Abstract
The phylogenetically closely related species Streptococcus salivarius and Streptococcus vestibularis are oral bacteria that are considered commensals, although they can also be found in human infections. The relationship between these two species and the relationship between strains isolated from carriers and strains responsible for invasive infections were investigated by multilocus sequence typing and additional sequence analysis. The clustering of several S. vestibularis alleles and the extent of genomic divergence at certain loci support the conclusion that S. salivarius and S. vestibularis are separate species. The level of sequence diversity in S. salivarius alleles is generally high, whereas that in S. vestibularis alleles is low at certain loci, indicating that the latter species might have evolved recently. Cluster analysis indicated that there has been genetic exchange between S. salivarius and S. vestibularis at three of the nine loci investigated. Horizontal gene transfer between streptococci belonging to the S. salivarius group and other oral streptococci was also detected at several loci. A high level of recombination in S. salivarius was revealed by allele index association and split decomposition sequence analyses. Commensal and infection-associated S. salivarius strains could not be distinguished by cluster analysis, suggesting that the pathogen isolates are opportunistic. Taken together, our results indicate that there is a high level of gene exchange that contributes to the evolution of two streptococcal species from the human oral cavity.
Streptococcus salivarius and Streptococcus vestibularis belong to the salivarius group of viridans streptococci (18). Many of the viridans streptococcal species are part of the normal microbial flora of humans. These commensal species are most prevalent in the oral cavity but also reside in the gastrointestinal and urogenital tracts. Viridans streptococci have emerged as important nosocomial pathogens, and they typically cause significant septicemia when mucosal lesions are present and host defense mechanisms are compromised (36, 53, 57). S. salivarius and S. vestibularis have been associated with severe human infections, such as, meningitis, endocarditis, and bacteremia (9, 11, 14, 31, 42, 47). Molecular taxonomy based on sequencing and comparison of the 16S rRNA or sodA genes demonstrated that these two species are closely related (34, 46).
The population structure of several oral streptococci has been studied previously (7, 19, 26, 38). Genetic typing of commensal or opportunistic pathogens has revealed considerable diversity, sometimes even within a single host. Data obtained in a Streptococcus mitis population analysis support the concept that the strains are transient, and this species appears to be maintained by clonal replacement of evolving strains rather than by stable strains (19, 26). Almost all isolates of Streptococcus mutans obtained either from 30 individuals or from a small group of families displayed distinctive restriction fragment length polymorphism or restriction endonuclease analysis patterns (7, 38). Fitzsimmons et al. suggested that the high degree of diversity observed in several mucosal bacteria may be a mechanism for avoiding immune elimination (19). S. mitis and Streptococcus oralis strains isolated from the blood of neutropenic cancer patients were also highly diverse, as they all had distinct fingerprint patterns (57). The population structure and genetic diversity of S. salivarius and S. vestibularis have been not extensively investigated, in spite of the fact that S. salivarius is the predominant oral species, especially during mouth colonization, and can be associated with caries (2, 35, 43).
Multilocus sequence typing (MLST), a method based on the nucleotide sequences of ∼500-bp internal fragments of multiple (usually about seven) housekeeping genes, has been widely used to study global epidemiology and bacterial population structure (15). MLST can reveal highly clonal populations, as well as freely recombining populations, and the latter are exemplified by Streptococcus uberis or Streptococcus pneumoniae populations (8, 16). Several reports have indicated that most streptococcal species have a highly recombinational population structure (33, 37, 58). The genetic relationships of group B, C, G, and A streptococcal isolates from asymptomatic carriers and from human infections were investigated by phylogenetic analysis of MLST data (3, 32, 33). Here we used the MLST method to investigate the genetic relationship between strains of S. salivarius and strains of S. vestibularis. Below we describe an analysis of data obtained from 27 S. salivarius strains and 9 S. vestibularis strains recovered from patients with septicemia or from the oral cavities of healthy individuals. Our results document the population structure of S. salivarius, the relationship between the two species, and the extent of gene exchange in the evolution of these oral streptococci.
(This work was presented in part at the 7th ASM Conference on Streptococcal Genetics, Saint Malo, France, June 2006.)
MATERIALS AND METHODS
Bacterial strains.
S. salivarius (n = 27) and S. vestibularis (n = 9) strains are listed in Table 1. These strains were isolated in different countries and from different sources over a 50-year period (1954 to 2004) and were obtained from the Institut Pasteur Collection (CIP), the Belgium Coordinated Collections of Microorganisms (LMG), Paris Cochin Hospital (CCH), and our laboratory collection (JIM). Strains were obtained from the oral cavity (n = 18), human blood (n = 10), and breast milk (n = 1). Commensal strains were isolated from different oral cavities; oral cavities 1 and 2 were the oral cavities of 3-year-old twin girls.
TABLE 1.
Strains and MLST data
| Strain | ST | Allele
|
Sourceb | Origin | Yr | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| glck | ddlA | pepO | ilvC | thrS | pyrE | dnaE | sodA | tkta | |||||
| S. salivarius strains | |||||||||||||
| CIP53.158 | 1 | 2 | 10 | 7 | 13 | 4 | 15 | 14 | 13 | NA | NV | Ithaca, NY | 1954 |
| CIP55.126 | 2 | 2 | 11 | 7 | 13 | 4 | 15 | 14 | 13 | NA | NV | France | 1955 |
| CIP102505 | 3 | 7 | 6 | 1 | 4 | 13 | 12 | 20 | 2 | NA | NV | New York, NY | 1971 |
| JIM8222 | 4 | 16 | 26 | 13 | 16 | 17 | 22 | 13 | 8 | NA | Oral cavity | Germany | 2000 |
