Abstract
Most secreted archaeal proteins are targeted to the membrane via a tripartite signal composed of a charged N terminus and a hydrophobic domain, followed by a signal peptidase-processing site. Signal peptides of archaeal flagellins, similar to class III signal peptides of bacterial type IV pilins, are distinct in that their processing sites precede the hydrophobic domain, which is crucial for assembly of these extracytoplasmic structures. To identify the complement of archaeal proteins with class III signal sequences, a PERL program (FlaFind) was written. A diverse set of proteins was identified, and many of these FlaFind positives were encoded by genes that were cotranscribed with homologs of pilus assembly genes. Moreover, structural conservation of primary sequences between many FlaFind positives and subunits of bacterial pilus-like structures, which have been shown to be critical for pilin assembly, have been observed. A subset of pilin-like FlaFind positives contained a conserved domain of unknown function (DUF361) within the signal peptide. Many of the genes encoding these proteins were in operons that contained a gene encoding a novel euryarchaeal prepilin-peptidase, EppA, homolog. Heterologous analysis revealed that Methanococcus maripaludis DUF361-containing proteins were specifically processed by the EppA homolog of this archaeon. Conversely, M. maripaludis preflagellins were cleaved only by the archaeal preflagellin peptidase FlaK. Together, the results reveal a diverse set of archaeal proteins with class III signal peptides that might be subunits of as-yet-undescribed cell surface structures, such as archaeal pili.
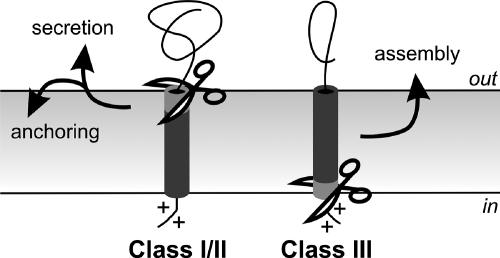
A diverse set of protein structures can decorate prokaryotic cell surfaces. They include the cell wall, flagella, and pili, which provide the cell with integrity, motility, adhesion, and the ability to transfer DNA. Prokaryotes have evolved distinct mechanisms to assemble subunits of such extracytoplasmic structures. For example, components of bacterial type IV pili require a dedicated membrane-associated machinery at the base of the growing pilus structure (12, 25, 27). Type IV pilins contain a conserved N-terminal hydrophobic stretch, and their interaction with each other provides a molecular scaffold for the helical assembly of the subunits into the pilus fiber (12, 13). This hydrophobic stretch is part of the signal peptide of the preprotein, which, unlike class I and II signal peptides, contains a signal peptidase cleavage site preceding the hydrophobic stretch (Fig. 1) (28). In addition to the prepilin peptidase, two conserved protein families are crucial for pilus biosynthesis: a VirB11-like ATPase (including GspE/TadA), which provides energy for the assembly and disassembly of the pilus, and a polytopic membrane protein (GspF/TadC) (34), which has been suggested to serve as an assembly platform for the pilus.
FIG. 1.
N-terminal signal peptide structures. Tripartite structure of class I (secretory) or class II (lipoprotein) signal peptides and class III (type IV pilin-like) signal peptide. Signal peptide cleavage by signal peptidases I and II, and prepilin peptidase, respectively, is symbolized by scissors; dark gray, hydrophobic region; light gray, cleavage region; +, positive charges.
In contrast to bacterial flagellar subunits, which are translocated using a specialized type III secretion apparatus (24), the secretion and assembly of archaeal flagellins resembles that of bacterial type IV pilins, as they possess class III signal peptides that are cleaved before the incorporation of the protein into the flagellar filament (9, 40). Moreover, several components of the archaeal flagellar assembly machinery are related to those of the type IV pilus biogenesis system, including a prepilin peptidase and the VirB11-like homologs FlaK and FlaI, respectively (8, 32). Additionally, the polytopic membrane protein FlaJ shows homology to TadC and might serve in a similar way as an assembly platform for the flagellum (31).
Interestingly, analysis of the predicted Sulfolobus solfataricus secretome revealed that certain membrane-bound substrate binding proteins (SBPs) of this crenarchaeon are also synthesized as preproteins with class III signal peptides (2). Consistent with this observation, the S. solfataricus prepilin peptidase homolog (PibD) could cleave both the flagellin subunit and the precursor of the glucose binding protein (4). While the biological roles of class III signal peptides associated with binding proteins are still unclear, it has been proposed that, similar to archaeal flagellins, these proteins also assemble into a cell surface structure (bindosome) upon secretion and signal peptide cleavage (1, 5). A function of the bindosome might be to locally increase the concentration of sugars for more efficient transport into the cell (5). Proteins with putative class III signal peptides were also observed in the Natronomonas pharaonis and Thermoplasma volcanium genomes (6, 17). The identification of archaeal nonflagellin proteins with class III signal peptides, which thus far have only been shown to be associated with subunits of cell surface structures (e.g., bacterial pili and archaeal flagella), suggests a diverse set of archaeal cell surface structures.
