Abstract
Nontypeable Haemophilus influenzae (NTHi) is a gram-negative bacterium and a common commensal organism of the upper respiratory tract in humans. NTHi causes a number of diseases, including otitis media, sinusitis, conjunctivitis, exacerbations of chronic obstructive pulmonary disease, and bronchitis. During the course of colonization and infection, NTHi must withstand oxidative stress generated by insult due to multiple reactive oxygen species produced endogenously by other copathogens and by host cells. Using an NTHi-specific microarray containing oligonucleotides representing the 1821 open reading frames of the recently sequenced NTHi isolate 86-028NP, we have identified 40 genes in strain 86-028NP that are upregulated after induction of oxidative stress due to hydrogen peroxide. Further comparisons between the parent and an isogenic oxyR mutant identified a subset of 11 genes that were transcriptionally regulated by OxyR, a global regulator of oxidative stress. Interestingly, hydrogen peroxide induced the OxyR-independent upregulation of expression of the genes encoding components of multiple iron utilization systems. This finding suggested that careful balancing of levels of intracellular iron was important for minimizing the effects of oxidative stress during NTHi colonization and infection and that there are additional regulatory pathways involved in iron utilization.
In the evolution of life, a delicate physiological balance has arisen to compensate for both an organism's need for oxygen and the attendant lethal consequences of oxygen's toxicity. For example, in the electron transport chain, electrons can be inadvertently transferred from redox-active proteins to oxygen, producing reactive oxygen species (ROS) such as superoxide and hydrogen peroxide (H2O2). The effects of ROS on cells can then be exacerbated by the presence of free iron which, via the Fenton reaction, can react with H2O2 to produce more highly reactive hydroxyl radicals (34, 40, 52). Within a host, this problem is compounded by the release of extracellular ROS by phagocytes, which again, can have deleterious effects on invading pathogens (13). Thus, bacteria have evolved an array of increasingly well-defined mechanisms to abrogate the effects of oxidative stress. Proteins involved in such processes include catalase, which decomposes H2O2, and members of both the AhpCF/TsaA family of alkylhydroperoxidases and the organic hydroperoxide resistance proteins (Ohr) that decompose organic peroxides. Also, DNA-binding ferritin-like proteins (Dps) sequester free iron and bind to DNA, thus protecting DNA from damage (27, 33, 45, 53, 59, 60, 67). In addition, many bacterial species possess genes encoding OxyR, a global regulator of antioxidant defenses (24, 66).
Protection against oxidative stress is especially important to nontypeable Haemophilus influenzae (NTHi). NTHi is one of three bacterial commensal organisms, the others being Streptococcus pneumoniae and Moraxella catarrhalis, which can ascend a virus-compromised eustachian tube and cause otitis media (OM) in the normally sterile middle ear space (5). During this process, the bacteria are subject to multiple innate host defenses. These include assault by antimicrobial peptides (28, 44, 49), as well as invasion of the middle ear by polymorphonuclear leukocytes, with the attendant possibility of insult by ROS (36, 37, 58). Also, S. pneumoniae produces extracellular H2O2 at concentrations that are bactericidal toward H. influenzae in vitro (64). Despite these attentions, the ability of NTHi to successfully infect the middle ear space suggests that NTHi have multiple robust mechanisms in place to withstand the effects of oxidative stress. However, candidate enzymes with roles in the decomposition of ROS and the repair of DNA or proteins damaged by oxidative stress have not been well characterized in Haemophilus spp. The sequence of the H. influenzae strain Rd genome initially indicated that strain Rd has few genes encoding predicted enzyme systems for protection against oxidative stress; htkE, which encodes catalase is the major exception. Subsequently, a chimeric peroxiredoxin-glutaredoxin (PGdx) was identified and characterized in strain Rd. Further potential mechanisms of the detoxification of ROS in clinical isolates of NTHi have been elucidated through our laboratory's investigations of strain differences between strain Rd and two NTHi isolates recovered from children with chronic OM (29, 55). Microarray comparisons between the genomes of strain Rd and that of the NTHi OM isolate 1885MEE revealed a homologue of the gene encoding the alkylhydroperoxidase TsaA in the OM isolate (55). Genes encoding PGdx, TsaA, catalase, ferritin-like Dps, manganese superoxide dismutase (MnSOD) and OxyR were identified in the genome of the NTHi OM isolate 86-028NP (29).
We have begun to define the defenses that NTHi strains utilize to combat oxidative stress. Strain 86-028NP and an oxyR mutant of strain 86-028NP were treated with H2O2, and then cDNAs were generated and used to interrogate an arrayed set of 70-mer oligonucleotides representing all of strain 86-028NP's known open reading frames (ORFs). We identified genes upregulated due to oxidative stress that were either dependent on or independent of OxyR in strain 86-028NP. These included genes encoding proteins with roles in protection against oxidative stress, in the repair of damaged DNA and proteins, and in the utilization of iron, allowing us to better understand the multiple mechanisms by which NTHi can protect itself against host defenses, both as a commensal organism and during the progress of infection.
MATERIALS AND METHODS
Bacterial strains and culture media used.
NTHi strain 86-028NP was recovered from the nasopharynx of a child with chronic otitis media. This strain has been well characterized both in vitro (8, 32) and in chinchilla models of OM (7, 47, 81). The genome sequence has been published (29).
For routine culturing, strain 86-028NP and the strain 86-028NP oxyR mutant were grown on chocolate II plates (Fisher Scientific, Pittsburgh, PA). For testing the viability of cells on protoporphyrin IX (PPIX) medium without supplemented iron, cells were grown on brain heart infusion (BHI) agar plates supplemented with 2 μg NAD/ml and either 20 μg PPIX/ml or 20 μg heme/ml. For liquid culture, all cells were grown in BHI supplemented with 2 μg NAD/ml and 2 μg heme/ml (sBHI). All growth was carried out without shaking at 37°C in a 5% CO2 atmosphere.
Construction of the 86-028NP oxyR::Tn5 mutant strain.
The oxyR mutant was identified in a random pool of Tn5 mutants that were constructed by in vitro mutagenesis using Transposomics reagents from Epicenter (Epicenter, Madison, WI) and a protocol similar to that described by Gwinn et al. (26). Briefly, chromosomal DNA from strain 86-028NP was isolated and mutagenized in vitro, and then the single strand breaks introduced by transposition were filled and ligated. DNA was transformed into strain 86-028NP, and kanamycin-resistant mutants were selected. Mutations in individual clones were mapped by DNA sequencing of amplicons generated using a single primer PCR method as described by Ducey and Dyer (18).
Complementation of the oxyR mutation in strain 86-028NP.
The oxyR gene, its 122-bp upstream intergenic region, and the first 23 bp of the 5′ end of the downstream gene were amplified by PCR, TA cloned into pGEM-T Easy (Promega Co., Madison, WI), and then moved into pSPEC1, a derivative of the Escherichia coli-Haemophilus shuttle vector pGZRS-39A that contains a spectinomycin resistance gene (6). The pSPEC1-oxyR construct was transformed into the 86-028NP oxyR::Tn5 mutant strain, and transformants were selected on chocolate agar containing 200 μg spectinomycin/ml. Complemented clones were identified due to restoration of growth on BHI agar plates supplemented with 2 μg NAD/ml and 20 μg PPIX/ml and further characterized by quantitative real-time PCR (QRT-PCR).
Generation of the 86-028NP strain-specific microarray probe set.
The array was manufactured using the commercial oligonucleotide set for H. influenzae, Rd KW20 (Operon Biotechnologies, Huntsville, AL), plus 548 additional custom-made 70-mer probes representing ORFs from the strain 86-028NP that were either absent in strain Rd or that had insufficient similarities to their Rd homologues.