| JIM8223 | 5 | 9 | 7 | 18 | 10 | 4 | 16 | 18 | 20 | NA | Oral cavity | Germany | 2000 |
| JIM8224 | 6 | 14 | 30 | 16 | 10 | 15 | 19 | 12 | 5 | NA | Oral cavity | Germany | 2000 |
| JIM8221 | 7 | 4 | 3 | 3 | 4 | 14 | 12 | 17 | 14 | NA | Oral cavity | Germany | 2000 |
| CIP102503T | 8 | 3 | 7 | 1 | 8 | 1 | 13 | 17 | 12 | NA | Human blood | New York, NY | 1950 |
| CIP104994 | 9 | 6 | 6 | 4 | 5 | 5 | 11 | 20 | 14 | NA | Human blood | Tourcoing, France | 1996 |
| LMG13104 | 10 | 1 | 15 | 17 | 14 | 2 | 14 | 22 | 10 | NA | NV | Munchen, Germany | 1983 |
| LMG13106 | 11 | 1 | 17 | 22 | 15 | 4 | 3 | 24 | 2 | NA | NV | Hospital, London, United Kingdom | 1983 |
| LMG13108 | 12 | 15 | 25 | 7 | 11 | 8 | 24 | 16 | 6 | NA | NV | Hospital, London, United Kingdom | 1983 |
| LMG13109 | 13 | 15 | 27 | 20 | 9 | 12 | 21 | 7 | 2 | NA | NV | Hospital, London, United Kingdom K | 1983 |
| LMG14652 | 14 | 5 | 14 | 3 | 5 | 5 | 15 | 21 | 14 | NA | Human blood | Ostersund, Sweden | 1993 |
| JIM8421 | 15 | 15 | 23 | 11 | 14 | 11 | 2 | 23 | 11 | NA | Breast milk | Finland | 2001 |
| JIM8775 | 16 | 2 | 16 | 6 | 12 | 4 | 15 | 17 | 7 | NA | Oral cavity 1 | France | 2004 |
| JIM8777 | 18 | 1 | 9 | 17 | 6 | 7 | 4 | 19 | 10 | NA | Oral cavity 1 | France | 2004 |
| JIM8776 | 17 | 2 | 12 | 12 | 7 | 6 | 10 | 15 | 9 | NA | Oral cavity 1 | France | 2004 |
| JIM8774 | 17 | 2 | 12 | 12 | 7 | 6 | 10 | 15 | 9 | NA | Oral cavity 2 | France | 2004 |
| JIM8773 | 21 | 17 | 4 | 21 | 9 | 1 | 20 | 9 | 4 | NA | Oral cavity 2 | France | 2004 |
| JIM8771 | 19 | 16 | 8 | 6 | 14 | 9 | 23 | 8 | 1 | NA | Oral cavity 2 | France | 2004 |
| JIM8772 | 20 | 8 | 28 | 2 | 8 | 10 | 25 | 17 | 7 | NA | Oral cavity 2 | France | 2004 |
| CCHSS1 | 22 | 16 | 29 | 14 | 17 | 20 | 18 | 11 | 3 | NA | Human blood | Hospital Cochin, France | 2003 |
| CCHSS2 | 23 | 14 | 1 | 8 | 10 | 16 | 19 | 10 | 2 | NA | Human blood | Cochin Hospital, France | 2001 |
| CCHSS3 | 24 | 5 | 13 | 19 | 5 | 4 | 1 | 6 | 14 | NA | Human blood | Cochin Hospital, France | 2002 |
| CCHSS4 | 25 | 4 | 24 | 5 | 5 | 5 | 11 | 20 | 2 | NA | human blood | Cochin Hospital, France | 2001 |
| CCHSS7 | 26 | 15 | 2 | 16 | 12 | 6 | 17 | 17 | 1 | NA | human blood | Cochin Hospital, France | 2004 |
| S. vestibularis strains | |||||||||||||
| CIP103363T | 1 | 12 | 19 | 10 | 3 | 3 | 9 | 2 | 16 | 5 | Oral cavity | Hospital, London, United Kingdom | 1987 |
| LMG17854 | 2 | 11 | 20 | 9 | 1 | 18 | 7 | 4 | 17 | 4 | Vestibular mucosa | Malmö, Sweden | 1966 |
| LMG14646 | 2 | 11 | 20 | 9 | 1 | 18 | 7 | 4 | 17 | 4 | Vestibular mucosa | Malmö, Sweden | 1966 |
| LMG14645 | 3 | 13 | 21 | 9 | 1 | 19 | 7 | 4 | 15 | 4 | Vestibular mucosa | Malmö, Sweden | 1966 |
| LMG14647 | 4 | 10 | 18 | 9 | 2 | 18 | 5 | 1 | 15 | 3 | Vestibular mucosa | Malmö, Sweden | 1966 |
| LMG17855 | 4 | 10 | 18 | 9 | 2 | 18 | 5 | 1 | 15 | 3 | Vestibular mucosa | Malmö, Sweden | 1966 |
| LMG17856 | 4 | 10 | 18 | 9 | 2 | 18 | 5 | 1 | 15 | 3 | Human dental plaque | Malmö, Sweden | 1966 |
| CCHSV5 | 5 | 10 | 22 | 15 | 1 | 21 | 8 | 5 | 18 | 1 | Human blood | Cochin Hospital, France | 2002 |
| CCHSV6 | 6 | 10 | 5 | 10 | 1 | 3 | 6 | 3 | 19 | 2 | Human blood | Cochin Hospital, France | 2004 |
NA, not amplified.
NV, not available.
Strains were grown overnight at 37°C on M17 broth medium containing glucose or lactose at final concentration of 1% in an anaerobic atmosphere. Samples were collected from oral cavities by scraping the tongue surface with sterile wooden tongue depressors that were used directly for isolation on M17 medium plates containing glucose. Both genetic identification and phenotypic identification were performed for all the strains. Phenotypic identification was performed with the rapid ID 32 Strep system (bioMérieux) or using a focused scheme for assessing hydrolysis of urea and acid production in lactose-, raffinose-, sorbitol-, or inulin-containing media. Hydrolysis of urea and a failure to ferment raffinose and inulin are characteristics of all S. vestibularis strains tested (10). Genetic identification at the species level was carried out by sequencing 16S rRNA and the sodA gene (46).
Multilocus sequence typing.
We searched for candidate MLST loci among housekeeping genes which were used previously for MLST of other gram-positive bacteria and which could be amplified with degenerate primers (ddlA, tkt, glcK, pyrE, and sodA) or whose sequences were available in a database for closely related species (ilvC, pepO, thrS, and dnaE). Furthermore, we checked to make sure that the genes were usually found in distant locations in previously described streptococcal genomes, especially the genome of the closely related species Streptococcus thermophilus. The nucleotide sequences of internal fragments of the following nine genes that were selected were determined: ilvC (encoding ketol-acid reductoisomerase), ddlA (encoding d-alanine d-alanine ligase), glcK (encoding glucose kinase), pepO (encoding endopeptidase), thrS (encoding threonyl-tRNA synthetase), tkt (encoding transketolase), pyrE (encoding orotate phosphoribosyltransferase), dnaE (encoding DNA polymerase III), and sodA (superoxide dismutase). For the sodA gene, we used primers and an amplification procedure described previously (46). Primers used in this study are listed in Table 2. One bacterial colony, freshly grown on an agar plate, was suspended in 50 μl of TES (10 mM Tris-HCl, 1 mM EDTA, 25% sucrose), and DNA was extracted by lysis in a thermocycler at 95°C for 10 min and at 4°C for 15 min. The DNA obtained was used immediately for PCR amplification performed with the GeneAmp 9700 PCR system (Perkin-Elmer) by using the following cycling parameters: 5 min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 50°C, and 1 min at 72°C. Amplified products (459 to 527 bp of internal gene fragments) were examined on a 1.5% agarose gel. They were used for direct sequencing by a fluorescent sequencing procedure with the same PCR primers that were used for the initial PCR amplification. The sequence of each fragment on both strands was determined with an Applied Biosystems 370A DNA sequencer by using a Bigdye Terminator sequencing kit according the manufacturer's protocol (Perkin-Elmer). To ensure the accuracy of the sequence, amplicons obtained from at least two bacterial colonies were used for sequence determination.
TABLE 2.
