Abstract
Bacteriophage T4 contains three self-splicing group I introns in genes in de novo deoxyribonucleotide biosynthesis (in td, coding for thymidylate synthase and in nrdB and nrdD, coding for ribonucleotide reductase). Their presence in these genes has fueled speculations that the introns are retained within the phage genome due to a possible regulatory role in the control of de novo deoxyribonucleotide synthesis. To study whether sequences in the upstream exon interfere with proper intron folding and splicing, we inhibited translation in T4-infected bacteria as well as in bacteria containing recombinant plasmids carrying the nrdB intron. Splicing was strongly reduced for all three T4 introns after the addition of chloramphenicol during phage infection, suggesting that the need for translating ribosomes is a general trait for unperturbed splicing. The splicing of the cloned nrdB intron was markedly reduced in the presence of chloramphenicol or when translation was hindered by stop codons inserted in the upstream exon. Several exon regions capable of forming putative interactions with nrdB intron sequences were identified, and the removal or mutation of these exon regions restored splicing efficiency in the absence of translation. Interestingly, splicing of the cloned nrdB intron was also reduced as cells entered stationary phase and splicing of all three introns was reduced upon the T4 infection of stationary-phase bacteria. Our results imply that conditions likely to be frequently encountered by natural phage populations may limit the self-splicing efficiency of group I introns. This is the first time that environmental effects on bacterial growth have been linked to the regulation of splicing of phage introns.
Bacteriophage T4 contains three self-splicing group I introns, which are situated in the genes coding for thymidylate synthase (td), the small subunit of the aerobic ribonucleotide reductase (nrdB), and the anaerobic ribonucleotide reductase (nrdD) (9, 17, 34, 44). Like many other group I introns, the td and nrdD introns each contain an open reading frame encoding a homing endonuclease that renders the introns mobile and they can be inserted into new phage genomes through the process of homing (27, 28). The nrdB intron is not mobile, due to a deletion in the intron-borne homing endonuclease gene (13). Horizontal transfer of the introns between phages through homing has played a significant role in the evolution of group I introns in T-even-like phages (4, 27, 29).
Much speculation has been raised about the potential benefits or costs of introns for phages and what effect they may have on the expression of intron-containing genes and phage viability, but few studies have addressed the question experimentally. Since all of the T4 introns are present in genes that encode proteins in the same biochemical pathway, de novo deoxyribonucleotide synthesis during phage infection, it was speculated early on that the introns may confer a selective advantage to the phage by offering the possibility of regulating the expression of the intron-containing genes by the regulation of splicing (15, 17, 33). One step towards such a regulatory role of splicing in T4 was the finding that, in the absence of translation of the upstream td exon, a specific interaction can be formed between exon sequences and the 3′ splice site of the td intron (31). This interaction prevents normal intron folding and inhibits splicing.
The catalytic structure of each of the T4 group I introns is made of several stem-loop pairings (called P1 through P12; for a schematic secondary-structure prediction, see reference 7) that are packed into a compact structure via tertiary RNA-RNA interactions (14, 22). The core of each intron is made of the contact region between the two separately folding P4 to P6 and P3 to P9 domains, with the active site located in the P7 helix (21). The 5′ splice site is determined by the position of the P1 stem-loop (5, 11), and the P9.0 interaction brings together the 3′ splice site and the active site, determining the position of the 3′ splice site (5). Alignment of the 5′ and 3′ splice sites at the active site for ligation of the two exons is facilitated via the P10 pairing between sequences in the P1 loop (L1) and sequences directly downstream of the 3′ splice site (10, 21). Since P9.0 and P10 are rather weak interactions and cannot be formed until the whole intron is transcribed, they are sensitive to misfoldings.
In this study, we show that splicing efficiency for all three T4 introns is strongly reduced if translation is reduced or abolished. Multiple upstream exon regions are predicted to be able to form interactions that interfere with normal intron folding for each intron-containing gene, and with nrdB as an example, we find that the lack of actively translating ribosomes in these exon regions inhibits splicing. Furthermore, a marked reduction of splicing of all three introns also occurs if the infected bacterium is in stationary phase, and this splicing reduction is coupled to the same regions of the upstream exon. These findings suggest that folding problems may be common to group I introns in the absence of efficient translation. This is the first time that environmental effects (like the growth phase of infected bacteria) that are probably frequently encountered by natural phage populations have been shown to affect the splicing of phage introns.
MATERIALS AND METHODS
Bacteria, phages, and plasmids.
Escherichia coli strain JM101 {supE thi Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15]} was used for all plasmid expression studies. Plasmid pBS5 (34) contains the wild-type nrdB gene from bacteriophage T4 cloned into the M13mp19 plasmid. Stop codon mutants were made in pBS5 by the PCR-based QuikChange site-directed mutagenesis method from Stratagene according to the manufacturer's protocols. E. coli B0 was used as host for all phage infections with T4D; both stocks have been maintained in our laboratory for many years.
Mutagenesis primers.
The primers used for mutagenesis were TAA10, 5′-CAGTTTTTAATACAAATCCATAAGATGTTTTGAATGAACCG; TAA23, 5′-CCGATGTTCTTTGGTTCTGGATAAGGTTTAGCTCGATATG; TAA44, 5′-GAACTCATTGAGCGGCAGTAAAGTTTTTTTTGGCGTCCTG; TAA100, 5′-CTTATGTCATTAATTTAAGACCCAAGCCTTG; TAA130, 5′-CATGCGAAATCTTTAAACTGATCCATCGAAGG; TAA166, 5′-GGTTAAAACCCGTTAATGGGAAAATGCTAAAGAC; TAA212, 5′-ATGCCTTATAAGCTATT (25); A49C, 5′-GATGTTTTGAATGAACCGCTGTTCTTTGGTTCTGG; T1259G, 5′-GCGAGCTCAATCGAACAGACGGTACCTTTAACTTCC; Δ12-15 5′-CCATAAGATGTTTTGAATGAATTTGGTTCTGGATTAGGTTTAGCTCG; Δ45-129 A (5′-CAGTATATTACTGCCGCTCAATGAG) and B (5′-CGGCAGTAATATACTGATCCATCGAAGG); ΔCry-P1 5′-TAATGCCTTAGAATAAATTCGTTTTGCGTAAAATGCGCCTTTAAACGG; ΔnrdB IVS A 5′-GTTAAAGGTACACGCAAAAGATACATAAAAACG and B; 5′-CTTTTGCGTGTACCTTTAACTTCCATAAG; and IVSmut, 5′-GCAAAACAAGGTTTAATAATTAGTCTTCGGACG. Underlining denotes nucleotide changes.
Total-RNA preparations.
Cells were grown in LB medium with or without antibiotics to an optical density at 600 nm (OD600) of 0.3. The expression of T4 nrdB mRNA from pBS5 or its derivatives was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM. Cells were harvested by adding equal volumes of bacteria and ice-cold RNAprotect bacterial reagent (QIAGEN), followed by immediate freezing in liquid nitrogen. Total RNA was extracted from the bacterial samples with the QIAGEN RNeasy miniprep spin kit according to the manufacturer's protocol.