Microarray analyses of the 86-028NP strain's response to H2O2.
NTHi, the 86-026NP strain, and the 86-028NP oxyR::Tn5 mutant strain were grown in sBHI to mid-exponential phase. The cultures were divided in two, with H2O2 added to one half of the culture to a final concentration of 250 μM, while an equivalent volume of medium was added to the second half of the culture. After incubating for 10 min at 37°C in a 5% CO2 atmosphere, 5-ml aliquots of cells were pelleted at 4°C for 5 min at 3,220 × g. Immediately, supernatant was aspirated from both the treated and untreated cultures, and total RNA was isolated using TRIZOL Reagent (Invitrogen Corporation, Carlsbad, CA) as outlined by Mason et al., (47). cDNA was synthesized and labeled using an indirect labeling method similar to that described by Hegde et al. (30). Full microarray manufacturing, hybridization protocols, and data analysis methods are shown in the supplementary material.
Quantitative RT-PCR.
Quantitative RT-PCR was used to confirm the relative expression of genes identified by microarray analyses. This assay utilized RNA samples obtained from both the 86-028NP and the 86-028NP oxyR mutant (untreated or treated with 250 μM H2O2) and the one-step QuantiTect SYBR Green RT-PCR kit (QIAGEN, Valencia, CA) as outlined by Mason et al. (47).
RESULTS AND DISCUSSION
Characterization of OxyR in strain 86-028NP.
The oxyR gene in strain 86-028NP encodes a protein that is 73% identical to the E. coli OxyR protein. Nineteen of twenty residues in the helix-turn-helix motif thought to be involved in DNA binding are identical (41). Further comparisons of the amino acid sequences of OxyR from strain 86-028NP and those from E. coli show that the other functional domains in the OxyR protein from both organisms are well conserved. These include domains involved in tetramerization, disulfide bond formation, and RNA polymerase binding (12, 41, 42, 83, 88, 90).
Characterization of the 86-028NP oxyR mutant.
The 86-028NP oxyR mutant strain was identified from a bank of random transposition mutants. The transposon insertion was between nucleotides 1242 and 1250 of the oxyR gene. Maciver and Hansen (46) showed that a mutation in oxyR inhibited the growth of a H. influenzae type b strain on medium in which heme was replaced with the heme precursor PPIX. Nakahigashi et al. showed that a ferrochelatase mutant of E. coli was sensitive to light. They postulated that ROS was generated as a result of PPIX accumulation (57). This suggested to Maciver and Hansen that the differential growth of an oxyR mutant on PPIX-containing medium was due to the generation of superoxide that, in turn, could spontaneously undergo dismutation to form H2O2. Addition of catalase to the PPIX-containing medium abrogated the oxyR mutant growth defect, further indicating the effect of H2O2 generation in media. In order to demonstrate that the truncated oxyR gene in strain 86-028NP did not encode a functional protein, the oxyR mutant was cultured on BHI containing PPIX instead of heme and no supplemental iron. Growth of the oxyR mutant on medium containing PPIX was completely abrogated, whereas the mutant grew on heme-containing media. The parent strain grew on both media. Complementation of the oxyR mutant returned the parental phenotype (data not shown).
Determination of strain 86-028NP and mutant strain 86-028NP oxyR sensitivities to hydrogen peroxide.
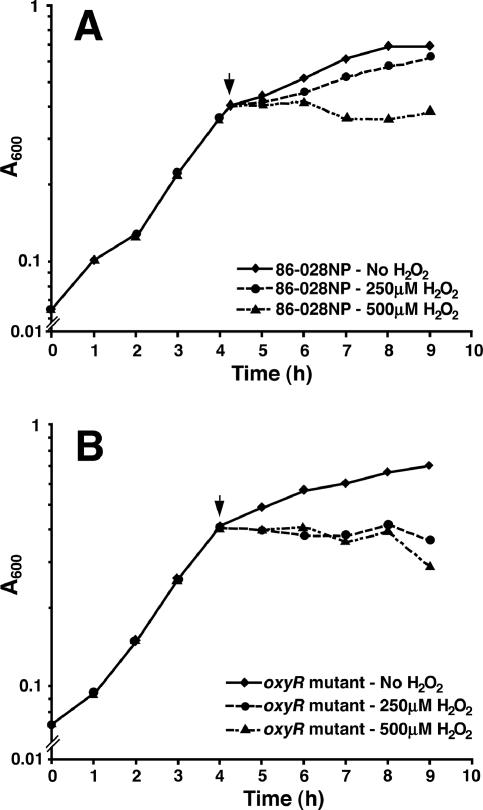
Strain 86-028NP and the oxyR mutant were grown without shaking in sBHI at 37°C in a 5% CO2 atmosphere to exponential phase, and then H2O2 was added to final concentrations of 250 μM and 500 μM. Growth was allowed to proceed, and the absorbance at 600 nm was measured at 1-hour intervals. Strain 86-028NP, in the presence of 250 μM H2O2, grew similarly to the untreated cells, but growth was inhibited at 500 μM H2O2. In contrast, the 86-028NP oxyR mutant was sensitive to both concentrations of H2O2 tested (Fig. 1). Also, the complemented strain grew similarly to the parent strain after the addition of 250 μm H2O2 to an exponentially growing culture showing restoration of OxyR function (data not shown). In order to determine whether the oxyR mutant could recover after treatment with 250 μM H2O2 for 10 min, cells were grown to exponential phase without shaking in sBHI at 37°C in a 5% CO2 atmosphere and then split into two aliquots. One aliquot was treated with H2O2 at a final concentration of 250 μM, while the second aliquot had an equivalent volume of medium added. After a 10-minute incubation, both aliquots were pelleted, washed, and resuspended in fresh sBHI. Absorbance at 600 nm was measured at 1-hour intervals to monitor growth (Fig. 2). Once H2O2 was removed from the cultures, growth resumed. These data indicate that incubating with 250 μM H2O2 for 10 min was an effective treatment to induce nonlethal oxidative stress.
FIG. 1.
The growth of NTHi 86-028NP strain (A) and the NTHi 86-028NP oxyR mutant strain (B) in increasing concentrations of H2O2. Both parent and mutant strains were grown to exponential phase, and then the designated concentration of H2O2 was added (arrowhead). Growth was then allowed to continue in the presence of H2O2.
FIG. 2.
Recovery of the NTHi 86-028NP strain and the NTHi 86-028NP oxyR mutant after a 10-min treatment with 250 μM H2O2. Both parent and mutant strains were grown to exponential phase and then treated with 250 μM H2O2 for 10 min (arrowhead indicates the point of H2O2 addition). The medium was then removed, and the cells were washed and resuspended in fresh medium. Growth was then allowed to continue.
DNA microarray analysis.
Cells were grown to exponential phase and split into two aliquots, and then H2O2 was added to one half of each culture to a final concentration of 250 μM. After 10 min, the cultures were harvested by centrifugation, and labeled cDNA was prepared as described in the supplemental material. For each comparison, a minimum of two sets of RNA was purified (biological replicates), while for each biological replicate, two slides were analyzed (technical replicates). This generated a minimum of 16 data points per gene for each comparison. Per-slide averaging and normalization were performed with Gene Traffic DUO, version 3.2-11 (Stratagene Corporation, La Jolla, CA). The mean 635 nm/532 nm ratios were generated from background-subtracted media intensities. Dye intensity normalization was applied by performing a LOWESS (subgrid) algorithm with all significant features including exogenous spike bacterial controls. Analysis of between-slide variance and replicate averaging was performed using the NIA/NIH analysis of variance (ANOVA) tool. A false discovery rate (FDR) of >0.05 was used to exclude false positives in reported genes.
