The experiment was performed in duplicate. Error bars, standard deviations.
Thin-section immunoelectron microscopy.
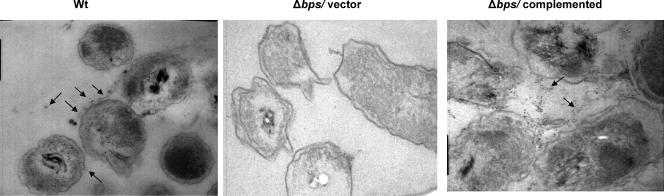
To further address the role of the bps-encoded polysaccharide in biofilm formation, we utilized immuno-TEM. The reactivities of the anti-PNAG sera with the matrix material of the biofilms formed by the wild-type and Δbps strains containing either the vector or the complementing plasmid were tested. The gold particles were found to be both cell associated and extracellular (Fig. 5) and were interpreted to be the antibody cross-reacting with the bps-specific polysaccharide constituting the biofilm matrix. This cross-reactivity was specific to the anti-PNAG antibody and was not observed when only the secondary antibody was used (data not shown). Additionally, consistent with the lack of the cross-reactive polysaccharide-like material in the immunoblot assays (Fig. 1), TEM of the biofilms formed by the Δbps strain harboring the vector plasmid (Fig. 5) or the Δbps strain by itself (data not shown) did not reveal the presence of any observable gold particles. Whereas the gold particles were mainly bound to the exopolysaccharide (EPS) material found on the outer surface of the wild-type strain, for the complemented strain they were also located in the intercellular spaces (Fig. 5). The presence of comparatively higher numbers of gold particles for the complemented strain (Fig. 5, right panel) is consistent with the immunoblot analyses, which demonstrate the overproduction of the Bps polysaccharide in this strain (Fig. 1).
FIG. 5.
Immunoelectron microscopy. Representative TEM micrographs of vertical thin sections of biofilms formed by wild-type and Δbps strains containing either the pBBR1MCS vector or the complementing plasmid pMM11 are shown. Arrows indicate colloidal gold beads.
Characterization of Bordetella biofilm development by confocal scanning laser microscopy.
The use of flow chambers combined with CSLM provides a nondestructive means of visualizing the complex architecture of live hydrated biofilms (25). Although biofilm formation in Bordetella has been demonstrated by utilizing microtiter dish assays and SEM (27, 44), no information is available on the 3-dimensional biofilm structures that Bordetella develops under live monitoring and hydrodynamic conditions. To determine whether Bordetella actually formed a 3-dimensional biofilm, the gfp-tagged wild-type strain RB50 was cultured planktonically to the mid-logarithmic phase and then injected into the flow cell. The cells were then incubated for 2 h to allow adherence to the glass coverslips of the flow cells. The flow of the growth medium was then resumed, and the dynamics of biofilm development was visualized by confocal microscopy every 24 h over a period of 4 days.
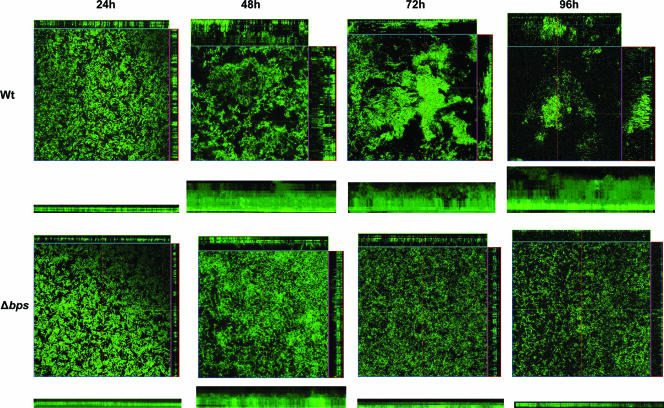
It was apparent from these analyses that at 24 h the wild-type strain adhered to the glass coverslip as a uniform monolayer (Fig. 6). However at this time point the biofilm lacked any apparent structural organization. By day 2, the wild-type strain, in addition to spreading horizontally, had also started to spread vertically, resulting in the appearance of some 3-dimensional multicellular structures in the form of pillars. At later time points (72 and 96 h), there was mainly vertical growth of the biofilm, giving rise to the characteristic pillar structures and water channels observed for other bacteria in continuous-flow systems (7). In addition, the attached wild-type cells existed mainly as discrete clusters or microcolonies at these later time-points.
