Abstract
In the denitrifying bacterium Thauera aromatica, the central intermediate of anaerobic aromatic metabolism, benzoyl-coenzyme A (CoA), is dearomatized by the ATP-dependent benzoyl-CoA reductase to cyclohexa-1,5-diene-1-carbonyl-CoA (dienoyl-CoA). The dienoyl-CoA is further metabolized by a series of β-oxidation-like reactions of the so-called benzoyl-CoA degradation pathway resulting in ring cleavage. Recently, evidence was obtained that obligately anaerobic bacteria that use aromatic growth substrates do not contain an ATP-dependent benzoyl-CoA reductase. In these bacteria, the reactions involved in dearomatization and cleavage of the aromatic ring have not been shown, so far. In this work, a characteristic enzymatic step of the benzoyl-CoA pathway in obligate anaerobes was demonstrated and characterized. Dienoyl-CoA hydratase activities were determined in extracts of Geobacter metallireducens (iron reducing), Syntrophus aciditrophicus (fermenting), and Desulfococcus multivorans (sulfate reducing) cells grown with benzoate. The benzoate-induced genes putatively coding for the dienoyl-CoA hydratases in the benzoate degraders G. metallireducens and S. aciditrophicus were heterologously expressed and characterized. Both gene products specifically catalyzed the reversible hydration of dienoyl-CoA to 6-hydroxycyclohexenoyl-CoA (Km, 80 and 35 μM; Vmax, 350 and 550 μmol min−1 mg−1, respectively). Neither enzyme had significant activity with cyclohex-1-ene-1-carbonyl-CoA or crotonyl-CoA. The results suggest that benzoyl-CoA degradation proceeds via dienoyl-CoA and 6-hydroxycyclohexanoyl-CoA in strictly anaerobic bacteria. The steps involved in dienoyl-CoA metabolism appear identical in all nonphotosynthetic anaerobic bacteria, although totally different benzene ring-dearomatizing enzymes are present in facultative and obligate anaerobes.
In recent years, a number of anaerobic bacteria have been identified that are capable of using aromatic growth substrates as the sole energy and carbon source (14). These bacteria comprise facultative (denitrifying or phototrophic bacteria) and obligate anaerobes (iron- or sulfate-reducing bacteria and fermenting bacteria, often in syntrophic association with an H2-consuming organism). So far, the overwhelming amount information about the biochemical processes involved in anaerobic aromatic metabolism has been derived from studies with facultative anaerobes (for recent reviews, see references 6-10, 14, and 25).
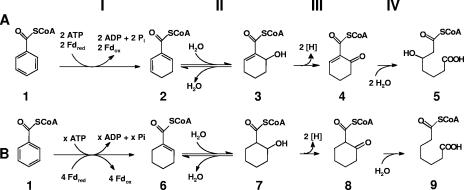
In both facultatively and obligately anaerobic bacteria, many aromatic compounds such as toluene, phenol, xylenes, cresols, phenylacetic acid, and benzoate are converted to the central intermediate benzoyl-coenzyme A (CoA). However, the subsequent steps resulting in dearomatization and cleavage of the benzene ring have so far only been studied in facultative anaerobes. The initial steps of benzoyl-CoA metabolism in the facultative anaerobic model organism Thauera aromatica, the so-called benzoyl-CoA pathway, can be summarized as follows (Fig. 1, pathway A). In reaction I, benzoyl-CoA (compound 1 in Fig. 1; compound numbers in the text refer to structures indicated in Fig. 1) becomes dearomatized by reduction in two single-electron transfer steps to the aromatic ring, yielding cyclohexa-1,5-diene-1-carbonyl-CoA (dienoyl-CoA; compound 2) (3, 5). The key enzyme, benzoyl-CoA reductase, couples this electron transfer from reduced ferredoxin to the aromatic ring with stoichiometric ATP hydrolysis (1 ATP/electron) (4). In reaction II, the dienoyl-CoA formed is hydrated to 6-hydroxycyclohex-1-ene-1-carbonyl-CoA (6-OH-cyclohexenoyl-CoA; compound 3) by cyclohexa-1,5-diene-1-carbonyl-CoA hydratase (dienoyl-CoA hydratase) (19). The subsequent two enzymatic steps, reactions III and IV, comprise the oxidation of the hydroxyl group by a specific alcohol dehydrogenase and ring cleavage by a hydrolase yielding 3-hydroxypimelyl-CoA (compound 5) (18). This compound becomes further metabolized by β-oxidation-like reactions to three acetyl-CoAs and one CO2. Notably, in the facultatively anaerobic organism Rhodopseudomonas palustris, benzoyl-CoA is reduced twice to cyclohex-1-ene-1-carbonyl-CoA (cyclohexenoyl-CoA; compound 6), each reduction involving the addition of two electrons (14, 15). As a consequence, the subsequent steps in R. palustris (Fig. 1, pathway B) differ from those in T. aromatica (22).
FIG. 1.
