Abstract
Spores of a Bacillus subtilis strain with a gerD deletion mutation (ΔgerD) responded much slower than wild-type spores to nutrient germinants, although they did ultimately germinate, outgrow, and form colonies. Spores lacking GerD and nutrient germinant receptors also germinated slowly with nutrients, as did ΔgerD spores in which nutrient receptors were overexpressed. The germination defect of ΔgerD spores was not suppressed by many changes in the sporulation or germination conditions. Germination of ΔgerD spores was also slower than that of wild-type spores with a pressure of 150 MPa, which triggers spore germination through nutrient receptors. Ectopic expression of gerD suppressed the slow germination of ΔgerD spores with nutrients, but overexpression of GerD did not increase rates of spore germination. Loss of GerD had no effect on spore germination induced by agents that do not act through nutrient receptors, including a 1:1 chelate of Ca2+ and dipicolinic acid, dodecylamine, lysozyme in hypertonic medium, a pressure of 500 MPa, and spontaneous germination of spores that lack all nutrient receptors. Deletion of GerD's putative signal peptide or change of its likely diacylglycerylated cysteine residue to alanine reduced GerD function. The latter findings suggest that GerD is located in a spore membrane, most likely the inner membrane, where the nutrient receptors are located. All these data suggest that, while GerD is not essential for nutrient germination, this protein has an important role in spores' rapid response to nutrient germinants, by either direct interaction with nutrient receptors or some signal transduction essential for germination.
The germination of spores of Bacillus species initiates the conversion of metabolically dormant spores into growing cells. Germination of Bacillus subtilis spores normally begins with the binding of specific nutrient germinants, either l-alanine or a mixture of l-asparagine, d-glucose, d-fructose, and potassium ions (AGFK), to specific receptors located in the spore's inner membrane (3, 18, 20, 30). Three functional nutrient receptors are found in B. subtilis spores, each encoded by the homologous tricistronic gerA, gerB, and gerK operons. The GerA nutrient receptor responds to l-alanine, while the GerB and GerK nutrient receptors cooperate in some fashion to respond to AGFK (18). Spores of gerA mutants are defective in germination with l-alanine, and spores of gerB or gerK mutants are defective in germination with AGFK. In addition, gerD and gerF mutants whose spores are affected in their germination with both l-alanine and AGFK have been identified (14, 18, 19, 20). Spores of gerD and gerF mutants have a defect early in germination, before the release of the spore's large depot of pyridine-2,6-dicarboxylic acid (dipicolinic acid [DPA]). GerF is a prelipoprotein diacylglycerol transferase necessary for nutrient receptor function (10, 11), but GerD function in spore germination is unknown.
GerD is an ∼20-kDa protein with homologs encoded in all sequenced genomes from Bacillus species (37) (Fig. 1), although not in the more distantly related Clostridium species. The GerD sequence is not homologous to that of any of the subunits of the spore's nutrient receptors. Transcription of gerD is at the same time as that of the gerA, gerB, and gerK operons and is induced only in the developing forespore compartment of the sporulating cell under the control of the forespore-specific sigma factor for RNA polymerase, σG (15). The precise location of GerD in the spore remains unclear, although there is a preliminary report that this protein is in the spore's outer layers (20). The amino acid sequence of GerD (37) includes a short N-terminal hydrophobic region that is likely a signal peptide (SP), followed by a recognition sequence for diacylglycerol addition to a specific cysteine residue (Fig. 1). The presence of these sequence features suggests that GerD is a membrane protein, and its forespore-specific expression is most consistent with its location being the spore's inner membrane.
FIG. 1.
Alignment of amino acid sequences of GerD from the various Bacillus species. The species compared were B. subtilis, Bacillus licheniformis, Geobacillus stearothermophilis, Bacillus anthracis, Bacillus thuringiensis, and Bacillus cereus. Gray shading indicates similar residues, and black shading indicates identical residues. The asterisk denotes the cysteine residue that is the putative site for diacylglycerol addition. The A and B over the sequences delineate the region of gerD coding sequence deleted and replaced with an antibiotic resistance gene in the ΔgerD strains used in this work.
As noted above, previous work indicates that gerD mutant spores are slow to germinate with both l-alanine and AGFK, although this is reported to be to different extents (14, 18, 35). The gerD mutant spores do germinate in l-alanine, albeit slower than wild-type spores; however, the decrease in the rate of gerD mutant spore germination compared to that of wild-type spores is much greater in AGFK (14, 18, 35). This finding, as well as the much-improved germination with l-alanine of gerD mutant spores prepared in a resuspension medium (RM), a defined minimal medium (21, 32), has led to the suggestion that GerD may not be an essential component of the spore germination apparatus (35). However, previous work on gerD mutant spores has used strains with either gerD point mutations or with a Tn917 insertion in the gerD coding sequence (35, 37). Since such mutants may retain partial GerD function, we have reexamined the gerD phenotype using strains in which a large portion of the gerD coding sequence has been deleted and replaced with an antibiotic resistance gene. We have also examined the importance of the likely SP and a putative site for diacylglycerol addition in GerD for the function of this protein.
MATERIALS AND METHODS
B. subtilis strains and their construction.
B. subtilis strains used in this study are listed in Table 1. All are isogenic derivatives of strain PS832, a prototrophic derivative of strain 168. New strains generated in this work were made via transformation with chromosomal or plasmid DNA (1). Mutations in various ger genes are deletions, with much of the genes' coding sequences replaced by antibiotic resistance genes (2, 10, 26). In the case of the ΔgerD strain, 35% of the gerD coding sequence has been deleted (Fig. 1) and replaced with a spectinomycin resistance (Spr) gene (10). Plasmid pVK73 (8), for replacing an Spr gene with a kanamycin resistance (Kmr) marker, was obtained from the B. subtilis Genetic Stock Center and was used to replace the Spr gene in strain PS3502 with a gene encoding Kmr, giving strain PS3946.
TABLE 1.