Limited primer extension.
Limited primer extension was performed either with labeled primer or with by-product labeling by the incorporation of [32P]dCTP. The two methods were used interchangeably and produced identical results. Total RNA was mixed with 25 pmol of unlabeled or 1 pmol of labeled primer in hybridization buffer (to a final concentration of 100 mM KCl, 1 mM EDTA, 50 mM Tris-HCl [pH 8]) in a final volume of 11 μl and denatured at 95°C for 2 min, followed by annealing at 42°C for 15 min. For product labeling, 6.5 μl of extension mix {3.5 μl avian myeloblastosis virus reverse transcriptase reaction buffer, 2.3 μl nucleotide mix (to a final concentration of 100 μM dATP, 100 μM dGTP, 250 μM ddTTP, 2.5 μM [32P]dCTP), 0.7 μl (15 U) of avian myeloblastosis virus reverse transcriptase (Roche Diagnostics)} was added to the annealing reaction and incubation continued at 42°C for 60 min. For the labeled primer reaction, the nucleotide mix contained equal amounts of dATP, dCTP, and dGTP (100 μM) and 250 μM ddTTP. To assay nrdD splicing, ddGTP was used as the terminating nucleotide and dTTP was included instead of dGTP in the nucleotide mixture accordingly. The primers used were RTnrdB25A (5′-CAT GTT CTT ATG GAA GTT AAA GGT A-3′) for nrdB, RTtdss (5′-TGA CGC AAT ATT AAA CGG TA-3′) for td, and RTnrdDss (5′-GAA TAA CGT GTT CAC TTC AT-3′) for nrdD. Lengths of products corresponding to pre-mRNA were 29 nucleotides (nt) (nrdB), 24 nt (td), and 25 nt (nrdD), and those corresponding to mRNA were 37 nt (nrdB), 35 nt (td), and 29 nt (nrdD). To remove unincorporated nucleotides, the RNA was degraded by the addition of RNase A (10 mg/ml) and incubation at 37°C for 60 min followed by the removal of nucleotides with the QIAGEN nucleotide removal kit. Samples were run on a 15% polyacrylamide gel. Gels were analyzed on a Fuji LAS3000 phosphorimager and quantified using Fuji Image Gauge 3.45 software. The product-labeling method incorporates stoichiometrically different amounts of label between spliced and unspliced product depending on the sequence between the primer and the terminating nucleotide. This was corrected for in calculations according to the following scheme: the ratio of td spliced/unspliced was 4/1 32P-C, that of nrdB spliced/unspliced was 3/2 32P-C, and that of nrdD spliced/unspliced was 2/1 32P-C.
Phage infection in the presence of chloramphenicol.
E. coli B0 was grown at 30°C in LB medium to an OD600 of 0.3, and the culture was divided into three parts, one with the addition of chloramphenicol 5 min prior to phage addition, one with the addition of chloramphenicol 2 min after infection, and one without the addition of chloramphenicol. The final concentration of chloramphenicol was 155 μM, a concentration that rapidly stops translation. Phage was added to a multiplicity of infection of approximately 5, samples were collected at 3, 7, and 12 min after infection as described above, and splicing was analyzed by limited primer extension.
Screen for interacting regions.
The upstream exons of the T4 td, nrdB, and nrdD genes were screened for regions with inverse complementarity to any region of their respective intron. Exon sequences were inverted and divided into blocks of 20 nucleotides with overlaps of 10 nucleotides between adjacent blocks and checked for complementarity to intron sequences, excluding homing endonuclease open reading frames (ORFs), by Wilbur-Lipman alignments (40). In the first round, any complementarity of seven bases or more, allowing for G-U base pairs and one mismatch or bulged nucleotide, was collected. In a second round of selection, positive sequences were mapped on the secondary structure of the respective intron and scored for the possibility to compete with intron structures depending on the strength of the alternative pairing. Only interactions with ΔG (46) values 1.5 times larger than that of the broken interaction were considered.
Pre-mRNA and mRNA stability.
To compare the stabilities of pre-mRNA and mRNA, one needs to use gene constructs that produce only one or the other RNA product, as splicing would otherwise interfere with the results. For measuring pre-mRNA stability, we used splicing-deficient constructs with four point mutations at the intron catalytic center and, for measuring mRNA stability, we used constructs with an exact deletion of the intron sequence. Transcription was induced as described above and stopped by the addition of rifampin to a final concentration of 500 μg/ml. Samples were collected, and total RNA was prepared as described above. The amount of transcript was assayed at three positions along the transcript simultaneously with the limited primer extension product-labeling method using three different primers: nrdBmRNA1 (5′-CAA CTG GAT TTG TAT TAA AAA CTG TA-3′) hybridizes 5 nt downstream of the start of nrdB with an extended product 28 of nt, nrdBmRNA2 (5′-GCT TCT AAG GCA TTA ATA ACG TGC-3′) hybridizes 26 nt upstream of the 5′ splice site with an extended product of 34 nt, and nrdBmRNA3 (5′-TGA ACT TCA TAA TCT TGG CAT TAC CTT C-3′) hybridizes 35 nt downstream of the 3′ splice site with an extended product of 31 nt. All products contain stoichiometrically equal amounts of 32P-C. Samples were analyzed as described above.
Chloramphenicol splicing experiment.
Cells were grown in LB medium with 100 μg/ml carbenicillin to an OD600 of 0.3, and chloramphenicol was added to a final concentration of 155 μM at the same time that transcription was induced with 1 mM IPTG. Samples were collected 10, 20, 30, 60, and 120 min after induction and analyzed by limited primer extension.
Phage infection of cells in different growth phases.
An E. coli B0 culture was grown at 37°C in LB medium. Upon infection, 20 ml of the growing culture was added to a prewarmed flask containing phage to a multiplicity of infection of approximately 5. Infections were made in mid-log phase, early stationary phase, and late stationary phase. Samples from infected cultures were collected as described above, and splicing was analyzed by limited primer extension.
Stationary-phase splicing experiment.
JM101 containing pBS5 or pBS5 Δ45-129;ΔCry-P1 was grown in LB medium with 100 μg/ml carbenicillin to an OD600 of 0.3, transcription was induced by adding IPTG to a final concentration of 1 mM, and growth was continued and monitored by measuring OD600. Samples were collected as described above. Growth was restarted by gently spinning down part of the stationary-phase cells and resolving them in fresh, prewarmed LB-medium containing carbenicillin and IPTG.
RESULTS
Splicing inhibition in the absence of translation is a general trait for all three T4 introns.