In this study, a PERL program (FlaFind [http://signalfind.org/]) was developed to screen archaeal genomes for proteins with class III signal peptides. In silico and in vivo analyses of FlaFind positives revealed the presence of a diverse set of proteins with class III signal peptides, including a subset of pilin-like proteins that are specifically cleaved by a novel prepilin peptidase. Colocalization of these FlaFind positives with bacterial type IV pilin assembly genes, as well as the structural resemblance of many of the FlaFind positives with homologs of bacterial pilin-like substrates, suggests that they may be subunits of archaeal cell surface structures. The identification of distinct classes of subunits of putative extracytoplasmic structures provides valuable data for future molecular and cell-biological investigations of archaeal cell surface structures, such as archaeal pili, which thus far have not been described in molecular detail.
MATERIALS AND METHODS
Sequence retrieval.
The protein sequences from the species listed in Table 1 were analyzed to identify putative class III signal sequence-containing proteins. The input data contained protein sequences from 22 different species downloaded from the NCBI website (http://www.ncbi.nlm.nih.gov/genomes/static/a.html).
TABLE 1.
Predicted archaeal class III signal peptide-containing proteins and homologs of type IV pilin-like biosynthesis components
| Organism | No. FlaFind positive | No. hypothetical | No. flagellin | No. SBP | No. DUF361 like | No. linkeda | No. FlaK | No. EppA | No. TadA | No. TadC |
|---|---|---|---|---|---|---|---|---|---|---|
| Crenarchaeota | ||||||||||
| Aeropyrum pernix K1 | 15 | 12 | 2 | 2 | 3 | 1 | 3 | 3 | ||
| Pyrobaculum aerophylum IM2 | 21 | 18 | 1 | 2 | 1c | 4 | 1 | |||
| Sulfolobus acidocaldarius DSM639 | 21 | 17 | 1 | 3 | 3 | 1 | 3 | 3 | ||
| Sulfolobus solfataricus P2 | 28 | 17 | 1 | 8 | 8 | 1 | 4 | 4 | ||
| Sulfolobus tokodaii strain 7 | 27 | 24 | 1 | 3 | 6 | 1 | 3 | 3 | ||
| Euryarchaeota | ||||||||||
| Archaeoglobus fulgidus DSM4304 | 17 | 13 | 2 | 6 | 1 | 4 | 3 | |||
| Haloarcula marismortui ATCC 43049 | 34 | 33 | 2b | 13 | 1 | 5 | 4 | |||
| Halobacterium sp. strain NRC-1 | 21 | 13 | 6b | 1 | 8 | 1 | 3 | 3 | ||
| Methanocaldococcus jannaschii DSM2661 | 15 | 10 | 3 | 1 | 7b | 8 | 2 | 1 | 4 | 4 |
| Methanococcus maripaludis S2 | 14 | 10 | 3b | 10 | 8 | 1 | 1 | 2 | 3 | |
| Methanopyrus kandleri AV19 | 6 | 4 | 2 | 3 | 1 | 1 | 2 | |||
| Methanosarcina acetivorans C2A | 31 | 19 | 3 | 6 | 8 | 4 | 6 | 6 | ||
| Methanosarcina mazei Go1 | 13 | 9 | 3 | 3 | 1 | 5 | 4 | |||
| Methanothermobacter thermautotrophicus DeltaH | 8 | 6 | 2b | 4 | 1 | 1 | 2 | |||
| Picrophilus torridus DSM9790 | 8 | 4 | 1 | |||||||
| Pyrococcus abyssi GE5 | 19 | 13 | 3 | 1 | 4 | 9 | 1 | 2 | 3 | |
| Pyrococcus furiosus DSM3638 | 18 | 11 | 2b | 1 | 3 | 7 | 1 | 2 | 3 | |
| Pyrococcus horikoshii OT3 | 20 | 15 | 5 | 2 | 3 | 10 | 1 | 2 | 3 | |
| Thermococcus kodakarensis KOD1 | 38 | 29 | 5 | 2 | 4 | 12 | 1 | 2 | 3 | |
| Thermoplasma acidophilum DSM1728 | 4 | 3 | 1 | 1 | 1c | 3 | 3 | |||
| Thermoplasma volcanium GSS1 | 7 | 3 | 2 | 1 | 1 | 1c | 3 | 3 | ||
| Nanoarchaeum equitans Kin-4-M | 3 | 3 | 2 | 2 | 2 |
Number of substrates linked to other FlaFind positives and/or genes involved in biosynthesis of type IV pilin-like structures.
Includes substrates that were likely erroneously annotated and thus were identified by FlaFind only upon reannotation.
These FlaK homologs show very low sequence conservation and were identified by examination of membrane topology predictions and the presence of conserved catalytic residues: Ta0254 (T. acidophilum), TVN1340 (T. volcanium), PAE1599 (P. aerophylum).
Class III signal peptide prediction.
