Abstract
Little is known of protein expression in Mycobacterium avium subsp. paratuberculosis and how this contributes to pathogenesis. In the present study, proteins from both membranes and cytosol were prepared from two strains of M. avium subsp. paratuberculosis, i.e., laboratory-adapted strain K-10 and a recent isolate, strain 187, obtained from a cow exhibiting clinical signs of Johne's disease. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of cytosol and membrane proteins from K-10 and 187 showed marked differences in protein expression. Relative levels of protein expression from both M. avium subsp. paratuberculosis strains were measured by using amine-reactive isobaric tagging reagents (iTRAQ) and tandem mass spectroscopy. Protein identification and relative expression data were obtained for 874 membrane and cytosolic proteins from the M. avium subsp. paratuberculosis proteome. These data showed a number of significant differences in protein expression between strain K-10 and clinical isolate 187. Examples of proteins expressed at higher levels in clinical isolate 187 compared to strain K-10 are AtpC, RpoA, and several proteins involved in fatty acid biosynthesis. In contrast, proteins such as AhpC and several proteins involved in nitrogen metabolism were expressed at higher levels in strain K-10 compared to strain 187. These data may provide insights into the proteins whose expression is important in natural infection but are modified once M. avium subsp. paratuberculosis is adapted to laboratory cultivation. Results from these studies will provide tools for developing a better understanding of M. avium subsp. paratuberculosis infection in the host and offer potential as diagnostic reagents and vaccine candidates.
Paratuberculosis (Johne's disease) is a significant economic problem in cattle and sheep worldwide (33). The causative agent of this chronic enteric disease is Mycobacterium avium subsp. paratuberculosis. M. avium subsp. paratuberculosis is a gram-positive intracellular pathogen that can persist and replicate within macrophages of the infected host (41). It is thought that neonatal cattle become infected by ingesting M. avium subsp. paratuberculosis shed in the feces of cattle that are in the clinical phase of the disease. Infections can persist for several years in a subclinical phase that is difficult to identify because of the absence of clinical signs. Once the disease enters the clinical phase, thickening of the intestinal wall leads to weight loss, decreased milk production, diarrhea, shedding of M. avium subsp. paratuberculosis in the feces, and eventual death (11, 16, 33). Diagnosis of Johne's disease is often complicated because of the slow progression of the disease and the prevalence of genetically similar mycobacterial species in the environment.
The recent sequencing and annotation of the K-10 strain of M. avium subsp. paratuberculosis has led to significant progress in the analysis of the genetic regulation of M. avium subsp. paratuberculosis (22). Microarray analysis has provided extensive data on gene expression in several species of mycobacteria, including M. avium subsp. paratuberculosis. Although single proteins have been identified as potential diagnostic tools for M. avium subsp. paratuberculosis, comprehensive surveys of the M. avium subsp. paratuberculosis proteome have been lacking until now. Recent studies have reported success in the use of methods such as two-dimensional electrophoresis and surface-enhanced laser desorption ionization-time of flight mass spectrometry (MS) to profile protein expression in M. avium subsp. paratuberculosis (10, 15). Direct comparison of gene expression between strains or within a single strain grown under differing conditions has proven useful in identifying mycobacterial growth and pathogenicity characteristics (14, 17). Analysis of the M. avium subsp. paratuberculosis proteome offers another level of regulation for study.
In postgenomic studies, high-throughput high-performance liquid chromatography coupled with MS has been used to identify greater than 25% of the predicted M. tuberculosis coding sequences (25). The identification of large sets of proteins like this is possible with extensive fractionation of the protein sample. Large-scale proteome analysis from multiple samples has thus far been limited by an inability to quantify MS results. Use of recently developed amine-labeled isobaric tags now allows the analysis of relative protein expression levels between samples (1). By using this method, we analyzed the levels of protein expression from two M. avium subsp. paratuberculosis strains.
Studies on M. avium subsp. paratuberculosis have shown that adaptation to laboratory growth conditions often is accompanied by changes in phenotype (5, 38). It is unknown how changes in phenotype correspond to protein expression profiles. The two strains used in this study were chosen to evaluate effects of in vitro propagation of M. avium subsp. paratuberculosis on protein expression. Comparing the proteome expression profiles of two M. avium subsp. paratuberculosis strains of different passage numbers may help us elucidate the roles of key components involved in M. avium subsp. paratuberculosis growth and pathogenesis. In the present study, our goals were to obtain a comprehensive survey of the M. avium subsp. paratuberculosis proteome and to demonstrate the differences in protein expression in two M. avium subsp. paratuberculosis strains, multiple-passage laboratory-adapted strain K-10 and low-passage strain 187 obtained from a cow with clinical Johne's disease.
MATERIALS AND METHODS
Culture strains and protein samples.
M. avium subsp. paratuberculosis strain K-10 was chosen as the reference strain because of its use in sequencing of the M. avium subsp. paratuberculosis genome and extensive study. Strain K-10 was passed 16 times in our laboratory, a value that does not account for the multiple passages performed in the laboratory of origin. M. avium subsp. paratuberculosis strain 187 was isolated from the ileum of a clinical cow. It was confirmed to be M. avium subsp. paratuberculosis by PCR IS900 (37; data not shown) and grown to passage 5. Twenty-milliliter volumes of actively growing cultures of both strains were individually transferred to 500 ml of Middlebrook 7H9 broth (pH 5.9) containing 0.05% Tween 80 and ferric mycobactin J (2 mg/liter; Allied Monitor, Fayette, MO). Culture growth was monitored, and cultures were harvested in log phase (A540 = 0.23 to 0.24). Expansion of each strain to log phase after passage took between 2 and 3 weeks because of the slow replication rate that is characteristic of M. avium subsp. paratuberculosis. Cultures were collected and washed three times in cold phosphate-buffered saline (PBS, pH 7.4). Cell sonicates were prepared as previously reported (4). Briefly, cells were resuspended in 5 ml of buffer (250 mM sucrose, 10 mM HEPES [pH 7.5], 1 mM EGTA) and sonicated three times for 10 min in an ice-water slurry. After each sonication step, the samples were cooled for 10 min prior to the start of the next sonication interval. The sonicates were centrifuged at 10,000 × g for 15 min to remove unbroken cells, and cellular debris and the supernatants were decanted. The supernatants were centrifuged at 100,000 × g for 1 h to pellet membranes. The supernatant was collected and served as the M. avium subsp. paratuberculosis cytosolic protein fraction. The pellet was resuspended in an equal volume of sonication buffer and served as the M. avium subsp. paratuberculosis membrane protein fraction. All samples were assayed for protein content by the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA) and stored at −20°C.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with precast 4 to 12% Bis-Tris buffered minigels in morpholinepropanesulfonic acid (MOPS) buffer (Invitrogen Corporation, Carlsbad, CA). Samples were diluted in 4× NuPAGE loading buffer and denatured at 95°C for 5 min. Silver staining was done with the SilverQuest silver staining kit (Invitrogen).
Immunoblot assays.