To determine whether the inhibition of splicing observed for the td intron in the absence of translation (31) is a general trait for all three T4 introns, we measured their splicing efficiencies in infected bacteria in the presence of the translational inhibitor chloramphenicol. The ratios of spliced transcript to unspliced transcript were assayed by limited primer extension (see Material and Methods). For these experiments, we needed to consider the different expression timing of the intron-containing genes. The early td promoter does not require any phage proteins, but the expression of nrdB and nrdD from middle-mode promoters requires the phage protein MotA (24, 36). Chloramphenicol could therefore be added prior to infection to measure td splicing but had to be added a few minutes after infection to measure nrdB and nrdD intron splicing to first allow for the expression of the MotA protein. In addition, all three intron-borne ORFs are expressed from late promoters (and for the nrdB ORF, also from a middle promoter) within the intron, thus adding transcripts containing only the 3′ half of the intron late in infection. This will produce nonspliceable RNA that artificially lowers the relative splicing efficiency at late times of infection. The ORF transcription is also affected by the addition of chloramphenicol, and we therefore compare splicing efficiencies at the peak of splicing to avoid these variations.
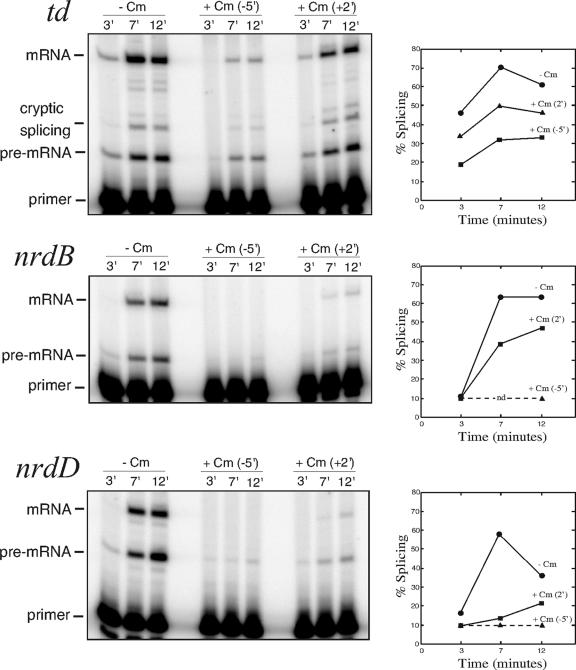
The addition of chloramphenicol clearly reduced the splicing efficiency of all three T4 introns (Fig. 1). For the td intron, splicing efficiency was reduced to 50% of the normal level at the peak of efficiency when chloramphenicol was added prior to infection and to 70% when it was added 2 min after infection, indicating that the addition of the antibiotic after infection perhaps does not show its full effect on splicing. Cryptic td splicing was increased in the presence of antibiotic from 3 to 7% of total splicing in both cases, indicating that compromised folding of the intron plays a part in the reduction of splicing. For the nrdB and nrdD introns, transcript levels are high in the absence of chloramphenicol but no or very little transcript can be seen when chloramphenicol was added 5 min prior to infection. When chloramphenicol instead was added 2 min after infection, both pre-mRNA and mRNA can be seen, although still at a level lower than that without antibiotic. Importantly, the addition of chloramphenicol reduces splicing to 75% of the normal level for nrdB, and for nrdD, where the effect is most pronounced, splicing is reduced to less than one-third of the normal level. Since chloramphenicol does not have a direct effect on group I intron splicing (38), these results (Fig. 1) indicate that, during phage infection, a reduction of translation has severe effects on the splicing capability of all three T4 introns.
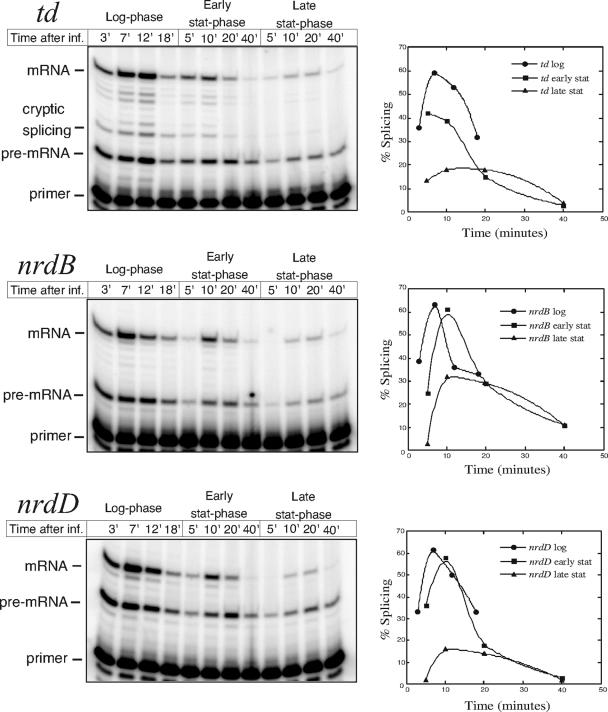
FIG. 1.
Effect of chloramphenicol on splicing during phage infection. E. coli B0 cells were grown at 30°C and infected with T4D in early log phase. Chloramphenicol was added either 5 min prior to infection (middle lanes) or 2 min after addition of phages (rightmost lanes). Splicing efficiencies at different time points are shown in the graph beside each panel. The peak of splicing efficiency of all three introns differed somewhat between experiments but was consistent relative to each other within experiments. Sampling at different time points determined the peaks to be 5 to 7 min for td, 7 to 10 min for nrdB, and 5 to 10 min for nrdD and the variation of splicing efficiency to be ±8.5% for td, ±5.1% for nrdB, and ±4.0% for nrdD. Dotted lines denote splicing efficiencies below the detection limit of 10% splicing. No nrdB transcript was detected before 12 min after infection when chloramphenicol was added 5 min prior to infection. −, absence of; +, presence of.
Several potential exon-intron interactions may disturb splicing of the T4 introns.