A PERL program (FlaFind) was written to identify substrates with putative class III signal peptides among the set of all annotated coding sequences from 22 completely sequenced archaeal genomes. The program receives as input the amino acid sequences in Fasta format and the results obtained from a TMHMM v2.0 analysis (22, 36) of each of the sequences. A sequence is FlaFind positive if (i) the sequence has one or two TMHMM-predicted hydrophobic segments, (ii) the first hydrophobic segment begins within the first 30 amino acids of the protein sequence, and (iii) the pattern [KR][GA][ALIFQMVED][ILMVTAS] is found preceding the hydrophobic segment but not more than 10 amino acids away from the beginning of the hydrophobic segment.
Sequence analyses.
The FlaFind-positive set was characterized using Pfam v19.0 with the default parameters and the Pfam_fs database (10). DUF361-like sequences were identified among FlaFind positives using a modified FlaFind program in which the motif was [KR][GA][Q][X] [STA][X][DE], where X is any amino acid. Pfam v19.0 was also used to scan the 22 genomes for the DUF361 domain.
Ortholog identification was done with OrthoMCL version 1.2, with the inflation parameter set to 1.01 and the cutoff P value for WU-BLAST set to 1e−10. OrthoMCL creates clusters of orthologous and paralogous protein sequences and, together with the Pfam analyses, can clarify the functions and relationships of the various FlaFind positives.
The chromosomal environment for FlaFind-positive genes was determined using Genomapper (http://www-archbac.u-psud.fr/Genomap/GenomapBrowser.html). This tool displays the location, frame, and direction of transcription of a given gene from completely sequenced microbial genomes. Genes in the genomic environment of FlaFind substrate genes were considered to be linked (i.e., in an operon) if the intergenic distance was less than 100 base pairs and the genes were transcribed in the same direction. An exception was made when a substrate gene met all the above criteria but was transcribed in the opposite direction to the remainder of the gene cluster. Putative operons were screened for the presence of genes encoding additional FlaFind substrates and/or type IV pilus biogenesis protein homologs, i.e., TadA-like ATPases, TadC-like membrane proteins, and type IV pilin-like signal peptidases.
To visualize sequence conservation in selected FlaFind substrates, N-terminal sequences were aligned manually (until residue +30 relative to the cleavage site) with the putative signal peptidase cleavage site as reference and analyzed using the Weblogo server (14). To quantify local sequence conservation in an alignment, a PERL script was written that uses the ClustalW consensus symbol output with an adjustable window size. The script quantifies “identical” and “conserved” consensus symbols and produces two output files, one containing the number of identical amino acids per window and one with identical plus similar amino acids. The results, using a window size of 10, were plotted as percent sequence conservation versus amino acid position (see Fig. S1 in the supplemental material).
Plasmid construction.
Genomic DNA of Methanococcus maripaludis S2 was a gift from John Leigh (University of Washington). The plasmids used in this study are listed in Table S3 in the supplemental material. MMP0233/epdA, MMP0237/epdC, and MMP1667/flaB2 open reading frames were amplified by PCR from M. maripaludis genomic DNA, with appropriate restriction sites in the primers and with the native stop codons deleted. The PCR fragments were cloned into NcoI/BamHI-cut pZA7 (39), which added a C-terminal hemagglutinin epitope tag, resulting in pZA10, pZA11, and pZA12, respectively. The MMP0232/eppA and MMP0555/flaK genes were amplified in a similar way and cloned into pSA4 (4), yielding pZA13 and pZA14, respectively. Precursor genes including epitope tags were transferred as NcoI/HindIII fragments into pBAD/Myc-His A (Invitrogen, Breda, The Netherlands). Plasmids suitable for coexpression of substrates and peptidases were constructed as follows. First, an NcoI/HindIII fragment of pZA13 or a BglII/HindIII fragment of pZA14 was transferred into the corresponding restriction sites of pUC18-pibD (unpublished data), a construct that contained an SphI cassette including a T7 promoter, the pibD open reading frame, a C-terminal six-histidine tag, and a T7 terminator. In this way, the pibD gene was replaced by the respective peptidase genes. From the resulting plasmids, the SphI cassette was transferred into the unique SphI restriction site of the pBAD/Myc-His A precursor constructs, resulting in coexpression plasmids with all combinations of precursor and peptidase genes (see Table S3 in the supplemental material).
Growth conditions and preparation of E. coli crude membranes.
BL21(DE3)(pLysS) was used in all overexpression studies. Bacterial strains were grown to an optical density at 600 nm of 0.6 to 0.8. Then, expression of the precursor genes was induced by addition of l-arabinose for 2 h. Full induction of the araBAD promoter often resulted in strong overexpression of substrate genes, leading to protein degradation. Therefore, the induction conditions were optimized and l-arabinose was added to final concentrations of 0.2% (constructs containing epdA), 0.004% (epdC), and 0.001% (flaB2). Subsequently, peptidase genes were induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 h. The culture was harvested by centrifugation, and the cell pellets were resuspended in 2 ml of buffer (50 mM Tris-HCl, pH 7.5, 1 mM EDTA). Crude membranes were isolated as described previously (4) and resuspended in 50 mM Tris-HCl, pH 7.5. Cleavage of substrates was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western immunoblot analysis of 5 μg (EpdC and FlaB2 membranes) or 10 μg (EpdA membranes) of crude membranes. Substrate proteins were detected using monoclonal anti-hemagglutinin antibodies (Sigma).