FIG. 6.
Confocal scanning laser micrographs of biofilms formed by the wild-type B. bronchiseptica strain RB50 (top panels) and the Δbps mutant (bottom panels). Strains were inoculated directly in the flow cell and visualized in situ every 24 h. For each micrograph, the middle panel represents the x-y plane, and the adjacent top and side panels represent the x-z and y-z planes, respectively. The panel underneath each micrograph represents the x-z reconstruction of each biofilm. For each strain, biofilm images were taken from at least five areas and the experiment was repeated at least four times. A representative CSLM image for each sample is shown.
The bpsABCD locus is important for the maintenance of the 3-dimensional structure of Bordetella biofilms.
To gain information on the role of the bps locus in contributing to the development of Bordetella biofilms, we compared the architecture of the biofilms formed by the wild-type and mutant strains in the flow cell. Initially, at 24 h, the Δbps mutant strain displayed the same pattern of attachment as the wild-type strain and existed as a uniform monolayer (Fig. 6). In contrast to the wild-type strain, which existed mainly as a heterogeneous community of microcolonies at later time points, the bps mutant continued to exist as single attached cells in the form of a monolayer across the coverslip; aggregates of the bps mutant were very rarely seen. Unlike those of the wild-type strain, the biofilms formed by the Δbps strain displayed an almost-complete absence of 3-dimensional structural features such as the pillars and the water channels (Fig. 6). In order to achieve a quantitative assessment of the observed differences in biofilm architecture, CSLM-generated images were analyzed by the COMSTAT computer software package. At 24 h and 48 h, statistical evaluation of the biofilms formed by the wild-type and Δbps strains revealed no significant differences in their biomass content or in their average thickness. At later time points (days 3 and 4), however, the biofilms formed by the Δbps strain consisted of significantly less biomass and biofilm thickness (Table 2). Thus, the results described above, taken together, indicate a role for bpsABCD in biofilm formation, particularly in forming and maintaining the complex architecture seen in wild-type biofilms.
TABLE 2.
COMSTAT analysis of CSLM-generated Z-series of the wild-type and Δbps mutant strainsa
| Time (h) | Strain | Biomass
|
Avg thickness
|
Maximum thickness
|
|||
|---|---|---|---|---|---|---|---|
| Valueb | Pc | Value (μm) | Pc | Value (μm) | Pc | ||
| 24 | wt | 0.41 (0.13) | 0.55 (0.22) | 4.9 (1.5) | |||
| Δbps | 0.72 (0.69) | 0.289 | 1.1 (1.1) | 0.197 | 4.6 (1.5) | 0.686 | |
| 48 | wt | 3.0 (0.8) | 6.2 (2.3) | 20.8 (4.8) | |||
| Δbps | 3.1 (0.95) | 0.905 | 5.4 (1.32) | 0.549 | 13.8 (1.3) | <0.05 | |
| 72 | wt | 1.7 (0.72) | 2.9 (1.5) | 12.48 (4.1) | |||
| Δbps | 0.33 (0.2) | <0.05 | 1.1 (1.8) | <0.05 | 3.8 (2.9) | <0.01 | |
| 96 | wt | 1.5 (1.7) | 2.4 (2.6) | 16.9 (8.9) | |||
| Δbps | 0.42 (0.38) | <0.01 | 0.51 (0.72) | <0.005 | 7.7 (3.5) | <0.0001 | |
Values are averages for five images from a representative experiment. Standard errors are given in parentheses. wt, wild type.
Given as μm3/μm2 of surface area covered by bacteria expressing GFP.
Determined using an unpaired two-tailed student's t-test.