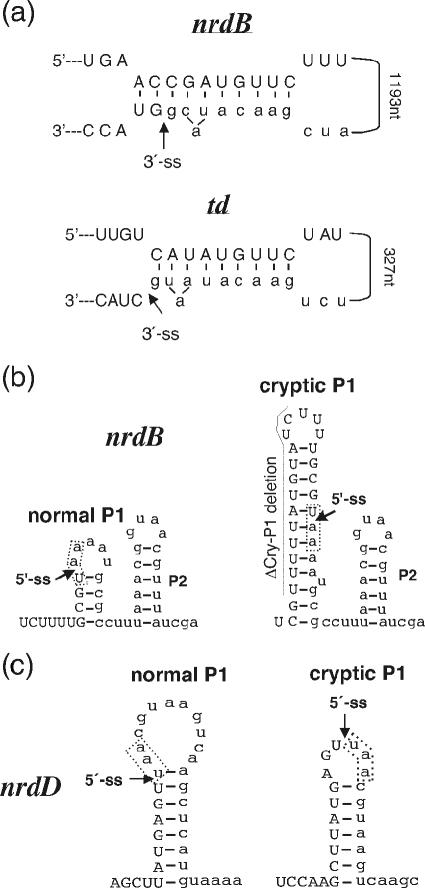
If the observed reduction of splicing of the T4 introns is due to interactions between exposed exon regions and intron sequences that interfere with correct intron folding, putative regions of complementarity between exon and intron sequences should exist. To identify such regions for experimental testing of their effects on splicing, we performed a computer-based screen for the complementarity of the upstream exon and the catalytic core sequences of the respective intron (i.e., excluding the endonuclease sequence since interactions formed between the exon and the looped-out open reading frame are not likely to affect splicing) as described in Material and Methods. Multiple putative interacting regions that would disrupt important secondary and tertiary folds of the introns were found for each of the three introns (Table 1 ). Our screen detected the exon-intron interaction found by Semrad and Schroeder to strongly interfere with td splicing (31). Most interestingly, one region of the nrdB exon (between codons 12 and 15) can form a putative structure almost identical to the td interaction, including an equivalent bulged adenosine located 2 nt from the 3′ splice site (Fig. 2a). In addition, several other putative interactions with potential to target the splice sites and other intron regions known to be vital for correct intron folding were found. For both the nrdB and the nrdD genes, we found potential cryptic P1 stems that would interfere with 5′ splice site selection (Fig. 2b and c). A cryptic P1 has already been identified for the T4 td intron (8).
TABLE 1.
Putative interactions between T4 exon and intron sequences
| Interacting exon codons | Interacting intron region | Distance (nt) | ΔG status (kcal/mol)a
|
|
|---|---|---|---|---|
| Formed | Βrokenb | |||
| nrdB | ||||
| 12-15 | P9.0 | 1,201 | −9.3 | ND |
| 50-53 | P2 | 504 | −7.6 | −4.8 |
| 82-85 | P7.2 to P3 and P12 | 881 | −10.1 | −6.0 |
| 106-108 | P5 and J3/4 and J6/7 | 377 | −7.8 | −2.4 |
| 133-135 | P9.0 | 837 | −7.2 | ND |
| 135-138 | P7 and J7/8 | 759 | −7.1 | −3.0 |
| 146-151 | P7 and J7/8 | 703 | −12.0 | −3.0 |
| 183-185 | P7 and J6/7 | 544 | −7.6 | −3.0 |
| 213-219 | Cry-P1 | −8.0 | −4.0 | |
| nrdD | ||||
| 39-43 | P9.0 | 1,439 | −4.9 | ND |
| 78-82 | P6 | 357 | −12.2 | −7.5 |
| 80-83 | P9.0 and P9.2 | 1,312 | −11.5 | −6.3 |
| 138-141 | L1 | 116 | −6.8 | |
| 156-158 | P6 | 134 | −8.0 | −4.1 |
| 177-181 | Cry-P1 | −5.3 | −7.5 | |
| td | ||||
| 27-31 | P9.0 | 1,462 | −6.8 | ND |
| 79-81 | L9.2 | 1,296 | −8.4 | ND |
| 111-116 | P9.0 and P10 | 1,221 | −10.4 | ND |
| 125-129 | L9.2 | 1,157 | −6.3 | −3.9 |
| 157-159 | P9.0 and P10 | 1,077c | −6.1 | ND |
| 171-175 | P9.0 and P10 | 1,036 | −6.2 | ND |
FIG. 2.
Putative interactions between exon and intron sequences. (a) Comparison of structures of the putative interaction found in nrdB and the interaction found to interfere with td splicing (31). Exon sequences are in uppercase letters, and intron sequences in lowercase letters. An arrow indicates the 3′ splice site. Distance between exon and intron regions in the two experimental systems is indicated. (b and c) Putative cryptic P1 pairings in the nrdB and nrdD introns. Exon sequences are in uppercase letters, and intron sequences are in lowercase letters. Arrows indicate the 5′ splice sites. The naturally occurring stop codon at the start of each intron is marked with a box and the extent of the ΔCry-P1 deletion in nrdB is indicated.
Splicing efficiency without translation of the T4 nrdB transcript.
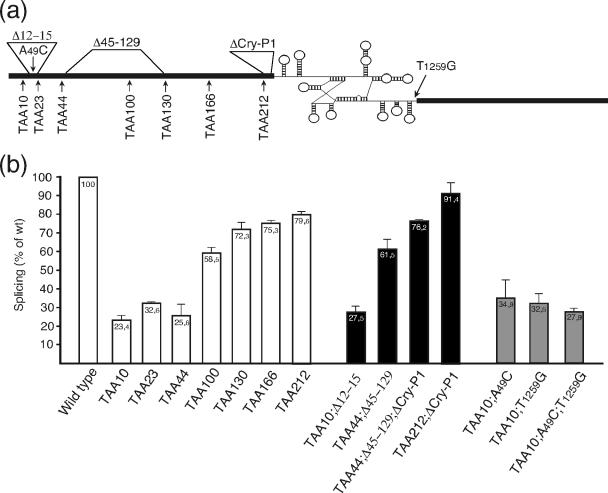
To determine whether the reduction of splicing found in the absence of translation really is due to our predicted exon-intron interactions, we chose to use the nrdB transcript as a model for assaying the importance of these predicted interactions. The nrdB transcript has a predicted interaction almost identical to the one previously shown to affect splicing in td (31) and a cryptic P1 as also found in td (8) and nrdD (this study). We introduced stop codons at several positions along the upstream exon of the nrdB gene (Fig. 3a). Stop codons will exclude translating ribosomes from downstream regions of the transcript and allow potential interactions between intron and exon regions. Indeed there was a clear reduction in splicing levels for all stop codon constructs (Fig. 3b).
FIG. 3.
Effect of stop codons in the upstream exon on splicing of the nrdB intron. (a) Positions of inserted stop codons, mutations, and deletions in the T4 nrdB gene. Thick lines represent exons. The intron is indicated with a schematic secondary fold. (b) Efficiency of nrdB splicing in the different constructs. Bars display the amount of spliced versus unspliced product of stop codon constructs (white) and those with deletions (black) and point mutations (gray) relative to the wild-type pBS5 level. Numbers are averages from two to five independent experiments. Error bars show the standard error of the mean.
The most pronounced reduction of splicing was found with stop codons inserted early in the exon. The TAA10 construct (numbers represent amino acid numbers from the start of the gene; exon 1 is 220 codons long) that would prevent ribosomes from reaching the putative interacting region between codons 12 and 15 and thus allow this to form shows only slightly more than 20% of the wild-type splicing efficiency (Fig. 3b). However, the two following stop codons constructs, TAA23 and TAA44, where ribosomes would disrupt the interaction, show only a very small increase in splicing levels, indicating that the putative interaction between codons 12 and 15 contributes only marginally to the reduction of splicing and that regions downstream of codon 44 are responsible for most of the interference of splicing.