RESULTS
To determine the diversity of archaeal proteins with class III signal peptides, a PERL program (FlaFind) was developed to detect proteins containing this class of N-terminal signals. The program predicts class III signal peptides based on the presence and position of the corresponding cleavage site (−2[KR] −1[GA] +1[ALIFQMVED] +2[MLIVTAS]), as well as the presence of a hydrophobic stretch following the cleavage site (see Materials and Methods). Analysis of the chromosomal localization of genes encoding FlaFind positives, identification of amino acid sequence similarities among proteins with predicted class III signal peptides, and preprotein-processing studies of FlaFind positives were carried out to validate and substantiate the significance of the FlaFind results.
In silico analysis identifies different classes of archaeal proteins with class III signal peptides. (i) Overview of FlaFind output.
FlaFind identified 388 proteins in 22 archaeal genomes, 102 of which were annotated as homologs of proteins with predicted functions (Table 1; see Table S1 in the supplemental material). Of these, 77 belonged to classes that had previously been shown to contain class III signal peptides, including 44 flagellins and 33 substrate binding proteins. The majority of the remaining 25 substrates belonged to different classes of extracytoplasmic proteins, including proteases and redox proteins. Only 4 of the 102 substrates were likely cytoplasmic proteins, suggesting that the rate of detection of proteins lacking a signal sequence is low (3.9%).
(ii) Chromosomal localization of FlaFind positives.
Bacterial type IV pilin-like structures consist of one major and several minor subunits. Genes encoding these subunits are often found in the same transcriptional unit, which also encodes proteins involved in the biosynthesis of bacterial type IV pilin-like structures (38). Consistent with their presence in cell surface structures, 120 FlaFind positives were predicted to be coregulated with additional FlaFind positives and/or genes coding for a TadA, TadC, and/or a type IV pilin peptidase homolog (Table 1 and Fig. 2; see Fig. S1 and Table S2 in the supplemental material). Moreover, in several cases, structural conservation of operons encoding homologs of FlaFind positives was observed among different organisms (Fig. 2 and Fig. 3). For example, the genes encoding the S. solfataricus FlaFind positives SSO0117 and SSO0118 are in an operon with tadA and tadC homologs, and this feature is conserved in the genomes of the three sequenced Sulfolobus species (Fig. 3). Interestingly, these operons, as well as an operon in each of the sequenced Pyrococcus strains that contained at least two FlaFind positives, were coregulated with an Lhr-like DNA helicase homolog, raising the possibility that these small substrates might be involved in DNA uptake or transfer (Fig. 3).
FIG. 2.
Cleavage of euryarchaeal pilin-like proteins by a dedicated signal peptidase. (A) Schematic representation of the conserved M. maripaludis and M. jannaschii operons containing genes that encode proteins with DUF361-like domains (black) and the novel subclass of euryarchaeal prepilin peptidase, eppA (white). Homologous genes are highlighted by identical shading. Note that the operon is split in M. jannaschii. (B) Transmembrane topologies of FlaK (left) and EppA (right) from M. maripaludis, predicted using the Phobius web server (21). Homologous regions in EppA and FlaK are in gray. A predicted four-transmembrane insertion in EppA is indicated in white. (C) Signal peptide cleavage of preproteins containing Duf361-like domains (EpdA and EpdC) and flagellin (FlaB2) from M. maripaludis by EppA or FlaK, as indicated. Control, E. coli expressing preprotein in the absence of an archaeal prepilin peptidase. Genes encoding precursor proteins under the arabinose-inducible promoter were expressed alone or in combination with IPTG-inducible peptidase genes. (D) Signal peptide cleavage of EpdA and EpdC with the flagellin cleavage site sequence or FlaB2 with the EpdA cleavage site sequence by EppA or FlaK. p, precursor, m, mature.
FIG. 3.
Structural conservation of operons encoding pilin-like FlaFind positives and a helicase; schematic representation of Sulfolobus and Pyrococcus operon structures. Genes coded for proteins with predicted functions, including FlaFind-positive proteins (green); proteins with a DUF361-like domain (hatched); an Lhr-like helicase (purple); a cell division GTPase/FtsZ3 (turquoise); COG5306, an uncharacterized conserved protein (light gray); COG4025, a predicted membrane protein (black); a TadA/VirB11-like ATPase (red); a TadC-like membrane protein (brown); an EndoIII-related endonuclease (COG0117, magenta); a transcriptional regulator (COG1474, cyan); a glycosyltransferase probably involved in cell wall biogenesis (COG1215, pink); a protein conserved in Sulfolobus (dark gray); and proteins conserved in Pyrococcus (white).
(iii) OrthoMCL analysis.
