Abstract
The Pseudomonas aeruginosa outer membrane is intrinsically impermeable to many classes of antibiotics, due in part to its relative lack of general uptake pathways. Instead, this organism relies on a large number of substrate-specific uptake porins. Included in this group are the 19 members of the OprD family, which are involved in the uptake of a diverse array of metabolites. One of these porins, OpdH, has been implicated in the uptake of cis-aconitate. Here we demonstrate that this porin may also enable P. aeruginosa to take up other tricarboxylates. Isocitrate and citrate strongly and specifically induced the opdH gene via a mechanism involving derepression by the putative two-component regulatory system PA0756-PA0757. Planar bilayer analysis of purified OpdH demonstrated that it was a channel-forming protein with a large single-channel conductance (230 pS in 1 M KCl; 10-fold higher than that of OprD); however, we were unable to demonstrate the presence of a tricarboxylate binding site within the channel. Thus, these data suggest that the requirement for OpdH for efficient growth on tricarboxylates was likely due to the specific expression of this large-channel porin under particular growth conditions.
The outer membrane of the opportunistic pathogen Pseudomonas aeruginosa is intrinsically poorly permeable to many classes of compounds, with a permeability coefficient that is 10- to 500-fold lower than that of Escherichia coli (17, 60). Despite the barrier presented by the outer membrane, P. aeruginosa is renowned for its metabolic versatility and is capable of importing and using such diverse compounds as carbohydrates, alcohols, organic acids, aromatic compounds, aliphatic compounds, and amino acids and other nitrogenous compounds as nutrient sources (44).
These hydrophilic nutrients enter the cell via outer-membrane-spanning, water-filled channels called porins. The major P. aeruginosa outer membrane porin is OprF. This protein belongs to a class of porins termed the slow porins, wherein one of two channel sizes is formed (31, 32, 46). The majority of OprF porins consist of small pores (0.36 nS) that provide rather inefficient uptake routes; however, a small fraction of OprF molecules (approximately 1 out of every 500) form larger channels (2 to 5 nS) that can facilitate the uptake of di-, tri-, and tetrasaccharides. Thus, it is believed that the majority of outer membrane transport occurs through substrate-specific porins (5, 11, 19).
Pseudomonas species possess a large collection of specific porins (48). The most extensively characterized specific porins of P. aeruginosa include the carbohydrate-specific porin OprB (58), the phosphate-specific porin OprP (18), the orthophosphate-specific porin OprO (42), and the basic amino acid-specific porin OprD (50). Generally, these porins tend to form narrow pores that are often occluded by long surface loops (27). The uptake of specific classes of molecules is facilitated through these channels by virtue of substrate-specific binding sites that orient substrates into favorable conformations as they traverse the channel (39, 59). It should be noted that certain compounds that do not recognize these binding sites are also able to traverse specific porins, increasing their net contribution to outer membrane permeability. A requirement for the passage of these nonspecific molecules is that they do not interact with the amino acids lining the interior of the channel. In P. aeruginosa, this nonspecific uptake activity by a specific porin has been observed with the basic amino acid-specific porin OprD, which enables the passive diffusion of gluconate (22).
OprD is the prototypical member of the recently characterized OprD family of porins, consisting of OprD and 18 homologues (45). In P. aeruginosa there are 19 members of this family, 8 of which have been shown to selectively take up diverse molecules such as amino acids, dipeptides, vanillate, pyroglutamate, and cis-aconitate (49). In this work, the functional and regulatory aspects of one OprD homologue, the cis-aconitate uptake porin OpdH, were investigated in greater detail. It is demonstrated that this porin is a highly regulated protein that may have a general role in tricarboxylate uptake. As well, OpdH appears to be unique among the Pseudomonas porins in forming a relatively large channel that does not contain a tricarboxylate-specific binding site. Based on the data presented here, we conclude that the regulation of OpdH expression, rather than its channel architecture, is the primary determinant of this porin's contribution to outer membrane permeability.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. A list of the primer sequences used in this study is available from the authors. All chemicals used were obtained from either Sigma or Fisher. Strains were routinely maintained on Luria-Bertani (LB) agar. Antibiotics for selection and maintenance were used at the following concentrations: tetracycline, 100 μg/ml for P. aeruginosa and 12.5 μg/ml for E. coli; gentamicin, 50 μg/ml for P. aeruginosa and 15 μg/ml for E. coli; carbenicillin, 500 μg/ml for P. aeruginosa; ampicillin, 100 μg/ml for E. coli. For cloning experiments, strains were grown in LB broth. For all other experiments, P. aeruginosa strains were grown in BM2 minimal medium (62 mM potassium phosphate buffer [pH 7], 0.5 mM MgSO4, 10 μM FeSO4) supplemented with specific carbon sources at the indicated concentrations. Overnight cultures were diluted to a starting optical density of 0.1 for growth experiments. For reporter gene assays, the cultures were inoculated with single colonies from LB plates. When required, the pH of the carbon sources was adjusted to 7. For experiments requiring low magnesium, the Mg2+ concentration was altered to 20 μM. High-iron conditions were achieved by adding FeSO4 to a final concentration of 100 μM to the medium. Low-iron conditions were created by culturing the bacteria in acid-washed glassware and adding dipyridyl to a final concentration of 0.3 mM.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source | ||
|---|---|---|---|---|
| Strains
|
||||
| P. aeruginosa
|
||||
| H103 | PAO wild-type strain | |||
| opdH′-4 | H103 opdH::xylE-Gmr/opdH | This study | ||
| opdH-10 | H103 opdH::xylE-Gmr | This study | ||
| PAK | PAK wild-type strain | 49 | ||
| opdH-26 | PAK opdH::mini-Tn5-Tcr | 49 | ||
| UW-PAO | PAO reference strain | 24 | ||
| UW-PA0756 | PAO PA0756::mini-Tn5-Tcr | 24 | ||
| UW-PA0757 | PAO PA0757::mini-Tn5-Tcr | 24 | ||
| UW-PA0756′ | UW-PA0756 complemented with PA0756 in pUCP20 | This study | ||
| UW-cbrA | PAO cbrA::mini-Tn5-Tcr | 24 | ||
| Crc | PAO crc::Tcr | 56 | ||
| H851 | H103 phoP::xylE-Gmr | 29 | ||
| PA0748 | H103 PA0748::luxCDABE-Tcr | 28 | ||
| RpoS | PAO rpoS::Tcr | 52 | ||
| E. coli
|
||||
| DH5α | General cloning strain | 16 | ||
| S17-1 | Mobilizing strain | 35 | ||
| Plasmids
|
||||
| pCR2.1 | TA cloning vector; Apr Kmr | Invitrogen | ||
| pEX18T | Suicide vector containing sacB gene; Tcr | 21 | ||
| pX1918GT | Source of xylE-Gmr cassette; Apr Gmr | 41 | ||
| pUCP20 | Broad-host-range expression vector; Cbr | 53 | ||
Genetic manipulations.
Routine cloning procedures were carried out as described by Sambrook et al. (36). All enzymes and cloning kits were purchased from Invitrogen (Carlsbad, CA), with the exception of Vent polymerase, which was from New England Biolabs (Ipswich, MA). An opdH knockout and transcriptional fusion was constructed by amplifying the gene from P. aeruginosa strain H103 with Vent polymerase in 1× reaction buffer, with 2 mM MgSO4, 5% dimethyl sulfoxide, 0.4 mM deoxynucleoside triphosphates (dNTPs), and 1 μM each primer. After an initial denaturation step at 94°C for 2 min, the amplification reaction mixture was incubated for 25 cycles at 94°C for 1 min, 63.5°C for 1 min, and 72°C for 1.5 min, followed by a final extension step at 72°C for 5 min. The resulting amplicon was treated with Taq polymerase, cloned into pCR2.1 TOPO by using the TA cloning kit, and transformed into electrocompetent E. coli DH5α. The opdH gene was excised from pCR2.1 TOPO by using HindIII and XbaI and was cloned into the corresponding sites in pEX18Tc. The opdH gene was then cut with SbfI, and a xylE-Gmr cassette from pX1918GT was cloned into this site. The orientation of the xylE-Gmr cassette was confirmed by restriction analysis. This plasmid was transformed into electrocompetent E. coli S17-1 and then mobilized into P. aeruginosa H103 by biparental mating followed by successive selection on gentamicin and 5% sucrose. The replacement of native opdH with opdH::xylE-Gmr in the resulting strain was confirmed by PCR analysis.