DISCUSSION
In this report, we have demonstrated that the Bordetella bpsABCD locus is involved in biofilm formation in Bordetella. The absence of the bps locus does not significantly affect the early steps of biofilm formation, in particular the initial attachment to abiotic surfaces. In this respect, the function of this locus is somewhat different from that of the homologous E. coli pgaABCD and S. aureus icaADBC loci, which have been shown to be required for initial surface attachment (23, 60). Both PGA and PIA have also been shown to promote cell-cell interaction (2, 23, 60), and based on our studies, it appears that this function is conserved for the Bordetella polymer encoded by the bps locus. Our findings suggest a role for the bps locus in microcolony formation and the development of the 3-dimensional structure of biofilms. The requirement for the bps-encoded protein products at a postattachment stage of biofilm formation is similar to that for a number of other bacterial loci or genes that have been implicated in the production of EPSs. For example, mutations that abolish the production of EPS in Vibrio cholerae and of colanic acid in E. coli K-12 do not block the abilities of these bacteria to attach to surfaces initially but do affect the development of matured biofilms (11, 62). Similarly, a mutation in the V. cholerae gene mbaA that led to an increase in the production of extracellular matrix material resulted in biofilms lacking the characteristic pillar structures and water channels (3).
The bps-dependent effect on biofilm formation was manifested in the presence of other Bordetella factors, particularly adhesins such as FHA (filamentous hemagglutinin), fimbriae, and pertactin (42). Recently, a positive role for FHA and fimbriae in biofilm formation on microtiter plates has been demonstrated (27). Our studies, in conjunction with others, suggest that there are at least two distinct steps in Bordetella biofilm development. Initial attachment to abiotic surfaces is probably mediated by adhesins such as FHA and fimbriae, whereas the bps locus subsequently participates in a postattachment step by contributing to the stability and the maintenance of the biofilm architecture.
The putative protein products encoded by the genes of the bpsABCD locus have high amino acid sequence similarity to the enzymes encoded by bacterial loci involved in the production of an extracellular polysaccharide, poly-β-1,6-N-GlcNAc (22, 33, 60). By comparing the extracts that were enriched in polysaccharides from the wild-type and bps mutant strains of B. bronchiseptica in an immunoblot assay, we have provided evidence that this locus is involved in the synthesis of a compound that is antigenically similar to S. aureus PNAG. Although we have not determined the precise chemical nature of the bps-dependent polysaccharide, the antibody utilized for immunoblotting was affinity purified on a column of immobilized deacetylated PNAG from S. aureus and has been shown to be specific for the products of the S. aureus ica locus (37). The inhibition of biofilm formation by the DspB enzyme and the dissolution of the matrix material from the biofilms in the presence of DspB further implicate poly-β-1,6 GlcNAc in contributing to the integrity of Bordetella biofilms. DspB has previously been demonstrated to display a β-hexosaminidase activity that is specific for the hydrolysis of glycosidic linkages of poly-β-1,6 GlcNAc and not the related β-1,4 linkages of chitin derivatives (28, 50). We are currently attempting to purify the specific polysaccharide produced by the bps locus for structural studies. Based on previous results, this may turn out to be technically challenging, since E. coli PGA was previously purified from a mutant strain that (i) was unable to synthesize colanic acid, (ii) harbored a mutation that disrupted csrA, which is a repressor of pgaABCD expression, and (iii) contained a high-copy-number plasmid containing the pga locus, thus allowing overproduction of PGA (28, 59). The expression of PIA in staphylococcal species is also under the control of multiple regulatory proteins and is regulated by diverse environmental factors (references 55 and 30 and references therein).