Bacillus subtilis strains used
| Strain | Genotype | Sourcea or reference |
|---|---|---|
| PS832 | Wild type | Laboratory stock |
| FB10 | gerBB* | 25 |
| FB62 | ΔgerD::spc | 10 |
| FB87 | ΔgerB::cat ΔgerK::erm | 26 |
| PS3476 | PsspD-gerA | 6 |
| PS3502 | PsspD-gerB* | 6 |
| PS3521 | gerBB* ΔgerA::neo | PS3609→FB10 |
| PS3609 | ΔgerA::neo | 10 |
| PS3651 | ΔgerA::neoΔgerK::erm | 2 |
| PS3665 | gerBB* ΔgerA::neo ΔgerK::erm | 2 |
| PS3926 | ΔgerA::neoΔgerD::spc | FB62→PS3609 |
| PS3927 | ΔgerB::cat ΔgerK::erm ΔgerD::spc | FB62→FB87 |
| PS3928 | ΔgerA::neo ΔgerB::cat ΔgerK::erm ΔgerD::spc | PS3609→PS3927 |
| PS3929 | gerBB* ΔgerD::spc | FB62→FB10 |
| PS3931 | gerBB* ΔgerA::neo ΔgerD::spc | FB62→PS3521 |
| PS3932 | gerBB* ΔgerA::neo ΔgerK::erm ΔgerD::spc | PS3657→PS3931 |
| PS3938 | ΔgerA::neo ΔgerK::erm ΔgerD::spc | PS3657→PS3931 |
| PS3940 | PsspD-gerA ΔgerD::spc | FB62→PS3476 |
| PS3945 | ΔgerA::neo ΔgerB::cat ΔgerK::erm | PS3609→FB87 |
| PS3946 | PsspD-gerB* | pVK73→PS3502 |
| PS3947 | PsspD-gerB* ΔgerD::spc | FB62→PS3946 |
| PS3980 | ΔgerD::cam | pΔgerD::cam→PS832 |
| PS3981 | gerD-spc | pgerDSpr→PS3980 |
| PS3982 | gerDC20A-spc | pgerDA20Spr→PS3980 |
| PS3983 | gerDΔSP-spc | pgerDΔSPSpr→PS3980 |
| PS3987 | ΔgerD amyE::gerD | pΔamyE::gerD→PS3980 |
| PS3988 | ΔgerD amyE::PsspB-gerD | pΔamyE::PsspB-gerD→PS3980 |
| PS3989 | amyE::gerD | pΔamyE::gerD→PS3981 |
| PS3990 | amyE::PsspB-gerD | pΔamyE::PsspB-gerD→PS3981 |
| PS3991 | amyE::gerDC20A | pΔamyE::gerDAla20→PS3981 |
| PS3992 | amyE::gerDΔSP | pΔamyE::gerDΔSP→PS3981 |
| PS3993 | gerDC20AamyE::gerD | pΔamyE::gerD→PS3982 |
| PS3994 | gerDC20AamyE::PsspB-gerD | pΔamyE::PsspB-gerD→PS3982 |
| PS3995 | gerDΔSPamyE::gerD | pΔamyE::gerD→PS3983 |
| PS3996 | gerDΔSPamyE::PsspB-gerD | pΔamyE::PsspB-gerD→PS3983 |
Strains made in this work were constructed by transformation of the strain to the right of the arrow with chromosomal or plasmid DNA from the strain to the left of the arrow. The abbreviation ΔSP denotes a gerD gene lacking the region coding for amino acids 7 to 17, which is a likely SP.
Preparation of B. subtilis strains expressing modified forms of GerD.
For construction of altered versions of gerD, a 1.2-kb fragment from 221 bp upstream to 421 bp downstream of the gerD translational start and stop codons, respectively, was PCR amplified (all primer sequences are available upon request) from B. subtilis PS832 genomic DNA. This fragment was digested with DraI (site upstream of gerD) and HindIII (site in extra nucleotides in the downstream primer) and cloned between the SmaI and HindIII sites in plasmid pUCΔ367 (12) (this plasmid is identical to pUC19 except that 367 bp between the SspI and NdeI sites has been deleted) in Escherichia coli JM109, giving plasmid pgerD. The 20th codon in gerD in this plasmid was then changed from TGC, encoding cysteine, to GCA, encoding alanine, by QuikChange II site-directed mutagenesis (Stratagene, Cedar Creek, TX), giving plasmid pgerDA20. A 266-bp fragment from bp 173 to 421 downstream of the gerD translation stop codon was PCR amplified from PS832 DNA and digested with SalI and HindIII (sites in the 5′ and 3′ PCR primers, respectively), and the fragment was cloned between the SalI and HindIII sites in plasmid pUC19, giving plasmid pBackgerD. A fragment containing bp 192 to 81 upstream of the gerD translation start codon was PCR amplified from pgerD. This fragment was digested with SacI and BamHI (SacI site originally from the multiple cloning site in pUC19; BamHI site in 3′ primer) and cloned between the SacI and BamHI sites in plasmid pBackgerD, giving plasmid pF/BgerD. This plasmid was digested with SalI and BamHI, and the small fragment removed was replaced with a 1.3-kb BamHI-SalI fragment containing a chloramphenicol resistance (Cmr) gene from plasmid pDG364 (9), giving plasmid pΔgerD::Cmr, in which the Cmr gene is flanked by sequences upstream and downstream of gerD but with the gerD coding sequence, promoter, and transcription terminator deleted.
To remove the hydrophobic region near the N terminus of GerD (amino acids 7 to 15) that is likely an SP, a 261-bp fragment containing the first 16 bp of the gerD coding sequence plus 195 bp of upstream sequence was PCR amplified from plasmid pgerD. This fragment was digested with SacI and HindIII (SacI site originally from the multiple cloning site in pUC19; HindIII site in the 3′ primer) and cloned in SacI-HindIII-digested pUCΔ367, giving plasmid pFrontSP. A 949-bp fragment encompassing bp 45 of the gerD coding sequence to 421 bp downstream of the gerD translation stop codon as well as extra residues at both ends, including appropriate restriction sites, was PCR amplified from plasmid pgerD, and after digestion with BglII and HindIII this fragment was cloned between the same sites in pFrontSP, giving plasmid pgerDΔSPR6, in which codons of gerD for 9 amino acids had been deleted while the sixth gerD codon had been mutagenized from a threonine codon to an arginine codon. Site-directed mutagenesis was then used to change this sixth codon to the original threonine codon, giving plasmid pgerDΔSP.
Plasmid pBackgerD was digested with BamHI and SalI, and a 1.1-kb BamHI-SalI fragment containing an Spr gene from plasmid pJL74 (16) was inserted, giving pBackgerDSpr. Plasmids pgerD, pgerDA20, and pgerDΔSP were used as substrates for PCR amplification of the gerD region from 192 bp upstream of the translation initiation codon to 144 bp downstream of the translation stop codon plus appropriate restriction sites in the PCR primers. These fragments were digested with SacI and BglII (SacI site originally from the multiple cloning site in pUC19; BglII site in the 3′ primer), and the resultant 0.9-kb fragments were cloned between the SacI and BamHI sites in plasmid pBackgerDSpr, giving plasmids pgerDSpr, pgerDA20Spr, and pgerDΔSPSpr, in which a 28-bp fragment beginning 75 bp downstream of gerD's putative transcription terminator had been removed and replaced with an Spr gene. For generation of B. subtilis strains carrying various gerD alleles at the gerD locus, strain PS3980 was first generated by transformation of strain PS832 to Cmr with ScaI-linearized plasmid pΔgerD::Cm. This strain was then transformed with the ScaI-linearized plasmids pgerDSpr, pgerDA20Spr, and pgerDΔSPSpr with selection for Spr, giving strains PS3981, PS3982, and PS3983. Spr transformants were further screened for those that had lost Cmr. In all cases, the appropriate chromosomal structure of the gerD locus in B. subtilis transformants was confirmed by PCR, and expected changes in the gerD sequence were confirmed by DNA sequencing.
Introduction of gerD and its variants at the amyE locus.