The opdH-deficient strain opdH-10 was complemented by creating a merodiploid. Essentially, a mating reaction between E. coli strain S17-1 harboring the opdH::xylE-Gmr construct and P. aeruginosa H103 was performed. The resulting clones were only selected on gentamicin, leaving a wild-type copy of the opdH gene in the resulting strain. The presence of both the wild-type and the insertionally inactivated copies of the opdH gene was confirmed by PCR analysis.
The PA0756 gene was cloned into the UW-PA0756 mutant strain by amplifying the gene as well as the 200-bp region upstream of it from UW-PAO with Platinum Pfx in 2× reaction buffer and 2× of the provided enhancer solution, with 1 mM MgSO4, 0.3 mM dNTPs, and 0.3 μM each primer. This mixture was incubated at 94°C for 5 min, followed by 25 cycles consisting of 15 s at 94°C, 30 s at 56.4°C, and 1 min at 68°C, and a final extension step at 68°C for 3 min. The resulting amplicon was cloned into pCR2.1 TOPO, excised with EcoRI and XbaI, and subcloned into pUCP20. The orientation of the gene was confirmed by restriction analysis, and the resulting construct was transformed into chemically competent UW-PA0756. The presence of the PA0756 gene in the complemented strain was confirmed by both restriction and PCR analysis.
Outer membrane isolation, protein electrophoresis, and sequencing.
Outer membranes were isolated from stationary-phase cultures as described previously (20). Cells were harvested, and the cell pellet was resuspended in an equal volume of cold 20% sucrose containing 100 μg/ml freshly prepared DNase I (Pharmacia, Somerset County, NJ) and incubated at room temperature for at least an hour. The cells were passed through a French pressure cell (American Instrument Co., Silver Spring, MD) twice at 1,500 lb/in2. The resulting lysate was layered onto a two-step sucrose gradient consisting of cold 70% and 50% sucrose, placed in a swinging bucket rotor (SW28; Beckman Instruments, Fullerton, CA), and spun in a Beckman Coulter Optima L-100XP ultracentrifuge at 28,000 rpm for 16 h at 4°C. The outer membrane fractionated at the interface between the 50% and 70% sucrose. This band was collected, diluted with distilled water, and centrifuged in a 70 Ti rotor (Beckman) at 43,000 rpm for 1 h at 4°C. Outer membranes were resuspended in 10 mM Tris-HCl (pH 8), and the protein concentration was determined as described in reference 37. Proteins were resolved in 11% sodium dodecyl sulfate (SDS)-acrylamide gels as described in reference 20.
The N-terminal sequence of the putative OpdH protein was determined by Edman degradation. Following electrophoresis, the proteins were transferred onto a polyvinylidene difluoride 1.0-μm-pore-size membrane (Millipore, Billerica, MA). The band corresponding to OpdH was excised and sequenced using an Applied Biosystems (Foster City, CA) 494 HT protein sequencer.
Reporter gene assays.
The catechol-2,3-dioxygenase (CDO) activities of the opdH::xylE-Gmr transcriptional fusion were determined as described previously (41). Cells were harvested at the indicated times, resuspended in 50 mM potassium phosphate buffer (pH 7.5) plus 10% acetone, and lysed by sonication. Unbroken cells were removed by centrifugation. The reaction buffer consisted of 50 mM potassium phosphate buffer (pH 7.5) and 0.3 mM catechol. Aliquots of the cell extracts were added to the reaction buffer, and the increase in absorbance at 375 nm was monitored on a dual-beam PYE Unicam PU8600 UV-visible light spectrophotometer (Philips, Amsterdam, The Netherlands). The change in absorbance was used to calculate the amount of 2-hydroxymuconic semialdehyde produced by using a molar extinction coefficient of 44,000. The protein content of the cellular extracts was determined by a modified Lowry method (37).
SQ-PCR.
Semiquantitative PCR (SQ-PCR) was carried out as described previously (49). Briefly, mRNA was isolated from exponential-phase cells with an RNeasy Mini RNA isolation kit (QIAGEN, Mississauga, Ontario, Canada). Residual genomic DNA was removed using a DNA-free kit (Ambion, Austin, TX). The RNA was quantified by determining its absorbance at 260 nm, and its quality was assessed by comparing its absorbance at 260 nm with its absorbance at 280 nm and by examining its appearance on a 2% agarose-Tris-acetate-EDTA gel. RNA aliquots were stored at −80°C.
Four micrograms of total RNA was combined with 750 ng of the random decamer (NS)5 and incubated at 70°C for 10 min, followed by 10 min at 25°C. A mixture containing 1× reverse transcription reaction buffer, 10 μM dithiothreitol, 0.5 μM dNTPs, 500 U/ml of Superase IN (Ambion), and 10,000 U/ml of Superscript reverse transcriptase II (Invitrogen) was added to the RNA and incubated for 1 h at 37°C and 2 h at 42°C. Following reverse transcription, the RNA was destroyed by the addition of 170 mM NaOH and incubation at 65°C for 15 min. The mixture was neutralized by the addition of 170 mM HCl.
One microliter of the resulting cDNA was used as the template for SQ-PCR using complementary designed to either the intergenic or the intragenic regions of the specified genes. The amplification reactions were carried out using Taq polymerase (Invitrogen) in 1× reaction buffer containing 2 mM MgCl2, 5% dimethyl sulfoxide, 0.4 mM dNTPs, and 40 nM forward and reverse primers. The reactions were cycled for 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C 20 or 25 times. The amplicons were resolved by electrophoresis on 2% agarose-Tris-acetate-EDTA gels stained with 50 μg/ml ethidium bromide. The intensities of the resulting bands on the agarose gels were quantified using ImageJ (http://rsb.info.nih.gov/ij/). All reported values were normalized to the levels of rpsL.
OpdH purification.
Outer membranes were isolated from stationary-phase cultures of an OprE-deficient mutant grown in BM2 minimal medium containing 10 mM citrate (pH 7) as described above. The outer membrane pellet was resuspended in 0.5% octyl-POE (Bachem, Torrance, CA) in 10 mM Tris-HCl (pH 8), placed on a rotating platform for 1 h at 37°C, and centrifuged at 43,000 rpm for 45 min at 4°C. The supernatant was reserved, and the procedure was repeated using 3% octyl-POE, followed by 3% octyl-POE plus 0.2 M NaCl and then 3% octyl-POE plus 5 mM EDTA (pH 8). The supernatant from this final solubilization step contained OpdH and was dialyzed for 2 days in 0.6% octyl-POE-10 mM EDTA-10 mM Tris-HCl (pH 8) at 4°C. The dialyzed protein mixture was fractionated using an ion-exchange fast protein liquid chromatography column (MonoQ; Pharmacia) that had been equilibrated with the dialysis buffer. The proteins were eluted by applying a linear gradient of NaCl from 0 mM to 100 mM. The protein-containing fractions were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), and those containing OpdH were pooled and dialyzed for 2 days in 0.6% octyl-POE and 25 mM histidine (pH 5.5). Further purification was achieved by loading the protein mixture onto an equilibrated chromatofocusing column (MonoP; Pharmacia) and eluting with 12% Polybuffer 7.4 (pH 3.5) (Pharmacia). The resultant fractions were analyzed by SDS-PAGE, and those containing OpdH were aliquoted and stored at −80°C.
Planar bilayer analysis.