Initial steps of benzoyl-CoA pathway in Thauera aromatica (A) and Rhodopseudomonas palustris (B). In T. aromatica, reaction I is catalyzed by ATP-dependent benzoyl-CoA reductase (A). Further reactions are catalyzed by (di)enoyl-CoA hydratase (reaction II), alcohol dehydrogenase (reaction III), and a ring-opening hydrolase (reaction IV). The following compounds are involved: compound 1, benzoyl-CoA; compound 2, dienoyl-CoA; compound 3, 6-OH-cyclohexenoyl-CoA; compound 4, 6-oxocyclohex-1-ene-1-carbonyl-CoA; compound 5, 3-hydroxypimelyl-CoA; compound 6, cyclohex-1-ene-1-carbonyl-CoA; compound 7, 6-hydroxycyclohexane-1-carbonyl-CoA; compound 8, 6-oxocyclohexane-1-carbonyl-CoA; and compound 9, pimelyl-CoA. These compound numbers are used in the text to refer to the individual structures shown here. Fdred and Fdox, reduced and oxidized ferredoxin, respectively.
Much less is known about the aromatic metabolism in obligate anaerobes. The initial step in benzoate metabolism is also catalyzed by benzoate-CoA ligases that have been purified from a syntrophic consortium (1), from the sulfate reducer Desulfococcus multivorans (23) and, after heterologous expression of the corresponding gene, from Geobacter metallireducens (28). However, the further steps of aromatic metabolism, including ring dearomatization and cleavage, have not been demonstrated so far in any obligate anaerobe.
In a recent proteomic approach, 44 benzoate-induced genes were identified in G. metallireducens; several of their products were identified by mass spectrometric analysis (11, 28). The genes are organized in two clusters comprising those putatively involved in the benzoyl-CoA degradation pathway (bam genes [benzoic acid metabolism]) and in β-oxidation reactions yielding acetyl-CoA and carbon dioxide. The bam genes include the structural gene of benzoate-CoA ligase (bamY). Amino acid sequence similarities of products from other genes (bamA, bamQ, and bamR) with enzymes from Thauera aromatica suggested that similar steps may be involved in the conversion of the dearomatized product to an aliphatic C7 compound (reactions II to IV in Fig. 1, pathway A). The gene products BamB to -I were considered to be involved in benzoyl-CoA dearomatization (reaction II in Fig. 1, pathway A). However, attempts to determine a benzoyl-CoA reductase activity in strict anaerobes have failed so far (23, 28); as a consequence, information is lacking about the product of benzene ring dearomatization and the further metabolism of the dearomatized product.
In the genome of G. metallireducens, the product of the bamR gene (gi 78223357) was annotated as an enoyl-CoA hydratase. However, BamR showed exceptionally high amino acid sequence identities (68 to 72%) to dienoyl-CoA hydratases from T. aromatica and Magnetospirillum species (28). In contrast, no gene with such high similarities to dienoyl-CoA hydratase of T. aromatica is present in the genome of the fermenting, benzoate-degrading Syntrophus aciditrophicus (NC_007759). In this organism, a product of a gene coding for a putative enoyl-CoA hydratase (gi 85860872) showed amino acid sequence identities (47%) to enoyl-CoA hydratases from aromatic compounds degrading Azoarcus species (20, 24). Notably, thermodynamic considerations argue that the amount of energy available to fermentative, iron-reducing and sulfate-reducing anaerobes is not sufficient to support an ATP-dependent, two-electron reduction of benzoyl-CoA to a dienoyl-CoA intermediate (26). For this reason, it has been proposed that energetically more favorable four-electron reduction reactions occur, forming cyclohex-1-enoyl-1-carboxyl-CoA from benzoyl-CoA (corresponding to pathway B in Fig. 1).
In order to investigate the benzoyl-CoA pathway in strict anaerobes for the first time, bamR from G. metallireducens (referred to bamRGeo) and the corresponding gene of S. aciditrophicus (referred to bamRSyn) were heterologously expressed with a His tag, purified, and characterized. The results clearly identified the gene products as highly specific, cyclohexa-1,5-diene-1-carbonyl-CoA hydratases. Dienoyl-CoA hydratase activities were also determined in extracts from cells of G. metallireducens, S. aciditophicus, and D. multivorans grown with an aromatic substrate. These results strongly suggest that—with the exception of R. palustris (Fig. 1B)—the benzoyl-CoA pathways are indeed identical in strictly and facultatively anaerobic bacteria metabolizing aromatic growth substrates, independent of the overall energy metabolism and the mode of benzoyl-CoA dearomatization.
MATERIALS AND METHODS
Growth of bacterial cells and preparation of cell extracts.
G. metallireducens (DSMZ-Nr. 7210) and D. multivorans (DSMZ-Nr. 2059) were obtained from Deutsche Sammlung von Mikroorganismen. S. aciditrophicus was from the culture collection of M. McInerney. G. metallireducens (21), S. aciditrophicus (13), and D. multivorans (23) were cultured anaerobically in a mineral salt medium as described previously. The cells were harvested in the exponential growth phase by centrifugation (10,000 × g) and were stored in liquid nitrogen. For the preparation of crude extracts, frozen cells were suspended in 20 mM triethanolamine hydrochloride-KOH buffer, pH 7.3 (1 g cells in 1.5 ml buffer), 10 mM MgCl2, 10% glycerol, and 0.1 mg of DNase I. Cell lysates were obtained by passage through a French pressure cell at 137 MPa. After centrifugation at 100,000 × g (1 h at 4°C), the supernatant was used for further studies.
Synthesis of CoA esters.