Differentially labeled cDNAs from either the 86-028NP strain (treated with or without H2O2; four biological replicates) or the 86-028NP strain and the oxyR mutant (both treated with H2O2; two biological replicates) were compared. Using a cutoff of greater than twofold induction, 40 genes were identified that were upregulated in the parent strain after H2O2 treatment compared to that of the untreated sample (Table 1). A further comparison of H2O2-treated parent cells and oxyR mutant cells identified 11 genes whose regulation was dependent on OxyR (Table 1, bold type). We further identified a subset of 15 genes whose expression was upregulated in strain 86-028NP after treatment with H2O2 relative to that of the similarly treated oxyR mutant but which did not exhibit significant responsiveness to H2O2 when the parent was analyzed in the presence or absence of H2O2 (data not shown). Among these genes were a number with roles in protein synthesis. Possibly, we were witnessing downregulation of expression of these genes in the oxyR mutant after insult with H2O2. This effect may be more striking in the oxyR mutant as it exhibited greater sensitivity to H2O2 (see Fig. 1B) and thus may be more prone to perturbations in protein synthesis as a response to cellular injury. Microarray analyses were confirmed by QRT-PCR of selected genes (Table 1).
TABLE 1.
Hydrogen peroxide-responsive genes in the NTHi 86-028NP strain
| NTHI no. | Gene name | Function | Fold increase in expression
|
||
|---|---|---|---|---|---|
| Parent (+H2O2)/parent | Parent (+H2O2)/mutant (+H2O2) | Parent (+H2O2)/complement (+H2O2) | |||
| NTHI1099a | hktE | Catalase | 61b(194)c | 46 (334) | 1c |
| NTHI0177 | hitA | Iron utilization periplasmic protein | 22 (7) | 1 | 1 |
| NTHI1169 | tbp2 | Transferrin-binding protein 2 | 18 (7) | 0.5 | 1 |
| NTHI1168 | tbp1 | Transferrin-binding protein 1 | 10 (8) | 0.7 | 1 |
| NTHI0782 | hgpB | Hemoglobin-haptoglobin binding protein B | 8 (5) | 1.5 | 2 |
| NTHI0481 | hfeA | Putative periplasmic chelated iron binding protein | 8 (4) | 4 (4) | 1 |
| NTHI0477 | hfeD | Putative ABC type chelated iron transport system, permease | 7 (6) | 3d | NDe |
| NTHI0479 | hfeB | Putative ABC type chelated iron transport system, ATPase | 7 (5) | 3 (3) | ND |
| NTHI0705 | pgdX | Peroxiredoxin/glutaredoxin | 7 (9) | 31 (35) | 1 |
| NTHI1794 | Pseudogene for conserved hypothetical protein | 7 | 0.5 | ND | |
| NTHI0199 | Conserved hypothetical protein | 6 (1) | 0.5 | ND | |
| NTHI0478 | hfeC | Putative ABC-type chelated iron transport system, permease | 6 (4) | 3 (3) | 1 |
| NTHI0736 | hgpD | Hemoglobin-haptoglobin binding protein D | 6 | 1 | ND |
| NTHI0179 | hitB | Iron(III) transport system permease protein | 5 (2) | 2d | ND |
| NTHI1817 | dps | Conserved hypothetical DNA-binding ferritin-like protein | 4 (1) | 7 (5) | 0.4 |
| NTHI0681 | gnd | 6-phosphogluconate dehydrogenase | 4 (4) | 6 (8) | 1 |
| NTHI0358 | tonB | Energizer of iron uptake systems | 4 | 1 | ND |
| NTHI1214 | Conserved putative gamma-carboxymuconolactone decarboxylase subunit | 4 | 0.4 | ND | |
| NTHI0359 | exbD | Transport protein ExbD | 4 | 1 | ND |
| NTHI0360 | exbB | Transport protein ExbB | 4 | 1 | ND |
| NTHI0369 | hxuC | Heme/hemopexin-binding protein C | 4 (2) | 1 | 1 |
| NTHI0180 | hitC | Iron utilization ATP-binding protein | 3 (2) | 2d | 0.4 |
| NTHI1171 | Conserved hypothetical protein | 3 | 0.6 | ND | |
| NTHI1632 | ftsK2 | DNA translocase | 3 | 0.1 | ND |
| NTHI0104 | Conserved hypothetical protein | 3 | 1 | ND | |
| NTHI0175 | Conserved hypothetical protein | 3 | 0.2 | ND | |
| NTHI0370 | hxuB | Heme/hemopexin-binding protein B | 3 (2) | 1 | 1 |
| NTHI0083 | recN | DNA repair protein RecN | 3 | 0.5 | ND |
| NTHI0202 | hemR | Hemin receptor | 3 | 0.5 | ND |
| NTHI0062 | Putative TRAP type C4-dicarboxylate transport system, periplasmic | 3 | 1 | ND | |
| NTHI1588 | impA | Error prone DNA polymerase | 3 | 0.3 | ND |
| NTHI1801 | pntB | NAD(P) transhydrogenase subunit beta | 3 | 4 | ND |
| NTHI1802 | pntA | NAD(P) transhydrogenase subunit alpha | 2 | 5 | ND |
| NTHI0371 | hxuA | Heme/hemopexin-binding protein A | 2 | 0.7 | ND |
| NTHI0063 | Conserved hypothetical zinc type alcohol dehydrogenase-like protein | 2 | 1 | ND | |
| NTHI0729 | recA | Recombinase | 2 | 1 | ND |
| NTHI0496 | Conserved hypothetical protein | 2 | 1 | ND | |
| NTHI0684 | Hypothetical protein | 2 (1) | 2 (2) | 0.1 | |
| NTHI0493 | hscA | Chaperone protein HscA | 2 | 1 | ND |
| NTHI1474 | lgtD | UDP-GlcNAc-lipooligosaccharide N-acetylglucosamine glycosyltransferase | 2 | 2d | ND |
Genes appearing in bold text were defined as members of the OxyR regulon.
Determined by microarray.
Determined by QRT-PCR.
FRD above 0.05 cutoff.
ND, not determined.
The transcriptional response of the complemented oxyR mutant to H2O2 treatment was also determined by microarray analysis. The 86-028NP strain, the oxyR mutant, and a complemented oxyR mutant were grown to exponential phase and treated with 250 μM H2O2 for 10 min, and then RNA was purified. Microarray analysis showing the transcriptional response of strain 86-028NP in the presence of H2O2 was similar to that previously observed. Importantly, the transcriptional response of the complemented oxyR mutant in the presence of H2O2 mirrored that of the parent, indicating restoration of OxyR function. This finding was confirmed by QRT-PCR analysis of significantly regulated genes (Table 1, column three).
Generally, the functions of the proteins encoded by the genes identified could be broadly divided into three classes: (i) proteins with roles in mitigating oxidative stress; (ii) proteins with roles in DNA and protein repair; or (iii) proteins with roles in iron utilization. Results will be discussed in terms of these three categories.
(i) Oxidative stress.
Proteins can have a direct role in controlling the effects of oxidative stress by detoxifying reactive oxygen species. NTHI1099 (htkE), which encodes catalase (10), was the most highly upregulated gene, with an approximate 60-fold induction in the H2O2-treated parent compared to that in the untreated parent. NTHI1099 was not induced in the H2O2-treated oxyR mutant, indicating that OxyR regulates the expression of NTHI1099.