Planar bilayer experiments were performed as described previously (7). Single channel conductance experiments were carried out using a Teflon cell containing two chambers connected by a small hole (diameter, 0.1 mm). The chambers were filled with an equal volume of buffered salt solution (pH 7), and a calomel electrode was placed in each. One of the electrodes was connected to a voltmeter, which was set to 20 mV, and the other was connected to a Keithley 427 current amplifier (Keithley Instruments, Cleveland, OH). Bilayers were formed by painting a solution of 1% diphytanoyl phosphatidylcholine (Avanti Polar Lipids, Alabaster, AL) in n-decane on either side of the hole. Once the bilayers had turned optically black, 1 μl of concentrated OpdH (∼100 ng) in a solution of 0.1% Triton X-100 was added to one side of the membrane. Single-channel insertion events were monitored on a digital storage oscilloscope (Tecktronix, Beaverton, OR) and recorded using a Houston Instruments strip chart recorder (Bausch and Lomb, Rochester, NY).
For the zero-current membrane potential measurements, the holes in the Teflon cells were much larger (diameter, 0.5 to 1 mm), 0.1 M KCl was used as the bathing solution, and one of the electrodes was connected to a Keithley electrometer instead of the current amplifier. Once the protein had been added, the membrane conductance increased rapidly for 15 to 20 min and then reached a virtually constant level. At this point, the applied voltage was turned off and the instrumentation was switched to measure zero-current membrane potentials. Aliquots of 3 M KCl were added to the cis side of the Teflon cell, and equal volumes of 0.1 M KCl were added to the trans side of the cell. The bathing solutions in the chambers were stirred, and once the zero-current membrane potential had stabilized, it was recorded and fit to the Goldman-Hodgkin-Katz equation (8) to determine the relative permeability of cations to anions.
Macroscopic conductance experiments, to assay for substrate binding, were performed as described for the zero-current potential measurements, with the following exception: once the membrane current was stationary, aliquots of the concentrated substrate solution (pH 7) were added to both sides of the membrane while the bathing solutions in the chambers were being stirred. Changes in the current level were recorded, and once it had stabilized, additional aliquots of substrate were added to the chambers.
MIC determinations.
Overnight cultures of the P. aeruginosa strain PAK and its isogenic opdH-deficient mutant, opdH-26, were serially diluted in fresh BM2 medium containing 20 mM citrate (pH 7) and a 10-fold excess of MgSO4 and FeSO4 to an inoculum size of 2 × 106 CFU/ml. Five microliters of diluted bacteria (approximately 104 cells) was then added to a 96-well culture dish containing BM2 plus twofold serial dilutions of the specified antibiotics. The MICs (the lowest antibiotic concentrations that inhibited cell growth) were determined after an 18-h incubation period at 37°C.
RESULTS
Genomic context of opdH.
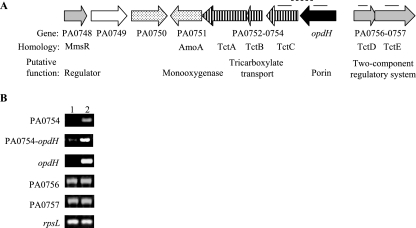
The opdH gene (PA0755) is upstream of three genes homologous (79% to 84% amino acid similarity) to the genes encoding the osmotic-shock-sensitive TctABC tricarboxylate transport system of Salmonella enterica serovar Typhimurium (Fig. 1A) (54). This system consists of a periplasmic tricarboxylate-binding protein, TctC, and two inner membrane protein components that have yet to be characterized. Unlike the arrangement in P. aeruginosa, the Salmonella serovar Typhimurium system lacks an outer membrane component. TctA and TctB have 4 and 12 predicted transmembrane segments, respectively. This transport system has recently been identified as the prototype of a large family of extracytoplasmic solute receptors termed the tripartite tricarboxylate transporters (www.tcdb.org/tcdb/index.php?tc=2.A.80) (2).
FIG. 1.
Genomic context and transcription of opdH. (A) Genomic context of opdH. Lines above the genes indicate the locations of the intragenic (solid lines) and intergenic (dashed lines) regions amplified by SQ-PCR. The putative functional classes of the genes are indicated as follows: stippled arrows, enzymes involved in carbon catabolism; open arrows, inner membrane transporter; solid arrows, porins; shaded arrows, two-component regulatory systems. (B) Transcription of regions within the opdH gene cluster. mRNA was isolated from exponential-phase cells grown in BM2 medium plus either 10 mM glucose (lane 1) or 10 mM citrate (pH 7) (lane 2) and was reverse transcribed into cDNA, which was amplified for 25 cycles by SQ-PCR. Results shown are representative of two separate experiments.
In Salmonella, the expression of the TctABC transport system is controlled by the divergently transcribed two-component regulatory system TctDE (55). The P. aeruginosa TctDE homologues, PA0756-PA0757, shared 73% and 56% amino acid similarity to their respective counterparts in Salmonella serovar Typhimurium and were also closely related to the PhoPQ and PmrAB two-component regulatory systems from both organisms (15, 29, 30).
The fifth gene in the putative opdH operon bore 70% amino acid similarity to the AmoA membrane-bound ammonia dioxygenase of Pseudomonas putida (13). Since the substrate specificities of these enzymes are quite broad (3), it is possible that PA0751 may be involved in the metabolism of compounds brought into the cell via OpdH and PA0752 to PA0754.
The previous demonstration of a role for OpdH in cis-aconitate uptake, and the high degree of similarity shared between the Salmonella serovar Typhimurium TctABCDE system and P. aeruginosa PA0752-PA0754 and PA0756-PA0757, suggested that this group of genes in P. aeruginosa may be involved in tricarboxylate uptake. SQ-PCR analysis of the levels of transcription of PA0754, opdH, and the region in between these two genes indicated that both genes were induced, possibly as a single transcriptional unit, on citrate but not on glucose (Fig. 1B). Applying a similar analysis to the genes for the putative two-component regulatory system PA0756-PA0757 indicated that these genes were transcribed in both glucose and citrate (Fig. 1B).
Construction and complementation of an opdH-deficient transcriptional fusion.
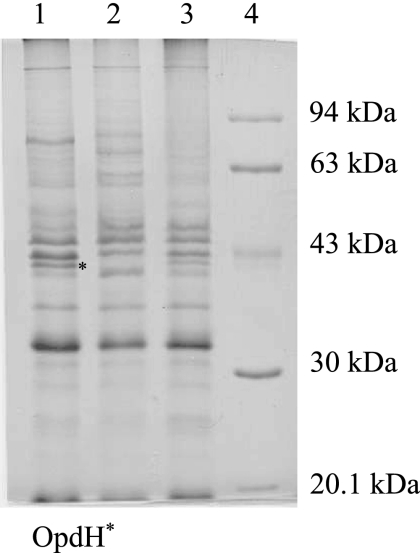
Previously, it was shown that an opdH transposon insertion mutant (opdH-26) was compromised in its ability to grow on cis-aconitate (49). This phenotype was specific to cis-aconitate, as both the opdH-26 mutant and its isogenic wild-type strain PAK grew equally well on citrate and not at all on isocitrate (46). To see if this phenotype was applicable in other strain backgrounds, an opdH mutant was constructed in a P. aeruginosa PAO background by inserting a copy of a xylE-Gmr cassette into the gene, thereby also creating a transcriptional fusion. Outer membranes isolated from the transcriptional fusion grown in BM2 minimal medium plus 10 mM citrate lacked a 40-kDa protein that was present in the wild-type strain (Fig. 2). The N-terminal amino acid sequence of the 40-kDa band from the wild-type strain, AXFLED, where X is unknown, corresponded to the first six amino acids of the mature OpdH protein (AGFLED).
FIG. 2.
Outer membrane profiles of wild-type P. aeruginosa, an opdH mutant, and an opdH-complemented strain. Outer membranes were isolated from stationary-phase cultures grown in BM2 medium plus 10 mM citrate (pH 7), and 20 μg was resolved on 11% SDS-PAGE gels. Lane 1, wild-type P. aeruginosa H103; lane 2, opdH-10; lane 3, opdH′-4; lane 4, Pharmacia low-range molecular weight marker.