Electrophoretic transfer of proteins to nitrocellulose (Invitrogen) was done with a Criterion blotting cell (Bio-Rad) in NuPAGE (Invitrogen) blotting buffer with 10% methanol for 30 min at 100 V. Membranes were washed for 5 min in PBS and blocked overnight at 4°C on a rotating platform with SuperBlock (Pierce Chemical Company, Rockford, IL) plus 0.05% Tween 20. Three M. avium subsp. paratuberculosis monoclonal antibodies were used in this study. MAP1643 codes for the AceAb protein, MAP3840 codes for the DnaK protein, and MAP2121c codes for the 35-kDa major membrane protein that is involved in M. avium subsp. paratuberculosis invasion of intestinal epithelial cells (3). Monoclonal antibodies were diluted in 15 ml of SuperBlock plus 0.05% Tween 20 at the following rates: anti-MAP1643, 1:300; anti-MAP3840, 1:200; anti-MAP2121c, 1:500. Blots were incubated with each antibody at room temperature for 1 h on a rocking platform. Following primary-antibody incubation, the blots received six 5-min washes in PBS containing 0.05% Tween 20. Next, the blots were incubated at room temperature on a rocking platform for 45 min with horseradish peroxidase-labeled anti-mouse secondary antibody (1:60,000 in 20 ml of SuperBlock plus 0.05% Tween 20). The blots were washed six times for 5 min in PBS plus 0.05% Tween 20. Specific banding was detected with the WestDura detection system (Pierce Immunochemicals, Rockford, IL).
iTRAQ labeling.
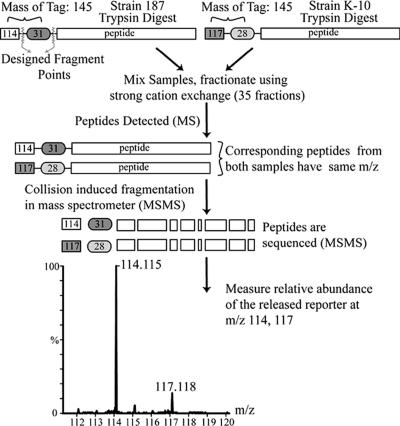
One hundred micrograms of protein from each sample was dried and processed with the iTRAQ kit (Applied Biosystems, Foster City, CA). M. avium subsp. paratuberculosis strain 187 was labeled with the 114 iTRAQ tag, and M. avium subsp. paratuberculosis strain K-10 was labeled with the 117 iTRAQ tag (Fig. 1). Peptides were digested with trypsin followed by labeling with the iTRAQ reagent. Subsequently, the peptides from the two samples were combined, dried, resuspended in 300 μl of 20 mM formic acid-20% acetonitrile (ACN), and further purified.
FIG. 1.
Schematic representation of iTRAQ protein expression analysis. The cytosolic protein fractions of strains 187 and K-10 were digested with trypsin, and the resulting peptides were labeled with appropriate iTRAQ tags. Samples were combined, and peptides were separated by strong cation exchange and analyzed with a Q-TOF Ultima API mass spectrometer. The ratio of peak areas between 114 and 117 was used to determine the relative abundance of proteins in the original sample. The procedure was repeated with the membrane-enriched fractions from both strains.
Strong cation exchange.
The iTRAQ samples were chromatographed on a cation-exchange column (Mono S PC 1.6/5; Amersham, Piscataway, NJ). The gradient solutions are mobile phase A (20 mM formic acid, 20% ACN, pH 2.7) and mobile phase B (20 mM formic acid, 20% ACN, 350 mM ammonium bicarbonate, pH 4.7). Sample fractions (0.5 ml) were collected over a 150-min gradient of 0 to 35% mobile phase B, followed by 5 min in 90% mobile phase B (the flow rate was 300 μl/min). Samples were dried and resuspended in 30 μl of 0.1% formic acid in 5% ACN and subjected to mass spectroscopy as follows.
High-performance chromatography and tandem mass spectroscopy of the samples.
Each strong-cation-exchange fraction was analyzed by capillary high-pressure liquid chromatography (CapLC; Waters, Milford, MA) in line with a Q-TOF Ultima API mass spectrometer (Waters, Milford, MA) (24). An Altantis C18 NanoEase column (75 μm by 100 mm) was used for peptide separation. The system was configured to concentrate and wash the injected sample on a Symmetry 300 C18 precolumn. Seven minutes after the start of sample loading, the precolumn was switched in line with the analytical column to allow the trapped peptides to be eluted onto the analytical column. Mobile phase A was 0.1% formic acid in 5% ACN. Mobile phase B was 0.1% formic acid in 95% ACN. The gradient was 95% A for 5 min and then ramped linearly to 60% A over 85 min. Over the next 2.5 min, it was ramped to 10% A and held for an additional 10 min before equilibration of the column. The flow rate was approximately 300 nl/min. The analytical column was connected to a Waters Lockspray-nanospray interface on the front of the mass spectrometer. The Lockspray used the peptides [Glu1]fibrinopeptide B and leucine enkephalin as standards (Sigma, St. Louis, MO). The capillary voltage was 3,500 V and was tuned for signal intensity. The five most intense ions with charge states between 2 and 4 were selected in each survey scan if they met the switching criteria. Three collision energies were used to fragment each peptide ion on the basis of its mass-to-charge (m/z) values. Each fraction was run four times, once collecting MS-MS data on the full range of parent masses, followed by runs collecting MS-MS data on parent masses in three mass ranges (400 to 635, 635 to 750, and 750 to 1,500) (32).
Bacterial processing.
All MS data files were processed into pkl files with Protein Lynx Global Server 2.0 (PLGS 2.0; Waters, Milford, MA) and analyzed with Mascot (Matrix Science, London, United Kingdom) with the NCBI nonredundant database with bacterial taxonomy. Peptide expression was determined by using the peak area of each of the iTRAQ labels, after the peaks were summed, smoothed, and centered. Data were organized to remove duplication of query assignments and duplicate proteins. All proteins without at least two MS-MS spectra and a probability of greater than 95% were removed. Only MS-MS spectra with abundance information for both iTRAQ labels were retained (23). Changes in protein expression were calculated as described by Ross and coworkers [187 label/(187 label + K-10 label)] (28). The range of this calculation is between 0 and 1, with a result of 0.5 meaning no change in protein expression. The average iTRAQ calculation for all MS-MS spectra in the membrane fraction was 0.569, with a standard deviation of 0.086. For the cytosol fraction, the average was 0.428, with a standard deviation of 0.057. A change of less than 2 standard deviations was deemed no change in protein expression.
RESULTS
Strains 187 and K-10 show different proteome expression profiles.
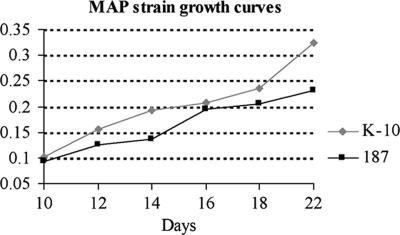
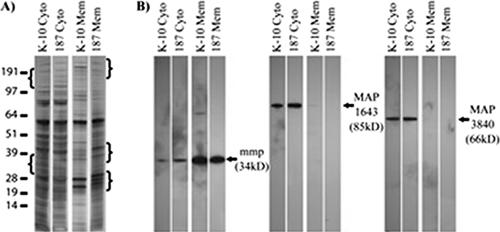
Differences in protein expression between M. avium subsp. paratuberculosis strains likely correlate with phenotypic traits such as differences in growth rates and virulence (2). To investigate to what extent this occurs in M. avium subsp. paratuberculosis, we analyzed two strains, low-passage M. avium subsp. paratuberculosis clinical isolate 187 and laboratory-adapted M. avium subsp. paratuberculosis strain K-10. Strain 187 grew at a slower rate than strain K-10 (Fig. 2). Protein samples from M. avium subsp. paratuberculosis strains 187 and K-10 showed different protein banding patterns on silver-stained SDS-PAGE gels (Fig. 3A). The regions with brackets show differences in banding patterns between the strains for both the cytosol and membrane-enriched fractions. Together, these data illustrate both phenotypic and proteomic profile differences between the two strains.