As noted above, several FlaFind positives could be classified into functional groups, i.e., flagellins and SBPs. To determine whether other, yet-unknown groups of conserved proteins were identified by FlaFind, OrthoMCL (23), a program designed to cluster proteins based on sequence similarity, was used to analyze all FlaFind-positive proteins. The program identified 47 groups, with 2 to 43 homologs in a given group (see Table S1 in the supplemental material). For example, of the 28 S. solfataricus FlaFind positives, 16 had at least one ortholog among the archaeal FlaFind positives. In fact, homologs of nine of these substrates were FlaFind positive in all three tested Sulfolobus species. Interestingly, in many cases, sequence homology was highest in the N-terminal portions of these proteins, a typical feature seen in subunits of bacterial type IV pili and proposed to be required for assembly (see Fig. S1 in the supplemental material) (12).
(iv) Pfam analysis.
Distinct from OrthoMCL, Pfam identifies small highly conserved domains within a protein (see Materials and Methods). To identify possible common themes among the large number of hypothetical proteins, all FlaFind positives were analyzed using Pfam (10). Consistent with the hypothesis that a diverse set of archaeal SBPs contains class III signal peptides, Pfam classified eight additional FlaFind positives as SBPs. Thus, in 14 of the 22 genomes, FlaFind identified at least one SBP, including (among others) sugar, dipeptide, and phosphate binding proteins (see Table S1 in the supplemental material).
Most striking, Pfam identified 19 euryarchaeal proteins with a domain of unknown function (DUF361), which is comprised of the amino acid motif QXSXEXXXL, where Q is the +1 position in the putative cleavage site of these proteins. Frequently, genes encoding DUF361-containing proteins were present in the same operon as genes encoding FlaFind positives, with a slightly varied domain sequence. In these “DUF361-like” domains, the serine was replaced by threonine or alanine, the glutamate was replaced by aspartate, and/or the leucine was replaced by a different hydrophobic amino acid (see Fig. S1 in the supplemental material). All FlaFind positives were screened for the presence of a DUF361-like domain with the modified FlaFind motif −2[KR] −1[GA] +1[Q] +2[X] +3[STA] +4[X] +5[DE]. This analysis revealed an additional 16 proteins, most of which were associated with the DUF361-containing proteins (Fig. 2A; see Table S1 in the supplemental material).
Interestingly, several genes encoding proteins with this conserved domain were found in operon structures, together with a gene encoding a novel subclass of euryarchaeal type IV prepilin peptidases, EppA. EppA, while homologous to FlaK, is substantially larger due to the presence of four additional predicted transmembrane segments (Fig. 2B). The chromosomal localization of eppA homologs and the fact that homologs of this prepilin peptidase were identified only in the eight euryarchaea that encoded DUF361-containing FlaFind positives strongly suggest a role as a specific signal peptidase for this class of preproteins (Table 1; see below).
EppA specifically cleaves proteins with DUF361-like domains.
FlaFind identified 14 substrates in M. maripaludis, including 3 flagellins and 10 DUF361-containing proteins. Three of these DUF361-containing proteins were coregulated with eppA (Fig. 2A). A similar operon is found in the genome of Methanocaldococcus jannaschii. However, there it is split and contains additional genes that are unique to the species (Fig. 2A). To determine whether the M. maripaludis EppA homolog specifically cleaves DUF361-containing proteins, either one of two genes encoding proteins with this conserved domain (MMP0233 and MMP0237) or a flagellin (MMP1667/flaB2) was coexpressed in E. coli with eppA from inducible promoters. In addition, the preproteins were coexpressed with flaK, the gene encoding the previously characterized M. maripaludis preflagellin peptidase (8). Processing of either of the two DUF361-containing proteins tested in E. coli could be observed only in cells that coexpressed EppA (Fig. 2C). Conversely, the novel peptidase was not able to cleave the flagellin subunit, strongly suggesting that the requirements for substrate recognition by the two subclasses of type IV pilin peptidases are distinct. Thus, here we will refer to MMP0233 and MMP0237 as EppA-dependent proteins (EpdA and EpdC, respectively).
The distinct substrate recognition characteristics of M. maripaludis FlaK and EppA sites were further demonstrated by an experiment in which the amino acids from positions −2 to +2 in EpdA were replaced with those of FlaB and vice versa (Fig. 2D). EppA was able to process the modified FlaB(RGQI) and did not cleave EpdA(KGAS) and EpdC(KGAS), indicating its requirement for the conserved glutamine at position +1. Conversely, FlaK was still able to cleave FlaB(RGQI), suggesting that it had a much broader cleavage site recognition capability and that its inability to cleave EpdA and EpdC was due to a distinct substrate recognition pattern. Consistent with this, FlaK was unable to process either of the EpdA(KGAS) and EpdC(KGAS) signal peptides despite the presence of the potential FlaK-processing site.