DNA fragments of ∼0.9 kb encompassing 203 bp upstream to 144 bp downstream of the gerD start and stop codons, respectively, were amplified by PCR from genomic DNA of B. subtilis strains PS832, PS3982, and PS3983. These fragments were digested with XhoI and BglII (sites within PCR primers), and the resultant ∼0.9-kb fragments were inserted between the XhoI and BamHI sites in plasmid pTI8a (11), giving plasmids pΔamyE::gerD, in which the gerD coding sequence with its promoter and putative transcription terminator sequences was inserted between the front and back regions of amyE, and plasmids pΔamyE::gerDAla20 and pΔamyE::gerDΔSP (where ΔSP denotes a gerD gene lacking the region coding for amino acids 7 to 17, which is a likely SP) which contain gerDC20A and gerDΔSP inserted in amyE, respectively. These plasmids were linearized with ScaI and used to transform B. subtilis strains PS3980 and PS3981 to Kmr, giving strains PS3987, PS3989, PS3991, PS3992, PS3993, and PS3995 (Table 1). For insertion of gerD at amyE but under the control of the strong forespore-specific promoter of the sspB gene (6), we utilized plasmid pTI8a (11), which contains the sspB promoter between segments of amyE. The full gerD coding sequence plus appropriate flanking restriction sites was PCR amplified from pgerD. This 724-bp fragment was digested with NdeI and BglII (sites in primers) and cloned between these sites in the 6.8-kb fragment from NdeI-BamHI-digested pTI8a, giving plasmid pΔamyE::PsspB-gerD. This plasmid was used to transform strains PS3980, PS3981, PS3982, and PS3983 to Kmr, giving strains PS3988, PS3990, PS3994, and PS3996, respectively. PCR and DNA sequencing confirmed the presence of the appropriate chromosomal structures at the amyE locus in these various transformants.
Spore preparation.
Spores were routinely prepared at 37°C on 2× SG medium agar plates without antibiotics (21). Since the composition of the sporulation medium has been reported to affect the GerD requirement for rapid spore germination with nutrients (35), spores were also prepared at 37°C on Spizizen's minimal medium (SMM) plates with 0.1% Casamino Acids or in liquid 2× SG medium, SMM plus 0.1% Casamino Acids, or RM (21, 31, 32). Spores were harvested, cleaned and stored as described previously (21, 22, 28). All spore preparations were >98% free of vegetative or sporulating cells, as determined by observation in a phase-contrast microscope.
Spore germination.
Prior to nutrient germination spores at an optical density at 600 nm (OD600) of 10 in water were heat activated at 70°C for 30 min and cooled on ice. Unless noted otherwise, germination was routinely at 37°C with spores at an initial OD600 of 1 in either Luria-Bertani (LB) medium (26), SMM (31) with either 10 mM l-alanine or AGFK (see below), or 2× YT medium (26) or in 25 mM Tris-HCl (pH 8.4) with either 10 mM l-alanine or a mixture of l-asparagine (2.5 mM), d-glucose (5 mg/ml), d-fructose (5 mg/ml), and KCl (50 mM) (AGFK). Alternatively, in one experiment l-alanine germination was in 11 mM l-alanine, 7 mg/ml of both KH2PO4 and Na2HPO4 (pH 7.25), 100 mM NaCl, and 200 mM KCl and AGFK germination was in 3 mM asparagine, 0.5 mg/ml glucose, 0.5 mg/ml fructose, and 50 mM KPO4 buffer (pH 7.4). Spore germination with l-alanine or AGFK was essentially identical under both the routine and alternative conditions. To examine the effect of pH on spore germination by l-alanine or AGFK the following buffers were used at 25 mM: succinate-NaOH (pH 6.0), Tris-HCl (pH 7.0 and 8.0), and glycine-NaOH (pH 9.0). In most cases spore germination was measured by monitoring the OD600 of spore cultures, which falls ∼60% upon complete spore germination (2). The extent of spore germination was also confirmed by phase-contrast microscopy. Spore germination by l-alanine and AGFK was also assessed by measuring DPA release as described below. The rate of spore germination was determined from the maximum rate of the fall of the OD600 of cultures as determined from the maximum slope of a plot of OD600 versus time (2, 6). All values reported are the averages of two experiments performed on two independent spore preparations, and individual values varied by less than 20% from average values shown. Note that since the rate of spore germination is expressed as percent/hour, if germination occurs rapidly, this value may be over 100%.
For assessment of very low rates of spore germination, glass microscope slides were coated with a thin layer of agar containing LB medium plus 10 mM l-alanine. Heat-activated spores (10-μl aliquots [∼105 spores]) containing kanamycin (50 μg/ml) to prevent cell growth were spotted on the slides, a glass coverslip was added and sealed with clear nail polish to prevent the slide from drying, and ∼100 spores on the slide were examined by phase-contrast microscopy during incubation at 37°C. Control experiments demonstrated that the addition of kanamycin did not affect rates of spore germination. RM spores were germinated similarly, but with the addition of only l-alanine or AGFK to the agar.
In some cases colony formation assays were used to assess spore germination, since mutations in gerD have no effects on cell growth rates (37) (data not shown). Spores at an OD600 of 1 (∼108 spores/ml) were heat activated as described above, and aliquots of various dilutions were spotted on LB medium agar plates, the plates were incubated at 37°C for 48 h, and colonies were counted. In another experiment, 200-μl aliquots of heat-activated spores containing ∼1.5 × 103 spores/ml were spread on LB medium agar plates, the plates were incubated at 37°C, and colonies were counted at various times. Spores were also germinated without heat activation at 37°C at an OD600 of 1 with Ca2+-DPA (60 mM CaCl2, 60 mM DPA [pH 8.0]) or at 37°C at an OD600 of 2 with dodecylamine (1 mM dodecylamine-20 mM KPO4 [pH 7.4]) as described previously (23, 28, 29). Spore germination with Ca2+-DPA was assessed by flow cytometry after staining with the fluorescent nucleic acid stain Syto 16 (Molecular Probes, Eugene, OR) (4). Spore germination with dodecylamine was analyzed by monitoring DPA release by measuring the OD270 of the supernatant fluid from germination incubations (29). Aliquots (1 ml) of germinating cultures were removed and centrifuged for 2 min in a microcentrifuge to obtain the supernatant fluid. The total DPA content of spores was determined by boiling samples for 30 min, centrifuging, and measuring the OD270 of the supernatant fluid. Previous work has shown that DPA comprises ∼85% of the material absorbing at 270 nm released from spores by boiling and >95% of the material absorbing at 270 nm released during spore germination (2, 6).
Germination was also performed on non-heat-activated spores at an OD600 of 1 in 50 mM Tris-HCl (pH 7.5) buffer either at 37°C with 150 MPa of pressure or at 50°C with 500 MPa of pressure (4, 5, 24, 36). The extent of spore germination due to pressure was analyzed by flow cytometry after staining with Syto 16 (4).
For germination with lysozyme, spores at an OD600 of 100 were first decoated for 30 min at 45°C in 0.1 M NaCl-0.1 M NaOH-0.5% sodium dodecyl sulfate-0.1 M dithiothreitol (34). The decoated spores were washed five to seven times with 0.15 M NaCl, resuspended in water at an OD600 of 10, and stored at 4°C. Washed decoated spores were resuspended at an OD600 of 1 in 1 ml hypertonic germination medium (0.3 M sucrose-10 mM Tris-HCl [pH 8.0]-10 mM MgCl2-10 mM CaCl2), germination was initiated by addition of 5 μl of 5 mg/ml lysozyme, and the suspension was incubated at 37°C (27). Germination was monitored by measuring the OD600 of the suspensions, as well as by phase-contrast microscopy.