The major drop in splicing efficiency (from 72 to 26% splicing compared to that of the wild type) occurs between stop codons 44 and 130, with the TAA100 construct showing intermediate levels. This drop in splicing efficiency indicates that ribosomal coverage of sequences between codons 44 and 130 is important for the efficiency of splicing. At least three potential interacting regions can be predicted between codons 44 and 130 (Table 1), one interacting with the P2 helix, one with the intron catalytic center, and one with the P5 helix extending to the triple-helix region J3/4 to J6/7.
The splicing activity appears to reach a plateau at a level of 75% of that of wild-type splicing with stop codons inserted after codon 130 (Fig. 3b), and the four predicted potential interactions located between codons 133 and 212 seem to have only a marginal effect on splicing. Three of these putative interactions target the catalytic site in the P7 helix. It is possible that the lack of effect is due to a high folding stability of the catalytic core of the intron. Since the last stop codon construct, TAA212, shows only 80% of the wild-type splicing activity, the translation of the 21-nt region between amino acid position 212 and the intron is important for efficient splicing to occur. Within this region, we can predict a strong putative 12-bp alternative P1 helix (Fig. 2b). Cryptic P1 stem formation has been shown to occur in other group I introns, including the T4 td intron (8), and ribosome coverage of the exon region closest to the intron will resolve such structures (31, 39). A cryptic P1 stem was also predicted for the T4 nrdD intron in our exon-intron complementarity screen (Table 1).
To make certain that mRNA stability is not affected in the stop codon mutants, thereby falsely giving the impression of a reduction in splicing efficiency, we deleted the intron and measured the mRNA half-lives of transcripts with and without stop codons (Table 2). Only the TAA100 construct had a significantly lower mRNA half-life than that of the wild-type gene. However, the pre-mRNA half-life of this construct was also reduced compared to that of the wild type and the mRNA/pre-mRNA stability ratio was therefore maintained (Table 2). In the presence of chloramphenicol, we observed a twofold increase for both mRNA and pre-mRNA stability (Table 2), which was in accordance with the stabilization of transcripts in the presence of this antibiotic found in previous studies (26, 30). The mRNA/pre-mRNA stability ratio was the same as that in the absence of chloramphenicol.
TABLE 2.
mRNA and pre-mRNA half-livesa
| Constructb | Transcriptc | t1/2 (min) |
|---|---|---|
| Wild type | mRNA | 8.3 ± 1.3 |
| Wild type | pre-mRNA | 7.6 ± 0.5 |
| TAA44 | mRNA | 7.2 ± 2.2 |
| TAA100 | mRNA | 6.1 ± 0.7 |
| TAA100 | pre-mRNA | 4.2 ± 0.8 |
| TAA166 | mRNA | 9.3 ± 0.9 |
| Wild type +Cm | mRNA | 20.5 ± 6.4 |
| Wild type +Cm | pre-mRNA | 15.9 ± 2.8 |
RNA stability was measured after the addition of rifampin to a final concentration of 500 μg/ml (see Materials and Methods). t1/2, half-life.
+Cm indicates the addition of chloramphenicol (155 μM final concentration) to the culture.
mRNA stability was measured in constructs having an exact deletion of the intron sequence, and pre-mRNA stability was measured in constructs with four point mutations at the catalytic center that inhibits splicing (see Materials and Methods).
Deletion of T4 nrdB exon sequences complementary to intron sequences restores splicing.
Three in-frame deletions were made to see what effect the removal of putative exon-intron interactions would have on splicing: a 9-bp deletion of codons 12 to 15 (Δ12-15), possibly interacting with the 3′ end of the intron; a 258-bp deletion of codons 45 to 129 (Δ45-129), removing several potential interactions in the region that shows the largest effect of the stop codon constructs; and a 12-bp deletion of codons 213 to 216, removing the potential cryptic P1 interaction (ΔCry-P1). All deletions were combined with the nearest upstream stop codon mutant to minimize interactions from exon regions upstream of the deletion. Splicing efficiencies of deletion constructs are shown in Fig. 3b.
The combination of TAA10 and Δ12-15 produces only a minor increase in splicing efficiency, from 23 to 28%. This is in line with the small increase in splicing efficiency between the TAA10 stop codon mutant (that would allow for the interaction to form) and the subsequent TAA23 and TAA44 mutants (where it would be disrupted by ribosomes) and confirms that this putative interaction in nrdB has only a minor role in the total decrease of splicing. In addition, the introduction of point mutations either in the exon (TAA10;A49C) or in the 3′ end of the intron (TAA10;T1259G) that would disrupt or weaken this interaction (Fig. 2a) results in the same small increase to 33 to 35% splicing efficiency (Fig. 3b), and restoring the base-pairing interaction by combining the two mutations (TAA10;A49C;T1259G) also reduces the splicing efficiency back to the initial level (Fig. 3b), again stressing the minor effect of the interaction shown in Fig. 2a for nrdB intron splicing.
In contrast, the deletion of codons 45 to 129, in combination with the TAA44 stop codon insertion (TAA44;Δ45-129), more than doubles the splicing efficiency (from 26 to 62%) (Fig. 3b) compared to that of the TAA44 mutation by itself, clearly showing that the region between codons 45 and 129 plays a major role in the reduction of splicing in the absence of translation. Finally, the deletion of the exon region with potential to form a cryptic-P1 interaction, in combination with TAA212, restores splicing to near wild-type levels (91%) (Fig. 3b).
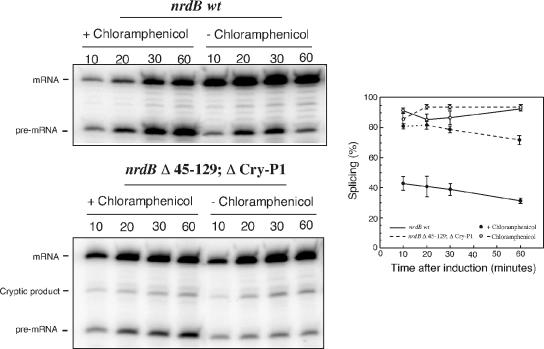
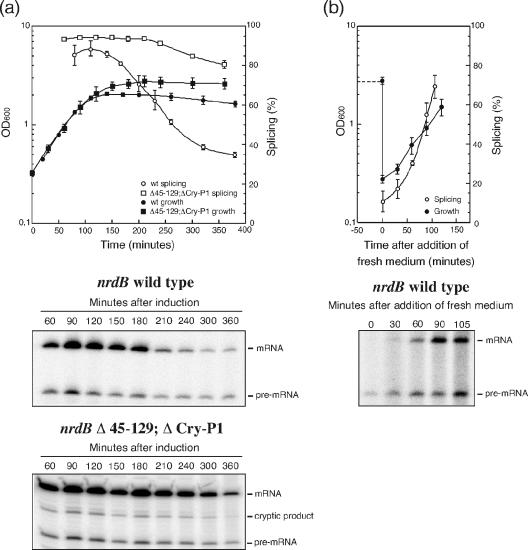
In agreement with the infection experiments, the splicing efficiency of the cloned nrdB intron was drastically reduced when cells were grown in the presence of chloramphenicol (Fig. 4). The reduction of splicing from 90 to 30% is also comparable to that seen with stop codons inserted early in the nrdB gene. One effect of the chloramphenicol treatment is exposure of upstream exon sequences that are normally covered by ribosomes, an effect similar to the introduction of stop codons. In support of the exon-intron interactions, the effect of chloramphenicol addition was almost abolished by the deletion of the 45-129 and Cry-P1 regions (Fig. 4).