Using similar analyses, the second member of the OxyR regulon identified in strain 86-028NP was NTHI0705 (pgdX), which encodes a glutathione-dependent, chimeric peroxidase. Originally identified in Rd, PGdx consists of an N-terminal peroxiredoxin domain and a C-terminal glutaredoxin domain. The peroxiredoxin domain of PGdx has a role in protection against H2O2 and alkylhydroperoxidase; the glutathione-dependent reducing action of the glutaredoxin domain allows the system to recycle (62, 63, 86, 87). The expression of PGdx was subsequently found to be upregulated during in vitro biofilm formation by multiple isolates of NTHi from patients with chronic obstructive pulmonary disease (56).
NTHI1817, encoding Dps, a conserved hypothetical DNA-binding ferritin-like protein, is a further member of the OxyR regulon in strain 86-028NP. Dps was identified in E. coli as a starvation response protein that binds DNA and protects against H2O2-induced oxidative stress. Subsequently, H2O2 was found to induce Dps expression in actively growing E. coli in an OxyR-dependent manner (3, 4, 92). Similarly, in Salmonella enterica serovar Typhimurium, Dps protects against H2O2-derived oxidative stress and is needed for bacterial survival after phagocytosis by macrophages, as shown in a mouse model of infection (27). Dps' protective function is derived from its ability to bind Fe2+ and abrogate H2O2-induced hydroxy radical production via the Fenton reaction; H2O2 treatment of Fe2+-DNA complexes, in the absence of Dps, leads to the total degradation of the DNA (89).
The final members of the OxyR regulon with a proposed role in mitigating oxidative stress in strain 86-028NP are NTHI1802 and NTHI1801. Both genes have FDRs higher than the set cutoff (0.06 and 0.09, respectively). However, they are significantly upregulated in the parent, compared with that in the oxyR mutant (fivefold and fourfold, respectively). Thus, we believe they are members of the OxyR regulon in strain 86-028NP. NTHI1802 and NTHI1801 are homologues of pntA and pntB which encode the α and β subunits of NAD(P) transhydrogenase, respectively. In E. coli, PntAB has a role in the generation of NADPH; treatment of E. coli with H2O2 induces both this activity and an increase in NADPH-dependent oxidase and peroxidase activity (11, 73). Both the thioredoxin and glutaredoxin systems utilize the reducing power of NADPH to reduce disulfide bonds, so this process will likely be important during periods of oxidative stress. Also, NADPH is significantly less efficient in reducing Fe3+ to Fe2+ than NADH. Brumaghim et al. suggest that H2O2 treatment depletes the NADH pool in favor of NADPH, thus minimizing the production of Fe2+ and thereby abrogating the bactericidal effect of the Fenton reaction (11).
Interestingly, the expression of the gene encoding the peroxiredoxin TsaA was found to be unresponsive to the H2O2 treatment of strain 86-028NP. This lack of upregulation was confirmed by quantitative real-time PCR. It should be further noted that experiments carried out to determine the affect of toxicity of H2O2 on strain 86-028NP showed that the susceptibility of the 86-028NP tsaA mutant to H2O2 was indistinguishable from that of the parent strain (results not shown). This is contrary to what was found for AhpC, E. coli's homologue of TsaA, which has a major role in the decomposition of H2O2 (75, 76). Homologues of TsaA are utilized in the decomposition of other ROS. For example, mutations in ahp in both E. coli and S. enterica serovar Typhimurium increase sensitivity to cumene hydroperoxide (80). Our data suggest that TsaA in strain 86-028NP may have a yet-unidentified role in protection against oxidative stress.
(ii) DNA and protein repair.
All genes with roles in DNA and protein repair after strain 86-028NP underwent insult with H2O2 were regulated in an OxyR-independent manner.
(iii) Iron utilization.
As with most bacteria, strain 86-028NP has an absolute requirement for extracellular sources of iron. Genes encoding members of the five major iron- or heme-uptake systems, identified through the annotation of the 86-028NP strain genome (29), were upregulated during H2O2 treatment. Of these, genes encoding the Hfe system were shown to be members of the OxyR regulon. ANOVA analysis of the regulation of gene response to H2O2 treatment in strain 86-028NP and the oxyR mutant show only the hfeA, hfeB, and hfeC genes as being upregulated in the parent, relative to the those in the oxyR mutant. However, several data points for the hfeD gene were aberrantly low, and these observations resulted in this gene's exclusion from the list due to an FDR higher than the cutoff value. As genes hfeA, -B, -C, and D are organized as an operon in strain 86-028NP, and genes hfeA, -B, and -C exhibited similar increases in transcription in the parent relative to those in the oxyR mutant, we concluded that hfeD was indeed a member of the OxyR regulon. This thesis was further supported by the identification of a putative OxyR-binding site upstream of hfeA, the first gene in the presumptive operon. Genes hfeABCD are homologues of genes encoding a chelated iron ABC transport system that was originally identified in Yersinia pestis (9).
Microarray analyses of H2O2-treated E. coli, Pseudomonas aeruginosa, and Neisseria gonorrhoeae have identified genes with roles in protection against oxidative stress and repair of oxidative damage, while only the last organism exhibits the induction of multiple genes with roles in iron utilization (71, 79, 92). Thus, identification of a major category of genes, whose expression was upregulated during H2O2 treatment, which encode proteins that have roles in the 86-028NP strain's iron utilization systems is of great interest.
Identification of the 86-028NP strain-specific OxyR consensus binding site.
The upstream intergenic regions of five putative OxyR-regulated genes (NTHI1099, NTHI0705, NTHI1817, NTHI0681, and NTHI0481, the first gene in the putative hfe operon) were searched for sequence homology to the E. coli OxyR consensus DNA binding site (83). The upstream regions of the two most highly upregulated genes (NTHI1099, and NTHI0705) showed obvious similarity. Because the presumed binding geometry of OxyR causes the binding site to be pseudopalindromic, we defined an initial 37-bp strain 86-028NP consensus from the NTHI1099 and NTHI0705 sites and their reverse-complement sequences. These sequences were used to define a position-specific scoring matrix (PSSM) for the entire motif, and the 86-028NP strain genome was searched for other occurrences of this pattern. High-scoring sequences that fell in the likely promoter regions of genes listed in Table 1 were added to the motif definition, the PSSM was regenerated, and the process was bootstrapped until no new genes from these tables were found. The final PSSM predicted OxyR targets upstream of NTHI1099, NTHI0705, NTHI1817, NTHI0251, and NTHI0177. Also, NTHI0179 possesses a weaker motif that overlaps the upstream NTHI0177 stop codon. The 86-028NP strain OxyR binding site possesses strong similarities to the repeated ATAGnt motif identified in E. coli (83). Also, the information content of the derived sequences displayed a small information peak offset from the 11-bp repeat of the presumed primary binding motifs. A similar phenomenon, suggestive of a minor-groove contact in the binding geometry, has been observed among E. coli OxyR binding sites (74). Of note, initially no similar patterns were identified upstream of the putative OxyR regulon members NTHI0681 and NTHI0481. However, if OxyR binding tolerates misspacing of the highly conserved presumed contact regions, NTHI0681 and NTHI0481 both possess potential OxyR binding sites, with that related to NTHI0681 containing one extra base in the 5′ subpseudopalindrome and that related to NTHI0481 being reduced by 1 base in each of the 5′ and 3′ subpseudopalindrome regions.
OxyR-independent genes.
Genes regulated independently of OxyR were defined as those whose change in expression, in the presence of H2O2, were similar in both the parent and the oxyR mutant. These OxyR-independent regulated genes were as follows:
(i) Oxidative stress.
All genes identified with direct roles in mitigating oxidative stress in strain 86-028NP were regulated by OxyR.
(ii) DNA and protein repair.