DISCUSSION
The majority of extracytoplasmic proteins are targeted to the prokaryotic cytoplasmic membrane by the presence of N-terminal tripartite signal peptides. However, subtle differences between these N-terminal sequences determine whether the protein is targeted to the membrane in a signal recognition particle-dependent or -independent manner (19), whether the substrate is targeted to the Sec or Twin arginine translocation pathway (33), and which signal peptidase processes the preprotein (28). The close resemblance of the signal peptide structures and the limited sequence conservation pose challenges to the ability to distinguish these signals from each other.
FlaFind, the program developed as part of this study, effectively identifies archaeal substrates containing class III signal peptides, as the program identified 41 of the 42 predicted archaeal flagellins and 19 of the 20 archaeal proteins containing a DUF361 domain, both classes of proteins that have been shown to be processed by a type IV prepilin-like peptidase (4, 8). In fact, all but one of the 14 M. maripaludis FlaFind positives were flagellins or DUF361-like substrates (3 and 10, respectively) suggesting that, certainly in this archaeon, the program successfully distinguishes class III signal peptides from other N-terminal signal peptides or transmembrane segments.
Consistent with the correct identification of class III signal peptide-containing proteins, the majority of nonflagellin FlaFind positives with annotated functions were SBPs, three of which had been shown experimentally to contain this class of signal peptide (3, 16). Moreover, only four of the annotated proteins were predicted cytoplasmic proteins, indicating that the rate of detection of proteins lacking a signal sequence is less than 4%. While the vast majority of FlaFind positives (75%) were annotated as hypothetical proteins, the chromosomal localization of many of the genes encoding these proteins, as well as the results of sequence homology and pattern searches among the FlaFind positives, strongly support the accurate identification of many of these proteins by FlaFind as class III signal peptide-containing proteins. It should be noted that, while Picrophilus torridus lacks any apparent TadA, TadC, or pilin peptidase homologs, FlaFind identified eight substrates in the archaeon. It is likely that, due to the lack of this peptidase, there is no selective pressure against the presence of a cleavage site-like pattern in secretory signal sequences and they are in fact false positives. Moreover, in different organisms, distinct consensus cleavage sequences may have evolved. For example, in Sulfolobus species, the +2 position is almost exclusively serine, while in other archaea, this position seems less important. Thus, it is unlikely that one will be able to define a perfect “global” consensus sequence for all archaeal class III signal peptides. However, our systematic genomic approach, in concert with additional in silico and in vivo analyses, has proven to yield valuable information about the diversity of predicted archaeal cell surface structures. To facilitate future studies on newly released genomes, the interactive version of FlaFind allows modification of the search pattern.
The substrates identified by FlaFind clustered into several distinct groups, including flagellins, SBPs, DUF361-containing proteins, and orthologous groups of small proteins, such as the Sulfolobus or Pyrococcus FlaFind positives that colocalized with a helicase (Fig. 3). The latter observation is particularly intriguing, as UV-induced exchange of genetic material between cells by a yet-unknown conjugational mechanism has been observed in Sulfolobus acidocaldarius and Haloferax volcanii (18, 26, 35).
Our data not only imply that the majority of FlaFind positives are indeed secreted proteins with N-terminal class III signal peptides, they are also consistent with the hypothesis that substrates with these signal peptides are subunits of cell surface structures, as (i) reminiscent of the coregulation of major and minor bacterial pilins and pseudopilins, genes encoding FlaFind positives were frequently cotranscribed with genes encoding additional FlaFind positives, and (ii) a significant number of FlaFind positives were encoded by genes located on an operon with homologs of genes encoding the pilin assembly components TadA and TadC. Moreover, a large number of FlaFind positives contain a negative charge at position +5, including 35 hypothetical proteins in which this charged amino acid is part of a conserved Pfam domain of unknown function, DUF361. The negative charge in DUF361-like proteins is embedded in a characteristic motif with the consensus sequence [RK][GA]QhShE (amino acid positions −2 to +5, with h being a hydrophobic residue). Similarly, a short characteristic motif was identified at the N terminus of a subclass of type IVb pilins (20). The presence of a negative charge at position +5 is a typical feature of bacterial type IV pilin-like subunits and is required for pilus assembly in Pseudomonas aeruginosa (30, 37). However, the absence of the +5 charge in many of the FlaFind positives does not suggest that these proteins lack the ability to form structures, as most archaeal flagellins do not possess a charge at this position. Distinct from poorly conserved amino acid sequences in the hydrophobic stretches of Tat and class I/II Sec signal sequences, the sequence of the hydrophobic stretch in the signal sequences of archaeal flagellins is highly conserved. This has also been observed for type IV pilins (12) and presumably allows optimal subunit-subunit interaction (11, 29). Consistent with the requirement for a highly conserved N-terminal hydrophobic “assembly domain,” several orthologous groups of FlaFind positives and substrates encoded by genes that cluster together share substantial sequence homology at their N termini (see Fig. S1 in the supplemental material).