FIG. 2.
Growth curves generated for M. avium subsp. paratuberculosis strains 187 and K-10 in Middlebrook 7H9 broth supplemented with Tween 80 and ferric mycobactin J. On the basis of their rates of growth, strain K-10 was harvested on day 18 (A540, 0.235) and strain 187 was harvested on day 22 (A540, 0.232).
FIG. 3.
Protein expression differences observed on both one- and two-dimensional silver-stained PAGE gels. (A) SDS-PAGE of cytosolic and membrane protein samples (0.5 μg/lane) from strains 187 and K-10. Differences in banding patterns between the strains are noted in the bracketed regions. The values on the left are molecular sizes in kilodaltons. (B) Immunoblot analysis of M. avium subsp. paratuberculosis strains 187 and K-10 with three available anti-M. avium subsp. paratuberculosis monoclonal antibodies. MAP1643 and MAP3840 are found predominantly in the cytosolic fractions, while MAP2121c is found mainly in the membrane fractions.
iTRAQ analysis identifies differentially expressed proteins.
Protein expression data were obtained for 550 proteins from the membrane fractions, of which 385 proteins were present only in the membranes and not the cytosol of the M. avium subsp. paratuberculosis strains (see the membrane protein data in the supplemental material). Of the 550 proteins identified in the M. avium subsp. paratuberculosis membranes, 266 of the proteins identified were hypothetical proteins in the SwissProt database. Within the membrane fractions of each strain, 37 proteins were expressed at higher abundance in strain K-10 than strain 187 (Table 1). In contrast, 35 M. avium subsp. paratuberculosis membrane proteins were expressed at higher abundance in strain 187 than in strain K-10 (Table 2). The incomplete and/or electronic-only annotation of the M. avium subsp. paratuberculosis genome limits the conclusions that can be made about the differentially regulated proteins observed in this study. It is of interest that membrane-associated proteins in clinical isolate 187 are generally upregulated to a greater extent (2.9- to 6.9-fold) than those in laboratory-adapted strain K-10 (Table 2). In contrast, upregulation of membrane-associated proteins in strain K-10 over those seen in strain 187 was more moderate (1.5- to 3.5-fold; Table 1). The greatest increase in protein expression was demonstrated for AtpC, a component of the ATP synthase complex, at 6.91-fold higher expression noted for strain 187 compared to strain K-10. The MVIN-like protein (MAP4336), a putative virulence factor in other organisms, was expressed 4.75-fold more in strain 187 than in strain K-10. Stress-associated proteins MAP4265 (GroEL1) and Map2281c (ClpP) were both expressed about threefold more in strain 187 than in strain K-10. Slower-growing clinical isolate 187 expressed 3.4-fold more of cell division protein MAP1894c (FtsZ) in the membrane fractions than was observed in laboratory-adapted strain K-10 (Table 2). Finally, a number of fatty acid metabolism proteins, FadE_2, InhA, FadE23, FadA1, FadE5, and FadB1 (2.9- to 4-fold), as well as RNA polymerase proteins RpoA and RpoB (3.6- to 3.9-fold; Table 2), were also upregulated in the recent clinical isolate (strain 187).
TABLE 1.
Proteins expressed at higher relative abundance in strain K-10 membrane fractions compared to strain 187 membrane fractions
| NCBI no. | Protein | Scorea | Descriptionb | Mean (SD) of 114/114 +117c | Fold increased | No. of unique peptidese | MS-MSf |
|---|---|---|---|---|---|---|---|
| gi 41407687 | MAP1589c | 171.26 | AhpC | 0.224 (0.012) | 3.46 | 2 | 2 |
| gi 41395410 | MAP0961c | 209.71 | Hypothetical protein | 0.251 (0.037) | 2.98 | 4 | 5 |
| gi 41399117 | MAP4185 | 254.35 | RpmD | 0.273 (0.092) | 2.66 | 2 | 8 |
| gi 41396278 | MAP1826c | 137.78 | Hypothetical protein | 0.291 (0.182) | 2.44 | 2 | 7 |
| gi 41408218 | MAP2120c | 599.40 | nifS-like protein | 0.301 (0.105) | 2.32 | 7 | 20 |
| gi 41397444 | MAP2987c | 345.10 | GlnB | 0.322 (0.170) | 2.11 | 7 | 9 |
| gi 41397250 | MAP2793 | 120.59 | Hypothetical protein | 0.325 (0.113) | 2.08 | 3 | 4 |
| gi 41398325 | MAP3396c | 110.75 | Hypothetical protein | 0.329 (0.136) | 2.04 | 2 | 3 |
| gi 41395411 | MAP0962c | 139.86 | Hypothetical protein | 0.342 (0.139) | 1.92 | 3 | 3 |
| gi 41395766 | MAP1316c | 223.80 | AppC | 0.346 (0.063) | 1.89 | 4 | 15 |
| gi 41399115 | MAP4183 | 70.03 | RplR | 0.349 (0.031) | 1.86 | 2 | 3 |
| gi 41398647 | MAP3717c | 181.06 | Hypothetical protein | 0.350 (0.161) | 1.86 | 3 | 4 |
| gi 41395491 | MAP1042 | 195.72 | Hypothetical protein | 0.350 (0.041) | 1.86 | 2 | 2 |
| gi 41395079 | MAP0631c | 174.39 | Hypothetical protein | 0.351 (0.041) | 1.85 | 4 | 5 |
| gi 41395078 | MAP0630c | 850.22 | Hypothetical protein | 0.356 (0.097) | 1.81 | 13 | 58 |
| gi 41394589 | MAP0143 | 257.33 | Hypothetical protein | 0.357 (0.026) | 1.80 | 4 | 6 |
| gi 41398430 | MAP3501 | 86.81 | Hypothetical protein | 0.360 (0.109) | 1.78 | 2 | 4 |
| gi 41395429 | MAP0980c | 395.83 | Hypothetical protein | 0.364 (0.097) | 1.75 | 6 | 8 |
| gi 41398294 | MAP3365c | 144.71 | Hypothetical protein | 0.368 (0.158) | 1.72 | 3 | 4 |
| gi 41394511 | MAP0233c | 163.76 | Hypothetical protein | 0.373 (0.050) | 1.68 | 2 | 3 |
| gi 41394679 | MAP1179 | 63.14 | CtaB | 0.374 (0.049) | 1.67 | 2 | 4 |
| gi 41395629 | MAP3171c | 116.67 | FtsX | 0.378 (0.041) | 1.64 | 2 | 3 |
| gi 41398089 | MAP1292c | 124.01 | Hypothetical protein | 0.379 (0.096) | 1.64 | 2 | 3 |
| gi 41395742 | MAP2939c | 160.68 | Hypothetical protein | 0.379 (0.158) | 1.64 | 4 | 4 |
| gi 41397396 | MAP0940 | 82.59 | Hypothetical protein | 0.381 (0.038) | 1.62 | 2 | 2 |
| gi 41395389 | MAP1940c | 292.78 | CtaC | 0.382 (0.037) | 1.62 | 4 | 4 |
| gi 41396393 | MAP2433 | 688.67 | Hypothetical protein | 0.382 (0.086) | 1.62 | 12 | 25 |
| gi 41396889 | MAP2988c | 112.85 | Amt_2 | 0.383 (0.052) | 1.61 | 2 | 3 |
| gi 41397445 | MAP3287 | 258.26 | Hypothetical protein | 0.384 (0.099) | 1.61 | 4 | 5 |
| gi 41398216 | MAP2057 | 656.43 | Hypothetical protein | 0.386 (0.071) | 1.59 | 14 | 26 |
| gi 41396510 | MAP3698c | 1656.99 | Hypothetical protein | 0.388 (0.064) | 1.58 | 27 | 72 |
| gi 41398628 | MAP3092 | 243.76 | FecB | 0.389 (0.130) | 1.56 | 3 | 4 |
| gi 41398010 | MAP1510 | 479.82 | Hypothetical protein | 0.393 (0.061) | 1.55 | 6 | 10 |
| gi 41395961 | MAP1738 | 223.21 | MmpL5 | 0.394 (0.075) | 1.54 | 4 | 5 |
| gi 41396190 | MAP3291c | 336.52 | Hypothetical protein | 0.400 (0.068) | 1.50 | 3 | 3 |
| gi 41398220 | MAP4169 | 242.16 | RpmC | 0.400 (0.087) | 1.50 | 5 | 5 |
Protein score assigned by Mascot.