Crotonyl-CoA was purchased from Fluka (Ulm, Germany). Benzoyl-CoA and cyclohexenoyl-CoA were enzymatically synthesized from the corresponding carboxylic acids and CoA by using purified His-tagged benzoate-CoA ligase from G. metallireducens (specific activity with benzoate was 16 μmol min−1 mg−1) (28). This enzyme catalyzes the following reaction: carboxylic acid + CoA + MgATP → carboxylic acid-CoA + MgAMP + PPi. Cyclohex-1-enecarboxylate was converted at 13% of the rate with benzoate. The assay, purification, and purity control of coenzyme A esters by preparative high-performance liquid chromatography (HPLC) are described elsewhere (16, 30).
Dienoyl-CoA and 6-OH-cyclohexenoyl-CoA were enzymatically synthesized from benzoate with an enriched benzoate-CoA ligase, benzoyl-CoA reductase, and dienoyl-CoA hydratase from T. aromatica as described previously (5, 16). The synthesis comprises the following two reaction steps: (i) benzoyl-CoA + 2 MgATP + 2 Ti(III)-citrate → dienoyl-CoA + 2 MgADP + 2 Pi + 2 Ti(IV)-citrate and (ii) reversible hydration of dienoyl-CoA to 6-OH-cyclohexenoyl-CoA. After benzoyl-CoA was completely converted, dienoyl-CoA and 6-OH-cyclohexenoyl-CoA were present in equal concentrations. Isolation and tests for purity of the coenzyme A esters were performed by preparative high-performance-liquid chromatography as described previously (18).
Cloning and expression of genes.
Standard protocols were used for DNA isolation and amplification (2).
The bamRGeo gene (gi 78223357) was amplified from G. metallireducens DNA by using the primer pair ATGAGCGAGAGCCCTCTCAA (forward primer) and GCGGTCTTGCCAGGCGGC (reverse primer); for the bamRSyn gene (gi 85860872), the primer pair ATGGGATTCAACACTATTCTTTTT (forward) and TTTGTCCTTGAACACCGGTTTTC (reverse) was used. Primers were designed in a way that the native stop codon was removed. Cloning positioned the gene of interest in frame with the DNA encoding a C-terminal peptide containing six histidines. Primers were synthesized by Biomers (Ulm, Germany). The following PCR program using Taq and Pfu polymerase (18:1) was applied for amplification: 30 cycles of 94°C for 30 s, 55°C for 45 s, and 72°C for 45 s (bamRGeo) and 30 cycles of 94°C for 1 min, 63°C for 1 min, and 72°C for 70 s (bamRSyn). The program started with 2 min at 94°C after the 30-cycle samples were incubated for a further 5 min at 72°C. The amplified genes were transferred into the pCRT7/CT-TOPO expression vector according to the manufacturer's instruction (Invitrogen Life Technologies, Karlsruhe, Germany). The constructs were transformed into One Shot TOP10F′ and One Shot BL21(DE3)pLysS cells for propagation, analysis, and maintenance, as well as expression of the fusion protein, respectively, according to the manufacturer's instructions. Expression of the gene was performed at 18°C (bamRGeo) or 25°C (bamRSyn) overnight; induction was carried out when the cells reached an optical density at 578 nm (OD578) of 0.5 by addition of 0.8 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The temperature was chosen in order to obtain the highest amount of soluble gene product. Cells were harvested at an OD578 of 1.8 (bamRGeo) or OD578 of 3.0 (bamRSyn), centrifuged at 13,700 × g for 15 min at 4°C, and stored at −80°C until further use.
Purification of His-tagged BamR proteins.
The overexpressed His-tagged BamRGeo and BamRSyn were purified at 4°C by Ni-chelating Sepharose Fast Flow chromatography (GE Healthcare) in a single step. The following purification procedure was applied. Four grams of E. coli strain One Shot BL21(DE3)pLysS cells (wet mass) containing the recombinant fusion protein was resuspended in 8 ml 20 mM Tris-HCl buffer, pH 7.9 (buffer A). Cell extracts were prepared as described above for G. metallireducens, after which 10% (by volume) glycerol was added, giving 5 ml cell lysate. After equilibration of an Ni-chelating Sepharose column (His-Trap HP; GE Healthcare [volume, 1 ml]) with buffer A containing 250 mM KCl and 10% glycerol (equilibration buffer), the cell extract was applied to the column at a flow rate of 0.2 ml min−1. The column was then washed with 6 column volumes of equilibration buffer plus 20 mM imidazole at a flow rate of 0.5 ml min−1, after which a linear gradient over 30 column volumes from 20 to 500 mM imidazole in equilibration buffer was applied (flow rate of 0.5 ml min−1). BamRGeo fusion protein eluted between 100 to 200 mM imidazole in 4 ml containing 18.4 mg protein; the BamRSyn eluted between 280 to 400 mM imidazole in 5 ml containing 9 mg protein. The purity of both BamR proteins was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Enzyme assays.