Direct defense mechanisms against oxidative stress cannot absolutely protect the cell against injury. Thus, a cell must have mechanisms in place that can repair damage due to ROS. Strain 86-028NP contained multiple homologues of such genes, whose expression was upregulated in the presence of H2O2 but in an OxyR-independent manner. NTHI0729 and NTHI0083 encode homologues of RecA and RecN. RecA and RecN have important roles in the repair of DNA damage which have been extensively characterized in UV damage experiments in many organisms, including E. coli and H. influenzae (43, 50, 51, 77, 82). Further repair mechanisms are potentially afforded by the gene products of NTHI0493 and NTHI0495, which encode homologues of the cochaperone proteins HscA and HscB, respectively, and that have roles in iron-sulfur cluster assembly (2, 31, 78). NTHI1588 is a homologue of a gene that encodes ImpA, an error-prone DNA polymerase originally identified in the incompatibility group plasmid TP110. Possession of this plasmid increases a cell's resistance to both UV and UV-mediated mutagenesis (25). In E. coli, impA is a member of the impCAB operon but, as with the H. influenzae Rd strain, strain 86-028NP lacks homologues of impB and impC. The sixth OxyR-independent gene in this group was NTHI1632, which is a homologue of a gene that encodes the DNA translocase FtsK. The role of FtsK in both DNA replication and cell division has been well established. In E. coli, FtsK is presumed to have a role in resolving chromosome dimers caused by crossovers during chromosome replication (17, 48).
(iii) Iron utilization.
The most strongly upregulated genes in this class were NTHI1168 and NTHI1169, which are homologues of genes encoding transferrin-binding proteins 1 and 2, respectively, (65). Other OxyR-independent upregulated genes encoding homologues of iron utilization proteins include NTHI0177, NTHI0179, and NTHI0180, which are homologues of genes encoding components of the Hit system, an ABC transport system that imports free iron into the cell (1, 72). Strain 86-028NP also possesses genes which are homologues of genes encoding hemoglobin and hemoglobin-haptoglobin binding proteins (HgpB, HgpC, and HgpD). H2O2 treatment upregulated the expression of the genes encoding HgpB and HgpD but not that of the gene encoding HgpC. The three encoded hemoglobin and hemoglobin-haptoglobin binding proteins in H. influenzae type b have overlapping functions (35, 54, 69), which suggests that if the upregulation of hemoglobin and hemoglobin-haptoglobin binding proteins is needed during oxidative stress, it is not vital that expression of all three proteins is upregulated. Finally, strain 86-028NP possesses genes which are homologues of the hxu system, the hemin receptor HemR, and components of the TonB-ExbBD system. All exhibit H2O2-induced, OxyR-independent upregulation of expression. The hxu system in H. influenzae type b encodes a heme utilization system that captures heme sequestered in a complex with hemopexin (14-16). The TonB-ExbBD system has a vital role in transducing the energy of the proton motor force to generate conformational changes in TonB-dependent outer membrane transporters (68). If the upregulation of outer membrane-associated iron uptake system proteins is functionally relevant to the conditions induced by treatment of strain 86-028NP with H2O2, then the concomitant upregulation of expression of the proteins needed to energize their function would be a logical step. The upregulation of iron utilization genes during periods of oxidative stress has been noted before, most recently in microarray analyses of H2O2-treated Neisseria meningitidis (79), although the number of iron utilization genes identified in N. meningitidis is fewer than that in strain 86-028NP. As is well established, the Fenton reaction is dependent on Fe2+ to react with H2O2 in the generation of hydroxyl radicals. It would thus seem counterintuitive for a bacterium under assault by H2O2 to upregulate proteins that would increase the intracellular concentration of iron. Three possible explanations could explain these observations. First, as suggested by Zheng et al. (92), the treatment of bacteria with H2O2 oxidizes intracellular pools of Fe2+, thus reducing the amount of available iron. The increased expression of genes involved in iron utilization in the presence of H2O2 may thus be unconnected to a response against oxidative stress. We may thereby be observing a response to iron limitation. However, bacteria were grown under aerobic conditions during which iron is already relatively insoluble. Subsequent oxidation by H2O2 may not induce a significant further reduction in iron solubility. Furthermore, the apparent control of expression of genes encoding components of the Hfe system by OxyR indicated that increased expression of genes with roles in iron utilization do have a role in response to oxidative stress. Second, an increase in intracellular iron could be utilized to repair 4Fe-4S cluster-containing proteins that have become damaged due to ROS insult. In E. coli, proteins containing 4Fe-4S clusters such as dihydroxy-acid dehydratase, 6-phosphoglucose dehydratase, aconitase, and quinolinate synthetase are particularly prone to damage due to superoxide-generated oxidative stress (19, 21-23). The superoxide radical oxidizes the clusters with the concomitant release of iron from the 4Fe-4S moiety, thus generating a nonfunctional cluster. In the case of dihydroxy-acid dehydratase, this does not produce a loss of protein or induce the synthesis of new protein. It appears that the damage is reversible, and the clusters can be repaired (19, 20, 39). Finally, the increased transcription of iron utilization genes in strain 86-028NP during a period of oxidative stress may also be related to 4Fe-4S cluster damage. The iron lost from 4Fe-4S clusters due to oxidative damage may impinge on DNA, causing further damage. Using an E. coli hpx mutant that lacks three major H2O2-scavenging enzymes, the use of iron chelators prevented both hydroxyl radical formation and DNA damage. An hpx dps double mutant also exhibited greater levels of DNA damage, again indicating both the damaging effects of iron and the role Dps plays in protecting against such damage (61). Binding of free iron may thus help abrogate the toxic effects of the Fenton reaction. In E. coli, Fe2+, a substrate for the Fenton reaction, may be generated by the reduction of Fe3+ by NADH (38). Thus, in the case of strain 86-028NP, the increased expression of homologues of proteins such as HitA may help mitigate the effects of the Fenton reaction due to a reduction in concentration of intracellular Fe3+ (1, 39).
OxyR regulon in strain 86-028NP.
In summary, by comparing the H2O2-induced increase in expression of genes in the parent with the expression of the same genes in the 86-028NP oxyR mutant, it was possible to identify genes whose upregulation was abrogated in cells lacking functional OxyR. These genes could thus be classified as being part of the 86-028NP strain OxyR regulon. Expectedly, the class of genes with roles in direct defenses against oxidative stress are all members of the OxyR regulon. By comparison, all genes with roles in direct defense against oxidative stress are also members of the OxyR regulon in E. coli (92). This shows the importance of OxyR in sensing oxidative stress and in promoting the expression of proteins that have a central role in mitigating the effects of such attacks. Conversely, genes encoding proteins involved in repairing DNA and protein damage generated by oxidative stress were expressed in an OxyR-independent manner. Interestingly, of the multiple iron utilization systems whose expression levels were upregulated in the presence of H2O2, only the Hfe system was regulated by OxyR. This result indicates alternative oxidative-stress-responsive regulation of these genes. The best-characterized global regulator of iron utilization genes is Fur (for a review of Fur in Neisseria, see Rohde and Dyer [70]), and the interrelatedness of Fur regulation and oxidative stress responses have been previously described. A fur mutant of E. coli grown at increased oxygen tensions exhibited oxidative DNA damage possibly due to excess free iron generated by derepression of iron uptake genes (85). Moreover, the promoter region of fldA, the cotranscribed gene upstream of fur in E. coli, exhibits a binding site for SoxS, the positive regulator of SOD (91). Furthermore, expression of both iron and manganese cofactored SOD is positively regulated by Fur in multiple species of bacteria (reviewed by Touati [84]). Finally, with regard to OxyR regulation of Fur, microarray analyses of hydrogen peroxide-treated E. coli indicated that fur expression was slightly upregulated compared to that of the untreated sample and that fur was a member of the OxyR regulon (92). This posits the question of whether Fur is regulating iron utilization genes in strain 86-028NP cells that are undergoing oxidative stress. Strain 86-028NP similarly possesses fldA upstream of fur, and their organization suggests that both genes are cotranscribed. However, strain 86-028NP does not possess a homologue of soxS. Also, the transcription of fur was not significantly altered in strain 86-028NP after H2O2 treatment. Thus, the regulation mechanisms that allows cross talk between the iron utilization and the oxidative stress resistance mechanisms in strain 86-028NP have yet to be elucidated.