Finally, our in vivo processing studies clearly demonstrated that DUF361-containing FlaFind positives were specifically cleaved by the novel subclass of prepilin peptidases, EppA, and that part of the “domain of unknown function” 361 is required for substrate recognition. While the involvement of FlaK in flagellum biogenesis has been demonstrated (7), future studies will reveal the specific function of EppA in the assembly of putative extracytoplasmic structures. However, it is tempting to speculate that the additional membrane-spanning segments in EppA are required for a function of this enzyme other than substrate cleavage, such as interaction with proteins involved in pilus assembly. Alternatively, the enzyme might exhibit an additional activity similar to bacterial prepilin peptidases that methylate the N terminus of the cleaved substrate. Also, the colocalization of eppA and substrate genes suggests coregulation. Recently, a second prepilin peptidase (FppA) was described in Pseudomonas aeruginosa (15). FppA is specific for a subclass of type IV pilins from the same organism, and it does not cleave the major pilin PilA, which in turn is a substrate of PilD. It is intriguing that two completely unrelated organisms apparently developed similar strategies to distinguish between various classes of pilin-like substrates.
The study described here opens many new directions for structural and genetic studies of archaeal extracytoplasmic structures. Future in vivo studies of the native archaeal hosts should provide validation of additional substrates (allowing refinement of the program) and identify additional archaeal components involved in the biosynthesis of extracellular structures, like the novel prepilin peptidase EppA. Additionally, data presented here raised countless intriguing questions, including the following. (i) What is the advantage of assembling SBPs into proposed cell surface structures? (ii) What are the functions of the FlaFind-positive hypothetical proteins? (iii) How do the distinct structures assemble? (iv) What is the significance of having two subclasses of prepilin peptidases if (as proposed earlier) cleavage of substrates occurs independently of their assembly?
Supplementary Material
Acknowledgments
We thank the Pohlschröder laboratory and Amy Decatur for critical reading and discussion of the manuscript. We also thank Brian C. Moore and John Leigh for providing M. maripaludis chromosomal DNA, invaluable for this analysis.
Support for this work was provided to M.P. by a National Science Foundation grant (reference no. MCB-0239215), to J.C.K. by an NIH/Fogarty International Center grant (reference no. 5D43TW007012-03), to S.-V.A. by VENI and VIDI grants from the Dutch Organization for Scientific Research (NWO), and to A.J.M.D. by the Van der Leeuw Programme of the Earth and Life Sciences Foundation (ALW), which is subsidized by NWO.
Footnotes
Published ahead of print on 17 November 2006.
Supplemental material for this article may be found at http://jb.asm.org.
REFERENCES
- 1.Albers, S. V., and A. J. Driessen. 2005. Analysis of ATPases of putative secretion operons in the thermoacidophilic archaeon Sulfolobus solfataricus. Microbiology 151:763-773. [DOI] [PubMed] [Google Scholar]
- 2.Albers, S. V., and A. J. M. Driessen. 2002. Signal peptides of secreted proteins of the archaeon Sulfolobus solfataricus: a genomic survey. Arch. Microbiol. 177:209-216. [DOI] [PubMed] [Google Scholar]
- 3.Albers, S. V., M. G. Elferink, R. L. Charlebois, C. W. Sensen, A. J. M. Driessen, and W. N. Konings. 1999. Glucose transport in the extremely thermoacidophilic Sulfolobus solfataricus involves a high-affinity membrane-integrated binding protein. J. Bacteriol. 181:4285-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albers, S. V., Z. Szabo, and A. J. Driessen. 2003. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J. Bacteriol. 185:3918-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albers, S. V., Z. Szabo, and A. J. Driessen. 2006. Protein secretion in the Archaea: multiple paths towards a unique cell surface. Nat. Rev. Microbiol. 4:537-547. [DOI] [PubMed] [Google Scholar]
- 6.Bardy, S. L., J. Eichler, and K. F. Jarrell. 2003. Archaeal signal peptides—a comparative survey at the genome level. Protein Sci. 12:1833-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bardy, S. L., and K. F. Jarrell. 2003. Cleavage of preflagellins by an aspartic acid signal peptidase is essential for flagellation in the archaeon Methanococcus voltae. Mol. Microbiol. 50:1339-1347. [DOI] [PubMed] [Google Scholar]
- 8.Bardy, S. L., and K. F. Jarrell. 2002. FlaK of the archaeon Methanococcus maripaludis possesses preflagellin peptidase activity. FEMS Microbiol. Lett. 208:53-59. [DOI] [PubMed] [Google Scholar]
- 9.Bardy, S. L., S. Y. Ng, and K. F. Jarrell. 2004. Recent advances in the structure and assembly of the archaeal flagellum. J. Mol. Microbiol. Biotechnol. 7:41-51. [DOI] [PubMed] [Google Scholar]
- 10.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang, S. L., R. K. Taylor, M. Koomey, and J. J. Mekalanos. 1995. Single amino acid substitutions in the N-terminus of Vibrio cholerae TcpA affect colonization, autoagglutination, and serum resistance. Mol. Microbiol. 17:1133-1142. [DOI] [PubMed] [Google Scholar]