The enoyl-CoA hydratase activity was determined by either a continuous or a discontinuous assay at 30°C. The continuous spectrophotometric assay is based on the difference in absorbance between dienoyl-CoA and 6-OH-cyclohexenoyl-CoA at 320 nm with Δɛ320 = 1,700 M−1 cm−1 (19); it was routinely used during enzyme purification and for Km value determinations. The typical assay mixture (500 μl) contained 100 mM Tris-HCl, pH 7.5, 0.5 mM of dienoyl-CoA or 6-OH-cyclohexenoyl-CoA, and 2 to 10 μl of a 1:500-fold dilution of enzyme preparation in the assay buffer corresponding to 0.1 to 0.5 μg of purified dienoyl-CoA hydratase. In the discontinuous method, 50-μl samples were taken from this assay at different time points. After addition of 5 μl 10% formic acid, the sample was centrifuged and the supernatant was analyzed by C18 reversed-phase high-performance liquid chromatography as described previously (19). In the HPLC assay, substrate consumption and product formation were directly analyzed by monitoring the absorption of the CoA esters at 260 nm and in addition by recording the typical UV/visible spectra of the individual substrates and products (Waters photodiode array detector model 996). The UV/visible spectra of dienoyl-CoA, 6-OH-cyclohexenoyl-CoA, cyclohexenoyl-CoA, 2-hydroxycyclohexane-1-carbonyl-CoA, crotonyl-CoA, and 3-hydroxybutyryl-CoA are described elsewhere (18, 19). The HPLC assay was used for product analysis, for determination of substrate specificity (concentration of the substrates used, 0.5 mM), and for testing the cyclohexenoyl-CoA hydration activity in crude extracts of G. metallireducens, S. aciditrophicus, and D. multivorans.
For the determination of Km values of dienoyl-CoA hydratase, 6-OH-cyclohexenoyl-CoA and dienoyl-CoA were added at final concentrations of 0.6, 0.3, 0.15, 0.075, 0.0375, and 0.018 mM to the assay mixture. Km values were determined by fitting the data to Michaelis-Menten curves using the Prism software package (GraphPad, San Diego, Calif.).
Determination of native molecular mass.
The native molecular masses of overexpressed BamR proteins were determined using an FPLC Superdex 200 HR 10/30 gel-filtration column (GE Healthcare; diameter, 10 mm; bed volume, 24 ml), which had been equilibrated with 10 mM Tris-HCl buffer, pH 7.5, containing 100 mM potassium chloride. About 100 μl of protein solution (18 mg ml−1) was applied at a flow rate of 0.2 ml min−1. In addition, native gels of different acrylamide concentrations were prepared for molecular mass determination by Ferguson blotting (12).
Further determinations.
SDS-PAGE (12.5% polyacrylamide) was performed as described by Laemmli (17). Proteins were visualized using Coomassie blue staining (29). Protein was routinely determined by the method of Bradford using bovine serum albumin as a standard (10). Sequence alignments for the formation of a phylogenetic tree of dienoyl-CoA hydratases was carried out with the MultAlign software (http://prodes.toulouse.inra.fr/multalin/).
RESULTS AND DISCUSSION
Cyclohexa-1,5-dienoyl-CoA hydratase activities in cell extracts of G. metallireducens, S. aciditrophicus, and D. multivorans.
A recent proteomic study with G. metallireducens suggested that steps II to IV in Fig. 1 are similar or identical in facultative and strict anaerobes (28). However, no experimental evidence for any of the enzymatic steps of the benzoyl-CoA pathway in strict anaerobes has been given so far. For this reason, the presence of dienoyl-CoA hydratases putatively catalyzing the second step in benzoyl-CoA degradation pathway in obligate anaerobes was studied in G. metallireducens, S. aciditrophicus, and D. multivorans. For this purpose, cell extracts of the individual organisms grown with benzoate were incubated with dienoyl-CoA (compound 2) and the time-dependent conversion to 6-OH-cyclohexenoyl-CoA (compound 3) was determined by HPLC analysis as described previously (16, 18, 19).
Cell extracts from G. metallireducens rapidly converted dienoyl-CoA to 6-OH-cyclohexenoyl-CoA (2.5 μmol mg−1 min−1) (Table 1) in a protein- and time-dependent manner. In a second phase, both compounds were then slowly converted, most probably to compounds 4 and 5 (reactions III and IV in Fig. 1) (data not shown). Upon addition of NAD+ (0.5 mM) to the assay, the conversion of 6-OH-cyclohexenoyl-CoA was sixfold increased, suggesting that NAD+ is the cosubstrate of 6-OH-cyclohexenoyl-CoA dehydrogenase catalyzing reaction III (Fig. 1). In contrast, virtually no conversion of cyclohexenoyl-CoA (compound 6) to more polar products was observed in extracts of cells grown with benzoate (Table 1). In addition, no significant conversion of dienoyl-CoA to 6-OH-cyclohexenoyl-CoA was observed in extracts of G. metallireducens cells grown with acetate (Table 1). The results are in accordance with the observed induction of bamR during growth on benzoate (28).
TABLE 1.
Hydratase activities in extracts of cells from strict anaerobes grown on different substrates
| Organism and growth substrate | Hydratase activity (U mg−1)a
|
|
|---|---|---|
| Dienoyl-CoA | Cyclohexenoyl-CoA | |
| G. metallireducens | ||
| Benzoate | 2.5 ± 0.3 | <0.01 |
| Acetate | <0.01 | <0.01 |
| S. aciditrophicus; benzoate | 1.8 ± 0.4 | ND |
| D. multivorans | ||
| Benzoate | 2 ± 0.4 | 1.7 ± 0.3 |
| Lactate | 0.1 | >2.5 |
The substrates tested for hydratase activities were cyclohex-1,5-diene-1-carbonyl-CoA (dienoyl-CoA; compound 2) and cyclohex-1-ene-1-carbonyl-CoA (cyclohexenoyl-CoA; compound 6). Activities with dienoyl-CoA were tested using the direct spectrophotometric assay; activity with cyclohexenoyl-CoA was tested by HPLC analysis of the substrate consumed/product formed. The mean value ± standard deviations from at least triple determinations are indicated. One unit refers to μmol min−1. ND, not determined.
