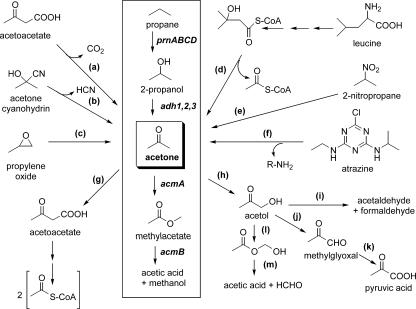
In this issue of the Journal of Bacteriology, Kotani et al. (11) elucidate the latter half of a new pathway for propane metabolism in Gordonia sp. strain TY-5, thus complementing their work on the earlier steps of the pathway from 3 years ago (10). Utilization of this gaseous three-carbon substrate, depicted in the boxed region of Fig. 1, provides new insights into the microbial metabolism of acetone—a central intermediate of the overall scheme. The initial step of propane utilization involves a four-component putative di-iron monooxygenase (encoded by prnABCD) that catalyzes the NADH-dependent subterminal hydroxylation of the alkane to form 2-propanol. Three distinct NAD+-dependent secondary alcohol dehydrogenases (encoded by adh1, adh2, and adh3) convert the hydroxylated intermediate to acetone. A flavin adenine dinucleotide (FAD)-dependent monooxygenase (encoded by acmA) catalyzes an NADPH-dependent Baeyer-Villiger reaction that inserts an oxygen atom into a carbon-carbon bond of acetone to produce methyl acetate. Finally, a hydrolase (encoded by acmB) splits the ester into acetic acid and methanol. Consistent with their proposed functions, these genes are induced by propane, 2-propanol, and acetone.
FIG. 1.
Overview of acetone metabolism. The boxed region depicts the novel pathway of propane metabolism uncovered in Gordonia sp. strain TY-5 (10, 11). Additional reactions that generate or degrade acetone are illustrated outside of the box.
The central position of acetone in the propane oxidation pathway is placed into a broader context by comparison to the other acetone-related reactions illustrated in Fig. 1. Many anaerobic bacteria, such as those in the genus Clostridium (4), produce acetone during fermentation by decarboxylation of acetoacetate (reaction a). Selected plants decompose cyanogenic glucosides to generate acetone cyanohydrin, which is metabolized by a lyase (reaction b) to form hydrogen cyanide and acetone (7). Epoxypropane, formed in certain bacteria by monooxygenation of propene, can undergo isomerization (reaction c) to acetone (18). Various members of the vibrio family possess a multistep pathway of leucine catabolism that produces 3-hydroxy-3-methylglutaryl-coenzyme A (15), which decomposes to yield this ketone (reaction d). Other microbes oxidize 2-nitropropane (reaction e), atrazine (reaction f), or other compounds to produce this molecule (13, 14). In the same manner, alternative pathways for acetone decomposition have been described. One well-characterized pathway involves the ATP-dependent, manganese-containing enzyme acetone carboxylase (reaction g) that forms acetoacetate, which is subsequently converted through multiple steps to form two molecules of acetyl-coenzyme A (1, 17). The conversion of acetone to acetol by cytochrome P450 has long been known (9), and other types of monooxygenases also may be capable of catalyzing such terminal hydroxylation (scheme h). Acetol cleavage to form acetaldehyde and formaldehyde (reaction i) has been proposed (5); however, no characterization of this enzyme (presumably containing thiamine pyrophosphate to facilitate this chemistry) has been reported. Acetol dehydrogenase (reaction j) and methylglyoxal dehydrogenase (reaction k) reactions have been characterized (3) and would convert acetol to pyruvate. Finally, evidence was presented 20 years ago for an NADPH-dependent acetol monooxygenase that catalyzes Baeyer-Villiger chemistry (reaction l) in Mycobacterium sp. strain P1 (8); the resulting hydroxymethylene acetate would spontaneously decompose (reaction m) to yield acetic acid and formaldehyde. In sum, the reactions shown in Fig. 1 highlight the position of acetone at a central metabolic crossroads.