Descriptions are from the SwissProt database.
Mean and standard deviation for all of the MS spectra for a given protein.
Fold increase compared to the other strain. A ratio of 0.5 is no change in protein expression.
Number of unique peptides identified for the protein.
MS-MS refers to the total number of MS spectra found for the given protein.
TABLE 2.
Proteins expressed at higher relative abundance in strain 187 membrane fractions compared to strain K-10 membrane fractions
| NCBI no. | Protein | Scorea | Descriptionb | Mean (SD) of 114/114 +117c | Fold increased | No. of unique peptidese | MS-MSf |
|---|---|---|---|---|---|---|---|
| gi 41408548 | MAP2450c | 95.36 | AtpC | 0.874 (0.046) | 6.91 | 2 | 3 |
| gi 41394941 | MAP0494 | 160.55 | Hypothetical protein | 0.852 (0.059) | 5.77 | 3 | 6 |
| gi 41397135 | MAP2679c | 145.98 | Hypothetical protein | 0.828 (0.112) | 4.82 | 2 | 5 |
| gi 41399268 | MAP4336 | 243.44 | Hypothetical protein | 0.826 (0.095) | 4.75 | 4 | 7 |
| gi 41395817 | MAP1367 | 530.25 | ArgG | 0.820 (0.093) | 4.55 | 8 | 16 |
| gi 41399164 | MAP4232 | 361.15 | RpsD | 0.805 (0.058) | 4.13 | 4 | 8 |
| gi 41395238 | MAP0790 | 1251.23 | FadB1 | 0.801 (0.099) | 4.02 | 17 | 50 |
| gi 41398624 | MAP3694c | 209.19 | FadE5 | 0.799 (0.117) | 3.99 | 3 | 4 |
| gi 41399062 | MAP4130 | 573.43 | RpoB | 0.797 (0.111) | 3.93 | 8 | 15 |
| gi 41407408 | MAP1310 | 87.39 | PykA | 0.793 (0.057) | 3.83 | 2 | 2 |
| gi 41395106 | MAP0658c | 647.22 | DesA1 | 0.791 (0.082) | 3.77 | 10 | 14 |
| gi 41399165 | MAP4233 | 429.61 | RpoA | 0.784 (0.150) | 3.63 | 7 | 13 |
| gi 41397348 | MAP2891c | 325.60 | GpsI | 0.784 (0.085) | 3.62 | 4 | 6 |
| gi 41395237 | MAP0789 | 282.86 | FadA1 | 0.779 (0.094) | 3.53 | 4 | 5 |
| gi 41396346 | MAP1894c | 545.69 | FtsZ | 0.774 (0.088) | 3.43 | 7 | 13 |
| gi 41396700 | MAP2246c | 130.37 | Hypothetical protein | 0.773 (0.115) | 3.41 | 2 | 3 |
| gi 41395960 | MAP1509 | 104.02 | Hypothetical protein | 0.773 (0.118) | 3.41 | 2 | 2 |
| gi 41397138 | MAP2682c | 81.60 | Hypothetical protein | 0.768 (0.041) | 3.32 | 2 | 5 |
| gi 41396537 | MAP2084 | 113.61 | Hypothetical protein | 0.768 (0.056) | 3.32 | 2 | 4 |
| gi 41399075 | MAP4143 | 884.65 | Tuf | 0.764 (0.120) | 3.23 | 8 | 37 |
| gi 41396008 | MAP1557c | 275.96 | Gnd | 0.762 (0.085) | 3.20 | 6 | 11 |
| gi 41396732 | MAP2787 | 161.39 | ClpX | 0.762 (0.078) | 3.20 | 3 | 3 |
| gi 41398576 | MAP3646 | 353.58 | PckA | 0.758 (0.115) | 3.14 | 9 | 11 |
| gi 41398107 | MAP3189 | 304.13 | FadE23 | 0.756 (0.085) | 3.09 | 4 | 5 |
| gi 41396735 | MAP2281c | 168.83 | ClpP | 0.755 (0.139) | 3.09 | 3 | 6 |
| gi 41395318 | MAP0870c | 89.73 | Hypothetical protein | 0.755 (0.062) | 3.08 | 2 | 2 |
| gi 41397978 | MAP3060c | 643.12 | FixB | 0.755 (0.074) | 3.08 | 8 | 17 |
| gi 41399185 | MAP4253 | 164.27 | GlmS | 0.754 (0.075) | 3.07 | 3 | 5 |
| gi 41399197 | MAP4265 | 601.67 | GroEL1 | 0.752 (0.075) | 3.04 | 7 | 16 |
| gi 41398243 | MAP3314c | 169.50 | Hypothetical protein | 0.752 (0.074) | 3.04 | 2 | 3 |
| gi 41407308 | MAP1210 | 168.78 | InhA | 0.751 (0.032) | 3.02 | 2 | 2 |
| gi 41397954 | MAP3036c | 343.67 | IlvC | 0.750 (0.075) | 3.00 | 6 | 9 |
| gi 41398500 | MAP3750c | 119.53 | FadE2 | 0.744 (0.094) | 2.91 | 2 | 2 |
| gi 41398385 | MAP3456c | 442.78 | Icd2 | 0.743 (0.069) | 2.89 | 8 | 17 |
| gi 41409122 | MAP3024c | 374.91 | hupB | 0.743 (0.091) | 2.89 | 6 | 32 |
Protein score assigned by Mascot.
Descriptions are from the SwissProt database.
Mean and standard deviation for all of the MS spectra for a given protein.
Fold increase compared to the other strain. A ratio of 0.5 is no change in protein expression.
Number of unique peptides identified for the protein.
MS-MS refers to the total number of MS spectra found for the given protein.