Attempts to complement the opdH-xylE transcriptional fusion by placing a cloned copy of the opdH gene into various expression vectors resulted in clones with the gene inserted in the inverse orientation to the plasmid-borne promoter, even on low-copy-number vectors, or with two copies of opdH inserted in tandem. We reasoned that overexpression of the gene might be lethal (43) and thus created a merodiploid by crossing the opdH::xylE-Gmr cassette into the chromosome and selecting for single-crossover events. The resulting strain, opdH′-4, contained normal levels of a 40-kDa outer membrane protein corresponding to the size of OpdH, as assessed by SDS-PAGE (Fig. 2).
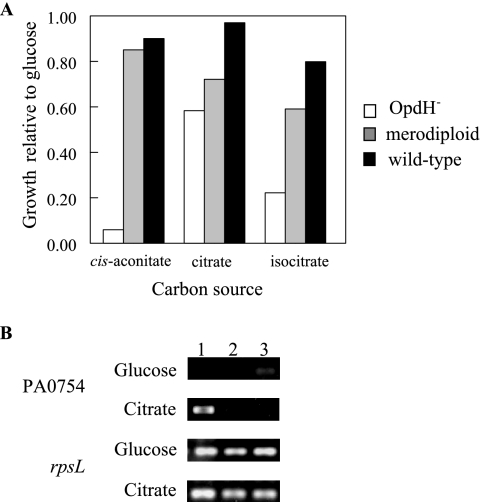
Growth curves in BM2 minimal medium plus 5 mM glucose, cis-aconitate, citrate, or isocitrate as the sole carbon source were performed using the three P. aeruginosa PAO1 strains (Fig. 3). In comparison to the P. aeruginosa PAO wild-type strain and the merodiploid, the opdH-deficient strain grew more slowly on cis-aconitate and isocitrate but only slightly more slowly on citrate. There were no differences in growth on succinate, pyruvate, fumarate, and α-ketoglutarate among the three strains (data not shown).
FIG. 3.
Growth phenotype of an opdH-deficient mutant. (A) Wild-type P. aeruginosa H103, the opdH-10 mutant, and the opdH′-4 merodiploid strain were grown in BM2 medium plus 5 mM cis-aconitate, citrate, or isocitrate. Average turbidity values obtained after 6.5 h from two separate experiments are reported. The growth of all three strains was normalized to their growth in BM2 medium plus 5 mM glucose due to the small-colony phenotype of the opdH-10 mutant. (B) Expression of PA0754 in wild-type P. aeruginosa H103 (lane 1), the opdH-10 mutant (lane 2), and the opdH′-4 merodiploid strain (lane 3). Strains were grown to exponential phase on BM2 medium plus 10 mM glucose or citrate. mRNA was extracted from these cultures, reverse transcribed, and used as the template for 25 rounds of SQ-PCR. Data are representative of two independent experiments.
The growth defect observed on cis-aconitate and isocitrate was attributable to the lack of OpdH alone, since SQ-PCR indicated that the downstream gene, PA0754, encoding the tricarboxylate-binding protein homologue, was not transcribed in either the opdH mutant or the merodiploid strain (Fig. 3B). This observation suggests the presence of another system responsible for transporting tricarboxylates across the cytoplasmic membrane.
Tricarboxylate-specific induction of OpdH.
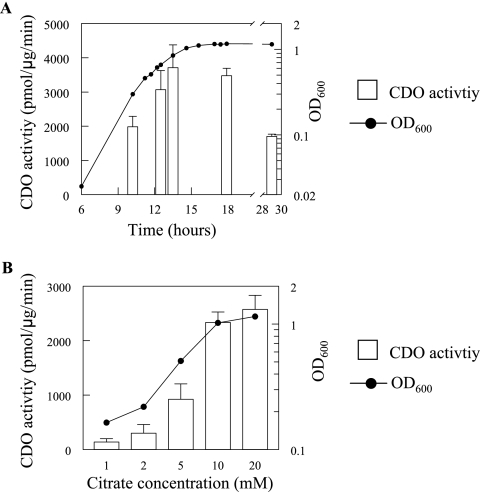
Since opdH was expressed on citrate (Fig. 1) and neither of the opdH-deficient mutants (opdH-10 and opdH-26) exhibited any major growth defects on citrate as a sole carbon source (Fig. 3), this compound was used to investigate the expression of opdH throughout the cell cycle (Fig. 4A). The CDO activity of the transcriptional fusion increased as the cell cycle progressed, reaching a maximal expression level during late-exponential phase (3,713 pmol/μg/min when the A600 was 0.8) that was maintained for approximately 4 h as the cells entered stationary phase (3,476 pmol/μg/min) and then dropped to 1,702 pmol/μg/min after overnight growth (total, 29 h).
FIG. 4.
Expression of OpdH in citrate. (A) Expression of OpdH as a function of the cell cycle. An opdH::xylE-Gmr transcriptional fusion was grown in BM2 medium plus 10 mM citrate (pH 7). Aliquots were taken at the indicated intervals for turbidity and CDO activity determinations. (B) Dose response of opdH to citrate. CDO activities were determined from stationary-phase cultures of the opdH::xylE-Gmr transcriptional fusion grown in BM2 medium plus 1, 2, 5, 10, or, 20 mM citrate (pH 7). Results shown are representative of two separate experiments performed in triplicate.
The decreased expression of opdH during late-stationary phase suggested that it may have been under the control of the RpoS stationary-phase sigma factor (51). To test this hypothesis, an rpoS mutant was grown in BM2 medium plus 10 mM glucose or citrate as the sole source of carbon. mRNA was extracted from both cultures and reverse transcribed into cDNA, which was then used as a template for SQ-PCR. The patterns and levels of opdH expression in the mutant and the corresponding wild-type strain were identical, suggesting that RpoS was not involved in the expression of this porin (see Fig. 7C).
FIG. 7.
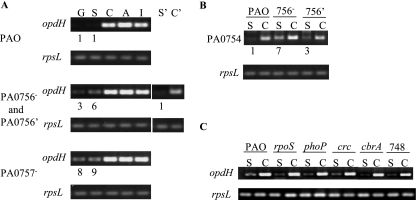
Regulation of opdH and PA0754 by the PA0756-PA0757 two-component regulatory system. Strains were grown in BM2 medium plus 10 mM of the indicated carbon source: G (glucose), S (succinate), C (citrate), A (cis-aconitate), or I (isocitrate). mRNA was extracted from exponential-phase cells, reverse transcribed, and used as the template for 25 rounds of SQ-PCR. Ratios of gene expression by the mutant to that by the wild-type strain are given below the gel images. All values were normalized to the levels of rpsL. Data are representative of at least two separate experiments. (A) Expression of opdH in wild-type P. aeruginosa UW-PAO (top panels), UW-PA0756 and UW-PA0756′ (center panels), and UW-PA0757 (lower panels). Lane S′, UW-PA0756′ complemented strain grown on succinate; lane C′, UW-PA0756′ complemented strain grown on citrate. (B) Expression of PA0754 in UW-PAO, UW-PA0756, and PA0756′ grown on BM2 medium plus succinate or citrate. (C) Expression of opdH in wild-type P. aeruginosa PAO, an rpoS mutant, a phoP mutant, a crc mutant, a cbrA mutant, and a mutant deficient in PA0748. Cultures were grown on BM2 medium plus succinate or citrate.
During the later stages of growth, the carbon source becomes limiting and may influence porin expression. To test if the expression of opdH was sensitive to the levels of citrate in the medium, the transcriptional fusion was grown in BM2 medium with 1, 2, 5, 10, or 20 mM citrate. The level of opdH expression increased with the citrate concentration, reaching a plateau at 10 mM citrate (Fig. 4B). Therefore, based on the results presented in Fig. 4, all subsequent induction assays were performed using early-stationary-phase cells grown in 10 mM carbon source.