Oligonucleotide primers for S. salivarius and S. vestibularis
| Locus | Accession no. | Primer function | Primer | Sequence (5′-3′) |
|---|---|---|---|---|
| ilvC | AF220670 | MLST | ilvC-up | GATCAGGTCACGATGTTAT |
| MLST | ilvC-dn | GGTGCATATCCAGCTTCAGT | ||
| ddlA | gbU69167 | MLST | ddlA-up | TCAAGTGTGGCTATGGA |
| MLST | ddlA-dn | GTAGATGGCTCCATCCTC | ||
| glcK | AF442552 | MLST | glcK-up | TGGGCAGAAACTCAAGA |
| MLST | glcK-dn | AACACCACCACCGATAAC | ||
| pyrE | CP000024 | MLST | pyrE-dn | CGCTTTACGGAGGAACAT |
| MLST | pyrE-up | GTCCGTCTGCAGTGATGT | ||
| thrS | CP000024 | MLST | thrS-up | ATCACTGAAGATGGAAGC |
| MLST | thrS-dn | CCAAGTTTACGGTGGTCA | ||
| dnaE | CP000024 | MLST | dnaE-up | GGACTGGGAGCCTGGGAT |
| MLST | dnaE-dn | ACTCCCTGCAGCAGACCC | ||
| pepO | AJ249396 | MLST | pepOup | AACTCTACCACCCTTATGA |
| MLST | pepOdo | GGTTTGTTCCACTTGCTCCAG | ||
| tkt | CP000024 | MLST (S. vestibularis) | tkt-up | GCAGCACCAATGGGTTAC |
| MLST (S. vestibularis) | tkt-dn | CCAAGTTTACGGTGGTCA | ||
| tkt-up2 | CTGGCCACCCTGGTGTGG | |||
| . | tkt-dn2 | GGAGCACCGTGAACACCG | ||
| tkt-up3 | AATGCTATTCGTTTTCTAGG | |||
| tkt-dn3 | TTGAAATCAGCATATACTT | |||
| tkt-up4 | CAGGTCATGGTTCAATGCT | |||
| tkt-dn4 | TGAATTACGAGTTGCTTG | |||
| tkt-dn5 | TTGATATCATTTGAATC | |||
| tkt-up6 | ATGGGTGCAGCACCAATGG | |||
| tkt-dn6 | ACCAATGACTGTCTTAAC | |||
| tkt region | Large region | tkt-dn7 | CTTAGAACCCCATTGACGG | |
| Large region | tkt-up8 | GGTGTACGTGAGTTTGCG | ||
| Large region | tkt-salup | CTTGGCATTGCTTGACTTCG | ||
| Large region | tkt-saldn | CCTTCGATTGAGATTAGTAAC | ||
| Large region | tkt-saldn8 | CAAAGGCAGCCTTGTATTCC | ||
| Large region | tkt-saldo9 | CAGAACCAGTCGCAATGA | ||
| Large region | tkt-vesup | CAAGATTATCTTGTCATCG | ||
| Large region | tkt-vesdo | GGCTGGAAAGTCCTTGCATAC | ||
| Large region | tkt-vesup9 | GGCAGCAAGTCTACCATGG | ||
| Large region | tkt-vesdo9 | GCTTGATACCATGCAGCC- | ||
| ilvC region | Large region | ilvB-up3 | GTGGACAGCTCAGTATTATC | |
| Large region | ilvC-dn3 | GCTTCTGCTACTGTGTAAG | ||
| Large region | ilvC-up5 | CTGAAGAAGATTTGTTTGGTG | ||
| Large region | tyrS-do4 | GGTAACATGACAGCAGGTAC | ||
| pepO region | Large region | pepO-up6 | GCAGGGTGCGTGCTCTTG | |
| Large region | pepO-up4 | ACGCTTACAAGATGATTT | ||
| Large region | pepO-dn6 | CTTCTGGAACGATGATTTTATC | ||
| Large region | pepO-dn4 | TCATAAGGGTGGTAGAGTT | ||
| Large region | pepO-dn5 | TACCAAATAATAACACGATC | ||
| Large region | pepO-dn7 | TAGTCATATGAAACTCCT | ||
| Large region | pepO-up5 | CTGGAGCAAGTGGAACAAACC | ||
| Large region | pepO-up7 | GAAATTTCAGTTGCTCATAG |
PCR amplification of ilvC, pepO, pyrE, and tkt genes for sequencing.
Primer pairs for the tkt locus did not amplify the DNA fragments of all S. salivarius isolates. Therefore, alternative sets of primers were designed using sequence information available in public databases. The primers listed in Table 2 (tkt-up2, tkt-dn2, tkt-up3, tkt-dn3, tkt-up4, tkt-dn4, tkt-dn5, tkt-up6, and tkt-dn6) were used in all possible up and dn combinations. The large region around the ilvC, pepO, and tkt genes was sequenced by primer walking, using primers designed on the basis of the regions sequenced. For amplification of the region containing the ilvB, ilvN, ilvC, and tyrS genes, we used ilvB-up3, ilvC-dn3, ilvC-up5, and tyrS-do4 (Table 2). For amplification of the region containing the dexS and pepO genes, we used pepO-up6, pepO-up4, pepO-dn6, pepO-dn4, pepO-dn5, pepO-dn7, pepO-up5, and pepO-up7 (Table 2). For amplification of the region containing the tkt gene, we used tkt-dn7 and tkt-up8 for both species, tkt-salup, tkt-saldn, tkt-saldn8, and tkt-saldo9 for S. salivarius, and tkt-vesup, tkt-vesdo, tkt-vesup9, and tkt-vesdo9 for S. vestibularis (Table 2).
For pyrE sequence alignment, the nucleotide sequence of the mobile element found in allele 11 (S. salivarius) was not taken into account.
Data treatment and statistical analysis.
For each locus, all the sequences were compared, and arbitrary allele numbers were assigned to the different sequences. The combination of alleles at each locus defined an allelic profile or sequence type (ST) for a strain. Strains with the same allelic profile were assigned to the same ST. The STs were identified by arbitrary numbers.
The number of polymorphic nucleotide sites and the maximal and average levels of nucleotide divergence of alleles (expressed as percentages) at a given locus were calculated using the MEGA software (version 3; http://www.megasoftware.net) (39). Phylogenetic analyses of the nucleotide sequences of each housekeeping gene separately and the concatenated sequence containing ddlA, thrS, pyrE, dnaE, and sodA were performed using the neighbor-joining (NJ) method and the same software. Because of their extensive nucleotide diversity, the glcK, ilvC, and pepO sequences could not be used in the MLST scheme, so we removed them from the concatenated sequence. A Kimura two-parameter distance model was used to estimate distances for nucleotide sequences. To determine the significance of the groups observed in trees constructed by the NJ method, a bootstrap analysis with 1,000 replicates was performed.
The split decomposition method was used to assess the degree of tree-like structure for alleles found for each locus (30). The sequence alignments were converted to NEXUS files, and the split decomposition analysis was performed with the SPLITSTREE 3.1 program (http://bibiserv.techfak.uni-bielefeld.de/splits/).
A statistical analysis based on the dN/dS ratio (41), which was the ratio of the number of substitutions that changed the amino acid sequence (dN is number of nonsynonymous substitutions per nonsynonymous site) to the number of silent substitutions (dS is the number of synonymous substitutions per synonymous site), was performed by using methods described with the START program (K. Jolley; http://mlst.zoo.ox.ac.uk/links/START). The standardized index of association (ISA) was calculated as described by Haubold and Hudson (24). In our study, the set of S. salivarius isolates analyzed for linkage equilibrium did not contain CIP55.126 and JIM8774, which are considered to be very closely related to CIP53.128 and JIM8776, respectively.
Nucleotide sequence accession numbers.
The sequences of all alleles have been deposited in the GenBank database under accession numbers DQ460524 to DQ460540 (ilvC fragment), DQ460609 to DQ460638 (ddlA fragment), DQ460507 to DQ460523 (glcK fragment), DQ460541 to DQ460562 (pepO fragment), DQ460588 to DQ460608 (thrS fragment), DQ460478 to DQ460482 (tkt fragment), DQ460563 to DQ460587 (pyrE fragment), DQ460483 to DQ460506 (dnaE fragment), and EF054776 to EF054795 (sodA fragment). The sequences of large regions have been deposited in the GenBank database under accession numbers DQ46995 (dexS and pepO of S. salivarius LMG13109), DQ464996 (dexS and pepO of S. vestibularis LMG14645), DQ464998 (ilvB, ilvN, ilvC, and tyrS of S. salivarius LMG13109), DQ464994 (ilvB, ilvN, ilvC, and tyrS of S. vestibularis LMG14645), DQ464997 (tkt and tRNAThr of S. salivarius LMG13109), and DQ464993 (orf1, tkt, and trkA2 of S. vestibularis LMG14645).
RESULTS
Development of an MLST scheme for S. salivarius and S. vestibularis.