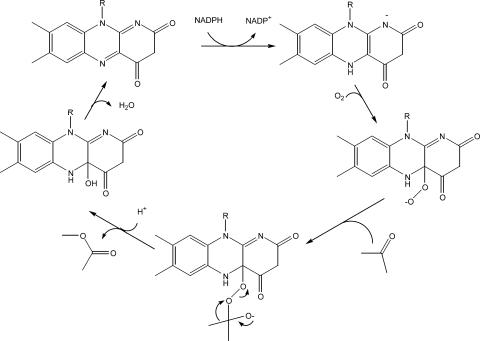
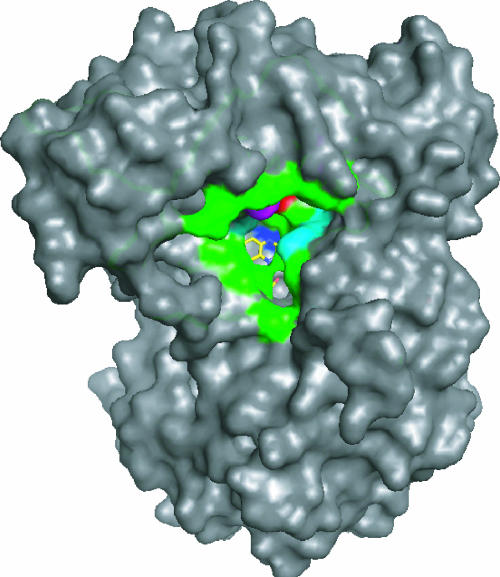
Although Baeyer-Villiger-type monooxygenases have been studied for decades, the Gordonia enzyme encoded by acmA appears to be unique in using the three-carbon compound acetone as an effective substrate. Indeed, a review of the literature reveals only slight activity with butanone as the smallest alternate substrate in one enzyme representative (2). The Baeyer-Villiger monooxygenases are flavoenzymes, and this is certainly the situation for AcmA. On the basis of its weak yellow color, this protein probably loses a portion of its FAD cofactor during purification—a situation known to occur during isolation of other family members (6, 12). Figure 2 depicts a reasonable FAD-dependent catalytic reaction for AcmA, using as a precedent the results of kinetic, mechanistic, and structural studies carried out with related enzymes. The figure is not meant to imply the order of substrate binding. For example, AcmA may exhibit a ter-ter kinetic mechanism, as described for cyclohexanone monooxygenase (16). Alternatively, the Gordonia enzyme might reduce its flavin by using NADPH, undergo a conformational change to react with oxygen, and then bind the substrate, as suggested by structural studies carried out with Thermobifida fusca phenylacetone monooxygenase (12). This structurally characterized family member is 43% identical in sequence to AcmA, raising questions concerning the structural basis of substrate specificity. Unfortunately, no structure is available for a substrate-bound form of any Baeyer-Villiger monooxygenase. Figure 3 depicts a surface representation of the T. fusca protein with its flavin illustrated in yellow. The residues lining its substrate-binding pocket are compared to the Gordonia enzyme and shown in green if conserved (Asp66, Leu153, Arg217, Thr218, Tyr331, Arg337, Gly388, Phe389, Ala391, and Ser500) or in other coloring when substituted (C65F, blue; S196A, teal; H220Q, magenta; K336H, cyan; I339P, red). Further comparisons of the sequences indicate a single amino acid deletion in AcmA, corresponding to Thr256 of the T. fusca enzyme, and three single amino acid insertions in the Gordonia protein (between Ile288 and Leu289, Glu375 and Arg376, and Ser441 and Ala442 of the phenylacetone monooxygenase). The first three of these changes are distant from the active site and unlikely to be important to substrate specificity or catalysis, whereas the latter insertion is predicted to occur near the FAD. Additional kinetic studies are needed to determine the catalytic efficiencies of the various substrates of AcmA, and mutagenic or structural investigations are required to discern the structural basis of the substrate specificity of these enzymes.
FIG. 2.
Mechanism of acetone monooxygenase AcmA, a Baeyer-Villiger-type monooxygenase. The order of substrate binding and product release is unknown; however, the reduced nicotinamide must be used to first reduce the flavin, the reduced flavin then reacts with oxygen, the peroxyflavin intermediate reacts with acetone, and a Criegee rearrangement leads to product formation.
FIG. 3.
Surface representation of phenylacetone monooxygenase and comparison of residues lining its substrate-binding pocket with those of AcmA. The structure of T. fusca phenylacetone monooxygenase (Protein Data Bank access code 1w4x) is shown in a surface representation (with the FAD depicted in yellow). Residues likely to be important for access to the active site or catalysis are compared to the corresponding residues of AcmA (green if conserved or other colors if they differ; see text).
Acknowledgments
I thank Jong Kyong Kim for comments on the text.
Research in my laboratory was supported by National Institutes of Health grant GM063584.