We have thus identified a complement of genes that are upregulated in NTHi strain 86-028NP as a consequence of oxidative stress induced by H2O2. We have further refined our analyses to identify those genes that are regulated by OxyR, a global regulator of cellular responses against oxidative stress. As expected, a number of these genes encoded proteins with direct roles in combating oxidative stress. More interestingly, we have also found that the expression of proteins that are members of the majority of the iron uptake systems in strain 86-028NP were also upregulated during H2O2 exposure. This raises the question as to whether the modulation of iron uptake by strain 86-028NP is a mechanism for combatting the effects of oxidative stress generated by the iron dependent Fenton reaction. Also, the upregulation of expression of the majority of the iron utilization genes occurred in an OxyR-independent manner, suggesting the possibility that additional regulation systems were in place. Elucidation of these additional systems will be of great interest as we further define the careful physiological balancing act NTHi cells are involved in, both as common commensal organisms and as major pathogens in the causation of multiple human respiratory tract diseases.
Supplementary Material
Acknowledgments
We acknowledge the technical assistance of Yingjie Zhang and Linda S. Johnson, who constructed and initially characterized the 86-028NP oxyR mutant.
This work was supported by NIH grant RO1-DC03915 (to L.O.B.).
Footnotes
Published ahead of print on 1 December 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adhikari, P., S. D. Kirby, A. J. Nowalk, K. L. Veraldi, A. B. Schryvers, and T. A. Mietzner. 1995. Biochemical characterization of a Haemophilus influenzae periplasmic iron transport operon. J. Biol. Chem. 270:25142-25149. [DOI] [PubMed] [Google Scholar]
- 2.Agar, J. N., C. Krebs, J. Frazzon, B. H. Huynh, D. R. Dean, and M. K. Johnson. 2000. IscU as a scaffold for iron-sulfur cluster biosynthesis: sequential assembly of [2Fe-2S] and [4Fe-4S] clusters in IscU. Biochemistry 39:7856-7862. [DOI] [PubMed] [Google Scholar]
- 3.Almiron, M., A. J. Link, D. Furlong, and R. Kolter. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6:2646-2654. [DOI] [PubMed] [Google Scholar]
- 4.Altuvia, S., M. Almiron, G. Huisman, R. Kolter, and G. Storz. 1994. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol. Microbiol. 13:265-272. [DOI] [PubMed] [Google Scholar]
- 5.Bakaletz, L. O. 2002. Otitis media, p. 259-298. In K. A. Brogden, and J. A. Guthmiller(ed.), Polymicrobial diseases. ASM Press, Washington, DC. [PubMed]
- 6.Bakaletz, L. O., B. D. Baker, J. A. Jurcisek, A. Harrison, L. A. Novotny, J. E. Bookwalter, R. Mungur, and R. S. Munson, Jr. 2005. Demonstration of type IV pilus expression and a twitching phenotype by Haemophilus influenzae. Infect. Immun. 73:1635-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakaletz, L. O., E. R. Leake, J. M. Billy, and P. T. Kaumaya. 1997. Relative immunogenicity and efficacy of two synthetic chimeric peptides of fimbrin as vaccinogens against nasopharyngeal colonization by nontypeable Haemophilus influenzae in the chinchilla. Vaccine 15:955-961. [DOI] [PubMed] [Google Scholar]
- 8.Bakaletz, L. O., B. M. Tallan, T. Hoepf, T. F. DeMaria, H. G. Birck, and D. J. Lim. 1988. Frequency of fimbriation of nontypeable Haemophilus influenzae and its ability to adhere to chinchilla and human respiratory epithelium. Infect. Immun. 56:331-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bearden, S. W., T. M. Staggs, and R. D. Perry. 1998. An ABC transporter system of Yersinia pestis allows utilization of chelated iron by Escherichia coli SAB11. J. Bacteriol. 180:1135-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishai, W. R., H. O. Smith, and G. J. Barcak. 1994. A peroxide/ascorbate-inducible catalase from Haemophilus influenzae is homologous to the Escherichia coli katE gene product. J. Bacteriol. 176:2914-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brumaghim, J. L., Y. Li, E. Henle, and S. Linn. 2003. Effects of hydrogen peroxide upon nicotinamide nucleotide metabolism in Escherichia coli: changes in enzyme levels and nicotinamide nucleotide pools and studies of the oxidation of NAD(P)H by Fe(III). J. Biol. Chem. 278:42495-42504. [DOI] [PubMed] [Google Scholar]
- 12.Choi, H., S. Kim, P. Mukhopadhyay, S. Cho, J. Woo, G. Storz, and S. Ryu. 2001. Structural basis of the redox switch in the OxyR transcription factor. Cell 105:103-113. [DOI] [PubMed] [Google Scholar]
- 13.Clark, R. A. 1990. The human neutrophil respiratory burst oxidase. J. Infect. Dis. 161:1140-1147. [DOI] [PubMed] [Google Scholar]
- 14.Cope, L. D., R. P. Love, S. E. Guinn, A. Gilep, S. Usanov, R. W. Estabrook, Z. Hrkal, and E. J. Hansen. 2001. Involvement of HxuC outer membrane protein in utilization of hemoglobin by Haemophilus influenzae. Infect. Immun. 69:2353-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cope, L. D., S. E. Thomas, Z. Hrkal, and E. J. Hansen. 1998. Binding of heme-hemopexin complexes by soluble HxuA protein allows utilization of this complexed heme by Haemophilus influenzae. Infect. Immun. 66:4511-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cope, L. D., R. Yogev, U. Muller-Eberhard, and E. J. Hansen. 1995. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J. Bacteriol. 177:2644-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donachie, W. D. 2002. FtsK: Maxwell's demon? Mol. Cell 9:206-207. [DOI] [PubMed] [Google Scholar]
- 18.Ducey, T. F., and D. W. Dyer. 2002. Rapid identification of EZ::TN transposon insertion sites in the genome of Neisseria gonorrhoeae. Epicentre Forum 9:6-7. [Google Scholar]
- 19.Flint, D. H., E. Smyk-Randall, J. F. Tuminello, B. Draczynska-Lusiak, and O. R. Brown. 1993. The inactivation of dihydroxy-acid dehydratase in Escherichia coli treated with hyperbaric oxygen occurs because of the destruction of its Fe-S cluster, but the enzyme remains in the cell in a form that can be reactivated. J. Biol. Chem. 268:25547-25552. [PubMed] [Google Scholar]
- 20.Flint, D. H., J. F. Tuminello, and M. H. Emptage. 1993. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J. Biol. Chem. 268:22369-22376. [PubMed] [Google Scholar]
- 21.Gardner, P. R., and I. Fridovich. 1991. Quinolinate synthetase: the oxygen-sensitive site of de novo NAD(P)+ biosynthesis. Arch. Biochem. Biophys. 284:106-111. [DOI] [PubMed] [Google Scholar]
- 22.Gardner, P. R., and I. Fridovich. 1991. Superoxide sensitivity of the Escherichia coli 6-phosphogluconate dehydratase. J. Biol. Chem. 266:1478-1483. [PubMed] [Google Scholar]
- 23.Gardner, P. R., and I. Fridovich. 1991. Superoxide sensitivity of the Escherichia coli aconitase. J. Biol. Chem. 266:19328-19333. [PubMed] [Google Scholar]
- 24.Georgiou, G. 2002. How to flip the (redox) switch. Cell 111:607-610. [DOI] [PubMed] [Google Scholar]