From M. avium subsp. paratuberculosis cytosolic fractions, protein expression data were obtained for 489 proteins, of which 324 proteins were identified only in the cytosolic fraction (see the cytosolic protein data in the supplemental material). Out of 489 proteins identified in M. avium subsp. paratuberculosis cytosol, 185 of the proteins identified were hypothetical proteins in the SwissProt database. Within the cytosolic fractions of each strain, 22 proteins were expressed at higher abundance in strain K-10 than in strain 187 (Table 3). In contrast, 18 cytosolic proteins were expressed at higher abundance in strain 187 than in strain K-10 (Table 4). The greatest increase in expression was demonstrated for Ffh, a signal recognition particle that participates in the secretory pathway for bacteria. Other major cytosolic proteins that were upregulated were GlnD, Icd2, and AtpC, all of which are involved in energy and nitrogen metabolism.
TABLE 3.
Proteins expressed at higher relative abundance in strain K-10 cytosolic fractions compared to strain 187 cytosolic fractions
| NCBI no. | Protein | Scorea | Descriptionb | Mean (SD) of 114/114 +117c | Fold increased | No. of unique peptidese | MS-MSf |
|---|---|---|---|---|---|---|---|
| gi 41397440 | MAP2983c | 87.30 | Ffh | 0.165 (0.176) | 5.06 | 2 | 3 |
| gi 41397443 | MAP2986c | 167.87 | GlnD | 0.180 (0.030) | 4.55 | 2 | 3 |
| gi 41394518 | MAP0072c | 266.55 | Hypothetical protein | 0.225 (0.076) | 3.44 | 5 | 8 |
| gi 41395953 | MAP1502 | 179.81 | Hypothetical protein | 0.234 (0.092) | 3.28 | 3 | 4 |
| gi 41398385 | MAP3456c | 367.30 | Icd2 | 0.238 (0.131) | 3.21 | 5 | 13 |
| gi 41398458 | MAP3529 | 72.89 | Hypothetical protein | 0.245 (0.110) | 3.08 | 2 | 4 |
| gi 41406377 | MAP0279 | 84.58 | Hypothetical protein | 0.246 (0.042) | 3.07 | 2 | 5 |
| gi 41399048 | MAP4116c | 182.64 | MmaA4 | 0.246 (0.091) | 3.07 | 3 | 7 |
| gi 41399128 | MAP4196 | 239.31 | Hypothetical protein | 0.262 (0.134) | 2.82 | 3 | 6 |
| gi 41399156 | MAP4224c | 126.33 | RmlC | 0.265 (0.070) | 2.78 | 2 | 3 |
| gi 41398646 | MAP3716c | 211.65 | FadE6 | 0.266 (0.098) | 2.75 | 4 | 13 |
| gi 41394854 | MAP0407c | 93.72 | Acs | 0.271 (0.031) | 2.70 | 2 | 2 |
| gi 41409122 | MAP3024c | 260.00 | hupB | 0.276 (0.038) | 2.63 | 4 | 20 |
| gi 41398803 | MAP3872 | 89.36 | Hypothetical protein | 0.276 (0.002) | 2.62 | 2 | 2 |
| gi 41398763 | MAP3832c | 83.46 | Hypothetical protein | 0.276 (0.091) | 2.62 | 2 | 6 |
| gi 41394457 | MAP0011 | 167.86 | PpiA | 0.280 (0.068) | 2.57 | 3 | 5 |
| gi 41396466 | MAP2013c | 87.44 | Hypothetical protein | 0.285 (0.089) | 2.51 | 2 | 3 |
| gi 41409145 | MAP3047 | 86.80 | Hypothetical protein | 0.286 (0.069) | 2.50 | 2 | 3 |
| gi 41396510 | MAP2057 | 150.58 | Hypothetical protein | 0.290 (0.034) | 2.44 | 3 | 6 |
| gi 41394882 | MAP0435c | 134.69 | Ppa | 0.295 (0.083) | 2.39 | 2 | 6 |
| gi 41395817 | MAP1367 | 516.06 | ArgG | 0.299 (0.074) | 2.35 | 6 | 9 |
| gi 41395429 | MAP0980c | 115.41 | Hypothetical protein | 0.310 (0.022) | 2.23 | 2 | 3 |
Protein score assigned by Mascot.
Descriptions are from the SwissProt database.
Mean and standard deviation for all of the MS spectra for a given protein.
Fold increase compared to the other strain. A ratio of 0.5 is no change in protein expression.
Number of unique peptides identified for the protein.
MS-MS refers to the total number of MS spectra found for the given protein.
TABLE 4.
Proteins expressed at higher relative abundance in strain 187 cytosolic fractions compared to strain K-10 cytosolic fractions
| NCBI no. | Protein | Scorea | Descriptionb | Mean of 114/114 +117c | SD of 114/114 +117c | Fold increased | No. of unique peptidese | MS-MSf |
|---|---|---|---|---|---|---|---|---|
| gi 41397329 | MAP2872c | 184.54 | FabG5_2 | 0.703 | 0.179 | 2.36 | 2 | 5 |
| gi 41397167 | MAP2710c | 357.16 | Hypothetical protein | 0.692 | 0.125 | 2.24 | 6 | 14 |
| gi 41395789 | MAP1339 | 82.60 | Hypothetical protein | 0.665 | 0.001 | 1.98 | 2 | 2 |
| gi 41394941 | MAP0494 | 383.10 | Hypothetical protein | 0.616 | 0.036 | 1.60 | 4 | 8 |
| gi 41397454 | MAP2997c | 351.82 | Hypothetical protein | 0.613 | 0.116 | 1.58 | 6 | 16 |
| gi 41408548 | MAP2450c | 113.18 | AtpC | 0.608 | 0.051 | 1.55 | 2 | 10 |
| gi 41395775 | MAP1325 | 563.23 | RpsA | 0.601 | 0.159 | 1.50 | 9 | 33 |
| gi 41396962 | MAP2506c | 117.11 | Hypothetical protein | 0.587 | 0.059 | 1.42 | 2 | 4 |
| gi 41396907 | MAP2451c | 657.81 | AtpD | 0.586 | 0.178 | 1.42 | 11 | 31 |
| gi 41396721 | MAP2267c | 194.66 | Rne | 0.578 | 0.128 | 1.37 | 3 | 3 |
| gi 41394596 | MAP0150c | 620.65 | FadE25_2 | 0.578 | 0.078 | 1.37 | 7 | 32 |
| gi 41398994 | MAP4063c | 153.40 | Hypothetical protein | 0.576 | 0.032 | 1.36 | 2 | 4 |
| gi 41394917 | MAP0470 | 98.56 | Hypothetical protein | 0.571 | 0.069 | 1.33 | 2 | 2 |
| gi 41395572 | MAP1123 | 117.28 | Gmk | 0.559 | 0.061 | 1.27 | 2 | 4 |
| gi 41396735 | MAP2281c | 209.32 | ClpP | 0.554 | 0.090 | 1.24 | 4 | 6 |
| gi 41394796 | MAP0349 | 180.79 | Hypothetical protein | 0.552 | 0.025 | 1.23 | 2 | 5 |
| gi 41398481 | MAP3551c | 178.96 | Hypothetical protein | 0.550 | 0.077 | 1.22 | 3 | 3 |
| gi 41399272 | MAP34340 | 150.17 | TrxC | 0.549 | 0.004 | 1.22 | 2 | 2 |
Protein score assigned by Mascot.
Descriptions are from the SwissProt database.
Mean and standard deviation for all of the MS spectra for a given protein.
Fold increase compared to the other strain. A ratio of 0.5 is no change in protein expression.
Number of unique peptides identified for the protein.
MS-MS refers to the total number of MS spectra found for the given protein.