FIG. 4.
Splicing of the nrdB intron in the presence of chloramphenicol. Cultures of JM101 containing either pBS5 or pBS5 Δ45-129;ΔCry-P1 were grown continuously at 37°C to an OD600 of 0.3, and half of each culture was treated with chloramphenicol 10 min prior to the induction of transcription. Samples were taken at the indicated time points after induction. The splicing efficiencies are displayed in the graph. Error bars show the standard error of the mean. −, absence of; +, presence of.
Taken together, these results show that, in the absence of translating ribosomes, a number of regions in the upstream exon have a strong collective negative effect on splicing efficiency of the nrdB intron.
Splicing efficiency of all three introns during T4 infection correlates with growth phase.
The rapid bacterial growth that occurs during laboratory culturing in rich medium is likely to be rare in nature. Since translation is known to be tightly regulated and the rate of translational initiation as well as elongation is reduced during starvation in E. coli (1, 35), we wanted to see whether the splicing of the T4 introns is also affected by infection in different bacterial growth phases. Aliquots of E. coli were infected with T4 phage during logarithmic as well as stationary growth, and samples were taken for RNA preparation at different time points during the infections. Splicing efficiencies of all three introns were measured. As described above for infections in the presence of chloramphenicol, the td gene is expressed immediately after infection, whereas the nrdB and nrdD genes are expressed several minutes later from middle-mode promoters (24, 36). In addition, the expression of nonspliceable transcripts from late endonuclease promoters (and for the nrdB ORF also from a middle promoter) within the intron will contribute to the pool of pre-mRNA transcripts, thus artificially lowering the relative splicing efficiency at late times of infection.
Upon infection of a log-phase culture where infection proceeds rapidly, all three introns have splicing efficiencies reaching at least 60% at the point of maximum gene expression (Fig. 5). In early stationary phase, the infection process is prolonged, the amount of transcript is reduced, and the maximal splicing levels drop slightly. In late stationary phase, there is a clear reduction of splicing efficiency for all three introns and a further reduction in transcript levels is also seen, possibly reflecting a lower transcriptional activity in the infected bacteria or a reduced infectivity of the phages. Splicing efficiency of the nrdB intron in late stationary phase shows a twofold reduction compared to splicing during logarithmic growth, while the efficiency of td and nrdD intron splicing is reduced more than threefold (Fig. 5). This clearly shows that the splicing of all three group I introns in T4 is affected by the growth of the infected bacterium. This is the first time that environmental conditions have been shown to affect self-splicing.
FIG. 5.
Effect of growth phase of infected cells on splicing. A culture of E. coli B0 was continuously grown in rich medium at 37°C. Upon infection (inf.), 20-ml samples were withdrawn and infected with T4D and samples were taken at the indicated time points. Note that sampling times were prolonged at stationary (stat)-phase infections to allow for the somewhat slower progression of infection. Graphs show splicing efficiencies for each intron at different growth phases.
Stationary-phase splicing reduction is due to exon-intron interactions for nrdB.
To assess the effect of growth phase on the efficiency of group I intron splicing in greater detail, bacteria containing the cloned nrdB gene were grown at 37°C for many hours and splicing was measured in the different growth phases. As can be seen in Fig. 6a and b, the splicing level is 80 to 90% during logarithmic growth but drops rapidly as the culture enters stationary phase and, after continued incubation, the ratio of spliced versus unspliced transcript drops to below 20%. Resuspension of the cells in fresh medium restored both growth rate and splicing efficiency (Fig. 6b). These results clearly show that splicing of the nrdB intron is strongly reduced by conditions encountered during stationary-phase growth.
FIG. 6.
Splicing of the nrdB intron in different growth phases. (a) Cultures of JM101 containing either pBS5 or pBS5 Δ45-129;ΔCry-P1 were grown at 37°C, and samples were taken at the indicated time points after induction of transcription. (b) After an overnight incubation, part of the pBS5 culture was centrifuged and resolved in fresh medium and incubation was continued. Splicing efficiency at the different time points was measured by limited primer extension. RNA was prepared from equal densities of cells for each sample. Open symbols in the graphs represent splicing efficiency, and closed symbols represent growth. Error bars show the standard error of the mean. wt, wild type.
Entry into stationary phase is characterized by a number of changes in the bacterial metabolism. A major response is the marked reduction of translation to about 10 to 20% of the level in exponentially growing cells (1, 35), further dropping during prolonged stationary growth (19). However, in addition to a reduction of translation, a variety of changes that occur upon entry into stationary phase might potentially affect splicing. To determine whether the reduction of splicing in stationary phase is due to interactions between exon and intron sequences, we repeated the growth phase experiment using the nrdB Δ45-129;ΔCry-P1 construct (deleting the major regions shown to interfere with nrdB splicing in the absence of translation). As predicted, splicing efficiency of the Δ45-129;ΔCry-P1 construct is only marginally affected by the entry into stationary phase compared to that of the wild-type gene, thereby supporting the possibility that the reduction of splicing in stationary phase is due to the same interfering folds as was seen with chloramphenicol addition or stop codon insertion.
DISCUSSION
Here we have demonstrated that the splicing of all three group I introns in bacteriophage T4 is reduced if translation of the upstream exon is abolished by the addition of translational inhibitors or by the insertion of stop codons that prevent further movement of ribosomes. A lack of actively translating ribosomes on the upstream exon of the td pre-mRNA was previously shown to permit the formation of a nonnative, long-range interaction between exonic and intronic sequences that interfered with the 3′ splice site recognition of the td intron (31, 39). In this work, we have found that multiple exon regions with complementarity to important regions in the respective intron exist for all three T4 group I introns and we have experimentally shown that several of these regions in the first nrdB exon interfere with the splicing of this intron in the absence of translation. Theoretically we predict that an interaction highly similar to the one described for the td intron (31) can potentially be formed between the first nrdB exon and the 3′ splice site of the nrdB intron. However, the effect of this interaction on splicing efficiency of the nrdB intron in the absence of translation was only marginal and, instead, several regions further downstream gave a strong collective interference with splicing. The exonic part of this putative interaction in nrdB is located at the beginning of the exon, while in td, it is close to the intron, i.e., a much longer exon sequence is present between the two interacting sites in nrdB compared to that in td. In addition, the td studies were performed with an artificially shortened intron (31). It is noteworthy that, of all self-splicing group I and group II introns found so far, introns in translated genes are found exclusively in organisms or organelles (mitochondria and chloroplasts) with coupled transcription/translation, while nuclear-encoded group I and group II introns are found in only nontranslated genes (rRNA and tRNA) (6). The splicing of introns within pre-rRNA transcripts can be affected by misfolding interactions with sequences upstream (42, 43) or downstream (41) of the intron, but the endogenous folding of the stable RNAs into defined structures suppresses these misfoldings in vivo (45). In comparison, mRNA exons are largely unstructured and the coupling of transcription and translation may be a general mechanism to avoid the interference of upstream exon sequences with intron folding before the 3′ end of the intron is transcribed. At the same time, all three T4 introns contain stop codons at the exon-intron junction or at the very 5′ region of the intron that keep the ribosomes from interfering with correct folding of the intronic parts (25).