- 25.Glazebrook, J. A., K. K. Grewal, and P. Strike. 1986. Molecular analysis of the UV protection and mutation genes carried by the I incompatibility group plasmid TP110. J. Bacteriol. 168:251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gwinn, M. L., A. E. Stellwagen, N. L. Craig, J.-F. Tomb, and H. O. Smith. 1997. In vitro Tn7 mutagenesis of Haemophilus influenzae Rd and characterization of the role of atpA in transformation. J. Bacteriol. 179:7315-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halsey, T. A., A. Vazquez-Torres, D. J. Gravdahl, F. C. Fang, and S. J. Libby. 2004. The ferritin-like Dps protein is required for Salmonella enterica serovar Typhimurium oxidative stress resistance and virulence. Infect. Immun. 72:1155-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris, R. H., D. Wilk, C. L. Bevins, R. S. Munson, Jr., and L. O. Bakaletz. 2004. Identification and characterization of a mucosal antimicrobial peptide expressed by the chinchilla (Chinchilla lanigera) airway. J. Biol. Chem. 279:20250-20256. [DOI] [PubMed] [Google Scholar]
- 29.Harrison, A., D. W. Dyer, A. Gillaspy, W. C. Ray, R. Mungur, M. B. Carson, H. Zhong, J. Gipson, M. Gipson, L. S. Johnson, L. Lewis, L. O. Bakaletz, and R. S. Munson, Jr. 2005. Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype d, strain KW20. J. Bacteriol. 187:4627-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hegde, P., R. Qi, K. Abernathy, C. Gay, S. Dharap, R. Gaspard, J. E. Hughes, E. Snesrud, N. Lee, and J. Quackenbush. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29:548-550, 552-554, 556. [DOI] [PubMed] [Google Scholar]
- 31.Hoff, K. G., D. T. Ta, T. L. Tapley, J. J. Silberg, and L. E. Vickery. 2002. Hsc66 substrate specificity is directed toward a discrete region of the iron-sulfur cluster template protein IscU. J. Biol. Chem. 277:27353-27359. [DOI] [PubMed] [Google Scholar]
- 32.Holmes, K. A., and L. O. Bakaletz. 1997. Adherence of non-typeable Haemophilus influenzae promotes reorganization of the actin cytoskeleton in human or chinchilla epithelial cells in vitro. Microb. Pathog. 23:157-166. [DOI] [PubMed] [Google Scholar]
- 33.Hong, Y., G. Wang, and R. J. Maier. 2006. Helicobacter hepaticus Dps protein plays an important role in protecting DNA from oxidative damage. Free Radic. Res. 40:597-605. [DOI] [PubMed] [Google Scholar]
- 34.Imlay, J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395-418. [DOI] [PubMed] [Google Scholar]
- 35.Jin, H., Z. Ren, P. W. Whitby, D. J. Morton, and T. L. Stull. 1999. Characterization of hgpA, a gene encoding a haemoglobin/haemoglobin-haptoglobin-binding protein of Haemophilus influenzae. Microbiology 145:905-914. [DOI] [PubMed] [Google Scholar]
- 36.Kawana, M., C. Kawana, and G. S. Giebink. 1992. Penicillin treatment accelerates middle ear inflammation in experimental pneumococcal otitis media. Infect. Immun. 60:1908-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawana, M., C. Kawana, T. Yokoo, P. G. Quie, and G. S. Giebink. 1991. Oxidative metabolic products released from polymorphonuclear leukocytes in middle ear fluid during experimental pneumococcal otitis media. Infect. Immun. 59:4084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keyer, K., A. S. Gort, and J. A. Imlay. 1995. Superoxide and the production of oxidative DNA damage. J. Bacteriol. 177:6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keyer, K., and J. A. Imlay. 1996. Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl. Acad. Sci. USA. 93:13635-13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korshunov, S., and J. A. Imlay. 2006. Detection and quantification of superoxide formed within the periplasm of Escherichia coli. J. Bacteriol. 188:6326-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kullik, I., J. Stevens, M. B. Toledano, and G. Storz. 1995. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for DNA binding and multimerization. J. Bacteriol. 177:1285-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kullik, I., M. B. Toledano, L. A. Tartaglia, and G. Storz. 1995. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J. Bacteriol. 177:1275-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, H. Y., A. Andalibi, P. Webster, S. K. Moon, K. Teufert, S. H. Kang, J. D. Li, M. Nagura, T. Ganz, and D. J. Lim. 2004. Antimicrobial activity of innate immune molecules against Streptococcus pneumoniae, Moraxella catarrhalis and nontypeable Haemophilus influenzae. BMC Infect. Dis. 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loprasert, S., W. Whangsuk, R. Sallabhan, and S. Mongkolsuk. 2004. DpsA protects the human pathogen Burkholderia pseudomallei against organic hydroperoxide. Arch. Microbiol. 182:96-101. [DOI] [PubMed] [Google Scholar]
- 46.Maciver, I., and E. J. Hansen. 1996. Lack of expression of the global regulator OxyR in Haemophilus influenzae has a profound effect on growth phenotype. Infect. Immun. 64:4618-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mason, K. M., R. S. Munson, Jr., and L. O. Bakaletz. 2003. Nontypeable Haemophilus influenzae gene expression induced in vivo in a chinchilla model of otitis media. Infect. Immun. 71:3454-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCool, J. D., and S. J. Sandler. 2001. Effects of mutations involving cell division, recombination, and chromosome dimer resolution on a priA2::kan mutant. Proc. Natl. Acad. Sci. USA. 98:8203-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGillivary, G., W. C. Ray, C. L. Bevins, R. S. Munson, Jr., and L. O. Bakaletz. A member of the cathelicidin family of antimicrobial peptides is produced in the upper airway of the chinchilla and its mRNA expression is altered by common viral and bacterial co-pathogens of otitis media. Mol. Immunol., in press. [DOI] [PMC free article] [PubMed]
- 50.McGrew, D. A., and K. L. Knight. 2003. Molecular design and functional organization of the RecA protein. Crit. Rev. Biochem. Mol. Biol. 38:385-432. [DOI] [PubMed] [Google Scholar]
- 51.Meddows, T. R., A. P. Savory, J. I. Grove, T. Moore, and R. G. Lloyd. 2005. RecN protein and transcription factor DksA combine to promote faithful recombinational repair of DNA double-strand breaks. Mol. Microbiol. 57:97-110. [DOI] [PubMed] [Google Scholar]
- 52.Messner, K. R., and J. A. Imlay. 1999. The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. J. Biol. Chem. 274:10119-10128. [DOI] [PubMed] [Google Scholar]
- 53.Mongkolsuk, S., W. Praituan, S. Loprasert, M. Fuangthong, and S. Chamnongpol. 1998. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J. Bacteriol. 180:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morton, D. J., P. W. Whitby, H. Jin, Z. Ren, and T. L. Stull. 1999. Effect of multiple mutations in the hemoglobin- and hemoglobin-haptoglobin-binding proteins, HgpA, HgpB, and HgpC, of Haemophilus influenzae type b. Infect. Immun. 67:2729-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Munson, R. S., Jr., A. Harrison, A. Gillaspy, W. C. Ray, M. Carson, D. Armbruster, J. Gipson, M. Gipson, L. Johnson, L. Lewis, D. W. Dyer, and L. O. Bakaletz. 2004. Partial analysis of the genomes of two nontypeable Haemophilus influenzae otitis media isolates. Infect. Immun. 72:3002-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murphy, T. F., C. Kirkham, S. Sethi, and A. J. Lesse. 2005. Expression of a peroxiredoxin-glutaredoxin by Haemophilus influenzae in biofilms and during human respiratory tract infection. FEMS Immunol. Med. Microbiol. 44:81-89. [DOI] [PubMed] [Google Scholar]