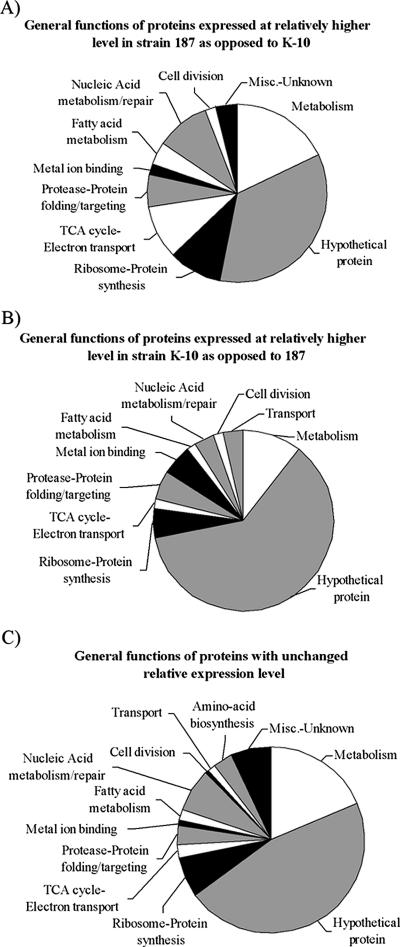
A total of 874 unique proteins were identified from M. avium subsp. paratuberculosis in this study, and 165 of these proteins were found in both the M. avium subsp. paratuberculosis membrane and cytosol preparations (see the supplemental material). Much of the protein function data available for M. avium subsp. paratuberculosis is inferred from electronic annotation and therefore must be used with that caveat in mind. Of the 58 proteins expressed at higher relative abundance in the strain K-10 membrane and cytosolic fractions compared to strain 187, 34 (57%) were hypothetical proteins (Fig. 4A). Of the 53 proteins expressed at higher relative abundance in the membrane and cytosol of strain 187 compared to strain K-10, 18 (34%) were hypothetical proteins (Fig. 4B). For the total M. avium subsp. paratuberculosis proteome, 763 proteins showed no expression changes and 374 (42%) were hypothetical proteins (Fig. 4C). Approximately 71% of the 4,350 annotated proteins from the sequencing of M. avium subsp. paratuberculosis are currently listed as hypothetical proteins (22). In each proteome grouping, hypothetical proteins were the largest category, followed by metabolism and ribosome-protein synthesis (Fig. 4A, B, and C). Clearly, as functions are attributed to the many hypothetical proteins identified, more important details will emerge from these data.
FIG. 4.
General functions of the proteins identified. (A) Fifty-three proteins were identified in strain 187 in both the membrane and cytosolic fractions at a significantly higher level compared to strain K-10 when grouped by function. The protein functions listed are from the SwissProt database and were inferred from electronic annotation. (B) Fifty-eight proteins were identified in strain K-10 in both the membrane and cytosolic fractions at a significantly higher level compared to strain 187 when grouped by function. (C) Seven hundred sixty-three proteins were identified in both strains whose relative expression level was found to be the same when grouped by function. Proteins identified in both the membrane and cytosolic fractions were counted one time. TCA, trichloroacetic acid; Misc., miscellaneous.
Immunoblot analysis confirms iTRAQ results.
Monoclonal antibodies to MAP1643, MAP2121c, and MAP3840 were used to confirm M. avium subsp. paratuberculosis iTRAQ expression data. None of these MAP proteins showed any change in expression in comparisons of M. avium subsp. paratuberculosis strains 187 and K-10 by either the iTRAQ method or immunoblotting (Fig. 3B; see the supplemental material). MAP1643 was only found in M. avium subsp. paratuberculosis cytosol, regardless of the method used. MAP3840 was identified in both the M. avium subsp. paratuberculosis membrane and cytosolic fractions by mass spectroscopy, but immunoblot data showed it to be present predominately within M. avium subsp. paratuberculosis cytosol preparations. In contrast, MAP2121c was identified only within the M. avium subsp. paratuberculosis membranes by mass spectroscopy but faint banding was observed in cytosolic fractions by immunoblotting (Fig. 3B; see the supplemental material). The correlation of immunoblot analysis and iTRAQ data with regard to both protein expression and subcellular localization supports the iTRAQ experimental results.
DISCUSSION
The aims of this study were to obtain a proteomic profile of M. avium subsp. paratuberculosis and to obtain information on the effects of in vitro growth on the relative protein expression data for two M. avium subsp. paratuberculosis strains. With just over 20% of potential M. avium subsp. paratuberculosis proteins identified by this shotgun MS-MS approach, we demonstrated that with the proper prefractionation, data sets large enough to look at global protein expression are obtainable for M. avium subsp. paratuberculosis. Moreover, this study is the most comprehensive proteomic survey of M. avium subsp. paratuberculosis performed to date. Of the 874 proteins found in both strains, 763 (87%) proteins showed no significant change in relative expression, indicating that both M. avium subsp. paratuberculosis strains expressed the majority of proteins at similar levels. As a screening protocol, the iTRAQ method proved capable of identifying 111 proteins with significantly different relative protein expression levels between the strains. The differences in protein expression ranged from no change to a sixfold increase in the expression of AtpC in strain 187 compared to strain K-10 membranes.
Since the membrane proteins are the first proteins presented to the host in an infection, it was of particular interest to us that MAP4336 was upregulated 4.7-fold in the membrane fraction of clinical isolate 187 over that seen in laboratory-adapted strain K-10. Since strain 187 was a recent isolate of M. avium subsp. paratuberculosis, it was anticipated that potential virulence proteins would be more highly expressed in strain 187 membranes. MAP4336 is an integral membrane protein from the MVIN family of proteins which, although biochemically uncharacterized, have been found to be virulence factors in other bacteria and therefore are important in bacterial pathogenesis (8, 29).
The higher expression of both GroEL1 and ClpP in clinical isolate 187 compared to strain K-10 may reflect the stress associated with the adaptation of strain 187 to laboratory culture medium. The function of GroEL1 is to promote the refolding and proper assembly of unfolded polypeptides generated under stress conditions (6). In association with this, the function of ClpP is in the degradation of misfolded proteins (21). Both of these proteins are upregulated in strain 187 to the same degree, and their complementary functions to prevent accumulation of incorrectly folded proteins make sense physiologically as this would likely occur during the transitional stress from the host environment to laboratory culture.
The slower growth of strain 187 in laboratory medium was not surprising since it was a recent isolate. But the high expression of a protein essential to cell division (FtsZ) is puzzling. Perhaps the higher expression of FtsZ in strain 187 can be explained by a slower completion of the cell division process due to the stress of movement to a laboratory environment. While in the preceding discussion the high expression of GroEL1 was attributed to stress, recent work has shown that GroEL1 associates with FtsZ in the cell division process (26). Therefore, this could explain part of the upregulation of GroEL1 seen in strain 187 compared to strain K-10.