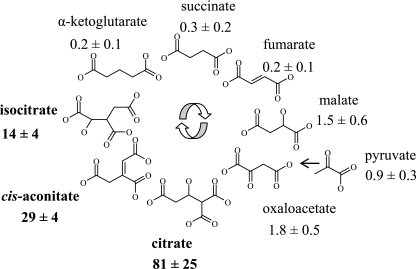
To investigate whether the citrate-mediated and the previously reported cis-aconitate-mediated induction of opdH (49) could be generalized to other di- and tricarboxylic acids, the opdH transcriptional fusion was grown in each of the Krebs cycle intermediates and then assayed for CDO activity. Cells grown in citrate demonstrated the highest CDO activity (1,326 pmol/μg/min), followed by cells grown in cis-aconitate (467 pmol/μg/min) and isocitrate (205 pmol/μg/min) (Fig. 5). Cells grown in oxaloacetate, malate, or pyruvate had low levels of opdH expression ranging from 14 pmol/μg/min to 30 pmol/μg/min. The CDO activities obtained from the cells grown in succinate, fumarate, and α-ketoglutarate were all less than 5 pmol/μg/min, which was considered insignificant.
FIG. 5.
Expression of OpdH in the Krebs cycle intermediates. Stationary-phase cultures of an opdH::xylE-Gmr transcriptional fusion were grown in minimal medium containing one of the Krebs cycle intermediates at a concentration of 10 mM (pH 7) as the sole carbon source. Results are from three independent experiments and are normalized to the transcription levels obtained in glucose, which was 16 ± 2 pmol/μg/min.
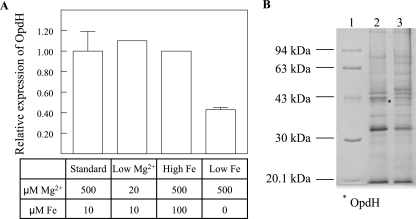
The effects of iron and magnesium on OpdH were also examined, since citrate is a known chelator of divalent cations (14). The opdH transcriptional fusion was grown on BM2 medium plus 10 mM citrate under standard, low-Mg2+, high-iron, or low-iron conditions, collected at stationary phase, and assayed for CDO activity. The growth of the low-iron culture was severely compromised, and it required 2 days to reach early-stationary phase. No significant effects on gene expression were detected in the presence of either excess iron or limiting concentrations of Mg2+. However, under low-iron conditions, the CDO activity was half of that observed under standard conditions (Fig. 6A). This result was confirmed by examining, by SDS-PAGE, the outer membrane profiles of the P. aeruginosa wild-type H103 strain grown under standard citrate conditions or citrate-low-iron conditions (Fig. 6B). The observed down-regulation of opdH was concomitant with the induction of several protein bands between 45 and 70 kDa.
FIG. 6.
Effects of metal ions on OpdH expression. (A) The opdH transcriptional fusion was grown in BM2 medium plus 10 mM citrate (pH 7) plus MgSO4 and FeSO4 at the indicated concentrations. Low-iron conditions were achieved by adding dypyridyl to the medium at a final concentration of 0.3 mM. Three experiments were performed for standard and low-iron conditions, and two for all others. (B) Outer membrane protein profiles of wild-type P. aeruginosa H103 grown in BM2 medium plus 10 mM citrate, pH 7 (lane 2), and in BM2 medium plus citrate plus low iron (lane 3). Lane 1, Pharmacia low-range molecular weight marker.
Regulation of opdH by PA0756-PA0757.
Mutants with insertions in the divergently transcribed putative two-component regulatory system PA0756-PA0757 were obtained to determine whether this system played a role in the tricarboxylate-induced expression of opdH. mRNA isolated from the wild-type P. aeruginosa PAO strain, the PA0756 mutant, and the PA0757 mutant, grown to exponential phase in BM2 medium plus 10 mM glucose, succinate, cis-aconitate, citrate, or isocitrate, was reverse transcribed into cDNA and used as the template for SQ-PCR with primers designed to a 100-bp region within the opdH gene.
In the wild-type strain, opdH was expressed at high levels in all three tricarboxylates tested relative to glucose and succinate (Fig. 7A). This pattern was conserved in both the regulator (PA0756) and sensor kinase (PA0757) mutants. The expression level of opdH did not differ significantly among the three strains when they were grown in any one of the tricarboxylates. A striking difference between the three strains was observed, however, when they were grown in either glucose or succinate (Fig. 7A, lanes G and S). In media containing these carbon sources, opdH was expressed at three- and sixfold-higher levels in the PA0756 mutant than in the parent strain and at eight- and ninefold-higher levels in the PA0757 mutant strain. The repression of opdH in succinate was restored when the PA0756 gene was overexpressed in the mutant strain (Fig. 7A, lane S′). PA0754, which is homologous to the periplasmic binding protein TctC of Salmonella serovar Typhimurium, was regulated by PA0756 in the same manner as opdH (Fig. 7B).
Because tricarboxylate-mediated induction of opdH was elevated in the PA0756-PA0757 mutant strains, the possible involvement of a transcriptional activator was investigated. Candidates for this role included the CbrAB two-component regulator and the Crc protein, which are involved in the metabolism of citrate and succinate, respectively (12, 33); the PhoPQ two-component regulatory system, which is homologous to PA0756-PA0757 (29); and the putative regulator PA0748, which is proximal to the 5-gene cluster containing opdH (Fig. 1A). The levels of opdH transcription in mutants lacking these regulators, however, were identical to those of the wild-type strain, suggesting that these systems do not have an impact on opdH expression (Fig. 7C).
Purification of OpdH.
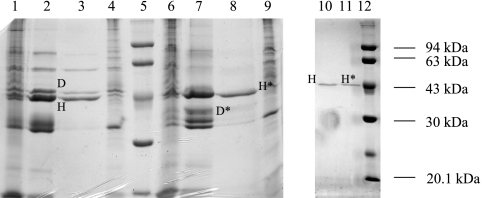
Given the induction of OpdH by tricarboxylates and its involvement in their uptake, we sought to determine whether it, like other specific porins, contained a binding site for its substrates by purifying the protein and analyzing its channel function in a planar bilayer apparatus. Based on its homology to OpdH and its similar mobility on SDS-PAGE gels, it seemed likely that OprE might contaminate preparations of OpdH; thus, a strain deficient in the production of OprE was used to purify OpdH. Both OprD and OpdH were soluble in octyl-POE plus EDTA, and they were the predominant proteins following ion-exchange chromatography (Fig. 8). Pure OpdH was obtained after passage through a chromatofocusing column (Fig. 8, lanes 10 and 11). The identity of OpdH was confirmed by comparing its mobility on an SDS-PAGE gel with those of the outer membrane proteins isolated from the wild type and an opdH mutant (Fig. 8, lanes 3 and 4). Also, unlike OprD, which demonstrates two different mobilities in SDS-PAGE gels depending on the temperature used to prepare the protein samples (20), OpdH was not heat modifiable. Therefore, the mobilities of heated and unheated OpdH protein samples were compared throughout the purification process to distinguish it from OprD (Fig. 8, lanes 3 and 8 and lanes 10 and 11).
FIG. 8.
Purification of OpdH. An 11% acrylamide gel showing the protein composition during various stages of OpdH purification. Each lane contains 20 μg of protein. Lanes 1 and 6, outer membranes isolated from a stationary-phase culture of an oprE mutant grown in BM2 medium plus 10 mM citrate; lanes 2 and 7, soluble protein fraction in 3% octyl-POE plus 5 mM EDTA; lanes 3 and 8, proteins eluted from the MonoQ column; lanes 4 and 9, outer membranes isolated from a stationary-phase culture of an opdH mutant grown in BM2 medium plus 10 mM citrate; lanes 10 and 11, proteins eluted from the MonoP column; lanes 5 and 12, molecular weight markers. Samples in lanes 1 to 5, 10, and 12 were heated at 88°C for 10 min; samples in lanes 6 to 9 and 11 were heated at 37°C for 10 min. The locations of OprD and OpdH in their heated and unheated (*) forms are shown.
Channel function of OpdH.