- 57.Nakahigashi, K., K. Nishimura, K. Miyamoto, and H. Inokuchi. 1991. Photosensitivity of a protoporphyrin-accumulating, light-sensitive mutant (visA) of Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 88:10520-10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nonomura, N., G. S. Giebink, S. K. Juhn, T. Harada, and D. Aeppli. 1991. Pathophysiology of Streptococcus pneumoniae otitis media: kinetics of the middle ear biochemical and cytologic host responses. Ann. Otol. Rhinol. Laryngol. 100:236-243. [DOI] [PubMed] [Google Scholar]
- 59.Ochsner, U. A., D. J. Hassett, and M. L. Vasil. 2001. Genetic and physiological characterization of ohr, encoding a protein involved in organic hydroperoxide resistance in Pseudomonas aeruginosa. J. Bacteriol. 183:773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park, M., S. T. Yun, S.-Y. Hwang, C.-I. Chun, and T. I. Ahn. 2006. The dps gene of symbiotic “Candidatus Legionella jeonii” in Amoeba proteus responds to hydrogen peroxide and phagocytosis. J. Bacteriol. 188:7572-7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park, S., X. You, and J. A. Imlay. 2005. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx− mutants of Escherichia coli. Proc. Natl. Acad. Sci. USA. 102:9317-9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pauwels, F., B. Vergauwen, and J. J. Van Beeumen. 2004. Physiological characterization of Haemophilus influenzae Rd deficient in its glutathione-dependent peroxidase PGdx. J. Biol. Chem. 279:12163-12170. [DOI] [PubMed] [Google Scholar]
- 63.Pauwels, F., B. Vergauwen, F. Vanrobaeys, B. Devreese, and J. J. Van Beeumen. 2003. Purification and characterization of a chimeric enzyme from Haemophilus influenzae Rd that exhibits glutathione-dependent peroxidase activity. J. Biol. Chem. 278:16658-16666. [DOI] [PubMed] [Google Scholar]
- 64.Pericone, C. D., K. Overweg, P. W. M. Hermans, and J. N. Weiser. 2000. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect. Immun. 68:3990-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perkins-Balding, D., M. Ratliff-Griffin, and I. Stojiljkovic. 2004. Iron transport systems in Neisseria meningitidis. Microbiol. Mol. Biol. Rev. 68:154-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pomposiello, P. J., and B. Demple. 2001. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19:109-114. [DOI] [PubMed] [Google Scholar]
- 67.Poole, L. B. 2005. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch. Biochem. Biophys. 433:240-254. [DOI] [PubMed] [Google Scholar]
- 68.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 69.Ren, Z., H. Jin, D. J. Morton, and T. L. Stull. 1998. hgpB, a gene encoding a second Haemophilus influenzae hemoglobin- and hemoglobin-haptoglobin-binding protein. Infect. Immun. 66:4733-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rohde, K. H., and D. W. Dyer. 2003. Mechanisms of iron acquisition by the human pathogens Neisseria meningitidis and Neisseria gonorrhoeae. Front. Biosci. 8:d1186-1218. [DOI] [PubMed] [Google Scholar]
- 71.Salunkhe, P., T. Töpfer, J. Buer, and B. Tümmler. 2005. Genome-wide transcriptional profiling of the steady-state response of Pseudomonas aeruginosa to hydrogen peroxide. J. Bacteriol. 187:2565-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanders, J. D., L. D. Cope, and E. J. Hansen. 1994. Identification of a locus involved in the utilization of iron by Haemophilus influenzae. Infect. Immun. 62:4515-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sauer, U., F. Canonaco, S. Heri, A. Perrenoud, and E. Fischer. 2004. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 279:6613-6619. [DOI] [PubMed] [Google Scholar]
- 74.Schneider, T. D. 1996. Reading of DNA sequence logos: prediction of major groove binding by information theory. Methods Enzymol. 274:445-455. [DOI] [PubMed] [Google Scholar]
- 75.Seaver, L. C., and J. A. Imlay. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seaver, L. C., and J. A. Imlay. 2001. Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. J. Bacteriol. 183:7182-7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Setlow, J. K., D. Spikes, and K. Griffin. 1988. Characterization of the rec-1 gene of Haemophilus influenzae and behavior of the gene in Escherichia coli. J. Bacteriol. 170:3876-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Silberg, J. J., T. L. Tapley, K. G. Hoff, and L. E. Vickery. 2004. Regulation of the HscA ATPase reaction cycle by the co-chaperone HscB and the iron-sulfur cluster assembly protein IscU. J. Biol. Chem. 279:53924-53931. [DOI] [PubMed] [Google Scholar]
- 79.Stohl, E. A., A. K. Criss, and H. S. Seifert. 2005. The transcriptome response of Neisseria gonorrhoeae to hydrogen peroxide reveals genes with previously uncharacterized roles in oxidative damage protection. Mol. Microbiol. 58:520-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Storz, G., F. S. Jacobson, L. A. Tartaglia, R. W. Morgan, L. A. Silveira, and B. N. Ames. 1989. An alkyl hydroperoxide reductase induced by oxidative stress in Salmonella typhimurium and Escherichia coli: genetic characterization and cloning of ahp. J. Bacteriol. 171:2049-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suzuki, K., and L. O. Bakaletz. 1994. Synergistic effect of adenovirus type 1 and nontypeable Haemophilus influenzae in a chinchilla model of experimental otitis media. Infect. Immun. 62:1710-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sweetman, W. A., E. R. Moxon, and C. D. Bayliss. 2005. Induction of the SOS regulon of Haemophilus influenzae does not affect phase variation rates at tetranucleotide or dinucleotide repeats. Microbiology 151:2751-2763. [DOI] [PubMed] [Google Scholar]
- 83.Toledano, M. B., I. Kullik, F. Trinh, P. T. Baird, T. D. Schneider, and G. Storz. 1994. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78:897-909. [DOI] [PubMed] [Google Scholar]
- 84.Touati, D. 2000. Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373:1-6. [DOI] [PubMed] [Google Scholar]
- 85.Touati, D., M. Jacques, B. Tardat, L. Bouchard, and S. Despied. 1995. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 177:2305-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vergauwen, B., M. Herbert, and J. J. Van Beeumen. 2006. Hydrogen peroxide scavenging is not a virulence determinant in the pathogenesis of Haemophilus influenzae type b strain Eagan. BMC Microbiol. 6:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vergauwen, B., F. Pauwels, M. Vaneechoutte, and J. J. Van Beeumen. 2003. Exogenous glutathione completes the defense against oxidative stress in Haemophilus influenzae. J. Bacteriol. 185:1572-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang, X., P. Mukhopadhyay, M. J. Wood, F. W. Outten, J. A. Opdyke, and G. Storz. 2006. Mutational analysis to define an activating region on the redox-sensitive transcriptional regulator OxyR. J. Bacteriol. 188:8335-8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao, G., P. Ceci, A. Ilari, L. Giangiacomo, T. M. Laue, E. Chiancone, and N. D. Chasteen. 2002. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J. Biol. Chem. 277:27689-27696. [DOI] [PubMed] [Google Scholar]
- 90.Zheng, M., F. Aslund, and G. Storz. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279:1718-1721. [DOI] [PubMed] [Google Scholar]
- 91.Zheng, M., B. Doan, T. D. Schneider, and G. Storz. 1999. OxyR and SoxRS regulation of fur. J. Bacteriol. 181:4639-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.