RESULTS
Response of gerD spores to nutrients.
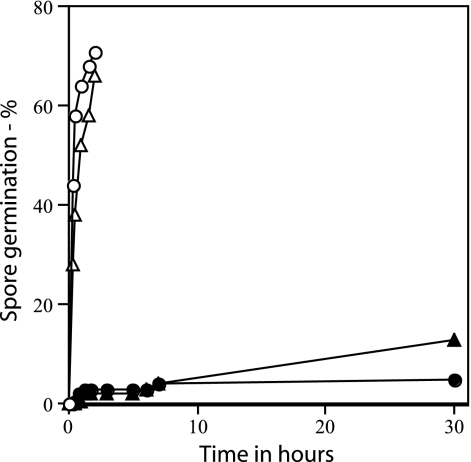
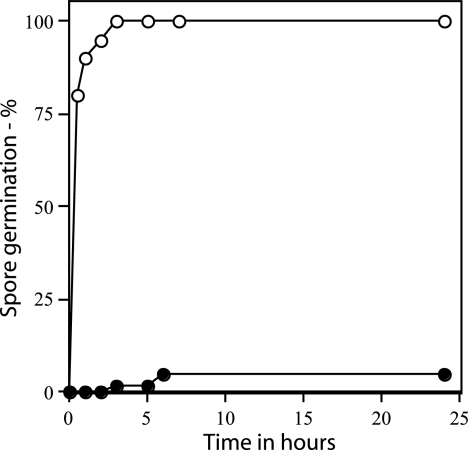
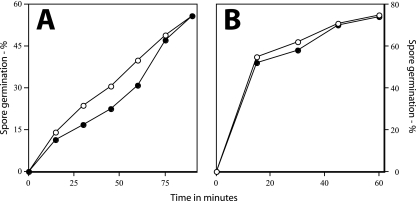
As found previously with spores of strains with point or insertion mutations in gerD, rates of germination of spores with a deletion mutation in gerD were much lower than that of wild-type spores using either AGFK or l-alanine to trigger spore germination (Table 2) (14, 18, 19, 35). The rate of germination of gerD spores in LB medium was also low (Table 2). The slow germination of ΔgerD spores with l-alanine or AGFK was observed not only when monitoring spore germination by measuring the OD600 of the culture but also by measuring DPA release (Fig. 2). To examine spore germination over even longer times in LB medium plus alanine, we monitored spore germination by phase-contrast microscopy. Again, while wild-type spores germinated completely in a few hours under these conditions, ΔgerD spores germinated more slowly (Fig. 3).
TABLE 2.
Germination of gerD spores with various genetic backgroundsa
| Genetic background (strainsb) | Germinant | Rate of germination (%/h)
|
|
|---|---|---|---|
| With gerD | Without gerD | ||
| Wild type (PS832/FB62) | Ala | 110 | <5 |
| Wild type (PS832/FB62) | AGFK | 60 | <5 |
| Wild type (PS832/FB62) | LB medium | 87 | <5 |
| ΔgerA (PS3609/PS3926) | Ala | <5 | <5 |
| ΔgerA (PS3609/PS3926) | AGFK | 61 | 6 |
| ΔgerA (PS3609/PS3926) | LB medium | <12 | <5 |
| ΔgerK ΔgerB (FB87/PS3927) | Ala | 105 | 6 |
| ΔgerK ΔgerB (FB87/PS3927) | AGFK | 6 | <5 |
| ΔgerK ΔgerB (FB87/PS3927) | LB medium | 73 | 12 |
| gerBB* (FB10/PS3929) | Ala | 239 | 15 |
| gerBB* (FB10/PS3929) | AGFK | 219 | 10 |
| gerBB* ΔgerA (PS3521/PS3931) | Ala | 83 | 5 |
| gerBB* ΔgerA (PS3521/PS3931) | AGFK | 271 | 14 |
| gerBB* ΔgerA ΔgerK (PS3665/PS3932) | Ala | 104 | 7 |
| gerBB* ΔgerA ΔgerK (PS3665/PS3932) | AGFK | 78 | 7 |
| PsspD-gerA (PS3476/PS3940) | Ala | 200 | 23 |
| PsspD-gerA (PS3476/PS3940) | AGFK | 28 | 5 |
| PsspD-gerB* (PS3946/PS3947) | Ala | 247 | 6 |
| PsspD-gerB* (PS3946/PS3947) | AGFK | 257 | <5 |
Spore germination was induced by addition of l-alanine (Ala), AGFK, or LB medium; germination was measured by monitoring the OD600 of cultures, and rates of spore germination were calculated as described in Materials and Methods.
The first strain listed contains gerD, and the second has the same genetic background but without gerD.
FIG. 2.
Rates of germination of wild-type and ΔgerD spores. Wild-type (PS832) and ΔgerD (FB62) spores were germinated with either l-alanine or AGFK, and germination was assessed by measuring DPA release as described in Materials and Methods. ○, wild-type spores with l-alanine; ▵, wild-type spores with AGFK; ▴, ΔgerD spores with l-alanine; •, ΔgerD spores with AGFK.
FIG. 3.
Rates of germination of wild-type and gerD spores measured by microscopy. Wild-type (PS832) and ΔgerD (FB62) spores were spotted on LB medium agar slides with l-alanine and kanamycin, and spore germination was assessed by examining ∼100 spores by phase-contrast microscopy as described in Materials and Methods. ○, wild-type spores; •, ΔgerD spores.
Colony formation by ΔgerD spores.
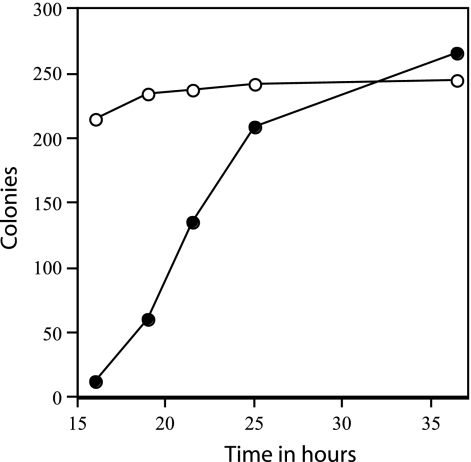
The slow germination of the ΔgerD spores noted above suggested that these spores might have a lower efficiency of colony formation than wild-type spores. However, this was not the case, as both wild-type and ΔgerD spores prepared in several different media had very similar efficiencies of colony formation, when colonies were counted after 48 h (Table 3). However, it was clear in this experiment that colonies from ΔgerD spores arose more slowly than from wild-type spores. The rate of appearance of colonies from heat-activated spores was determined by counting the colonies that appeared at various times from approximately identical numbers of spores, both ΔgerD and wild type, that were spread on agar plates and incubated at 37°C. While the number of colonies from wild-type spores was almost maximal after ≤16 h of incubation, the rate of appearance of colonies from ΔgerD spores was much lower (Fig. 4). The rates of appearance of colonies from ΔgerD spores were similar whether or not a heat activation step was applied, and this was also the case with wild-type spores (data not shown).