Footnotes
Published ahead of print on 1 December 2006.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Boyd, J. M., H. Ellsworth, and S. A. Ensign. 2004. Bacterial acetone carboxylase is a manganese-dependent metalloenzyme. J. Biol. Chem. 279:46644-46651. [DOI] [PubMed] [Google Scholar]
- 2.Britton, L. N., and A. J. Markovetz. 1977. A novel ketone monooxygenase from Pseudomonas cepacia. Purification and properties. J. Biol. Chem. 252:8561-8566. [PubMed] [Google Scholar]
- 3.Cooper, R. A. 1984. Metabolism of methylglyoxal in microorganisms. Annu. Rev. Microbiol. 38:49-68. [DOI] [PubMed] [Google Scholar]
- 4.Dürre, P., R. J. Fischer, A. Kuhn, K. Lorenz, W. Schreiber, B. Sturzenhofecker, S. Ullman, K. Winzer, and U. Sauer. 1995. Solventogenic enzymes of Clostridium acetobutylicum: catalytic properties, genetic organization, and transcriptional regulation. FEMS Microbiol. Rev. 17:251-262. [DOI] [PubMed] [Google Scholar]
- 5.Ensign, S. A., F. J. Small, J. R. Allen, and M. K. Sluis. 1998. New roles for CO2 in the microbial metabolism of aliphatic epoxides and ketones. Arch. Microbiol. 169:179-187. [DOI] [PubMed] [Google Scholar]
- 6.Griffin, M., and P. W. Trudgill. 1976. Purification and properties of cyclohexanone oxygenase of Pseudomonas NCBI 9872. Eur. J. Biochem. 63:199-209. [DOI] [PubMed] [Google Scholar]
- 7.Gruber, K., G. Gartler, B. Krammer, H. Schwab, and C. Kratky. 2004. Reaction mechanism of hydroxynitrile lyases of the α/β-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of Lys236. J. Biol. Chem. 279:20501-20510. [DOI] [PubMed] [Google Scholar]
- 8.Hartmans, S., and J. A. M. de Bont. 1986. Acetol monooxygenase from Mycobacterium Py1 cleaves acetol into acetate and formaldehyde. FEMS Microbiol. Lett. 36:155-158. [Google Scholar]
- 9.Koop, D. R., and J. P. Casazza. 1985. Identification of ethanol-inducible P-450 isozyme 3a as the acetone and acetol monooxygenase of rabbit microsomes. J. Biol. Chem. 260:13607-13612. [PubMed] [Google Scholar]
- 10.Kotani, T., T. Yamamoto, H. Yurimoto, Y. Sakai, and N. Kato. 2003. Propane monooxygenase and NAD+-dependent secondary alcohol dehydrogenase in propane metabolism by Gordonia sp. strain TY-5. J. Bacteriol. 185:7120-7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotani, T., H. Yurimoto, N. Kato, and Y. Sakai. 2007. Novel acetone metabolism in a propane-utilizing bacterium, Gordonia sp. strain TY-5. J. Bacteriol. 189:886-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malito, E., A. Alfieri, M. W. Fraaije, and A. Mattevi. 2004. Crystal structure of a Baeyer-Villiger monooxygenase. Proc. Natl. Acad. Sci. USA 101:13157-13162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagpal, A., M. P. Valley, P. F. Fitzpatrick, and A. M. Orville. 2006. Crystal structures of nitroalkane oxidase: insights into the reaction mechanism from a covalent complex of the flavoenzyme trapped during turnover. Biochemistry 45:1138-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagy, I., F. Compernolle, K. Ghys, J. Vanderleyden, and R. De Mot. 1995. A single cytochrome P-450 system is involved in degradation of the herbicides EPTC (S-ethyl dipropylthiocarbamate) and atrazine by Rhodococcus sp. strain NI86/21. Appl. Environ. Microbiol. 61:2056-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nemecek-Marshall, M., C. Wojciechowski, W. P. Wagner, and R. Fall. 1999. Acetone formation in the vibrio family: a new pathway for bacterial leucine catabolism. J. Bacteriol. 181:7493-7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryerson, C. C., D. P. Ballou, and C. T. Walsh. 1982. Mechanistic studies on cyclohexanone oxygenase. Biochemistry 21:2644-2655. [DOI] [PubMed] [Google Scholar]
- 17.Sluis, M. K., and S. A. Ensign. 1997. Purification and characterization of acetone carboxylase from Xanthobacter strain Py2. Proc. Natl. Acad. Sci. USA 94:8456-8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Small, F. J., J. K. Tilley, and S. A. Ensign. 1995. Characterization of a new pathway for epichlorohydrin degradation by whole cells of Xanthobacter strain Py2. Appl. Environ. Microbiol. 61:1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]