Extracts from D. multivorans and S. aciditrophicus cells grown with benzoate also showed a high level of dienoyl-CoA hydratase activity (Table 1). As described above, 6-OH-cyclohexenoyl-CoA was further converted to more polar products. In D. multivorans, the dienoyl-CoA hydratase activity in extracts from cells grown on lactate was only 5% of that in extracts of cells grown on benzoate (Table 1). In contrast to G. metallireducens, cell extracts of D. multivorans also catalyzed the hydration of cyclohexenoyl-CoA. However, the cyclohexenoyl-CoA hydration activity was even higher in extracts from cells grown with lactate (Table 1). Thus, this activity does not appear to be specifically associated with anaerobic aromatic acid metabolism.
The presence of benzoate-induced dienoyl-CoA hydratases in three different obligately anaerobic bacteria strongly suggests that dienoyl-CoA is an intermediate of the aromatic acid metabolism in strictly anaerobic bacteria, making it unlikely that the reduction of benzoyl-CoA occurs by a four-electron reaction as has been suggested for theoretical considerations for Syntrophus gentianae (26).
Expression of bamRGeo and bamRSyn and purification of the gene products.
Results from the in vitro dienoyl-CoA assays indicated that cells from G. metallireducens, S. aciditrophicus, and D. multivorans grown with benzoate contain benzoate-induced dienoyl-CoA hydratases. In a recent study, the benzoate-induced bamR gene (gi 78223357), annotated as an enoyl-CoA hydratase, has been suggested to code for a putative dienoyl-CoA hydratase (28). In contrast, in the recently sequenced genome of the fermenting, benzoate-degrading S. aciditrophicus (NC_007759), no open reading frame with high similarities to dienoyl-CoA hydratase of T. aromatica is present. Instead, an open reading frame coding for a putative enoyl-CoA hydratase (gi 85860872) showed amino acid identities (47%) to benzoate-induced putative dienoyl-CoA hydratases from Azoarcus species (20, 24). Both genes (referred to as bamRGeo and bamRSyn) were amplified with a C-terminal His6 tag by PCR, and the DNA obtained was cloned and overexpressed in Escherichia coli.
Expression of bamRGeo and bamRSyn in E. coli yielded soluble proteins of approximately 30 kDa (Fig. 2). The molecular mass corresponded to that predicted from the gene sequence (30.4 kDa for BamRGeo and 29.9 kDa for BamRSyn [including His tags]). Each protein was purified from the soluble protein fraction of recombinant E. coli cell extracts by Ni-chelating chromatography in a single step. BamRGeo was eluted between 100 to 200 mM imidazole, and BamRSyn was eluted between 280 and 400 nM imidazole. Almost all of the soluble protein of E. coli eluted with the equilibration buffer (Fig. 2).
FIG. 2.
SDS-PAGE analysis of protein fractions obtained during heterologous expression and purification of His-tagged BamRGeo and BamRSyn. Lanes 1 and 5, molecular mass standards; lanes 2 and 3, insoluble (lane 2) and soluble (lane 3) protein fractions of E. coli cells containing a plasmid with bamRGeo after induction by IPTG; lane 4, His-tagged BamRGeo (approximately 30 kDa) after purification by Ni-chelating affinity chromatography; lanes 6 and 7, soluble proteins of E. coli containing a plasmid with bamRSyn before (lane 6) and after (lane 7) induction by IPTG; lane 8, His-tagged BamRSyn (approximately 30 kDa) after purification by Ni-chelating affinity chromatography. Proteins were visualized by staining with Coomassie blue.
Properties of BamRGeo and BamRSyn.
The native molecular masses were determined by gel filtration, giving 92 ± 15 kDa for BamRGeo and 115 ± 10 kDa for BamRSyn. The mass of BamRGeo suggested an unusual α3 composition; for this reason, an additional Ferguson blot analysis (12) was carried out, giving a mass of 70 ± 10 kDa. The data suggest the composition of BamRSyn to be α4, whereas the composition of BamRGeo is suggested to be either α2 or α3. The subunit architecture of BamRGeo corresponds to that of dienoyl-CoA hydratase of T. aromatica, which may result from the higher amino acid sequence similarities between BamRGeo and dienoyl-CoA hydratase from T. aromatica (19).
To test whether BamRGeo and BamRSyn code for dienoyl-CoA hydratases, the substrate preferences of the enzymes were investigated in view of their possible roles in benzoate metabolism. For this purpose, the putative hydratase activities of the produced proteins were monitored in a discontinuous assay followed by HPLC analysis of the substrate consumed or product formed. The putative substrates or products dienoyl-CoA (compound 2) and 6-OH-cyclohexenoyl-CoA (compound 3) were enzymatically synthesized by the combined action of enriched benzoate-CoA ligase, benzoyl-CoA reductase, and dienoyl-CoA hydratase from T. aromatica.