TABLE 3.
Efficiency of colony formation by wild-type and ΔgerD sporesa
| Sporulation medium | Avg no. of colonies
|
|
|---|---|---|
| Wild type | ΔgerD | |
| 2× SG | 2.1 × 108 | 1.9 × 108 |
| SMM plus 0.1% Casamino Acids | 1.5 × 108 | 1.2 × 108 |
| RM | 6 × 107 | 6.9 × 107 |
Spores of strains PS832 (wild type) and FB62 (ΔgerD) were prepared using various media. The spores were purified, and aliquots of various dilutions of spore suspensions with an OD600 of 1 (1 × 108 to 2 × 108 spores/ml) were spotted on LB medium agar plates. The plates were incubated for 48 h at 37°C, and colonies were counted. Values shown are the averages of duplicate determinations.
FIG. 4.
Rates of appearance of colonies from wild-type and ΔgerD spores. Two hundred fifty to 300 spores of strains PS832 (wild type) or FB62 (ΔgerD) were spread on an LB medium agar plate without antibiotics, the plates were incubated at 37°C, and colonies were counted after various times of incubation. ○, wild-type spores; •, ΔgerD spores.
Germination of ΔgerD spores lacking or with elevated levels of nutrient germinant receptors.
The gerD mutation reduced the rate of nutrient germination not only of spores with an otherwise wild-type background but also in spores lacking the GerA nutrient receptor, or both the GerB and GerK nutrient receptors (Table 2). The gerD mutation also reduced the rate of germination of spores carrying a point mutation in the gerBB cistron of the gerB operon, termed the gerBB* mutation. This mutation generates a GerB* nutrient receptor that functions even in the absence of the GerK nutrient receptor to trigger spore germination in response to either l-alanine or l-asparagine alone (2, 25).
Since a gerD mutation decreased spore germination triggered by any of the spore's nutrient receptors, a possible mechanism of GerD action is that this protein either positively regulates nutrient receptor formation or increases the function of individual nutrient receptors. If either of these explanations is correct, increasing the levels of nutrient receptors in spores might suppress the effects of a gerD mutation. In previous work we have constructed strains that have 10- to 20-fold-higher levels of GerA or GerB* receptors in spores by placing the gerA and gerB* operons under the control of the moderately strong forespore-specific promoter of the sspD gene (6). Although overexpression of these receptors increased the rates of spore germination with appropriate nutrients, as shown previously (6), this did not suppress the effects of the gerD mutation on spore germination (Table 2). Thus it is unlikely that GerD acts by increasing either the function or level of individual nutrient receptors.
Spores lacking all three functional nutrient receptors do not respond to nutrient germinants (26). However, the spores of such strains, termed ger3 spores, exhibit a very low rate of spontaneous germination (0.01 to 0.1%/day), and this low rate of germination continues for at least a week (26). It was thus of interest to examine the effect of a gerD mutation on this spontaneous spore germination. Similar to results found previously (26), ∼0.03% of the spores of a ger3 strain (PS3945) germinated per day at 37°C, while approximately the same percentage (0.06%) of spores of an isogenic strain also lacking gerD (PS3928; ger3 ΔgerD) germinated per day under the same conditions (data not shown). Thus a gerD mutation did not slow the rate of spontaneous germination of spores lacking all functional nutrient receptors.
Germination of ΔgerD spores prepared in different media or under different conditions.
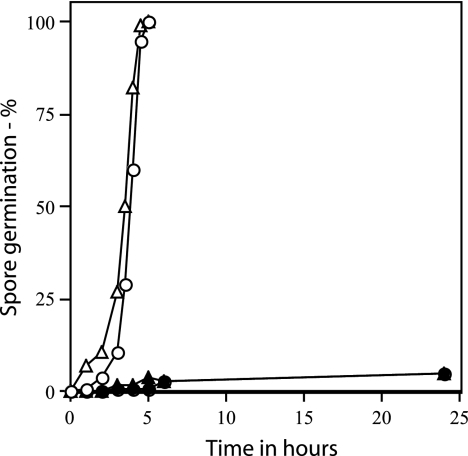
Previous studies have suggested that sporulation conditions may affect the ability of gerD mutant spores to germinate, as sporulation of gerD point mutants in RM yields spores that germinate with l-alanine at rates much closer to those of wild-type spores (35, 37). However, we observed a severe germination defect in l-alanine with ΔgerD spores that had been prepared in RM (Table 4). This was also true of ΔgerD spores prepared in SMM with 0.1% Casamino Acids, another more minimal medium (Table 4). While initial experiments with spores prepared in RM measured spore germination by monitoring the OD600 of cultures (Table 4), we also examined the nutrient germination of these spores over a longer time period using phase-contrast microscopy; again there was only slow germination of the ΔgerD spores prepared in RM (Fig. 5). In addition to modifications in the sporulation medium, we also examined the effects of preparation of ΔgerD spores in liquid 2× SG medium or SMM plus 0.1% Casamino Acids, as well as on solid 2× SG medium at temperatures from 23 to 44°C, as these changes have been found to affect spore properties, including rates of spore germination (17, 29). However, again these changes did not suppress the defect in the germination of ΔgerD spores with nutrients (data not shown).
TABLE 4.
Germination of ΔgerD and wild-type spores prepared and germinated in various mediaa
| Germinant | Rate of spore germination (%/h) for indicated strainb in:
|
|||||
|---|---|---|---|---|---|---|
| 2× SG
|
RM
|
SMM + CAA
|
||||
| PS832 | FB62 | PS832 | FB62 | PS832 | FB62 | |
| LB medium | 87 | <5 | NDc | ND | ND | ND |
| Ala | ND | ND | 34 | <5 | ND | ND |
| AGFK | ND | ND | 56 | <5 | ND | ND |
| SMM plus Ala | ND | ND | ND | ND | 64 | <5 |
| SMM plus AGFK | ND | ND | ND | ND | 90 | <5 |
Spores were prepared in 2× SG medium, RM, or SMM plus 0.1% Casamino Acids (CAA), and spore germination was carried out with either LB medium, L-alanine (Ala), AGFK, or SMM with 10 mM L-alanine or with AGFK. Spore germination was measured by monitoring the OD600 of cultures as described in Materials and Methods. Rates of germination are given as the percentages of spores that germinate in 1 hour as described in Materials and Methods.
PS832 is the wild type, and FB62 lacks gerD.
ND, not done.
FIG. 5.
Rates of germination of wild-type and ΔgerD spores prepared in RM. Wild-type (PS832) and ΔgerD (FB62) spores prepared in RM were spotted on agar slides with kanamycin and either l-alanine or AGFK, and spore germination was assessed by examining ∼100 spores under phase-contrast microscopy as described in Materials and Methods. ○, wild-type spores with l-alanine; ▵, wild-type spores with AGFK; •, ΔgerD spores with l-alanine; ▴, ΔgerD spores with AGFK.
Effects of monovalent cations, pH, and temperature on ΔgerD spore germination.