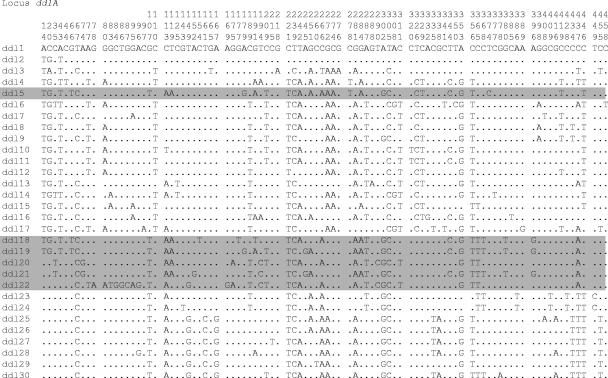
Twenty-seven S. salivarius strains and nine S. vestibularis strains isolated from different sources (oral cavity, breast milk, and human blood) and in different countries over a 50-year period were collected (Table 1). Misclassification due to the natural intraspecies phenotypic variability and the high level of 16S rRNA sequence identity (99%) between S. salivarius and S. vestibularis was corrected by characterization of the sodA gene sequence. The type strains of S. salivarius (CIP102503 [= NCTC8618]) and S. vestibularis (CIP103363 [= NCTC 12166]) were used as standards. The nucleotide sequences of 404- to 527-bp internal fragments were determined for eight and nine loci in S. salivarius and S. vestibularis, respectively, as tkt could not be amplified for S. salivarius with the first set of primers. The polymorphic sites for the ddlA alleles are shown in Fig. 1 and for the eight other loci in Fig. S1 in the supplemental material. The average levels of sequence diversity for the S. salivarius and S. vestibularis alleles were 5.8% and 2%, respectively.
FIG. 1.
Polymorphic nucleotide sites of ddlA alleles of S. salivarius and S. vestibularis: variable sites in each allele of the ddlA locus from the 27 S. salivarius strains and 9 S. vestibularis strains. The site numbers at the top are in vertical format, and S. vestibularis alleles are shaded.
In the S. salivarius population, a high number of unique alleles was found for each gene, and there was no particularly predominant allele. The number of alleles per locus for the 27 S. salivarius strains ranged from 13 (glcK) to 24 (ddlA). The highest level of nucleotide sequence divergence for a locus ranged from 4.9% (sodA) to 45.3% (pepO) (Table 3). The dN/dS ratio was calculated as a measure of the degree of selection in the population. For S. salivarius, the dN/dS ratios for glcK, pyrE, ilvC, sodA, and dnaE were <0.25. In contrast, the dN/dS ratio for the thrS, ddlA, and pepO loci were >1, indicating that there was a lower degree of selection against amino acid change (32).
TABLE 3.
Genetic diversity at S. salivarius and S. vestibularis loci
| Locus | Length of sequence (bp) | No. of alleles
|
No. of variable sites
|
Maximal % divergence
|
||||
|---|---|---|---|---|---|---|---|---|
| S. salivarius (27 isolates) | S. vestibularis (9 isolates) | S. salivarius (27 isolates) | S. vestibularis (9 isolates) | Within S. salivarius | Within S. vestibularis | Between S. salivarius and S. vestibularis | ||
| glcK | 470 | 13 | 4 | 65 | 4 | 11.9 | 0.9 | 11.7 |
| pepO | 460 | 19 | 3 | 193 | 138 | 45.3 | 38.2 | 41.2 |
| ilvC | 492 | 14 | 3 | 82 | 3 | 15.5 | 0.6 | 16 |
| sodA | 404 | 15 | 5 | 29 | 3 | 4.9 | 0.7 | 11.6 |
| thrS | 497 | 17 | 4 | 60 | 27 | 6.1 | 4 | 6.8 |
| pyrE | 519 | 20 | 5 | 69 | 3 | 7.3 | 0.6 | 7.3 |
| dnaE | 480 | 19 | 5 | 76 | 34 | 6.7 | 6.3 | 11.1 |
| ddlA | 459 | 24 | 6 | 75 | 50 | 8.1 | 8.8 | 9.3 |
| tkta | 527 | 5 | 4 | 0.6 | ||||
The tkt locus was amplified only in S. vestibularis isolates.
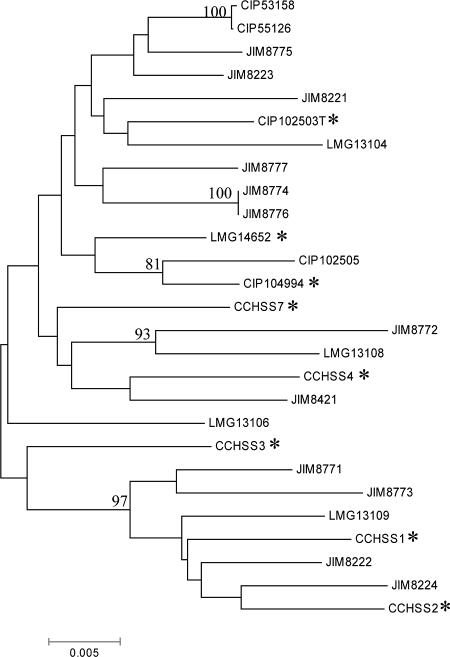
Table 1 summarizes the allelic profiles of the 27 S. salivarius strains used in this study. Each unique combination of allele numbers represents one allelic profile or ST. For S. salivarius, 26 different allelic profiles were found (ST1 to ST26), which allowed discrimination of all but two strains in our collection (ST17) (Table 1). Most STs differed at at least three of seven loci; the exceptions were ST1 and ST2, which differed at only one locus (Table 1). ST1 and ST2 were represented by two strains from the Institut Pasteur Collection (CIP53.158 and CIP55.126) and differed by only one nucleotide in the ddlA allele (Fig. 1 and Table 1). This is in sharp contrast to the high levels of variability observed for most other isolates and suggests that CIP53.158 and CIP55.126 originated from the same strain. The two ST17 strains, JIM8776 and JIM8774, are probably independent isolates of the same strain originating from the oral cavities of twin girls (Table 1). However, analysis of multiple isolates from the same two oral cavities revealed three different STs in one oral cavity and four different STs in the other, showing that S. salivarius isolates from the same individual are genetically heterogeneous. For the commensal and disease-associated S. salivarius isolates, the concatenated ddlA, thrS, pyrE, dnaE, and sodA sequences (total length, 2,359 bp) did not allow us to clearly distinguish the two classes, as judged from the tree shown in Fig. 2.
FIG. 2.
Phylogenetic tree based on concatenated sequences of five housekeeping genes (ddlA, thrS, pyrE, dnaE, and sodA). The tree was constructed using the neighbor-joining method. Bootstrap values that are ≥80% are indicated at nodes. Scale bar = 0.005 nucleotide substitution per site. Disease-related isolates are indicated by an asterisk.
For the set of nine S. vestibularis strains, the maximal level of nucleotide sequence divergence ranged from 0.6% (pyrE) to 38.2% (pepO) (Table 3), and six different allelic profiles were found, (ST1 to ST6) (Table 1). Two and three strains belonged to ST2 and ST4, respectively. All of these strains were isolated from vestibular mucosa at the same time and place (1966 in Malmö, Sweden). The strains with the same ST might correspond to multiple isolates of the same strains (Table 1). Because of the low number of independent isolates in our set, we could not use S. vestibularis in the MLST scheme.
Evidence of intraspecific recombination in S. salivarius obtained by statistical analysis.
Two types of statistical analysis were used to characterize intragenic recombination at S. salivarius loci analyzed by MLST.
(i) Standardized index of association.
The index of association has been widely used to analyze the degree of linkage disequilibrium between alleles in MLST and multilocus enzyme electrophoresis data (49). Haubold and Hudson described the standardized index of association, which does not depend on the number of loci analyzed (24). An ISA value of zero is expected for alleles in linkage equilibrium, indicating that alleles are distributed independent of each other because of free recombination (24). The ISA value calculated for our S. salivarius data was close to zero (0.057), suggesting that there was free recombination and thus a high level of gene exchange.
(ii) Split decomposition analysis.