In addition to the role in biofilm formation, production of extracellular PIA/PNAG-like polysaccharides has also been linked to the disease processes and the pathogenesis of bacteria. Previous studies have demonstrated a critical role for PNAG/PIA in resisting the components of innate and adaptive immune systems and in contributing to the virulence of S. aureus and S. epidermidis in laboratory animals (34, 35, 37, 38, 56, 58). The hmsHFRS locus of Y. pestis, which is a homologue of bpsABCD, has been linked to the production of an extracellular polysaccharide that appears to be involved in efficient transmission of the bacteria from the fleas to mammals (12, 29). Bordetellae are primarily extracellular pathogens and have developed multiple strategies to avoid the innate and adaptive immune responses. We speculate that the bpsABCD locus may play an important role in Bordetella virulence, especially in mediating resistance to host defenses. In this context, it is noteworthy that the human pathogens B. pertussis and B. parapertussis contain an intact homologue of the bpsABCD locus and produce a PNAG-like material. Even though B. bronchiseptica is considered the evolutionary progenitor of both B. pertussis and B. parapertussis, there are drastic differences in genome size and gene expression patterns among the three species (47). In particular, B. pertussis contains a smaller genome and has lost a large number of genes. B. pertussis and B. parapertussis contain a large number of pseudogenes, many of which have been inactivated by insertion of insertion elements, in-frame stop codons, and frameshift mutations (47). Thus, it seems that the human-adapted species of Bordetella are evolving by genome decay to express only factors that will be important for their growth and/or virulence (5, 10, 47). Further studies are under way to determine whether the bpsABCD locus contributes to the virulence of Bordetella.
Acknowledgments
We thank Dan Wozniak, Ed Swords, Neelima Sukumar, and Cheraton Love for critical reading of the manuscript. We are grateful to Gerry B. Pier for a gift of the PNAG-specific antibody. We thank Ken Grant of the Wake Forest Microscopy Core Facility for help with microscopy.
Research in the laboratory of R.D. is supported by funds from Wake Forest University Health Sciences and by grants from the USDA (CSREES-USDA grant 2006-35604-16874) and NIH (R21 AI071054). T.R. is funded by the National Institutes of Health (GM066794).
Kane Biotech, Inc., may develop applications related to the findings herein. T.R. serves as chief scientific advisor for, owns equity in, and may receive royalties from this company. The terms of this arrangement have been reviewed and approved by Emory University in accordance with its conflict-of-interest policies.
Footnotes
Published ahead of print on 17 November 2006.
REFERENCES
- 1.Agladze, K., D. Jackson, and T. Romeo. 2003. Periodicity of cell attachment patterns during Escherichia coli biofilm development. J. Bacteriol. 185:5632-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agladze, K., X. Wang, and T. Romeo. 2005. Spatial periodicity of Escherichia coli K-12 biofilm microstructure initiates during a reversible, polar attachment phase of development and requires the polysaccharide adhesin PGA. J. Bacteriol. 187:8237-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bomchil, N., P. Watnick, and R. Kolter. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 185:1384-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Branda, S. S., A. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 13:20-26. [DOI] [PubMed] [Google Scholar]
- 5.Brinig, M. M., C. A. Cummings, G. N. Sanden, P. Stefanelli, A. Lawrence, and D. A. Relman. 2006. Significant gene order and expression differences in Bordetella pertussis despite limited gene content variation. J. Bacteriol. 188:2375-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, J. A., G. J. Davies, V. Bulone, and B. Henrissat. 1998. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 329:719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappinscott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 8.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 9.Cotter, P. A., and J. F. Miller. 1994. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect. Immun. 62:3381-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cummings, C. A., M. M. Brinig, P. W. Lepp, S. van de Pas, and D. A. Relman. 2004. Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J. Bacteriol. 186:1484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darby, C., S. L. Ananth, L. Tan, and B. J. Hinnebusch. 2005. Identification of gmhA, a Yersinia pestis gene required for flea blockage, by using a Caenorhabditis elegans biofilm system. Infect. Immun. 73:7236-7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative Eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deora, R. 2002. Differential regulation of the Bordetella bipA gene: distinct roles for different BvgA binding sites. J. Bacteriol. 184:6942-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drenkard, E. 2003. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 5:1213-1219. [DOI] [PubMed] [Google Scholar]