One model for spore germination has suggested that spores require a pool of K+ ions before germination can occur (35). Indeed work using spores with point mutations in gerD found that addition of monovalent cations to the germination medium suppresses the gerD mutant spore's germination defect (35, 37). However with ΔgerD spores addition of KCl, NaCl, LiCl, or NH4Cl at concentrations up to 250 mM did not suppress the germination defect with either l-alanine or AGFK (data not shown). We also checked whether other changes in germination conditions might alter the GerD requirement for rapid spore germination with nutrients. However, varying the germination pH from 6 to 9 or the temperature from 23 to 44°C did not suppress the germination defect of the ΔgerD spores (data not shown). Increasing the l-alanine concentration from 10 to 100 mM to determine if ΔgerD spores have decreased sensitivity to nutrient germinants or removing the spore coats to determine if ΔgerD spores are defective in permeation of nutrient germinants (3) also did not suppress the nutrient germination defect of ΔgerD spores (data not shown). Under all of these conditions, the rates of germination of ΔgerD spores with either alanine or AGFK remained at ≤10% of those of wild-type spores (data not shown).
Germination of ΔgerD spores with nonnutrient germinants.
While a gerD deletion mutation slowed spore germination triggered by nutrients, there are many nonnutrient agents that can trigger spore germination, including cationic surfactants such as dodecylamine, lysozyme, Ca2+-DPA, and high pressures (4, 5, 23, 24, 26, 27, 29, 30). Many of these agents (dodecylamine, lysozyme, and Ca2+-DPA) trigger spore germination by pathways different from that for nutrient germination. Examining the effects of this group of nonnutrient germinants showed that rates of spore germination with dodecylamine as measured by DPA release were nearly identical for both wild-type and ΔgerD spores (Fig. 6A). This was also the case for germination in hypertonic medium with lysozyme (data not shown) as well as with Ca2+-DPA (Fig. 6B).
FIG. 6.
Rates of germination of wild-type and ΔgerD spores with (A) dodecylamine or (B) Ca2+-DPA. Wild-type (PS832) and ΔgerD (FB62) spores were germinated with 1 mM dodecylamine (A) or with Ca2+-DPA (B), and spore germination was measured either by following DPA release (dodecylamine germination) or flow cytometry after staining with Syto 16 (Ca2+-DPA germination), as described in Materials and Methods. ○, wild-type spores; •, ΔgerD spores.
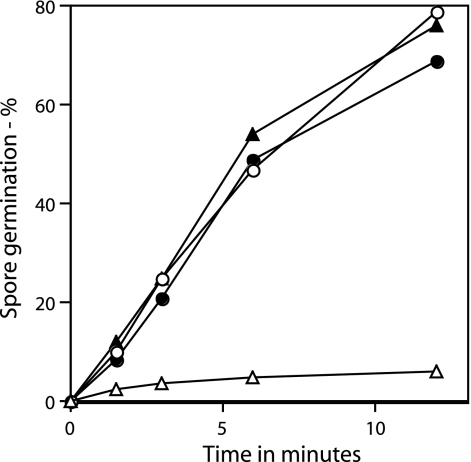
Previous work has established that different high pressures trigger spore germination via different pathways (4, 5, 24, 36). Pressures of about 150 MPa induce germination primarily via the nutrient receptor-mediated pathway, while pressures of about 500 MPa induce germination primarily via mechanisms independent of the nutrient receptors. Since ΔgerD spores are slowed in nutrient receptor-mediated germination, we would expect that germination of ΔgerD spores at 150 MPa of pressure would also be slow, which is what was found (Fig. 7). In contrast, since spore germination with 500 MPa of pressure does not proceed through the nutrient receptors, we would expect to see nearly wild-type spore germination rates with ΔgerD spores under these conditions, and this is again what was observed (Fig. 7). These results are consistent with a previous pressure germination study with spores of a gerD point mutant (36).
FIG. 7.
Germination of wild-type and ΔgerD spores by high pressures. Wild-type (PS832) and ΔgerD (FB62) spores were germinated with pressure, and spore germination was assessed by measuring DPA release as described in Materials and Methods. ○, wild-type spores germinated with 150 MPa of pressure; •, wild-type spores germinated with 500 MPa of pressure; ▵, ΔgerD spores germinated with 150 MPa of pressure; ▴, ΔgerD spores germinated with 500 MPa of pressure.
Effects of modifications in the GerD level or amino acid sequence on spore germination.
As noted above, GerD contains a likely SP followed by a putative recognition site for diacylglycerol addition, including the cysteine residue to which the diacylglycerol is added. One subunit of the spore's nutrient receptors also contains both of these sequence features, and the change of the diacylglycerylated cysteine to an alanine eliminates the function of the GerA nutrient receptor and greatly reduces GerB nutrient receptor function (10). Thus it was of obvious interest to determine the importance of these GerD sequence features. Perhaps not surprisingly, deletion of GerD's likely SP or the change of its putative diacylglycerylated cysteine to an alanine destroyed GerD function in spore germination with either l-alanine or AGFK (Table 5). However, when the latter mutant genes were at the gerD locus, expression of wild-type GerD from the amyE locus restored GerD function, as did expression of gerD from amyE in a strain with a gerD deletion at the gerD locus (Table 5). As expected, expression of the altered gerD genes from the amyE locus did not interfere with germination of spores that had wild-type gerD at the gerD locus (Table 5).
TABLE 5.
Germination of spores of strains with different gerD genes in various locationsa
| Strain | Relevant genotypeb | Relative rate of spore germinationc with:
|
|
|---|---|---|---|
| L-Alanine | AGFK | ||
| PS3980 | ΔgerD | ≤8 | ≤8 |
| PS3981 | Wild type | 100 | 100 |
| PS3982 | gerDC20A | ≤8 | ≤4 |
| PS3983 | gerDΔSP | ≤8 | ≤4 |
| PS3987 | ΔgerD amyE::gerD | 105 | 85 |
| PS3988 | ΔgerD amyE::PsspB-gerD | 105 | 85 |
| PS3989 | amyE::gerD | 95 | 95 |
| PS3990 | amyE::PsspB-gerD | 100 | 95 |
| PS3991 | amyE::gerDC20 | 105 | 110 |
| PS3992 | amyE::gerDΔ7-15 | 100 | 110 |
| PS3993 | gerDC20AamyE::gerD | 105 | 105 |
| PS3994 | gerDC20AamyE::PsspB-gerD | 100 | 110 |
| PS3995 | gerDΔSPamyE::gerD | 100 | 95 |
| PS3996 | gerDΔSPamyE::PsspB-gerD | 105 | 80 |
Spores prepared at 37°C on 2× SG plates were germinated using the alternative conditions with either L-alanine or AGFK, and the rates of spore germination were determined by measuring the rate of DPA release as described in Materials and Methods. All values reported are the averages of results with two preparations of spores and have been rounded off to the nearest 5, except for values less than 10.
The abbreviation ΔSP denotes a gerD gene lacking the region coding for amino acids 7 to 17, which is a likely SP.
All values are expressed relative to that for spores of strain PS3981, which was set at 100.