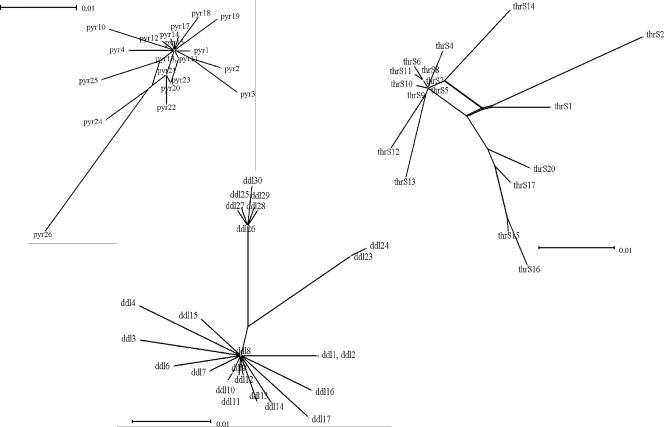
Evidence of recombination was obtained by split decomposition analysis using the SPLITSTREE 3.1 program. The algorithm used in this software is able to display conflicting results in the phylogenetic descent of sequences. A tree-like structure is created when the descent is clonal, but an interconnecting network-like structure appears when recombination plays a role in the evolutionary history of the population analyzed (30, 52). The results obtained with unique S. salivarius alleles are shown in Fig. 3 and in Fig. S2 in the supplemental material. All but one split graph have network-like structures consistent with a recombinational population structure. The sole exception is the ddlA split graph, which is closer to a bush-like structure.
FIG. 3.
Split decomposition analysis of alleles present in 27 S. salivarius strains: split graphs for thrS, pyrE, and ddlA alleles. In some graphs several alleles are connected to each other by multiple pathways, forming an interconnected network and suggesting that there were recombination events. The numbers are allele numbers. Split graphs for ilvC, pepO, glcK, dnaE, and sodA alleles are shown in Fig. S2 in the supplemental material.
Clustering S. salivarius and S. vestibularis allele trees.
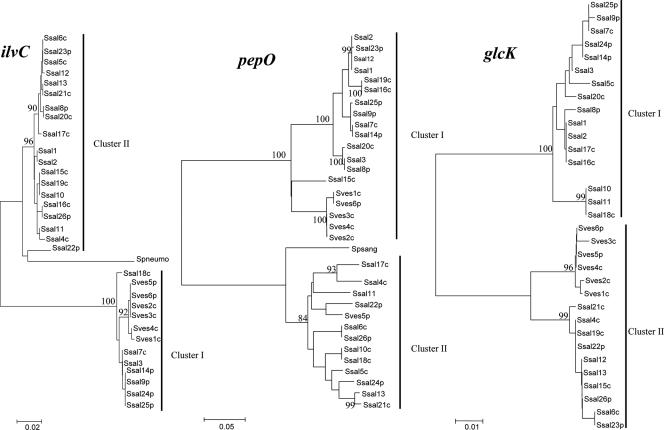
The relationship between S. salivarius and S. vestibularis strains was analyzed further by using the nucleotide sequences of the alleles at each MLST locus. Allele trees that included all S. salivarius and S. vestibularis alleles were constructed for each of the seven loci by using the NJ method (48) (Fig. 4). On the one hand, as expected for a separate species, S. vestibularis alleles are clustered together in the glcK, pyrE, dnaE, sodA, and ilvC trees, and this clustering is supported by bootstrap values of 100%. On the other hand, the ddlA, pepO, and thrS allele trees do not allow discrimination between S. vestibularis and S. salivarius alleles (Fig. 4). For example, in the case of pepO, the S. vestibularis pepO15 allele (see Fig. S1 in the supplemental material) clearly clusters with S. salivarius alleles such as pepO14 (5.2% divergence), while it exhibits more than 38% divergence with other S. vestibularis alleles. In the same way, the ddlA and thrS alleles of both species are mixed in different branches (Fig. 1; see Fig. S1 in the supplemental material).
FIG. 4.
Phylogenetic relationships among 27 S. salivarius strains and 9 S. vestibularis strains. Trees are shown for glcK, ilvC, and pepO loci. Ssal, S. salivarius; Sves, S. vestibularis; p, pathogen; c, commensal; Spneumo, S. pneumoniae; Spsang, S. parasanguinis. The strain numbers correspond to ST numbers shown in Table 1. The trees were constructed by using the neighbor-joining method. Clusters and bootstrap values that are ≥80% are indicated. The scale bars indicate the number of nucleotide substitutions per site.
Origin of S. salivarius and S. vestibularis genes.
In S. salivarius, the maximal level of divergence of nucleotide sequences ranged from less than 10% (4.9 to 8.1%) for five loci to more than 10% for three loci (11.9 to 43.5%; pepO, ilvC, and glcK) (Table 3). In the same way, in S. vestibularis the maximal level of divergence of nucleotide sequences was 38.2% for the pepO locus (Table 3). Interestingly, the pepO, ilvC, and glcK trees were each divided into two clusters strongly supported by a bootstrap value of 100% (Fig. 4). None of the clusters corresponded to known features, such as species or origin (commensal versus pathogen or country). For each locus, the average levels of divergence were similar within the cluster, whereas they were three- to fourfold higher between the clusters (Table 4). Interestingly, the pepO cluster II sequences were more closely related to the Streptococcus parasanguinis pepO sequence (GenBank accession no. AF116532) than to cluster I sequences (Fig. 4). In the same way, ilvC cluster II sequences were more closely related to S. thermophilus (GenBank accession no. AE008420 and AE007356) and S. pneumoniae (GenBank accession no. AF220670) ilvC sequences than to cluster I sequences (Fig. 4).
TABLE 4.
Genetic diversity of clusters at the glcK, ilvC, and pepO loci
| Locus | No. of alleles
|
Maximal % divergence
|
|||
|---|---|---|---|---|---|
| Cluster I | Cluster II | Within cluster I | Within cluster II | Between cluster I and cluster II | |
| glcK | 7 | 9 | 3.9 | 2.6 | 11.9 |
| ilvC | 11 | 6 | 2.1 | 4.2 | 16 |
| pepO | 11 | 11 | 13.2 | 12.9 | 45.3 |
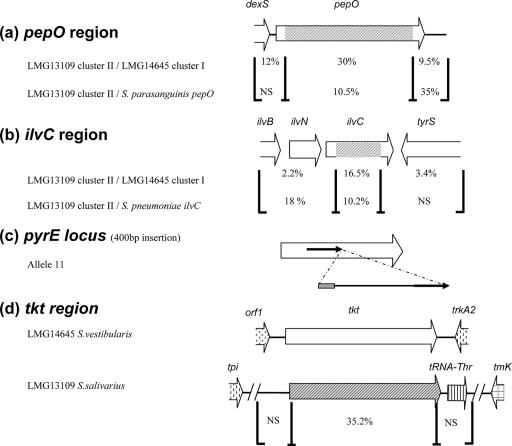
The levels of divergence for pepO and ilvC regions in strains having alleles that belong to different clusters are summarized in Fig. 5. The 1,783-bp cluster II pepO sequence exhibited 10.5% divergence from S. parasanguinis pepO, compared with 30% divergence from the cluster I pepO allele (Fig. 5a). Similarly, the 725-bp cluster II ilvC sequence exhibited 5.1% and 10.2% divergence from S. thermophilus and S. pneumoniae ilvC sequences, respectively, compared with 16.5% divergence from cluster I ilvC alleles (Fig. 5b). These results suggest that there was horizontal gene transfer (HGT) of the pepO and ilvC alleles from different streptococcal species. A similar process seems to take place at the glcK locus, as the alleles are split in two well-supported clusters. However, neither of the clusters appears to be closely related to known sequences (Table 4 and Fig. 4), suggesting that there was HGT from an uncharacterized streptococcal species.
FIG. 5.