Periods of slow growth due to limiting nutrient supplies are likely to be commonly encountered by enteric bacteria in nature. Upon entry into stationary phase, the total amount of translating ribosomes in E. coli is known to drop drastically (1, 2, 19). In addition, the rate of translational elongation is reduced due to a lack of amino acid-charged tRNAs resulting in stalled ribosomes on the transcripts (1). There are multiple examples of how this is used by bacteria for regulation of gene expression by attenuation mechanisms (18). It is also intriguing that a wide range of different bacteriophages have group I introns inserted in genes involved in crucial steps of DNA metabolism (3, 6, 12, 16, 23, 37). Could the regulatory potential of the coupling between translation and intron splicing provide any selective advantage to the phage? It is known that T4 infectivity (injection of phage DNA) of bacteria in stationary phase is quite efficient (20). However, T4 does not lyse the infected bacteria in stationary phase. Instead, the infection stalls but is not terminated; it remains dormant inside the bacterium and can be resumed, and progeny phage can be produced if new nutrients are added to the medium (20). In such a situation, the restriction of the use of energy and reducing equivalents by limiting the de novo production of deoxyribonucleotides would leave more energy to cell survival and prolong the period that the phage can remain inside the bacterium. Such a regulatory potential of the T4 introns was speculated on already 20 years ago when the introns were first discovered (17, 32, 33). An attempt to determine how introns affect phage viability showed that deletion of the introns in T4 does not yield any detectable difference in burst size during logarithmic growth with different nutrient sources (12), but to our knowledge, no studies have been performed comparing the prolonged survivals of intron-containing and intronless phages in stationary phase. Our findings here of the reduction of splicing in stationary-phase infections merit such additional investigations to see whether introns can affect the viability of the phage under different growth conditions. Although the molecular biology and genetics of T4 have been substantially studied, very little is known about its natural growth conditions and habitats, and whether there are instances in the T4 life cycle in nature where introns are advantageous or detrimental to the phage is still an enigma.
Acknowledgments
This work was supported by the Swedish Science Research Council.
Footnotes
Published ahead of print on 22 November 2006.
REFERENCES
- 1.Albertson, N. H., and T. Nyström. 1994. Effects of starvation for exogenous carbon on functional mRNA stability and rate of peptide chain elongation in Escherichia coli. FEMS Microbiol. Lett. 117:181-187. [DOI] [PubMed] [Google Scholar]
- 2.Albertson, N. H., T. Nyström, and S. Kjelleberg. 1990. Macromolecular synthesis during recovery of the marine Vibrio sp. S14 from starvation. J. Gen. Microbiol. 136:2201-2207. [Google Scholar]
- 3.Bechhofer, D. H., K. K. Hue, and D. A. Shub. 1994. An intron in the thymidylate synthase gene of Bacillus bacteriophage beta 22: evidence for independent evolution of a gene, its group I intron, and the intron open reading frame. Proc. Natl. Acad. Sci. USA 91:11669-11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell-Pedersen, D., S. Quirk, J. Clyman, and M. Belfort. 1990. Intron mobility in phage T4 is dependent upon a distinctive class of endonucleases and independent of DNA sequences encoding the intron core: mechanistic and evolutionary implications. Nucleic Acids Res. 18:3763-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke, J. M. 1989. Selection of the 3′-splice site in group I introns. FEBS Lett. 250:129-133. [DOI] [PubMed] [Google Scholar]
- 6.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. Bioinformatics 3:2. [Online.] [DOI] [PMC free article] [PubMed]
- 7.Cech, T. R., S. H. Damberger, and R. R. Gutell. 1994. Representation of the secondary and tertiary structure of group I introns. Nat. Struct. Biol. 1:273-280. [DOI] [PubMed] [Google Scholar]
- 8.Chandry, P. S., and M. Belfort. 1987. Activation of a cryptic 5′ splice site in the upstream exon of the phage T4 td transcript: exon context, missplicing, and mRNA deletion in a fidelity mutant. Genes Dev. 1:1028-1037. [DOI] [PubMed] [Google Scholar]
- 9.Chu, F. K., G. F. Maley, F. Maley, and M. Belfort. 1984. Intervening sequence in the thymidylate synthase gene of bacteriophage T4. Proc. Natl. Acad. Sci. USA 81:3049-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, R. W., R. B. Waring, J. A. Ray, T. A. Brown, and C. Scazzocchio. 1982. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature 300:719-724. [DOI] [PubMed] [Google Scholar]
- 11.Davies, R. W., R. B. Waring, and P. Towner. 1987. Internal guide sequence and reaction specificity of group I self-splicing introns. Cold Spring Harbor Symp. Quant. Biol. 52:165-171. [DOI] [PubMed] [Google Scholar]
- 12.Eddy, S. 1992. Introns in the T-even bacteriophages. Ph.D. thesis. University of Colorado, Boulder.
- 13.Eddy, S. R., and L. Gold. 1991. The phage T4 nrdB intron: a deletion mutant of a version found in the wild. Genes Dev. 5:1032-1041. [DOI] [PubMed] [Google Scholar]
- 14.Golden, B. L., A. R. Gooding, E. R. Podell, and T. R. Cech. 1998. A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science 282:259-264. [DOI] [PubMed] [Google Scholar]
- 15.Goodrich-Blair, H., V. Scarlato, J. M. Gott, M. Q. Xu, and D. A. Shub. 1990. A self-splicing group I intron in the DNA polymerase gene of Bacillus subtilis bacteriophage SPO1. Cell 63:417-424. [DOI] [PubMed] [Google Scholar]
- 16.Goodrich-Blair, H., and D. A. Shub. 1994. The DNA polymerase genes of several HMU-bacteriophages have similar group I introns with highly divergent open reading frames. Nucleic Acids Res. 22:3715-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gott, J. M., D. A. Shub, and M. Belfort. 1986. Multiple self-splicing introns in bacteriophage T4: evidence from autocatalytic GTP labeling of RNA in vitro. Cell 47:81-87. [DOI] [PubMed] [Google Scholar]
- 18.Henkin, T. M., and C. Yanofsky. 2002. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays 24:700-707. [DOI] [PubMed] [Google Scholar]
- 19.Kolter, R., D. A. Siegele, and A. Tormo. 1993. The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47:855-874. [DOI] [PubMed] [Google Scholar]
- 20.Kutter, E., E. Kellenberger, K. Carlson, S. Eddy, J. Neitzel, L. Messinger, J. North, and B. Guttman. 1994. Effect of bacterial growth conditions and physiology on T4 infection, p. 406-418. In J. D. Karam J. W. Drake, K. N. Kreuzer, G. Mosig, D. H. Hall, F. A. Eiserling, L. W. Black, E. K. Spicer, E. Kutter, K. Carlson, and E. S. Miller (ed.), Molecular biology of bacteriophage T4. ASM Press, Washington, DC.