The fact that wild-type gerD expressed at amyE could restore GerD function indicated that functional GerD was expressed from gerD at amyE. Previous work has shown that overexpression of various nutrient receptors can have significant effects on both sporulation and spore germination (6, 11). However, when gerD was expressed at amyE under the control of either the gerD promoter or the very strong forespore-specific sspB promoter, the resultant strains sporulated normally (data not shown) and the spores of these strains exhibited rates of germination very similar to those of wild-type spores (Table 5). While we do not know the degree to which GerD was overexpressed in spores of the strain with gerD under PsspB control, previous work has found that placing the gerB operon under PsspB control results in a 200-fold increase in levels of GerBA (27).
DISCUSSION
The results with a ΔgerD mutation presented in this work revealed a greater effect of GerD on spore germination than observed previously, as our ΔgerD spores were very slow to germinate with all nutrients tested. This was the case irrespective of the medium used for spore preparation or the presence or absence of various cations in the germination solutions. The smaller effect(s) on spore germination of gerD mutations seen in earlier work, in particular with l-alanine as the germinant (18, 35, 37), is perhaps due to the retention of some function by the products of the mutant gerD genes used previously, as has been suggested for the gerD point mutants (37). The differences in the behavior of the different gerD mutations could also be due to differences in the genetic backgrounds of the B. subtilis 168 strains used in different laboratories.
It was somewhat surprising that the rate of germination of ΔgerD spores inferred from the rate of appearance of colonies from ΔgerD spores appeared to be significantly higher than the rate of ΔgerD spore germination seen either by monitoring DPA release or phase-contrast microscopy. While the reason for the differences between these different experiments is not clear, these experiments were carried out under significantly different conditions. One such difference is the much higher spore concentrations in experiments measuring germination by monitoring DPA release or phase-contrast microscopy compared to monitoring colony formation. It has been reported with spores of several species that the spore concentration can have a significant effect on the rate of germination, although there has usually been more rapid germination by spores at higher concentrations (7, 38). However, this phenomenon has not been studied with B. subtilis spores, and perhaps higher spore concentrations of B. subtilis ΔgerD spores result in inhibition of spore germination. Another significant difference between the various experiments is that in measuring spore germination by phase-contrast microscopy the spores were on only a thin layer of agar, and again this may have resulted in a lower rate of spore germination. Whatever the precise explanation for the differences in the rates of germination of ΔgerD spores in various experiments, it is clear that the rate of ΔgerD spore germination is significantly lower than that of wild-type spores, although within ∼48 h ΔgerD spores germinate as well as wild-type spores.
Since GerD appears to play a significant, yet not absolutely essential role in spore germination, the important question is how GerD functions to stimulate the rate of spore germination. Knowledge of proteins involved in spore germination is currently confined largely to (i) the nutrient receptors that bind nutrient germinants, with this binding leading to subsequent germination events (30); (ii) the redundant enzymes that digest the spore's peptidoglycan cortex in response to some signal from the nutrient germinant receptors (30); (iii) the GerP proteins that may be responsible for allowing nutrient germinants to pass through the spore's outer layers to reach their receptors (3); and (iv) the SpoVA proteins that may be involved in Ca2+-DPA release upon activation of the spore's nutrient receptors (33). Since gerD mutants are blocked in or before Ca2+-DPA release and cortex hydrolysis is not required for Ca2+-DPA release (30), GerD must act before cortex hydrolysis and either on nutrient access to their receptors, on the activity of or the signal transduction from nutrient receptors, or on Ca2+-DPA release. However, it seems unlikely that GerD is involved in allowing access of nutrients to their receptors, since l-alanine concentrations >100-fold higher than needed for maximal rates of spore germination did not suppress the effect of the gerD mutation. Similarly, decoating of spores, a process that suppresses effects of gerP mutations on spore germination (3), also did not suppress the effects of the gerD mutation.
It also seems unlikely that GerD functions directly in the release of Ca2+-DPA that takes place early in germination, since Ca2+-DPA release also takes place during germination induced by nonnutrient germinants such as dodecylamine, exogenous Ca2+-DPA, and a pressure of 500 MPa, yet the rates of germination of ΔgerD spores with these agents were essentially equal to those of wild-type spores. We are left then with GerD function involving the spore's nutrient receptors in some fashion. That GerD does affect the nutrient receptors or their signaling in some way is consistent with the finding that germination with all agents that require these receptors is greatly slowed by the gerD mutation, while germination with all agents that do not require these receptors is not. How then may GerD function to increase the rate of germination caused by activation of nutrient receptors? As noted above, this seems unlikely to be due to allowing easier access of nutrient germinants to their receptors in the spore's inner membrane. Indeed, spore germination due to a pressure of 150 MPa, a process that requires no nutrient germinant yet proceeds through the nutrient receptors (4, 24, 36), is greatly slowed by the gerD mutation. It also seems unlikely that GerD increases the level of functional nutrient receptors, since overexpression of nutrient receptors did not suppress the effects of the gerD mutation. However, we cannot rule out the possibility that GerD somehow either modifies the nutrient receptors to make them more sensitive to their ligands or alters the sporulation process itself such that the responsiveness of the nutrient receptors is increased.
Analysis of the precise effects of GerD on nutrient germination strongly suggests that GerD is not essential for spore germination via the nutrient receptors, as ΔgerD spores germinate, outgrow, and eventually form colonies on plates; this process is just slower than that with wild-type spores. The difference between ΔgerD and wild-type spores in their germination with nutrients appears to be in the duration of the lag time after exposure of spores to nutrients and before initiation of obvious germination processes such as Ca2+-DPA release, with this lag time being much longer with ΔgerD spores. The lag times for ΔgerD spores also are extremely variable for individual spores, as colonies from ΔgerD spores appeared over a long time period, much longer than that for wild-type spores. While the presence of a lag period after exposure of spores to nutrients and prior to measurable germination events has been seen with spores of a number of Bacillus species, the events taking place in this lag period are largely unknown and factors that determine the length of this lag period are not understood. However, one event determining the duration of this lag period could be the efficiency of transduction of signals from ligand-activated nutrient receptors to components acting later in the spore germination process, such as enzymes that degrade the peptidoglycan cortex. Thus it is attractive to adopt as at least a working model that GerD is involved in some fashion in the transduction of a signal of some sort between the nutrient receptors and some other component of the spore germination apparatus.
If GerD is involved in the latter signal transduction process, it seems likely that GerD would be located near the nutrient receptors in the spore's inner membrane. The presence of an essential SP and a cysteine that is probably diacylglycerylated suggests that GerD is likely a membrane protein, probably a peripheral membrane protein since most of the protein is hydrophilic (37). Since gerD is transcribed only in the developing forespore, GerD is most likely present in the spore's inner membrane, and attempts to definitively localize GerD in the spore are in progress. If, as seems likely, GerD is in the inner membrane, then a more intriguing question is what other proteins physically interact with GerD. The answer to this question may provide crucial information not only about GerD function but also about the overall signal transduction pathway in spore germination.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (GM19698) to P. Setlow and by a grant from the USDA to D. Hoover and P. Setlow.