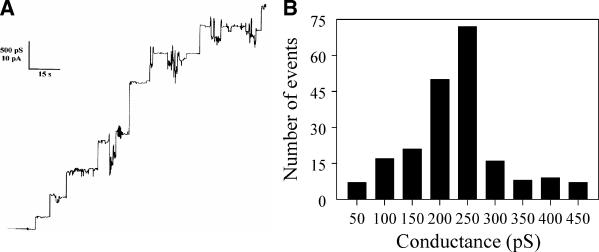
When the purified OpdH was analyzed in a planar bilayer apparatus, stepwise increases in conductance were observed (Fig. 9A). These steps were not observed in control experiments where detergent alone was added and therefore likely corresponded to the channel activity of OpdH. After two or three channel insertion events, the insertion of subsequent channels caused an increase in the background channel noise, and the bilayer had to be re-formed in order to identify channel events unambiguously. More than 200 single-channel events were observed in 1 M KCl, and the average conductance of these events was 230 pS (Fig. 9B), which is considerably lower (approximately 10-fold) than that of the major porins of the E. coli outer membrane (9). The single-channel conductance in KCl was a linear function of the electrolyte concentration, suggesting that the channel interior did not contain a binding site for either K+ or Cl− (Table 2).
FIG. 9.
Channel properties of OprD. (A) Single-channel chart recording of OpdH. Chart recording demonstrates the stepwise increases in conductance formed upon the insertion of OpdH into a bilayer composed of 1% diphytanoyl phosphatidylcholine and bathed in 1 M KCl (pH 7). (B) Histogram of the single-channel insertion events of OpdH in 1 M KCl. The average single-channel conductance of 207 channel insertion events was 230 pS.
TABLE 2.
Single-channel conductance of purified OpdH
| Salt solution | Conductance (pS) | No. of events |
|---|---|---|
| 0.01 M KCl | 9 | 79 |
| 0.03 M KCl | 40 | 205 |
| 0.1 M KCl | 73 | 109 |
| 0.3 M KCl | 190 | 82 |
| 1 M KCl | 230 | 207 |
| 3 M KCl | 889 | 89 |
| 1 M LiCl | 81 | 65 |
| 1 M K acetate | 238 | 52 |
To investigate the ion selectivity of OpdH further, the single channel conductance values of the porin in a variety of electrolyte solutions were determined. Table 2 shows the single-channel conductance of OpdH in the presence of different salt solutions. First chloride was replaced by acetate. This change had only a minor influence on the conductance of the channel. The influence of the cationic species was much more substantial: the average single-channel conductance decreased from 230 to 81 pS when K+ was replaced with Li+ (Table 2), a finding consistent with a weakly cation-selective channel. The ratio of the permeability of K+ and Cl− ions was calculated to be 4.6.
The addition of concentrated solutions of citrate, isocitrate, or cis-aconitate, either alone or in complex with metal ions, to the OpdH bilayer did not result in an associated decrease in membrane conductance, suggesting that these agents do not block the passage of ions through OpdH by binding within the pore.
Antibiotic susceptibility of an OpdH deficient mutant.
Given the relatively large size of the OpdH channel, we reasoned that it may serve as a conduit for antibiotics. Previous results indicated that this channel was not involved in antibiotic uptake in rich medium (49); therefore, MIC assays were performed under conditions known to induce OpdH expression. Compared to its isogenic wild-type strain, PAK, the opdH-26 mutant demonstrated an eightfold increase in resistance (from a MIC of 0.125 to a MIC of 1 μg/ml) to the bulky cephalosporin antibiotic ceftazidime when grown in 20 mM citrate. This resistance was specific to ceftazidime (molecular weight [MW], 637) but was not observed with other, smaller cephalosporins, including cefotaxime (MW, 477), cefepime (MW, 481), cefpirome (MW, 515), cefsoludin (MW, 555), or ceftriaxone (MW, 555), or with other classes of antibiotics, including imipenem (MW, 317), gentamicin (MW, 478), and enoxacin (MW, 320).
DISCUSSION
OpdH belongs to the OprD family of specific porins in P. aeruginosa. Previous work with this family showed that several of these channels are involved in the uptake of specific compounds, including OpdH, which was implicated in cis-aconitate uptake. Here we elaborate on that work and show that in P. aeruginosa the growth of an opdH-deficient mutant is also compromised on isocitrate. No significant growth defect was observed when citrate or other structurally related dicarboxylates were used as sole carbon sources, indicating the possibility that these compounds traverse the outer membrane through an alternate or multiple porin(s) such as any of the other 18 members of the OprD family of porins (47, 49).
A purified preparation of OpdH demonstrated a single-channel conductance in 1 M KCl of 230 pS, which is relatively large compared to that of the basic amino acid-specific porin OprD (∼20 pS). However, despite its homology with OprD and the seemingly specific nature of its uptake capabilities, we were unable to demonstrate the presence of a tricarboxylate binding site in the channel. If, like other specific porins, OpdH possessed a binding site and is a single-file channel (1, 6, 10), the addition of either citrate or cis-aconitate to the planar bilayer apparatus would have blocked the pore and caused a decrease in conductance rather than a slight increase. In addition, the observed cation selectivity of the channel was inconsistent with the expected presence of a tricarboxylate-specific binding site within the channel. Although there is a possibility that the OpdH substrate may be a metal-tricarboxylate chelate, the data presented indicate that the channel functions as a general pore that supports the nonspecific diffusion of tricarboxylates. It is worth noting that such a general pore function has also been demonstrated for OprD in gluconate uptake and for other substrate-specific porins.
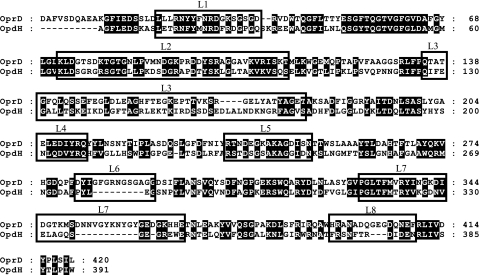
Our results do not eliminate the possibility that binding sites exist among the extracellular loops of OpdH. Extracellular binding sites of other porins are predicted to be involved in bringing substrates to the vicinity of the channel and thus might not impact transport rates through the porin (26, 59). A comparison of the amino acid sequences of OprD and OpdH revealed that loops 3, 6, 7, and 8 demonstrated the greatest variability (Fig. 10) and would thus be likely candidates for containing tricarboxylate binding sites in OpdH. Further studies of these loop regions and their impact on OpdH function are required in order to clarify whether this porin binds to its substrate or not.
FIG. 10.
Alignment of OprD and OpdH. Signal peptides were predicted using SignalP and were removed from the protein sequences, which were then aligned using ClustalX. The locations of the surface loops are based on the published topology map of OprD (outlined).
It is probable that differences in the loop arrangements of OprD and OpdH may explain the disparity observed in their channel sizes. A protein sequence alignment of these two porins revealed that OprD has deletions in three of its C-terminal loops (Fig. 10). The mouth of OprD is known to be constricted by the presence of loops 5, 7, and 8, resulting in an average single-channel conductance of 20 pS in 1 M KCl. When an 8-amino-acid deletion is introduced into loop 5 of OprD, the single-channel conductance of this porin rises to 675 pS (23). It is believed that this situation is analogous to that of OpdH, which, because of the decreased length of its loops 6, 7, and 8, is expected to have a channel mouth that is relatively unobstructed and open.
Although the single-channel conductance of OpdH was on average 10 times greater than those of other characterized specific porins of Pseudomonas species, it is considered small compared to those of OmpF and OmpC, the major porins of E. coli (0.23 nS versus 1 to 2 nS) (9, 23, 25, 38, 57). Also, of the many compounds that were tested as potential substrates for OpdH, significant growth defects were observed only for two compounds: isocitrate and cis-aconitate. This observation demonstrates that despite the presence of OpdH in the outer membrane, the low permeability of the Pseudomonas outer membrane is still maintained (60). Nevertheless, this channel does appear to have a role in susceptibility to the important antipseudomonal cephalosporin ceftazidime under opdH-inducing conditions, consistent with nonspecific porin activity.