Both BamRGeo and BamRSyn converted dienoyl-CoA to a more polar product in a time- and enzyme-dependent fashion (data not shown [but see Fig. 3 for the reverse reaction]). Coelution with a standard and the characteristic UV spectrum (19) clearly indicated that the product formed was 6-OH-cyclohexenoyl-CoA. Both BamR enzymes also catalyzed the reverse reaction, the dehydration of 6-OH-cyclohexenoyl-CoA to dienoyl-CoA (Fig. 3). Again, the product from the conversion of 6-OH-cyclohexenoyl-CoA was identified as dienoyl-CoA by coelution with the conjugated 1,5-dienoyl-CoA compound and by the typical UV spectrum (19). With the spectrophotometric assay, following the hydration of dienoyl-CoA to 6-OH-cyclohexenoyl-CoA (0.5 mM each), rates of 550 μmol min−1 mg−1 (BamRGeo) and 350 μmol min−1 mg−1 (BamRSyn) were determined; the rate in the reverse reaction was 730 μmol min−1 mg−1 for BamRGeo (Table 2). The apparent Km values for dienoyl-CoA were 85 ± 30 μM (BamRGeo) and 30 ± 10 μM (BamRSyn); the apparent Km for 6-OH-cyclohexenoyl-CoA for BamRGeo was 50 ± 15 μM. The equilibrium concentration ratio between the substrate and the corresponding product was nearly 1:1 (Fig. 3). The discontinuous HPLC assay was also used to monitor a possible hydration of cyclohexenoyl-CoA (compound 6) or crotonyl-CoA (each at 1 mM) by both BamR enzymes. Neither substrate was converted at a significant rate by BamRGeo and BamRSyn.
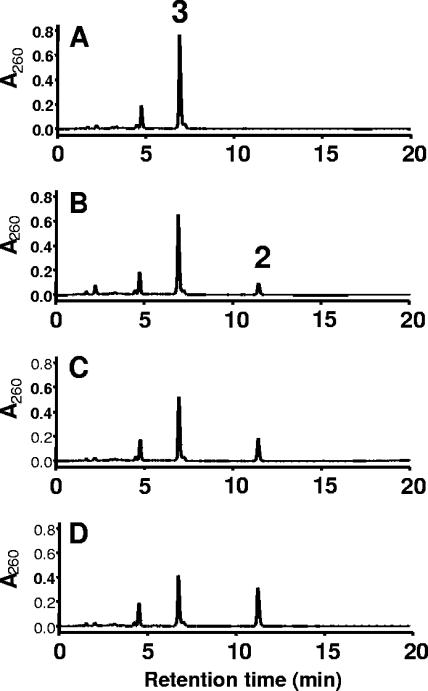
FIG. 3.
Conversion of 6-OH-cyclohexa-1-ene-1-carbonyl-CoA and cyclohex-1-ene-1-carbonyl-CoA (0.5 mM each) by purified BamRGeo. The numbers of the peaks refer to the structures shown in Fig. 1. Shown is the time-dependent dehydration of 6-OH-cyclohexenoyl-CoA (peak 3) to dienoyl-CoA (peak 2) from HPLC analysis of samples taken at 0 s (A), 15 s (B), 45 s (C), and 180 s (D). The reaction leveled off after 3 min by reaching the 1:1 equilibrium between substrate and product. Detection of all compounds was carried out by a UV monitor at 260 nm.
TABLE 2.
Properties of cyclohexa-1,5-diene-1-carbonyl-CoA hydratases from G. metallireducens (BamRGeo) and S. aciditrophicus (BamRSyn)
| Hydratase | Substrate used
|
Substrates not used | Km (μM) | Sp act (μmol min−1 mg−1)
|
Catalytic no. for hydration (s−1) | Subunit compositiona | ||
|---|---|---|---|---|---|---|---|---|
| Hydration | Dehydration | Hydration | Dehydration | |||||
| BamRGeo | Dienoyl-CoA | 6-OH-cyclohexenoyl-CoA | Cyclohex-1-ene-1-carbonyl-CoA, crotonyl-CoA | 85 ± 20 (dienoyl-CoA), 50 ± 20 (6-OH-cyclohexenoyl-CoA) | 550 | 730 | 280 | α2 or α3 |
| BamRSyn | Dienoyl-CoA | 6-OH-cyclohexenoyl-CoA | Cyclohex-1-ene-1-carbonyl-CoA, crotonyl-CoA | 30 ± 10 (dienoyl-CoA) | 350 | 175 | α4 | |
The subunit was 30 kDa.
The results obtained clearly indicate that BamRGeo and BamRSyn code for highly specific dienoyl-CoA hydratases catalyzing the second step in the benzoyl-CoA pathway in G. metallireducens and S. aciditrophicus. This result is in accordance with the activity measurements in cell extracts.
Phylogenetic analysis of dienoyl-CoA hydratases.