HGT events detected in S. salivarius pepO, ilvC, pyrE, and tkt genes. Regions where there are high and low levels of diversity are indicated by brackets, and the level of diversity is shown for each region. NS, no significant homology. (a) In the pepO region, 2,095-bp nucleotide sequences of LMG13109 cluster II and LMG14645 cluster I were compared. The regions were compared with the S. parasanguinis pepO region. (b) In the ilvC region, 2,765-bp nucleotide sequences of LMG13109 cluster II and LMG14645 cluster I were compared. The regions were compared with the S. pneumoniae ilvC region. (c) In allele 11 of the pyrE locus, a 234-bp DNA insertion containing 30 bp of S. salivarius IS1139 (gray box) and 204 bp of DNA whose function and origin are unknown is present and is flanked by a 157-bp duplication (arrows). (d) In the tkt region, 2,506-bp nucleotide sequences of S. salivarius LMG13109 and S. vestibularis LMG14645 were compared. orf1 exhibits the highest level of homology to str0310 from S. thermophilus.
Genomic features that distinguish S. salivarius and S. vestibularis.
The maximal level of nucleotide divergence was high (4.9 to 45%) at all loci in S. salivarius. In contrast, several S. vestibularis loci (glcK, ilvC, tkt, sodA, and pyrE) diverged very little (maximal divergence, <0.9% [Table 3]). These differences in the levels of allele divergence indicate that these loci evolved differently and might be used to develop rapid test to differentiate the two species.
Strikingly, the tkt loci were significantly different in S. salivarius and S. vestibularis as the S. salivarius gene could not be amplified with several oligonucleotide pairs. Successful amplification of an internal fragment was achieved only with oligonucleotides tkt-up6 and tkt-dn6, and the fragment was sequenced. The level of divergence of the S. salivarius and S. vestibularis tkt gene sequences was 35.2% (Fig. 5d). Phylogenetic analysis showed that two types of tkt genes are present in streptococci (Fig. 6). The tkt gene of S. salivarius belongs to the first group (highest level of identity with Streptococcus pyogenes and Streptococcus agalactiae genes, 78%), whereas the tkt gene of S. vestibularis belongs to the second group (highest level of identity with S. thermophilus gene, 98%). Further amplification by inverse PCR and sequencing of the flanking regions showed that the genetic organization in the tkt region was different in the two species (Fig. 5d). The S. vestibularis tkt gene is flanked by an ABC transporter gene (highest level of homology to str0310 from S. thermophilus) and trkA2 (encoding potassium uptake protein); a similar organization occurs in S. thermophilus. By contrast, the S. salivarius tkt gene is followed by a tRNAThr gene and is situated in a region flanked by tpi (encoding triose phosphate isomerase) and tmk (thymidylate kinase), which are adjacent in the S. thermophilus genome (data not shown). PCR experiments showed that the genetic organization of the tkt region is typical for each species (S. salivarius and S. vestibularis) (data not shown). The trkA2 and tpi loci are located 170 kb apart in the S. thermophilus genome, indicating that a major rearrangement occurred during differentiation of the S. vestibularis and S. salivarius loci.
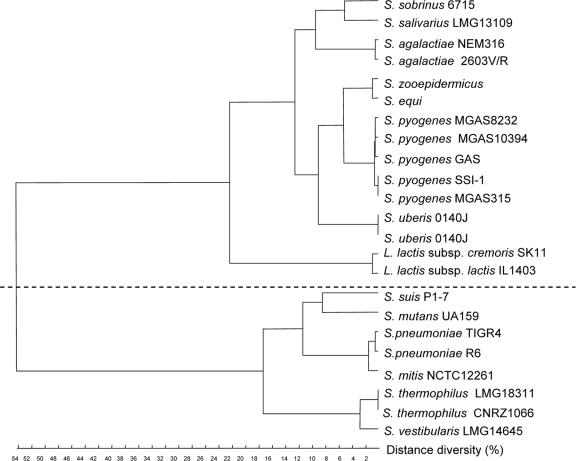
FIG. 6.
Additive distance tree for tkt genes from streptococci and lactococci. Alignment was performed with ClustalW, and the tree was constructed by the flexible method. The levels of identity between the two clusters range from 54% to 58%, whereas the levels of identity are more than 73% for the genes within each cluster.
DISCUSSION
In the present study, we carried out a comparative sequence analysis of several loci to explore the population structure of S. salivarius and S. vestibularis. Due to the small number of S. vestibularis isolates, an MLST scheme was obtained only for S. salivarius. The analysis allowed us (i) to investigate the genetic diversity within S. salivarius and S. vestibularis isolates and the relationships between them, (ii) to evaluate the relationships of commensal and pathogen strains, and (iii) to characterize extensive HGT in the two populations.
MLST of S. salivarius.
As a first step in developing an MLST method, the internal fragments of eight loci (dnaE, thrS, pyrE, ilvC, pepO, ddlA, sodA, and glcK) were amplified from all strains tested. One locus, tkt, could not be amplified from S. salivarius strains. The eight loci were polymorphic, and the maximal levels of divergence of three genes, ilvC, pepO, and glcK, were more than 12%. These high levels of divergence, which could have resulted from interspecific HGT events (see below), prevented use of these three loci in an MLST scheme for S. salivarius.
An analysis of alleles and STs showed that the diversity in S. salivarius is very high. The variability is comparable to the maximal sequence divergence (5 to 32%) and the mean nucleotide diversity (16.6%) reported for group C and G streptococci and for S. oralis, respectively (33, 55). Like S. salivarius, these streptococci are considered to be commensals, even though they are sometimes associated with human disease. The level of S. salivarius diversity is significantly higher than the levels of diversity of group A streptococci (maximal divergence, 1.4 to 6.1% [17]) and group B streptococci (1.2 to 2.5% variable nucleotide sites [32]). We found that S. salivarius isolates from the same individual exhibit a high degree of genetic diversity, as observed previously for several other mucosal streptococci (S. mitis, S. mutans, and S. oralis) (19, 38, 57). The phylogenetic tree of concatenated sequences of S. salivarius did not allow us to cluster strains isolated from human blood specifically. We concluded that commensal and potentially pathogenic strains do not belong to distinct S. salivarius populations. The lack of clear separation of commensal and clinical S. salivarius isolates resembles the findings reported for several other commensal streptococci, such as group C and G streptococci (S. mitis and S. oralis), which also exhibit opportunistic pathogenicity (33, 55).
Genetic relationship of S. salivarius and S. vestibularis isolates.
S. salivarius and S. vestibularis are included in the salivarius group of viridans streptococci (18). In 1988, S. vestibularis was described as a new oral species (56), while further analysis showed that it is very closely related to S. salivarius (34, 46). In this work, we found that the levels of relatedness between S. vestibularis and S. salivarius are very different at different genetic loci.
Two findings support the notion that S. salivarius and S. vestibularis should be considered separate species: (i) the strongly supported clusters of S. vestibularis alleles at the glcK, pyrE, dnaE, sodA, and ilvC loci and (ii) the extent of genomic divergence in particular loci, such as the divergence observed for tkt. Incidentally, the great differences in the sequence and genetic organization of the tkt locus could probably be used to develop a rapid test for discriminating these two bacteria.
By contrast, the lack of resolution of S. vestibularis and S. salivarius at three loci (ddlA, pepO, and thrS) suggests that there is frequent gene exchange between these two species. Considering the mosaic structure of these three S. vestibularis genes and the low levels of divergence found at five other loci (glcK, ilvC, pyrE, sodA, and tkt), we suggest that S. vestibularis may be a recently emerging population which evolves by interspecific gene exchange. However, analysis of a larger sample of S. vestibularis strains is necessary to test this hypothesis more stringently and to better define the evolutionary scheme of S. vestibularis.
Intraspecific recombination in S. salivarius and S. vestibularis populations.
Analysis of different loci of S. salivarius and S. vestibularis indicated that the loci have different evolutionary histories. Extensive intraspecific recombinational exchanges should have occurred to explain the features observed.