The apparent specificity of tricarboxylate transport through OpdH may have arisen, in part, from interactions between OpdH and the putative tricarboxylate transport system encoded by PA0754 to PA0752. The putative periplasmic binding protein PA0754 was cotranscribed and coregulated with opdH. Since the intergenic regions between PA0754, PA0753, and PA0752 are not very large and the homologous tricarboxylate transport system in Salmonella serovar Typhimurium forms an operon, it seems probable that all three genes are coregulated and form an operon with opdH. It is worthy of mention, though, that in the strain PAK opdH-26 knockout, PA0754 is still transcribed, albeit in a manner independent of citrate, suggesting that there may be differences between PAO and PAK (data not shown). Transport systems similar to the tct operon have been conserved in both sequence and arrangement in many bacterial species (2), but the inclusion of a porin gene within these putative operons appears to be unique to the pseudomonads. Fractionation studies have demonstrated that the TctC periplasmic binding protein of Salmonella serovar Typhimurium interacts with both the outer and inner membranes and may funnel substrates from one membrane to the other (54). Thus, it is possible that the P. aeruginosa PA0754 protein acts in an analogous manner, although further studies would be needed to confirm this hypothesis.
The selective uptake of tricarboxylates by OpdH may also be a consequence of its tight regulation. Of the nine di- and tricarboxylates tested, only the latter group of compounds led to high levels of opdH production. The level of opdH transcription in oxaloacetate, malate, and pyruvate was quite low (around 10% of the amount observed on the tricarboxylates), and it was negligible in fumarate, succinate, α-ketoglutarate, or other structurally diverse compounds including amino acids and pyroglutamate. An inspection of the chemical structures of these compounds revealed that at least three carbonyl groups were required for induction of efficient opdH transcription. Compounds without three carbonyl groups but possessing a high density of carbonyl or alcohol groups (i.e., fewer than 2 carbon atoms/functional group) led to low levels of opdH transcription. In the presence of compounds lacking these structural features, there was virtually no opdH transcription, limiting the range of environments where this channel is expected to be functional.
The expression profile of OpdH differed markedly from that of OprD. OprD is expressed on a broad range of carbon sources and is induced to higher levels in response to its substrate, arginine, via the ArgR regulator (34). OpdH, however, was expressed only in the presence of its substrates and, rather than being induced in response to these compounds, was normally repressed by the PA0756-PA0757 two-component regulatory system. These differences in regulation may be a reflection of the differences in channel architecture between the two porins. In 1 M KCl, the average single-channel conductance of OpdH was 12 times larger than that of OprD. The constitutive presence of such a large pore in the outer membrane would significantly increase its permeability to a variety of noxious agents, typified here by the bulky cephalosporin antibiotic ceftazidime; thus, limiting its expression might be of considerable advantage to P. aeruginosa. E. coli employs a similar strategy with its sucrose-specific porin ScrY and with RafY, a general porin involved in the uptake of saccharides. Both channels are relatively large, 1.4 nS and 2.9 nS, respectively, and their expression is tightly controlled by repressor proteins, thereby preventing the promiscuous uptake of potentially dangerous compounds into the cell (4, 40).
Acknowledgments
We thank P. V. Phibbs for the crc mutant and the University of Washington Genome Center for providing us with PA0756, PA0757, cbrA, and cbrB mutants.
This work was funded by grants from the Canadian Institutes of Health Research, the Canadian Cystic Fibrosis Foundation, the Deutsche Forschungsgemeinschaft, and the Fonds der Chemischen Industrie. S.T. received a CCFF studentship. R.E.W.H. holds a Canada Research Chair.
Footnotes
Published ahead of print on 17 November 2006.
REFERENCES
- 1.Andersen, C., B. Rak, and R. Benz. 1999. The gene bglH present in the bgl operon of Escherichia coli, responsible for uptake and fermentation of β-glucosides encodes for a carbohydrate-specific outer membrane porin. Mol. Microbiol. 31:499-510. [DOI] [PubMed] [Google Scholar]
- 2.Antoine, R., F. Jacob-Dubuisson, H. Drobecq, E. Willery, S. Lesjean, and C. Locht. 2003. Overrepresentation of a gene family encoding extracytoplasmic solute receptors in Bordetella. J. Bacteriol. 185:1470-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arp, D. J., L. A. Sayavedra-Soto, and N. G. Hommes. 2002. Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch. Microbiol. 178:250-255. [DOI] [PubMed] [Google Scholar]
- 4.Aslanidis, C., I. Muiznieks, and R. Schmitt. 1990. Successive binding of raf repressor to adjacent raf operator sites in vitro. Mol. Gen. Genet. 223:297-304. [DOI] [PubMed] [Google Scholar]
- 5.Bellido, F., N. L. Martin, R. J. Siehnel, and R. E. W. Hancock. 1992. Reevaluation, using intact cells, of the exclusion limit and role of porin OprF in Pseudomonas aeruginosa outer membrane permeability. J. Bacteriol. 174:5196-5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benz, R., C. Egli, and R. E. W. Hancock. 1993. Anion transport through the phosphate-specific OprP channel of the Pseudomonas aeruginosa outer membrane: effects of phosphate, di- and tribasic anions and of negatively-charged lipids. Biochim. Biophys. Acta 1149:224-230. [DOI] [PubMed] [Google Scholar]
- 7.Benz, R., and R. E. W. Hancock. 1981. Properties of the large ion-permeable pores formed from protein F of Pseudomonas aeruginosa in lipid bilayer membranes. Biochim. Biophys. Acta 646:298-308. [DOI] [PubMed] [Google Scholar]
- 8.Benz, R., K. Janko, and P. Lauger. 1979. Ionic selectivity of pores formed by the matrix protein (porin) of Escherichia coli. Biochim. Biophys. Acta 551:238-247. [DOI] [PubMed] [Google Scholar]
- 9.Benz, R., A. Schmid, and R. E. W. Hancock. 1985. Ion selectivity of gram-negative bacterial porins. J. Bacteriol. 162:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benz, R., A. Schmid, and G. H. Vos-Scheperkeuter. 1987. Mechanism of sugar transport through the sugar-specific LamB channel of Escherichia coli outer membrane. J. Membr. Biol. 100:21-29. [DOI] [PubMed] [Google Scholar]
- 11.Brinkman, F. S. L., M. Bains, and R. E. W. Hancock. 2000. The amino terminus of Pseudomonas aeruginosa outer membrane protein OprF forms channels in lipid bilayer membranes: correlation with a three-dimensional model. J. Bacteriol. 182:5251-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collier, D. N., P. W. Hager, and P. V. Phibbs, Jr. 1996. Catabolite repression control in the Pseudomonads. Res. Microbiol. 147:551-561. [DOI] [PubMed] [Google Scholar]
- 13.Daum, M., W. Zimmer, H. Papen, K. Kloos, K. Nawrath, and H. Bothe. 1998. Physiological and molecular biological characterization of ammonia oxidation of the heterotrophic nitrifier Pseudomonas putida. Curr. Microbiol. 37:281-288. [DOI] [PubMed] [Google Scholar]
- 14.Gautier-Luneau, I., C. Merle, D. Phanon, C. Lebrun, F. Biaso, G. Serratrice, and J. L. Pierre. 2005. New trends in the chemistry of iron(III) citrate complexes: correlations between X-ray structures and solution species probed by electrospray mass spectrometry and kinetics of iron uptake from citrate by iron chelators. Chemistry 11:2207-2219. [DOI] [PubMed] [Google Scholar]