- 16.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 17.Fetherston, J. D., P. Schuetze, and R. D. Perry. 1992. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol. Microbiol. 6:2693-2704. [DOI] [PubMed] [Google Scholar]
- 18.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid Rk2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furukawa, S., S. L. Kuchma, and G. A. O'Toole. 2006. Keeping their options open: acute versus persistent infections. J. Bacteriol. 188:1211-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fux, C. A., J. W. Costerton, P. S. Stewart, and P. Stoodley. 2005. Survival strategies of infectious biofilms. Trends Microbiol. 13:34-40. [DOI] [PubMed] [Google Scholar]
- 21.Gerke, C., A. Kraft, R. Sussmuth, O. Schweitzer, and F. Gotz. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273:18586-18593. [DOI] [PubMed] [Google Scholar]
- 22.Gotz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 23.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 24.Heininger, U., P. A. Cotter, H. W. Fescemyer, G. M. de Tejada, M. H. Yuk, J. F. Miller, and E. T. Harvill. 2002. Comparative phenotypic analysis of the Bordetella parapertussis isolate chosen for genomic sequencing. Infect. Immun. 70:3777-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heydorn, A., B. K. Ersboll, M. Hentzer, M. R. Parsek, M. Givskov, and S. Molin. 2000. Experimental reproducibility in flow-chamber biofilms. Microbiology 146:2409-2415. [DOI] [PubMed] [Google Scholar]
- 26.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 27.Irie, Y., S. Mattoo, and M. H. Yuk. 2004. The Bvg virulence control system regulates biofilm formation in Bordetella bronchiseptica. J. Bacteriol. 186:5692-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itoh, Y., X. Wang, B. J. Hinnebusch, J. F. Preston III, and T. Romeo. 2005. Depolymerization of β-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 187:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarrett, C. O., E. Deak, K. E. Isherwood, P. C. Oyston, E. R. Fischer, A. R. Whitney, S. D. Kobayashi, F. R. Deleo, and B. J. Hinnebusch. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190:783-792. [DOI] [PubMed] [Google Scholar]
- 30.Jefferson, K. K., D. B. Pier, D. A. Goldmann, and G. B. Pier. 2004. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 186:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jesaitis, A. J., M. J. Franklin, D. Berglund, M. Sasaki, C. I. Lord, J. B. Bleazard, J. E. Duffy, H. Beyenal, and Z. Lewandowski. 2003. Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J. Immunol. 171:4329-4339. [DOI] [PubMed] [Google Scholar]
- 32.Kaplan, J. B., C. Ragunath, N. Ramasubbu, and D. H. Fine. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous β-hexosaminidase activity. J. Bacteriol. 185:4693-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaplan, J. B., K. Velliyagounder, C. Ragunath, H. Rohde, D. Mack, J. K. Knobloch, and N. Ramasubbu. 2004. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J. Bacteriol. 186:8213-8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly-Quintos, C., L. A. Cavacini, M. R. Posner, D. Goldmann, and G. B. Pier. 2006. Characterization of the opsonic and protective activity against Staphylococcus aureus of fully human monoclonal antibodies specific for the bacterial surface polysaccharide poly-N-acetylglucosamine. Infect. Immun. 74:2742-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly-Quintos, C., A. Kropec, S. Briggs, C. L. Ordonez, D. A. Goldmann, and G. B. Pier. 2005. The role of epitope specificity in the human opsonic antibody response to the staphylococcal surface polysaccharide poly-N-acetyl glucosamine. J. Infect. Dis. 192:2012-2019. [DOI] [PubMed] [Google Scholar]