- 21.Michel, F., M. Hanna, R. Green, D. P. Bartel, and J. W. Szostak. 1989. The guanosine binding site of the Tetrahymena ribozyme. Nature 342:391-395. [DOI] [PubMed] [Google Scholar]
- 22.Michel, F., A. Jacquier, and B. Dujon. 1982. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie 64:867-881. [DOI] [PubMed] [Google Scholar]
- 23.Mikkonen, M., and T. Alatossava. 1995. A group I intron in the terminase gene of Lactobacillus delbrueckii subsp. lactis phage LL-H. Microbiology 141:2183-2190. [DOI] [PubMed] [Google Scholar]
- 24.Öhman, M. 1993. Analysis of regulatory RNA in accessory elements of Escherichia coli. Antisense RNA control in plasmid R1 and self-splicing introns in bacteriophage T4. Ph.D. thesis. Stockholm University, Stockholm, Sweden.
- 25.Öhman-Heden, M., A. Åhgren-Stålhandske, S. Hahne, and B. M. Sjöberg. 1993. Translation across the 5′-splice site interferes with autocatalytic splicing. Mol. Microbiol. 7:975-982. [DOI] [PubMed] [Google Scholar]
- 26.Pato, M. L., P. M. Bennett, and K. von Meyenburg. 1973. Messenger ribonucleic acid synthesis and degradation in Escherichia coli during inhibition of translation. J. Bacteriol. 116:710-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quirk, S. M., D. Bell-Pedersen, and M. Belfort. 1989. Intron mobility in the T-even phages: high frequency inheritance of group I introns promoted by intron open reading frames. Cell 56:455-465. [DOI] [PubMed] [Google Scholar]
- 28.Quirk, S. M., D. Bell-Pedersen, J. Tomaschewski, W. Rüger, and M. Belfort. 1989. The inconsistent distribution of introns in the T-even phages indicates recent genetic exchanges. Nucleic Acids Res. 17:301-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandegren, L., and B. M. Sjöberg. 2004. Distribution, sequence homology and homing of group I introns among T-even like bacteriophages: evidence for recent transfer of old introns. J. Biol. Chem. 279:22218-22227. [DOI] [PubMed] [Google Scholar]
- 30.Schneider, E., M. Blundell, and D. Kennell. 1978. Translation and mRNA decay. Mol. Gen. Genet. 160:121-129. [DOI] [PubMed] [Google Scholar]
- 31.Semrad, K., and R. Schroeder. 1998. A ribosomal function is necessary for efficient splicing of the T4 phage thymidylate synthase intron in vivo. Genes Dev. 12:1327-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shub, D. A., J. M. Gott, M. Q. Xu, B. F. Lang, F. Michel, J. Tomaschewski, J. Pedersen-Lane, and M. Belfort. 1988. Structural conservation among three homologous introns of bacteriophage T4 and the group I introns of eukaryotes. Proc. Natl. Acad. Sci. USA 85:1151-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shub, D. A., M. Q. Xu, J. M. Gott, A. Zeeh, and L. D. Wilson. 1987. A family of autocatalytic group I introns in bacteriophage T4. Cold Spring Harbor Symp. Quant. Biol. 52:193-200. [DOI] [PubMed] [Google Scholar]
- 34.Sjöberg, B. M., S. Hahne, C. Z. Mathews, C. K. Mathews, K. N. Rand, and M. J. Gait. 1986. The bacteriophage T4 gene for the small subunit of ribonucleotide reductase contains an intron. EMBO J. 5:2031-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svitil, A. L., M. Cashel, and J. W. Zyskind. 1993. Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. J. Biol. Chem. 268:2307-2311. [PubMed] [Google Scholar]
- 36.Tseng, M. J., P. He, J. M. Hilfinger, and G. R. Greenberg. 1990. Bacteriophage T4 nrdA and nrdB genes, encoding ribonucleotide reductase, are expressed both separately and coordinately: characterization of the nrdB promoter. J. Bacteriol. 172:6323-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Sinderen, D., H. Karsens, J. Kok, P. Terpstra, M. H. Ruiters, G. Venema, and A. Nauta. 1996. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Mol. Microbiol. 19:1343-1355. [DOI] [PubMed] [Google Scholar]
- 38.von Ahsen, U., J. Davies, and R. Schroeder. 1992. Non-competitive inhibition of group I intron RNA self-splicing by aminoglycoside antibiotics. J. Mol. Biol. 226:935-941. [DOI] [PubMed] [Google Scholar]
- 39.Waldsich, C., K. Semrad, and R. Schroeder. 1998. Neomycin B inhibits splicing of the td intron indirectly by interfering with translation and enhances missplicing in vivo. RNA 4:1653-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilbur, W. J., and D. J. Lipman. 1983. Rapid similarity searches of nucleic acid and protein data banks. Proc. Natl. Acad. Sci. USA 80:726-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woodson, S. A. 1992. Exon sequences distant from the splice junction are required for efficient self-splicing of the Tetrahymena IVS. Nucleic Acids Res. 20:4027-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woodson, S. A., and T. R. Cech. 1991. Alternative secondary structures in the 5′ exon affect both forward and reverse self-splicing of the Tetrahymena intervening sequence RNA. Biochemistry 30:2042-2050. [DOI] [PubMed] [Google Scholar]
- 43.Woodson, S. A., and V. L. Emerick. 1993. An alternative helix in the 26S rRNA promotes excision and integration of the Tetrahymena intervening sequence. Mol. Cell. Biol. 13:1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young, P., M. Öhman, M. Q. Xu, D. A. Shub, and B. M. Sjöberg. 1994. Intron-containing T4 bacteriophage gene sunY encodes an anaerobic ribonucleotide reductase. J. Biol. Chem. 269:20229-20232. [PubMed] [Google Scholar]
- 45.Zaug, A. J., M. M. McEvoy, and T. R. Cech. 1993. Self-splicing of the group I intron from Anabaena pre-tRNA: requirement for base-pairing of the exons in the anticodon stem. Biochemistry 32:7946-7953. [DOI] [PubMed] [Google Scholar]
- 46.Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244:48-52. [DOI] [PubMed] [Google Scholar]