- 12.Craig, L., M. E. Pique, and J. A. Tainer. 2004. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2:363-378. [DOI] [PubMed] [Google Scholar]
- 13.Craig, L., R. K. Taylor, M. E. Pique, B. D. Adair, A. S. Arvai, M. Singh, S. J. Lloyd, D. S. Shin, E. D. Getzoff, M. Yeager, K. T. Forest, and J. A. Tainer. 2003. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol. Cell 11:1139-1150. [DOI] [PubMed] [Google Scholar]
- 14.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Bentzmann, S., M. Aurouze, G. Ball, and A. Filloux. 2006. FppA, a novel Pseudomonas aeruginosa prepilin peptidase involved in assembly of type IVb pili. J. Bacteriol. 188:4851-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elferink, M. G., S. V. Albers, W. N. Konings, and A. J. Driessen. 2001. Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC transporters. Mol. Microbiol. 39:1494-1503. [DOI] [PubMed] [Google Scholar]
- 17.Falb, M., F. Pfeiffer, P. Palm, K. Rodewald, V. Hickmann, J. Tittor, and D. Oesterhelt. 2005. Living with two extremes: conclusions from the genome sequence of Natronomonas pharaonis. Genome Res. 15:1336-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grogan, D. W. 1996. Exchange of genetic markers at extremely high temperatures in the archaeon Sulfolobus acidocaldarius. J. Bacteriol. 178:3207-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber, D., D. Boyd, Y. Xia, M. H. Olma, M. Gerstein, and J. Beckwith. 2005. Use of thioredoxin as a reporter to identify a subset of Escherichia coli signal sequences that promote signal recognition particle-dependent translocation. J. Bacteriol. 187:2983-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kachlany, S. C., P. J. Planet, R. DeSalle, D. H. Fine, D. H. Figurski, and J. B. Kaplan. 2001. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 40:542-554. [DOI] [PubMed] [Google Scholar]
- 21.Kall, L., A. Krogh, and E. L. Sonnhammer. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027-1036. [DOI] [PubMed] [Google Scholar]
- 22.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 23.Li, L., C. J. Stoeckert, Jr., and D. S. Roos. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 25.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 26.Mevarech, M., and R. Werczberger. 1985. Genetic transfer in Halobacterium volcanii. J. Bacteriol. 162:461-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nudleman, E., and D. Kaiser. 2004. Pulling together with type IV pili. J. Mol. Microbiol. Biotechnol. 7:52-62. [DOI] [PubMed] [Google Scholar]
- 28.Paetzel, M., A. Karla, N. C. Strynadka, and R. E. Dalbey. 2002. Signal peptidases. Chem. Rev. 102:4549-4580. [DOI] [PubMed] [Google Scholar]
- 29.Park, H. S., M. Wolfgang, J. P. van Putten, D. Dorward, S. F. Hayes, and M. Koomey. 2001. Structural alterations in a type IV pilus subunit protein result in concurrent defects in multicellular behaviour and adherence to host tissue. Mol. Microbiol. 42:293-307. [DOI] [PubMed] [Google Scholar]
- 30.Pasloske, B. L., D. G. Scraba, and W. Paranchych. 1989. Assembly of mutant pilins in Pseudomonas aeruginosa: formation of pili composed of heterologous subunits. J. Bacteriol. 171:2142-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peabody, C. R., Y. J. Chung, M. R. Yen, D. Vidal-Ingigliardi, A. P. Pugsley, and M. H. Saier, Jr. 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149:3051-3072. [DOI] [PubMed] [Google Scholar]
- 32.Planet, P. J., S. C. Kachlany, R. DeSalle, and D. H. Figurski. 2001. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. USA 98:2503-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pohlschroder, M., E. Hartmann, N. J. Hand, K. Dilks, and A. Haddad. 2005. Diversity and evolution of protein translocation. Annu. Rev. Microbiol. 59:91-111. [DOI] [PubMed] [Google Scholar]
- 34.Py, B., L. Loiseau, and F. Barras. 2001. An inner membrane platform in the type II secretion machinery of Gram-negative bacteria. EMBO Rep. 2:244-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt, K. J., K. E. Beck, and D. W. Grogan. 1999. UV stimulation of chromosomal marker exchange in Sulfolobus acidocaldarius: implications for DNA repair, conjugation and homologous recombination at extremely high temperatures. Genetics 152:1407-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences, p. 175-182. In J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen (ed.), Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA. [PubMed]
- 37.Strom, M. S., and S. Lory. 1991. Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J. Biol. Chem. 266:1656-1664. [PubMed] [Google Scholar]
- 38.Strom, M. S., and S. Lory. 1993. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47:565-596. [DOI] [PubMed] [Google Scholar]
- 39.Szabo, Z., S. V. Albers, and A. J. Driessen. 2006. Active-site residues in the type IV prepilin peptidase homologue PibD from the archaeon Sulfolobus solfataricus. J. Bacteriol. 188:1437-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas, N. A., S. L. Bardy, and K. F. Jarrell. 2001. The archaeal flagellum: a different kind of prokaryotic motility structure. FEMS Microbiol. Rev. 25:147-174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.