Laboratory-adapted and low-passage strains of bacteria have been shown to possess differing characteristics (35). Of particular interest are membrane proteins which serve as the host interaction sites for these intracellular pathogens (12, 39). Low passage number strains of M. tuberculosis have been shown to retain protein expression profiles that include important antigenic markers that are not expressed in higher passage number laboratory attenuated strains (2). This is critical information suggesting that a loss of virulence genes may occur during in vitro propagation of the bacteria, which could potentially result in misdirected searches for immunogenic proteins. Fifty years ago, Segal and Bloch noted that M. tuberculosis grows in vivo by metabolizing fatty acids as opposed to carbohydrates in vitro (7, 30). The M. avium subsp. paratuberculosis membrane proteins FadE_2, InhA, FadE23, FadA1, FadE5, and FadB1 and cytosolic proteins FadE25_2 and FabG5_2 are all involved in fatty acid metabolism as determined by homology to proteins in the KEGG database (19, 20). All eight of these proteins were expressed at significantly higher levels in M. avium subsp. paratuberculosis clinical isolate 187 compared to in vitro-adapted M. avium subsp. paratuberculosis strain K-10. Laboratory medium for cultivation of M. avium subsp. paratuberculosis is supplemented with fatty acids for optimal growth. It has been shown that M. avium subsp. paratuberculosis cultured in medium lacking fatty acids shows distinct morphological changes, reduced resistance to acid conditions, and altered protein expression (35). These data are also indicative that these proteins are important for the survival of M. avium subsp. paratuberculosis within the host, enabling optimal assimilation of fatty acids for generation of the cell wall and cellular membrane. Consequently, the proteins involved may be missed or underrepresented during screening of in vitro-adapted M. avium subsp. paratuberculosis to obtain potential targets for identification of M. avium subsp. paratuberculosis-infected animals.
The RNA polymerase proteins RpoA and RpoB were both expressed at higher levels in strain K-10 compared to strain 187. RpoB, in particular, has been intensely scrutinized in M. tuberculosis as mutations in RpoB confer resistance to the antibiotic rifampin (13, 36). With a relative expression of RpoB greater than sixfold higher in strain 187 than in the laboratory-adapted strain, RpoB may prove an interesting candidate for study in M. avium subsp. paratuberculosis. Whether this represents an adaptation toward optimized growth in the laboratory strain remains to be investigated. GlnB, GlnD, and the nifs-like protein are all involved in nitrogen metabolism and were found at higher relative abundance in strain K-10 compared to strain 187, perhaps a reflection of a readily assessable nitrogen source for growth (18, 34). However, this indicates once again that strain-specific protein expression differences may also lead to an overemphasis on proteins that could be laboratory growth artifacts.
Although a necessary cofactor, excess iron can lead to oxidative stress in bacteria (9). It is thought that in response to excess iron, bacteria may induce the synthesis of proteins involved in iron binding in order to reduce iron-mediated oxidative damage (31). M. tuberculosis has more than 40 enzymes for which iron is a necessary cofactor (40). Although several iron-binding proteins were identified in the present survey, only FecB was expressed at a higher level in the strain K-10 membrane fraction. Alkyl hydroperoxide reductase C (AhpC) reduces organic hyperoxides and was present at threefold higher levels in the K-10 membrane fraction. Together, these two proteins may represent part of the adaptation response of M. avium subsp. paratuberculosis strain K-10 to oxidative stress (12). Additionally, AhpC has proven useful as an immunological identifier to distinguish M. avium from M. avium subsp. paratuberculosis, as antibodies against AhpC were found in the sera of M. avium subsp. paratuberculosis-infected goats (27). However, if field strains of M. avium subsp. paratuberculosis show lower expression of the protein in cattle, this may limit the usefulness of AhpC to screen for infection.
The results from this study demonstrate the potential for analyzing relative protein levels from large data sets. In addition to being the most comprehensive proteome to date for M. avium subsp. paratuberculosis, 451 hypothetical proteins inferred from the nucleotide sequence have been identified. Proteins expressed at relatively higher levels in the low-passage strain may prove useful in the development of diagnostic techniques, while proteins expressed at relatively higher levels in the high-passage strain need to be analyzed with that in mind. Although the present study is based upon a limited number of high- and low-passage M. avium subsp. paratuberculosis strains, we have identified differences in relative protein levels that both confirm expectations and challenge previous findings. It is clear that laboratory adaptation of M. avium subsp. paratuberculosis through continuous passage in complete medium will alter the growth pattern and protein expression profile, but how this directly affects the virulence of the bacterium remains to be tested. The broad scope of proteomic analysis coupled with the added power of direct comparison of individual proteins holds the potential for applications similar to that found in nucleic acid chip technology.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 1 December 2006.
REFERENCES
- 1.Aggarwal, K., L. H. Choe, and K. H. Lee. 2006. Shotgun proteomics using the iTRAQ isobaric tags. Brief. Funct. Genomics Proteomics 5:112-120. [DOI] [PubMed] [Google Scholar]
- 2.Bahk, Y. Y., S. A. Kim, J. S. Kim, H. J. Euh, G. H. Bai, S. N. Cho, and Y. S. Kim. 2004. Antigens secreted from Mycobacterium tuberculosis: identification by proteomics approach and test for diagnostic marker. Proteomics 4:3299-3307. [DOI] [PubMed] [Google Scholar]
- 3.Bannantine, J. P., J. F. J. Huntley, E. Miltner, J. R. Stabel, and L. E. Bermudez. 2003. The Mycobacterium avium subsp. paratuberculosis 35 kDa protein plays a role in invasion of bovine epithelial cells. Microbiology 149:2061-2069. [DOI] [PubMed] [Google Scholar]
- 4.Bannantine, J. P., and J. R. Stabel. 2001. Identification of two Mycobacterium avium subspecies paratuberculosis gene products differentially recognised by sera from rabbits immunised with live mycobacteria but not heat-killed mycobacteria. J. Med. Microbiol. 50:795-804. [DOI] [PubMed] [Google Scholar]
- 5.Begg, D. J., R. O'Brien, C. G. Mackintosh, and J. F. Griffin. 2005. Experimental infection model for Johne's disease in sheep. Infect. Immun. 73:5603-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Zvi, A. P., and P. Goloubinoff. 2001. Review: mechanisms of disaggregation and refolding of stable protein aggregates by molecular chaperones. J. Struct. Biol. 135:84-93. [DOI] [PubMed] [Google Scholar]
- 7.Bloch, H., and W. Segal. 1956. Biochemical differentiation of Mycobacterium tuberculosis grown in vivo and in vitro. J. Bacteriol. 72:132-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carsiotis, M., B. A. Stocker, D. L. Weinstein, and A. D. O'Brien. 1989. A Salmonella typhimurium virulence gene linked to flg. Infect. Immun. 57:3276-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castagnetto, J. M., S. W. Hennessy, V. A. Roberts, E. D. Getzoff, J. A. Tainer, and M. E. Pique. 2002. MDB: the Metalloprotein Database and Browser at The Scripps Research Institute. Nucleic Acids Res. 30:379-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, D., N. Sung, and M. T. Collins. 2006. Identification of proteins of potential diagnostic value for bovine paratuberculosis. Proteomics 6:5785-5794. [DOI] [PubMed] [Google Scholar]