- 15.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 17.Hancock, R. E. W. 1997. The bacterial outer membrane as a drug barrier. Trends Microbiol. 5:37-42. [DOI] [PubMed] [Google Scholar]
- 18.Hancock, R. E. W., and R. Benz. 1986. Demonstration and chemical modification of a specific phosphate binding site in the phosphate-starvation-inducible outer membrane porin protein P of Pseudomonas aeruginosa. Biochim. Biophys. Acta 860:699-707. [DOI] [PubMed] [Google Scholar]
- 19.Hancock, R. E. W., and F. S. L. Brinkman. 2002. Function of Pseudomonas porins in uptake and efflux. Annu. Rev. Microbiol. 56:17-38. [DOI] [PubMed] [Google Scholar]
- 20.Hancock, R. E. W., and A. M. Carey. 1979. Outer membrane of Pseudomonas aeruginosa: heat and 2-mercaptoethanol-modifiable proteins. J. Bacteriol. 140:902-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 22.Huang, H., and R. E. W. Hancock. 1993. Genetic definition of the substrate selectivity of outer membrane porin protein OprD of Pseudomonas aeruginosa. J. Bacteriol. 175:7793-7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, H., and R. E. W. Hancock. 1996. The role of specific surface loop regions in determining the function of the imipenem-specific pore protein OprD of Pseudomonas aeruginosa. J. Bacteriol. 178:3085-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaouen, T., L. Coquet, L. Marvin-Guy, N. Orange, S. Chevalier, and E. De. 2006. Functional characterization of Pseudomonas fluorescens OprE and OprQ membrane proteins. Biochem. Biophys. Res. Commun. 346:1048-1052. [DOI] [PubMed] [Google Scholar]
- 26.Klebba, P. E. 2002. Mechanism of maltodextrin transport through LamB. Res. Microbiol. 153:417-424. [DOI] [PubMed] [Google Scholar]
- 27.Koebnik, R., K. P. Locher, and P. Van Gelder. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37:239-253. [DOI] [PubMed] [Google Scholar]
- 28.Lewenza, S., R. K. Falsafi, G. Winsor, W. J. Gooderham, J. B. McPhee, F. S. L. Brinkman, and R. E. W. Hancock. 2005. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res. 15:583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macfarlane, E. L., A. Kwasnicka, M. M. Ochs, and R. E. W. Hancock. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34:305-316. [DOI] [PubMed] [Google Scholar]
- 30.McPhee, J. B., S. Lewenza, and R. E. W. Hancock. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50:205-217. [DOI] [PubMed] [Google Scholar]
- 31.Nestorovich, E. M., E. Sugawara, H. Nikaido, and S. M. Bezrukov. 2006. Pseudomonas aeruginosa porin OprF: properties of the channel. J. Biol. Chem. 281:16230-16237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishijyo, T., D. Haas, and Y. Itoh. 2001. The CbrA-CbrB two-component regulatory system controls the utilization of multiple carbon and nitrogen sources in Pseudomonas aeruginosa. Mol. Microbiol. 40:917-931. [DOI] [PubMed] [Google Scholar]
- 34.Ochs, M. M., C. D. Lu, R. E. W. Hancock, and A. T. Abdelal. 1999. Amino acid-mediated induction of the basic amino acid-specific outer membrane porin OprD from Pseudomonas aeruginosa. J. Bacteriol. 181:5426-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parke, D. 1990. Construction of mobilizable vectors derived from plasmids RP4, pUC18 and pUC19. Gene 93:135-137. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 37.Sandermann, H., Jr., and J. L. Strominger. 1972. Purification and properties of C55-isoprenoid alcohol phosphokinase from Staphylococcus aureus. J. Biol. Chem. 247:5123-5131. [PubMed] [Google Scholar]
- 38.Saravolac, E. G., N. F. Taylor, R. Benz, and R. E. W. Hancock. 1991. Purification of glucose-inducible outer membrane protein OprB of Pseudomonas putida and reconstitution of glucose-specific pores. J. Bacteriol. 173:4970-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schirmer, T., T. A. Keller, Y. F. Wang, and J. P. Rosenbusch. 1995. Structural basis for sugar translocation through maltoporin channels at 3.1 Å resolution. Science 267:512-514. [DOI] [PubMed] [Google Scholar]
- 40.Schmid, K., R. Ebner, J. Altenbuchner, R. Schmitt, and J. W. Lengeler. 1988. Plasmid-mediated sucrose metabolism in Escherichia coli K12: mapping of the scr genes of pUR400. Mol. Microbiol. 2:1-8. [DOI] [PubMed] [Google Scholar]
- 41.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 42.Siehnel, R. J., C. Egli, and R. E. W. Hancock. 1992. Polyphosphate-selective porin OprO of Pseudomonas aeruginosa: expression, purification and sequence. Mol. Microbiol. 6:2319-2326. [DOI] [PubMed] [Google Scholar]
- 43.Siehnel, R. J., R. S. L. Wong, H. Huang, and R. E. W. Hancock. 1994. Cloning of outer membrane protein genes and molecular genetic methods for studying structure function relationships. Methods Mol. Genet. 3:311-324. [Google Scholar]
- 44.Stanier, R. Y., N. J. Palleroni, and M. Doudoroff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 45.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 46.Sugawara, E., E. M. Nestorovich, S. M. Bezrukov, and H. Nikaido. 2006. Pseudomonas aeruginosa porin OprF exists in two different conformations. J. Biol. Chem. 281:16220-16229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamber, S., and R. E. Hancock. 2006. Involvement of two related porins, OprD and OpdP, in the uptake of arginine by Pseudomonas aeruginosa. FEMS Microbiol. Lett. 260:23-29. [DOI] [PubMed] [Google Scholar]
- 48.Tamber, S., and R. E. W. Hancock. 2004. The outer membranes of pseudomonads, p. 575-601. In J. L. Ramos (ed.), Pseudomonas, vol. 1. Kluwer Academic, New York, NY. [Google Scholar]
- 49.Tamber, S., M. M. Ochs, and R. E. W. Hancock. 2006. Role of the novel OprD family of porins in nutrient uptake in Pseudomonas aeruginosa. J. Bacteriol. 188:45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trias, J., and H. Nikaido. 1990. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J. Biol. Chem. 265:15680-15684. [PubMed] [Google Scholar]
- 51.Venturi, V. 2003. Control of rpoS transcription in Escherichia coli and Pseudomonas: why so different? Mol. Microbiol. 49:1-9. [DOI] [PubMed] [Google Scholar]
- 52.Vijgenboom, E., J. E. Busch, and G. W. Canters. 1997. In vivo studies disprove an obligatory role of azurin in denitrification in Pseudomonas aeruginosa and show that azu expression is under control of rpoS and ANR. Microbiology 143:2853-2863. [DOI] [PubMed] [Google Scholar]
- 53.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]
- 54.Widenhorn, K. A., W. Boos, J. M. Somers, and W. W. Kay. 1988. Cloning and properties of the Salmonella typhimurium tricarboxylate transport operon in Escherichia coli. J. Bacteriol. 170:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Widenhorn, K. A., J. M. Somers, and W. W. Kay. 1989. Genetic regulation of the tricarboxylate transport operon (tctI) of Salmonella typhimurium. J. Bacteriol. 171:4436-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolff, J. A., C. H. MacGregor, R. C. Eisenberg, and P. V. Phibbs, Jr. 1991. Isolation and characterization of catabolite repression control mutants of Pseudomonas aeruginosa PAO. J Bacteriol. 173:4700-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wylie, J. L., C. Bernegger-Egli, J. D. O'Neil, and E. A. Worobec. 1993. Biophysical characterization of OprB, a glucose-inducible porin of Pseudomonas aeruginosa. J. Bioenerg. Biomembr. 25:547-556. [DOI] [PubMed] [Google Scholar]
- 58.Wylie, J. L., and E. A. Worobec. 1995. The OprB porin plays a central role in carbohydrate uptake in Pseudomonas aeruginosa. J. Bacteriol. 177:3021-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye, J., and B. van den Berg. 2004. Crystal structure of the bacterial nucleoside transporter Tsx. EMBO J. 23:3187-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshimura, F., and H. Nikaido. 1982. Permeability of Pseudomonas aeruginosa outer membrane to hydrophilic solutes. J. Bacteriol. 152:636-642. [DOI] [PMC free article] [PubMed] [Google Scholar]