The noncongruence between allele phylogenetic trees and the low index of association of MLST alleles can be explained by frequent recombination events. The extent of the conflicting phylogenetic results is illustrated by the fact that a strain which has a certain position in one allele tree can occupy a very different position in another allele tree, with both positions strongly supported by bootstrap analysis. The conflicting phylogenetic signals found for seven S. salivarius loci in split decomposition analysis, coupled with a low association index value (ISA, 0.057), indicate that intraspecific recombination occurs frequently in S. salivarius and plays a major role in generating sequence diversity between strains. A specific gene transfer event is revealed by the presence of pyrE allele 11 in two strains isolated from human blood 5 years apart (CIP104994 isolated in 1996 and CCHSS4 isolated in 2001). This allele contains a specific DNA insertion consisting of 30 bp of S. salivarius insertion sequence IS1139 (40), followed by 204 bp of DNA whose origin is unknown (Fig. 5c). The events required to generate this allele were unlikely to occur independently twice, suggesting that the allele was exchanged horizontally. In S. vestibularis, split decomposition analysis suggested that intragenic recombination occurred at four of the nine loci (not shown). Taken together, these results support the conclusion that intraspecific recombination occurs extensively in the S. salivarius population and probably in S. vestibularis.
HGT between oral streptococci.
The presence of longer branches in ilvC, glcK, and pepO S. salivarius split graphs (see Fig. S2 in the supplemental material) are consistent with the importation of divergent genes from other species (27). This hypothesis is supported by the higher levels of homology of cluster II alleles of ilvC and pepO loci with S. pneumoniae and S. parasanguinis alleles, respectively, than with the other S. salivarius allele clusters. S. pneumoniae and S. parasanguinis are oral streptococci which might come in contact with S. salivarius and S. vestibularis in the buccal cavity. Evaluation of the extent of HGT in ilvC and pepO genes by sequence analysis indicated that exchange of 725- and 1,783-bp internal gene fragments, respectively, took place (Fig. 5a and b). Differences in chromosomal localization and high levels of nucleotide divergence between tkt genes from S. salivarius and S. vestibularis are also consistent with an HGT event. Detailed cluster analysis of these sequences and the presence of multiple variants in clusters suggest that the transfers are ancient. For example, the transfer of the ilvC divergent allele, possibly from the mitis group, likely preceded the emergence of the food species S. thermophilus in the salivarius group (4, 28). Notably, alleles of each cluster are still maintained in the population.
The extent of recombination and HGT in S. salivarius and S. vestibularis populations suggests that these bacteria have efficient mechanisms for gene acquisition and recombination, although they are not known to be naturally competent (6). Natural transformation has been described for S. pneumoniae and several viridans streptococci, such as S. mutans and Streptococcus sanguinis (1, 20, 44), and plays an important role in generating the high levels of genotypic diversity in these species. Recombinational events are responsible for the dissemination of genes encoding virulence factors, such as antibiotic resistance markers, quinolone resistance, immunoglobulin A1 protease, and competence-stimulating peptide (5, 21, 25, 45, 54). Altered penicillin binding protein genes are directly involved in penicillin resistance, and DNA sequences closely related to them appear to have been distributed horizontally between S. pneumoniae and viridans streptococci (12, 13, 22). Gene flow from human pathogens to commensal streptococci, described previously for groups A, C, and G (33, 50) and for the oral mitis group (55) and also observed here, may be involved in the evolutionary model of the commensal streptococci, which can be associated with bacterial infections. Recently, phylogenetic analyses of more than 20 streptococcal species revealed several examples of interspecific homologous recombination in housekeeping genes of viridans streptococci (29). Evidence for HGT and recombination events in this study associated S. salivarius and S. vestibularis with this group of oral streptococci, which have great potential to be receptors of foreign genes.
The absence of clustering of S. salivarius strains isolated from blood suggests that these strains are opportunistic pathogens. However, the high potential of S. salivarius and S. vestibularis for acquisition of new alleles at loci involved in general functioning of the cell and the possible acquisition of antibiotic resistance determinants (23, 51) raise the possibility that strains isolated from human infections acquired DNA that promoted their pathogenic behavior. Further studies are required to examine this possibility.
Supplementary Material
Acknowledgments
We thank Per S. J. Saris and Christina Jerusalem for providing S. salivarius and S. vestibularis strains.
Supplemental material for this article may be found at http://jb.asm.org/.
Footnotes
Published ahead of print on 3 November 2006.
REFERENCES
- 1.Avery, O. T., C. M. MacLeod, and M. McCarty. 1995. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyribonucleic acid fraction isolated from Pneumococcus type III. 1944. Mol. Med. 1:344-365. [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisharat, N., D. W. Crook, J. Leigh, R. M. Harding, P. N. Ward, T. J. Coffey, M. C. Maiden, T. Peto, and N. Jones. 2004. Hyperinvasive neonatal group B Streptococcus has arisen from a bovine ancestor. J. Clin. Microbiol. 42:2161-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolotin, A., B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyprides, B. Purnelle, D. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau, and P. Hols. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bracco, R. M., M. R. Krauss, A. S. Roe, and C. M. Macleod. 1957. Transformation reactions between Pneumococcus and three strains of Streptococci. J. Exp. Med. 106:247-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckley, N. D., C. Vadeboncoeur, D. J. LeBlanc, L. N. Lee, and M. Frenette. 1999. An effective strategy, applicable to Streptococcus salivarius and related bacteria, to enhance or confer electroporation competence. Appl. Environ. Microbiol. 65:3800-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caufield, P. W., and T. M. Walker. 1989. Genetic diversity within Streptococcus mutans evident from chromosomal DNA restriction fragment polymorphisms. J. Clin. Microbiol. 27:274-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffey, T. J., G. D. Pullinger, R. Urwin, K. A. Jolley, S. M. Wilson, M. C. Maiden, and J. A. Leigh. 2006. First insights into the evolution of Streptococcus uberis: a multilocus sequence typing scheme that enables investigation of its population biology. Appl. Environ. Microbiol. 72:1420-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conte, A., P. Chinello, R. Civljak, A. Bellussi, P. Noto, and N. Petrosillo. 2005. Streptococcus salivarius meningitis and sphenoid sinus mucocele. Case report and literature review. J. Infect. 52:27-30. [DOI] [PubMed] [Google Scholar]
- 10.Coykendall, A. L. 1989. Classification and identification of the viridans streptococci. Clin. Microbiol. Rev. 2:315-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunliffe, N. A., and A. J. Jacob. 1997. Streptococcus vestibularis bacteraemia. J. Infect. 34:85. [DOI] [PubMed] [Google Scholar]
- 12.Dowson, C. G., T. J. Coffey, C. Kell, and R. A. Whiley. 1993. Evolution of penicillin resistance in Streptococcus pneumoniae; the role of Streptococcus mitis in the formation of a low affinity PBP2B in S. pneumoniae. Mol. Microbiol. 9:635-643. [DOI] [PubMed] [Google Scholar]
- 13.Dowson, C. G., A. Hutchison, N. Woodford, A. P. Johnson, R. C. George, and B. G. Spratt. 1990. Penicillin-resistant viridans streptococci have obtained altered penicillin-binding protein genes from penicillin-resistant strains of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 87:5858-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyuk, E., O. J. Ormerod, and I. C. Bowler. 2002. Native valve endocarditis due to Streptococcus vestibularis and Streptococcus oralis. J. Infect. 45:39-41. [DOI] [PubMed] [Google Scholar]