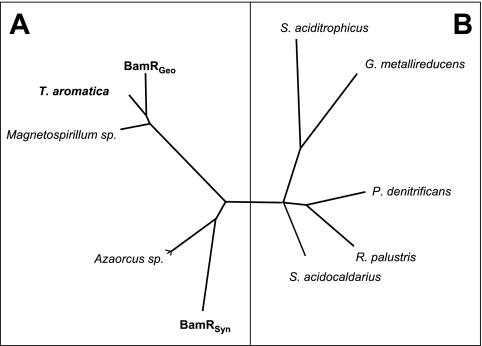
The unambiguous identification of the function of the two dienoyl-CoA hydratases allowed a phylogenetic analysis and comparison with other enoyl-CoA hydratases. Obviously dienoyl-CoA hydratases can be distinguished from other enoyl-CoA hydratases by their amino acid sequence (Fig. 4). The results obtained suggest that there are two phylogenetic clusters of dienoyl-CoA hydratases: one including the enzymes from T. aromatica, G. metallireducens, and deduced gene products from Magnetospirillum species and a second with the enzyme from S. aciditrophicus and deduced gene products from Azoarcus species. It is very likely that the deduced putative dienoyl-CoA hydratases from Magnetospirillum magnetotacticum (ZP 00053821) and Magnetospirillum sp. strain TS-1 (BAD 42370), both showing 68% and 67% amino acid sequence identities to BamRGeo, and the enzymes from Azoarcus sp. (Azoarcus sp. strain EbN1, YP 160034; Azoarcus evansii, CAD 21363; and Azoarcus sp. strain CIB, AAQ08815) are also specific dienoyl-CoA hydratases catalyzing the second step in anaerobic benzoate metabolism in both facultative anaerobes.
FIG. 4.
Phylogenetic tree of dienoyl-CoA hydratases (A) and other enoyl-CoA hydratases (B). Enzymes from organisms in boldface have been characterized as dienoyl-CoA hydratases. The putative dienoyl-CoA hydratases from Magnetospirillum and Azoarcus species are deduced from the amino acid sequence. In panel A, all putative dienoyl-CoA hydratases from anaerobic bacteria growing with aromatic compounds are presented. In panel B, only some representative enoyl-CoA hydratases are shown from bacteria that are not capable growing on aromatics along with enoyl-CoA hydratases of unknown function from the genomes of G. metallireducens and S. aciditrophicus.
It is interesting to note that the phylogenetic dienoyl-CoA hydratase clusters represent neither the phylogenetic relationship nor a common overall energy metabolism of the organisms (facultative versus obligate anaerobes) but rather suggest that distribution of dienoyl-CoA hydratase genes occurred by horizontal gene transfer. The high and distinguishing amino acid sequence similarities of dienoyl-CoA hydratases, especially of the Thauera/Geobacter type, may allow the construction of gene probes for conserved regions. With such gene probes, both facultatively and obligately anaerobic bacteria with the capacity to degrade aromatic compounds could be detected in less defined samples (enrichment cultures, environmental samples, etc.). So far gene probes for the ATP-dependent dearomatizing benzoyl-CoA reductases only detect facultatively anaerobic aromatic compounds degrading bacteria (27).
Conclusions.
In this work, evidence was obtained that with exception of R. palustris, dienoyl-CoA hydratases are involved in the aromatic acid metabolism of all facultatively and strictly anaerobic bacteria, independent of the overall energy metabolism (nitrate-, iron-, and sulfate-reducing and -fermenting bacteria) and the nature of the benzoyl-CoA dearomatizing enzyme. This result has an impact on the question how the aromatic ring is dearomatized in strictly anaerobic bacteria. A hydroxylation of the ring or a reduction by more than two electrons would not require the expression of a highly specific dienoyl-CoA hydratase during growth on aromatic compounds and can now be ruled out. Despite the fact that for unknown reasons a benzoyl-CoA-reducing activity could not be determined so far in any strictly anaerobic bacterium, the results provide a first biochemical evidence that the central benzoyl-CoA pathway proceeds via identical intermediates in nearly all anaerobes.
Acknowledgments
We are grateful to S. Wischgoll (Freiburg) for help in cultivation of G. metallireducens.
This work was funded by the Deutsche Forschungsgemeinschaft. The Yamada Science Foundation (Osaka, Japan) gave Y.S. financial support for this study.
Footnotes
Published ahead of print on 22 November 2006.
REFERENCES
- 1.Auburger, G., and J. Winter. 1992. Purification and characterization of benzoyl-CoA ligase from a syntrophic, benzoate-degrading, anaerobic mixed culture. Appl. Microbiol. Biotechnol. 37:789-795. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 3.Boll, M., and G. Fuchs. 1995. Benzoyl-CoA reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP-dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K 172. Eur. J. Biochem. 234:921-933. [DOI] [PubMed] [Google Scholar]
- 4.Boll, M., S. J. P. Albracht, and G. Fuchs. 1997. Benzoyl-CoA reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. A study of adenosintriphosphatase activity, ATP stoichiometry of the reaction and EPR properties of the enzyme. Eur. J. Biochem. 244:840-851. [DOI] [PubMed] [Google Scholar]
- 5.Boll, M., D. Laempe, W. Eisenreich, A. Bacher, T. Mittelberger, J. Heinze, and G. Fuchs. 2000. Non-aromatic products from anoxic conversion of benzoyl-CoA with benzoyl-CoA reductase and cyclohexa-1,5-diene-1-carbonyl-CoA hydratase. J. Biol. Chem. 275:21889-21895. [DOI] [PubMed] [Google Scholar]
- 6.Boll, M., G. Fuchs, and H. Heider. 2002. Anaerobic oxidation of aromatic compounds and hydrocarbons. Curr. Opin. Chem. Biol. 6:604-611. [DOI] [PubMed] [Google Scholar]
- 7.Boll, M. 2005. Key enzymes of anaerobic aromatic metabolism catalyzing Birch-like reductions. Biochim. Biophys. Acta 1707:34-50. [DOI] [PubMed] [Google Scholar]
- 8.Boll, M., and G. Fuchs. 2005. Unusual reactions involved in anaerobic metabolism of phenolic compounds. Chem. Biol. 386:989-997. [DOI] [PubMed] [Google Scholar]
- 9.Boll, M. 2006. Dearomatizing benzene ring reductases. J. Mol. Microbiol. Biotechnol. 10:132-142. [DOI] [PubMed] [Google Scholar]
- 10.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 11.Carmona, M., and E. Diaz. 2005. Iron-reducing bacteria unravel novel strategies for the anaerobic catabolism of aromatic compounds. Mol. Microbiol. 58:1210-1215. [DOI] [PubMed] [Google Scholar]
- 12.Coligan, J. E., B. M. Dunn, D. W. Speicher, and P. T. Wingfield. 1995. Current protocols in protein science, volume 2. John Wiley & Sons, Inc., New York, NY.