Footnotes
Published ahead of print on 22 November 2006.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:74-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atluri, S., K. Ragkousi, D. E. Cortezzo, and P. Setlow. 2006. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J. Bacteriol. 188:28-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behravan, J., H. Chirakkal, A. Masson, and A. Moir. 2000. Mutations in the gerP locus of Bacillus subtilis and Bacillus cereus affect access of germinants to their targets in spores. J. Bacteriol. 182:1987-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, E. P., K. Koziol-Dube, D. Guan, J. Wei, B. Setlow, D. E. Cortezzo, D. Hoover, and P. Setlow. 2005. Factors influencing germination of Bacillus subtilis spores via activation of nutrient receptors by high pressure. Appl. Environ. Microbiol. 71:5879-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, E. P., J. Wei, S. Atluri, D. E. Cortezzo, K. Koziol-Dube, D. G. Hoover, and P. Setlow. 2006. Analysis of factors influencing the rate of germination of spores of Bacillus subtilis by very high pressure. J. Appl. Microbiol. 102:65-76. [DOI] [PubMed] [Google Scholar]
- 6.Cabrera-Martinez, R.-M., F. Tovar-Rojo, V. R. Vepachedu, and P. Setlow. 2003. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J. Bacteriol. 185:2457-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caipo, M. L., S. Duffy, L. Zhao, and D. W. Schaffner. 2002. Bacillus megaterium spore germination is influenced by inoculum size. J. Appl. Microbiol. 92:879-884. [DOI] [PubMed] [Google Scholar]
- 8.Chary, V. C., E. I. Amaya, and P. J. Piggot. 1997. Neomycin- and spectinomycin-resistance replacement vectors for Bacillus subtilis. FEMS Microbiol. Lett. 153:135-139. [DOI] [PubMed] [Google Scholar]
- 9.Cutting, S. C., and P. B. Vander Horn. 1990. Genetic analysis, p. 27-60. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 10.Igarashi, T., B. Setlow, M. Paidhungat, and P. Setlow. 2004. Effects of a gerF mutation on the germination of spores of Bacillus subtilis. J. Bacteriol. 186:2984-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Igarashi, T., and P. Setlow. 2005. Interaction between individual protein components of the GerA and GerB nutrient receptors that trigger germination of Bacillus subtilis spores. J. Bacteriol. 187:2513-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Igarashi, T., and P. Setlow. 2006. Transcription of the Bacillus subtilis gerK operon, which encodes a spore germinant receptor, and comparison with that of operons encoding other germinant receptors. J. Bacteriol. 188:4131-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irie, R., T. Okamoto, and Y. Fujita. 1982. A germination mutant of Bacillus subtilis deficient in response to glucose. J. Gen. Appl. Microbiol. 28:345-354. [Google Scholar]
- 14.Irie, R., T. Okamoto, and Y. Fujita. 1986. Characterization and mapping of Bacillus subtilis gerD mutants. J. Gen. Appl. Microbiol. 32:303-315. [Google Scholar]
- 15.Kemp, E. H., R. L. Sammons, A. Moir, D. Sun, and P. Setlow. 1991. Analysis of transcriptional control of the gerD spore germination gene of Bacillus subtilis 168. J. Bacteriol. 173:4646-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LeDeaux, J. R., and A. L. Grossman. 1995. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J. Bacteriol. 177:166-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melly, E., P. C. Genest, M. E. Gilmore, S. Little, D. L. Popham, A. Driks, and P. Setlow. 2002. Analysis of the properties of spores of Bacillus subtilis prepared at different temperatures. J. Appl. Microbiol. 92:1105-1115. [DOI] [PubMed] [Google Scholar]
- 18.Moir, A., and D. A. Smith. 1990. The genetics of bacterial spore germination. Annu. Rev. Microbiol. 44:531-553. [DOI] [PubMed] [Google Scholar]
- 19.Moir, A., E. Lafferty, and D. A. Smith. 1979. Genetic analysis of spore germination mutants of Bacillus subtilis 168: the correlation of phenotype with map location. J. Gen. Microbiol. 111:165-180. [DOI] [PubMed] [Google Scholar]
- 20.Moir, A., E. H. Kemp, C. Robinson, and B. M. Corfe. 1994. The genetic analysis of bacterial spore germination. J. Appl. Bacteriol. 76:9S-16S. [PubMed] [Google Scholar]
- 21.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 22.Paidhungat, M., B. Setlow, A. Driks, and P. Setlow. 2000. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J. Bacteriol. 182:5505-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paidhungat, M., K. Ragkousi, and P. Setlow. 2001. Genetic requirements for induction of germination of spores of Bacillus subtilis by Ca2+-dipicolinate. J. Bacteriol. 183:4886-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paidhungat, M., B. Setlow, W. B. Daniels, D. Hoover, E. Papafragkou, and P. Setlow. 2002. Mechanisms of initiation of germination of spores of Bacillus subtilis by pressure. Appl. Environ. Microbiol. 68:3172-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paidhungat, M., and P. Setlow. 1999. Isolation and characterization of mutations in Bacillus subtilis that allow spore germination in the novel germinant d-alanine. J. Bacteriol. 181:3341-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paidhungat, M., and P. Setlow. 2000. Role of Ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 182:2513-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popham, D. L., J. Helin, C. E. Costello, and P. Setlow. 1996. Analysis of the peptidoglycan structure of Bacillus subtilis endospores. J. Bacteriol. 178:6451-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ragkousi, K., P. Eichenberger, C. van Ooij, and P. Setlow. 2003. Identification of a new gene essential for germination of Bacillus subtilis spores with Ca2+-dipicolinate. J. Bacteriol. 185:2315-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Setlow, B., A. E. Cowan, and P. Setlow. 2003. Germination of spores of Bacillus subtilis with dodecylamine. J. Appl. Microbiol. 95:637-648. [DOI] [PubMed] [Google Scholar]
- 30.Setlow, P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550-556. [DOI] [PubMed] [Google Scholar]
- 31.Spizizen, J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA 44:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tovar-Rojo, F., M. Chander, B. Setlow, and P. Setlow. 2002. The products of the spoVA operon are involved in dipicolinic acid uptake into developing spores of Bacillus subtilis. J. Bacteriol. 184:584-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vary, J. C. 1973. Germination of Bacillus megaterium spores after various extraction procedures. J. Bacteriol. 116:797-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warburg, R. J., A. Moir, and D. A. Smith. 1985. Influence of alkali metal cations on the germination of spores of wild-type and gerD mutants of Bacillus subtilis. J. Gen. Microbiol. 131:221-230. [Google Scholar]
- 36.Wuytack, E. Y., J. Soons, F. Poschet, and C. W. Michiels. 2000. Comparative study of pressure- and nutrient-induced germination of Bacillus subtilis spores. Appl. Environ. Microbiol. 66:257-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yon, J. R., R. L. Sammons, and D. A. Smith. 1989. Cloning and sequencing of the gerD gene of Bacillus subtilis. J. Gen. Microbiol. 135:3434-3445. [DOI] [PubMed] [Google Scholar]
- 38.Zhao, L., T. J. Montville, and D. W. Schaffner. 2000. Inoculum size of Clostridium botulinum 56A spores influences time-to-detection and percent growth-positive samples. J. Food Sci. 65:1369-1375. [Google Scholar]