- 36.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop, and K. M. Peterson. 1994. Pbbr1Mcs: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 37.Kropec, A., T. Maira-Litran, K. K. Jefferson, M. Grout, S. E. Cramton, F. Gotz, D. A. Goldmann, and G. B. Pier. 2005. Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for virulence in murine models of systemic infection. Infect. Immun. 73:6868-6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, H. L., L. Xu, J. P. Wang, Y. M. Wen, C. Vuong, M. Otto, and Q. Gao. 2005. Conversion of Staphylococcus epidermidis strains from commensal to invasive by expression of the ica locus encoding production of biofilm exopolysaccharide. Infect. Immun. 73:3188-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mah, T. F., B. Pitts, B. Pellock, G. C. Walker, P. S. Stewart, and G. A. O'Toole. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306-310. [DOI] [PubMed] [Google Scholar]
- 41.Maira-Litran, T., A. Kropec, D. A. Goldmann, and G. B. Pier. 2005. Comparative opsonic and protective activities of Staphylococcus aureus conjugate vaccines containing native or deacetylated staphylococcal poly-N-acetyl-β-(1-6)-glucosamine. Infect. Immun. 73:6752-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattoo, S., and J. D. Cherry. 2005. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin. Microbiol. Rev. 18:326-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKenney, D., J. Hubner, E. Muller, Y. Wang, D. A. Goldmann, and G. B. Pier. 1998. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect. Immun. 66:4711-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mishra, M., G. Parise, K. D. Jackson, D. J. Wozniak, and R. Deora. 2005. The BvgAS signal transduction system regulates biofilm development in Bordetella. J. Bacteriol. 187:1474-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 46.Ouahrani-Bettache, S., F. Porte, J. Teyssier, J. P. Liautard, and S. Kohler. 1999. pBBR1-GFP: a broad-host-range vector for prokaryotic promoter studies. BioTechniques 26:620-622. [DOI] [PubMed] [Google Scholar]
- 47.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. G. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 48.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 49.Perry, R. D., A. G. Bobrov, O. Kirillina, H. A. Jones, L. Pedersen, J. Abney, and J. D. Fetherston. 2004. Temperature regulation of the hemin storage (Hms+) phenotype of Yersinia pestis is posttranscriptional. J. Bacteriol. 186:1638-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramasubbu, N., L. M. Thomas, C. Ragunath, and J. B. Kaplan. 2005. Structural analysis of dispersin B, a biofilm-releasing glycoside hydrolase from the periodontopathogen Actinobacillus actinomycetemcomitans. J. Mol. Biol. 349:475-486. [DOI] [PubMed] [Google Scholar]
- 51.Relman, D., E. Tuomanen, S. Falkow, D. T. Golenbock, K. Saukkonen, and S. D. Wright. 1990. Recognition of a bacterial adhesin by an integrin: macrophage Cr3 (αMβ2, Cd11b/Cd18) binds filamentous hemagglutinin of Bordetella pertussis. Cell 61:1375-1382. [DOI] [PubMed] [Google Scholar]
- 52.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spoering, A. L., and M. S. Gilmore. 2006. Quorum sensing and DNA release in bacterial biofilms. Curr. Opin. Microbiol. 9:133-137. [DOI] [PubMed] [Google Scholar]
- 54.Stainer, D. W., and M. J. Scholte. 1970. A simple chemically defined medium for the production of phase I Bordetella pertussis. J. Gen. Microbiol. 63:211-220. [DOI] [PubMed] [Google Scholar]
- 55.Vuong, C., J. B. Kidder, E. R. Jacobson, M. Otto, R. A. Proctor, and G. A. Somerville. 2005. Staphylococcus epidermidis polysaccharide intercellular adhesin production significantly increases during tricarboxylic acid cycle stress. J. Bacteriol. 187:2967-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vuong, C., S. Kocianova, J. M. Voyich, Y. F. Yao, E. R. Fischer, F. R. Deleo, and M. Otto. 2004. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 279:54881-54886. [DOI] [PubMed] [Google Scholar]
- 57.Vuong, C., and M. Otto. 2002. Staphylococcus epidermidis infections. Microbes Infect. 4:481-489. [DOI] [PubMed] [Google Scholar]
- 58.Vuong, C., J. M. Voyich, E. R. Fischer, K. R. Braughton, A. R. Whitney, F. R. Deleo, and M. Otto. 2004. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 6:269-275. [DOI] [PubMed] [Google Scholar]
- 59.Wang, X., A. K. Dubey, K. Suzuki, C. S. Baker, P. Babitzke, and T. Romeo. 2005. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 56:1648-1663. [DOI] [PubMed] [Google Scholar]
- 60.Wang, X., J. F. Preston III, and T. Romeo. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186:2724-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watnick, P., and R. Kolter. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Webb, J. S., M. Givskov, and S. Kjelleberg. 2003. Bacterial biofilms: prokaryotic adventures in multicellularity. Curr. Opin. Microbiol. 6:578-585. [DOI] [PubMed] [Google Scholar]
- 64.Yeh, S. H. 2003. Pertussis: persistent pathogen, imperfect vaccines. Expert Rev. Vaccines 2:113-127. [DOI] [PubMed] [Google Scholar]