- 13.Elshahed, M. S., and M. J. McInerney. 2001. Benzoate fermentation by the anaerobic bacterium Syntrophus aciditrophicus in the absence of hydrogen-using microorganisms. Appl. Environ. Microbiol. 67:5520-5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson, J., and C. S. Harwood. 2002. Metabolic diversity in aromatic compound utilization anaerobic microbes. Annu. Rev. Microbiol. 56:345-369. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, K. J., and J. Gibson. 1992. Potential early intermediates in anaerobic benzoate degradation by Rhodopseudomonas palustris. Appl. Environ. Microbiol. 58:696-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koch, J., W. Eisenreich, A. Bacher, and G. Fuchs. 1993. Products of enzymatic reduction of benzoyl-CoA, a key reaction in anaerobic aromatic metabolism. Eur. J. Biochem. 211:649-661. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Laempe, D., M. Jahn, and G. Fuchs. 1999. 6-Hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase and 6-oxocyclohex-1-ene-1-carbonyl-CoA hydrolase, enzymes of the benzoyl-CoA pathway of anaerobic aromatic metabolism in the denitrifying bacterium Thauera aromatica. Eur. J. Biochem. 263:420-429. [DOI] [PubMed] [Google Scholar]
- 19.Laempe, D., W. Eisenreich, A. Bacher, and G. Fuchs. 1998. Cyclohexa-1, 5-diene-1-carbonyl-CoA hydratase, an enzyme involved in anaerobic metabolism of benzoyl-CoA in the denitrifying bacterium Thauera aromatica. Eur. J. Biochem. 255:618-627. [DOI] [PubMed] [Google Scholar]
- 20.López-Barragán, M. J., M. Carmona, M. T. Zamarro, B. Thiele, M. Boll, G. Fuchs, J. L. García, and E. Díaz. 2004. The bzd gene cluster coding for the anaerobic benzoate catabolism in Azoarcus sp. strain CIB. J. Bacteriol. 186:5762-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovley, D. R., and E. J. P. Phillips. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron and manganese. Appl. Environ. Microbiol. 54:1472-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pelletier, D. A., and C. S. Harwood. 2000. 2-Hydroxycyclohexanecarboxyl coenzyme A dehydrogenase, an enzyme characteristic of the anaerobic benzoate degradation pathway used by Rhodopseudomonas palustris. J. Bacteriol. 182:2753-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters, F., M. Rother, and M. Boll. 2004. Selenocysteine-containing proteins in anaerobic benzoate metabolism of Desulfococcus multivorans. J. Bacteriol. 186:2156-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rabus, R., M. Kube, J. Heider, A. Beck, K. Heitmann, F. Widdel, and R. Reinhardt. 2005. The genome sequence of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Arch. Microbiol. 183:27-36. [DOI] [PubMed] [Google Scholar]
- 25.Schink, B., B. Philipp, and J. Müller. 2000. Anaerobic degradation of phenolic compounds. Naturwissenschaften 87:12-23. [DOI] [PubMed] [Google Scholar]
- 26.Schöcke, L., and B. Schink. 1999. Energetics and biochemistry of fermentative benzoate degradation by Syntrophus gentianae. Arch. Microbiol. 171:331-337. [DOI] [PubMed] [Google Scholar]
- 27.Song, B., and B. B. Ward. 2005. Genetic diversity of benzoyl-coenzyme A reductase genes detected in denitrifying isolates and estuarine sediment communities. Appl. Environ. Microbiol. 71:2036-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wischgoll, S., D. Heintz, F. Peters, A. Erxleben, E. Sarnighausen, R. Reski, A. van Dorsselaer, and M. Boll. 2005. Gene clusters involved in anaerobic benzoate degradation of Geobacter metallireducens. Mol. Microbiol. 58:1238-1252. [DOI] [PubMed] [Google Scholar]
- 29.Zehr, B. D., T. J. Savin, and R. E. Hall. 1989. A one-step, low background Coomassie staining procedure for polyacrylamide gels. Anal. Biochem. 182:157-159. [DOI] [PubMed] [Google Scholar]
- 30.Ziegler, K., R. Buder, J. Winter, and G. Fuchs. 1989. Activation of aromatic acids and aerobic aromatic metabolism in a denitrifying Pseudomonas strain. Arch. Microbiol. 151:171-176. [Google Scholar]