- 11.Cocito, C., P. Gilot, M. Coene, M. de Kesel, P. Poupart, and P. Vannuffel. 1994. Paratuberculosis. Clin. Microbiol. Rev. 7:328-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fandiño, A. S., I. Rais, M. Vollmer, H. Elgass, H. Schagger, and M. Karas. 2005. LC-nanospray-MS/MS analysis of hydrophobic proteins from membrane protein complexes isolated by blue-native electrophoresis. J. Mass Spectrom. 40:1223-1231. [DOI] [PubMed] [Google Scholar]
- 13.Felmlee, T. A., Q. Liu, A. C. Whelen, D. Williams, S. S. Sommer, and D. H. Persing. 1995. Genotypic detection of Mycobacterium tuberculosis rifampin resistance: comparison of single-strand conformation polymorphism and dideoxy fingerprinting. J. Clin. Microbiol. 33:1617-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, Q., K. Kripke, Z. Arinc, M. Voskuil, and P. Small. 2004. Comparative expression studies of a complex phenotype: cord formation in Mycobacterium tuberculosis. Tuberculosis 84:188-196. [DOI] [PubMed] [Google Scholar]
- 15.Gumber, S., D. L. Taylor, and R. J. Whittington. 17 August 2006. Protein extraction from Mycobacterium avium subsp. paratuberculosis: comparison of methods for analysis by sodium dodecyl sulphate polyacrylamide gel electrophoresis, native PAGE and surface enhanced laser desorption/ionization time of flight mass spectrometry. J. Microbiol. Methods (Epub ahead of print) doi: 10.1016/j.mimet.2006.07.003. [DOI] [PubMed]
- 16.Harris, N. B., and R. G. Barletta. 2001. Mycobacterium avium subsp. paratuberculosis in veterinary medicine. Clin. Microbiol. Rev. 14:489-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huntley, J. F., J. R. Stabel, M. L. Paustian, T. A. Reinhardt, and J. P. Bannantine. 2005. Expression library immunization confers protection against Mycobacterium avium subsp. paratuberculosis infection. Infect. Immun. 73:6877-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakoby, M., R. Kramer, and A. Burkovski. 1999. Nitrogen regulation in Corynebacterium glutamicum: isolation of genes involved and biochemical characterization of corresponding proteins. FEMS Microbiol. Lett. 173:303-310. [DOI] [PubMed] [Google Scholar]
- 19.Kanehisa, M. 1997. A database for post-genome analysis. Trends Genet. 13:375-376. [DOI] [PubMed] [Google Scholar]
- 20.Kanehisa, M., and S. Goto. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krüger, E., E. Witt, S. Ohlmeier, R. Hanschke, and M. Hecker. 2000. The Clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins. J. Bacteriol. 182:3259-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, L., J. P. Bannantine, Q. Zhang, A. Amonsin, B. J. May, D. Alt, N. Banerji, S. Kanjilal, and V. Kapur. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. USA 102:12344-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lippolis, J. D., B. D. Peterson-Burch, and T. A. Reinhardt. 2006. Differential expression analysis of proteins from neutrophils in the periparturient period and neutrophils from dexamethasone-treated dairy cows. Vet. Immunol. Immunopathol. 111:149-164. [DOI] [PubMed] [Google Scholar]
- 24.Lippolis, J. D., and T. A. Reinhardt. 2005. Proteomic survey of bovine neutrophils. Vet. Immunol. Immunopathol. 103:53-65. [DOI] [PubMed] [Google Scholar]
- 25.Mawuenyega, K. G., C. V. Forst, K. M. Dobos, J. T. Belisle, J. Chen, E. M. Bradbury, A. R. Bradbury, and X. Chen. 2005. Mycobacterium tuberculosis functional network analysis by global subcellular protein profiling. Mol. Biol. Cell 16:396-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogino, H., M. Wachi, A. Ishii, N. Iwai, T. Nishida, S. Yamada, K. Nagai, and M. Sugai. 2004. FtsZ-dependent localization of GroEL protein at possible division sites. Genes Cells 9:765-771. [DOI] [PubMed] [Google Scholar]
- 27.Olsen, I., L. J. Reitan, G. Holstad, and H. G. Wiker. 2000. Alkyl hydroperoxide reductases C and D are major antigens constitutively expressed by Mycobacterium avium subsp. paratuberculosis. Infect. Immun. 68:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross, P. L., Y. N. Huang, J. N. Marchese, B. Williamson, K. Parker, S. Hattan, N. Khainovski, S. Pillai, S. Dey, S. Daniels, S. Purkayastha, P. Juhasz, S. Martin, M. Bartlet-Jones, F. He, A. Jacobson, and D. J. Pappin. 2004. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3:1154-1169. [DOI] [PubMed] [Google Scholar]
- 29.Rudnick, P. A., T. Arcondeguy, C. K. Kennedy, and D. Kahn. 2001. glnD and mviN are genes of an essential operon in Sinorhizobium meliloti. J. Bacteriol. 183:2682-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segal, W., and H. Bloch. 1957. Pathogenic and immunogenic differentiation of Mycobacterium tuberculosis grown in vitro and in vivo. Am. Rev. Tuberc. 75:495-500. [DOI] [PubMed] [Google Scholar]
- 31.Smith, J. L. 2004. The physiological role of ferritin-like compounds in bacteria. Crit. Rev. Microbiol. 30:173-185. [DOI] [PubMed] [Google Scholar]
- 32.Spahr, C. S., M. T. Davis, M. D. McGinley, J. H. Robinson, E. J. Bures, J. Beierle, J. Mort, P. L. Courchesne, K. Chen, R. C. Wahl, W. Yu, R. Luethy, and S. D. Patterson. 2001. Towards defining the urinary proteome using liquid chromatography-tandem mass spectrometry. I. Profiling an unfractionated tryptic digest. Proteomics 1:93-107. [DOI] [PubMed] [Google Scholar]
- 33.Stabel, J. R. 1998. Johne's disease: a hidden threat. J. Dairy Sci. 81:283-288. [DOI] [PubMed] [Google Scholar]
- 34.Sun, J., A. Van Dommelen, J. Van Impe, and J. Vanderleyden. 2002. Involvement of glnB, glnZ, and glnD genes in the regulation of poly-3-hydroxybutyrate biosynthesis by ammonia in Azospirillum brasilense Sp7. Appl. Environ. Microbiol. 68:985-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sung, N., and M. T. Collins. 2003. Variation in resistance of Mycobacterium paratuberculosis to acid environments as a function of culture medium. Appl. Environ. Microbiol. 69:6833-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, M. J. Colston, L. Matter, K. Schopfer, and T. Bodmer. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 37.Vary, P. H., P. R. Andersen, E. Green, J. Hermon-Taylor, and J. J. McFadden. 1990. Use of highly specific DNA probes and the polymerase chain reaction to detect Mycobacterium paratuberculosis in Johne's disease. J. Clin. Microbiol. 28:933-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whittington, R. J., I. Marsh, S. McAllister, M. J. Turner, D. J. Marshall, and C. A. Fraser. 1999. Evaluation of modified BACTEC 12B radiometric medium and solid media for culture of Mycobacterium avium subsp. paratuberculosis from sheep. J. Clin. Microbiol. 37:1077-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong, Y., M. J. Chalmers, F. P. Gao, T. A. Cross, and A. G. Marshall. 2005. Identification of Mycobacterium tuberculosis H37Rv integral membrane proteins by one-dimensional gel electrophoresis and liquid chromatography electrospray ionization tandem mass spectrometry. J. Proteome Res. 4:855-861. [DOI] [PubMed] [Google Scholar]
- 40.Yellaboina, S., S. Ranjan, V. Vindal, and A. Ranjan. 2006. Comparative analysis of iron regulated genes in mycobacteria. FEBS Lett. 580:2567-2576. [DOI] [PubMed] [Google Scholar]
- 41.Zurbrick, B. G., and C. J. Czuprynski. 1987. Ingestion and intracellular growth of Mycobacterium paratuberculosis within bovine blood monocytes and monocyte-derived macrophages. Infect. Immun. 55:1588-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.