- 15.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 16.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 17.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Facklam, R. 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin. Microbiol. Rev. 15:613-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fitzsimmons, S., M. Evans, C. Pearce, M. J. Sheridan, R. Wientzen, G. Bowden, and M. F. Cole. 1996. Clonal diversity of Streptococcus mitis biovar 1 isolates from the oral cavity of human neonates. Clin. Diagn. Lab. Immunol. 3:517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaustad, P. 1979. Genetic transformation in Streptococcus sanguis. Distribution of competence and competence factors in a collection of strains. Acta Pathol. Microbiol. Scand. Sect. B 87:123-128. [PubMed] [Google Scholar]
- 21.Gonzalez, I., M. Georgiou, F. Alcaide, D. Balas, J. Linares, and A. G. de la Campa. 1998. Fluoroquinolone resistance mutations in the parC, parE, and gyrA genes of clinical isolates of viridans group streptococci. Antimicrob. Agents Chemother. 42:2792-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hakenbeck, R., A. Konig, I. Kern, M. van der Linden, W. Keck, D. Billot-Klein, R. Legrand, B. Schoot, and L. Gutmann. 1998. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level beta-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J. Bacteriol. 180:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartley, D. L., K. R. Jones, J. A. Tobian, D. J. LeBlanc, and F. L. Macrina. 1984. Disseminated tetracycline resistance in oral streptococci: implication of a conjugative transposon. Infect. Immun. 45:13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haubold, B., and R. R. Hudson. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Linkage analysis. Bioinformatics 16:847-848. [DOI] [PubMed] [Google Scholar]
- 25.Havarstein, L. S., R. Hakenbeck, and P. Gaustad. 1997. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J. Bacteriol. 179:6589-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hohwy, J., J. Reinholdt, and M. Kilian. 2001. Population dynamics of Streptococcus mitis in its natural habitat. Infect. Immun. 69:6055-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes, E. C., R. Urwin, and M. C. Maiden. 1999. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol. Biol. Evol. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 28.Hols, P., F. Hancy, L. Fontaine, B. Grossiord, D. Prozzi, N. Leblond-Bourget, B. Decaris, A. Bolotin, C. Delorme, S. Dusko Ehrlich, E. Guedon, V. Monnet, P. Renault, and M. Kleerebezem. 2005. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 29:435-463. [DOI] [PubMed] [Google Scholar]
- 29.Hoshino, T., T. Fujiwara, and M. Kilian. 2005. Use of phylogenetic and phenotypic analyses to identify nonhemolytic streptococci isolated from bacteremic patients. J. Clin. Microbiol. 43:6073-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huson, D. H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 31.Idigoras, P., A. Valiente, L. Iglesias, P. Trieu-Cout, and C. Poyart. 2001. Meningitis due to Streptococcus salivarius. J. Clin. Microbiol. 39:3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M. S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalia, A., M. C. Enright, B. G. Spratt, and D. E. Bessen. 2001. Directional gene movement from human-pathogenic to commensal-like streptococci. Infect. Immun. 69:4858-4869. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Kawamura, Y., X. G. Hou, F. Sultana, H. Miura, and T. Ezaki. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 35.Kazor, C. E., P. M. Mitchell, A. M. Lee, L. N. Stokes, W. J. Loesche, F. E. Dewhirst, and B. J. Paster. 2003. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 41:558-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kennedy, H. F., D. Morrison, D. Tomlinson, B. E. Gibson, J. Bagg, and C. G. Gemmell. 2003. Gingivitis and toothbrushes: potential roles in viridans streptococcal bacteraemia. J. Infect. 46:67-70. [DOI] [PubMed] [Google Scholar]
- 37.King, S. J., J. A. Leigh, P. J. Heath, I. Luque, C. Tarradas, C. G. Dowson, and A. M. Whatmore. 2002. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J. Clin. Microbiol. 40:3671-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulkarni, G. V., K. H. Chan, and H. J. Sandham. 1989. An investigation into the use of restriction endonuclease analysis for the study of transmission of mutans streptococci. J. Dent. Res. 68:1155-1161. [DOI] [PubMed] [Google Scholar]
- 39.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 40.Lortie, L. A., G. Gagnon, and M. Frenette. 1994. IS1139 from Streptococcus salivarius: identification and characterization of an insertion sequence-like element related to mobile DNA elements from gram-negative bacteria. Plasmid 32:1-9. [DOI] [PubMed] [Google Scholar]
- 41.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 42.Partridge, S. M. 2000. Prosthetic valve endocarditis due to Streptococcus vestibularis. J. Infect. 41:284-285. [DOI] [PubMed] [Google Scholar]
- 43.Pearce, C., G. H. Bowden, M. Evans, S. P. Fitzsimmons, J. Johnson, M. J. Sheridan, R. Wientzen, and M. F. Cole. 1995. Identification of pioneer viridans streptococci in the oral cavity of human neonates. J. Med. Microbiol. 42:67-72. [DOI] [PubMed] [Google Scholar]
- 44.Perry, D., and H. K. Kuramitsu. 1981. Genetic transformation of Streptococcus mutans. Infect. Immun. 32:1295-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poulsen, K., J. Reinholdt, C. Jespersgaard, K. Boye, T. A. Brown, M. Hauge, and M. Kilian. 1998. A comprehensive genetic study of streptococcal immunoglobulin A1 proteases: evidence for recombination within and between species. Infect. Immun. 66:181-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poyart, C., G. Quesne, S. Coulon, P. Berche, and P. Trieu-Cuot. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J. Clin. Microbiol. 36:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruoff, K. L., S. I. Miller, C. V. Garner, M. J. Ferraro, and S. B. Calderwood. 1989. Bacteremia with Streptococcus bovis and Streptococcus salivarius: clinical correlates of more accurate identification of isolates. J. Clin. Microbiol. 27:305-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 49.Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sriprakash, K. S., and J. Hartas. 1996. Lateral genetic transfers between group A and G streptococci for M-like genes are ongoing. Microb. Pathog. 20:275-285. [DOI] [PubMed] [Google Scholar]
- 51.Stadler, C., and M. Teuber. 2002. The macrolide efflux genetic assembly of Streptococcus pneumoniae is present in erythromycin-resistant Streptococcus salivarius. Antimicrob. Agents Chemother. 46:3690-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suerbaum, S., M. Lohrengel, A. Sonnevend, F. Ruberg, and M. Kist. 2001. Allelic diversity and recombination in Campylobacter jejuni. J. Bacteriol. 183:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tunkel, A. R., and K. A. Sepkowitz. 2002. Infections caused by viridans streptococci in patients with neutropenia. Clin. Infect. Dis. 34:1524-1529. [DOI] [PubMed] [Google Scholar]
- 54.Whatmore, A. M., V. A. Barcus, and C. G. Dowson. 1999. Genetic diversity of the streptococcal competence (com) gene locus. J. Bacteriol. 181:3144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whatmore, A. M., A. Efstratiou, A. P. Pickerill, K. Broughton, G. Woodard, D. Sturgeon, R. George, and C. G. Dowson. 2000. Genetic relationships between clinical isolates of Streptococcus pneumoniae, Streptococcus oralis, and Streptococcus mitis: characterization of “atypical” pneumococci and organisms allied to S. mitis harboring S. pneumoniae virulence factor-encoding genes. Infect. Immun. 68:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whiley, R. A., and J. M. Hardie. 1988. Streptococcus vestibularis sp. nov. from the human oral cavity. Int. J. Syst. Bacteriol. 38:335-339. [DOI] [PubMed] [Google Scholar]
- 57.Wisplinghoff, H., R. R. Reinert, O. Cornely, and H. Seifert. 1999. Molecular relationships and antimicrobial susceptibilities of viridans group streptococci isolated from blood of neutropenic cancer patients. J. Clin. Microbiol. 37:1876-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zadoks, R. N., Y. H. Schukken, and M. Wiedmann. 2005. Multilocus sequence typing of Streptococcus uberis provides sensitive and epidemiologically relevant subtype information and reveals positive selection in the virulence gene pauA. J. Clin. Microbiol. 43:2407-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.