Abstract
Streptococcus mutans secretes and utilizes a 21-amino-acid signaling peptide pheromone to initiate quorum sensing for genetic competence, biofilm formation, stress responses, and bacteriocin production. In this study, we designed and synthesized a series of truncated peptides and peptides with amino acid substitutions to investigate their structure-activity relationships based on the three-dimensional structures of S. mutans wild-type signaling peptide UA159sp and C-terminally truncated peptide TPC3 from mutant JH1005 defective in genetic competence. By analyzing these peptides, we demonstrated that the signaling peptide of S. mutans has at least two functional domains. The C-terminal structural motif consisting of a sequence of polar hydrophobic charged residues is crucial for activation of the signal transduction pathway, while the core α-helical structure extending from residue 5 to the end of the peptide is required for receptor binding. Peptides in which three or more residues were deleted from the C terminus did not induce genetic competence but competitively inhibited quorum sensing activated by UA159sp. Disruption of the amphipathic α-helix by replacing the Phe-7, Phe-11, or Phe-15 residue with a hydrophilic residue resulted in a significant reduction in or complete loss of the activity of the peptide. In contrast to the C-terminally truncated peptides, these peptides with amino acid substitutions did not compete with UA159sp to activate quorum sensing, suggesting that disruption of the hydrophobic face of the α-helical structure results in a peptide that is not able to bind to the receptor. This study is the first study to recognize the importance of the signaling peptide C-terminal residues in streptococcal quorum sensing.
Many bacteria are now known to regulate diverse physiological processes through a mechanism called quorum sensing, in which bacterial cells communicate with each other for coordinated activities (6, 13, 23). During quorum sensing, bacteria produce and detect small diffusible signal molecules, called autoinducers, to sense the population density and regulate the gene expression to optimize their physiology (6, 13, 37). Bacteria produce autoinducing molecules at basal levels. When the population density increases, the concentration of autoinducing molecules also increases and reaches a threshold that activates a signal transduction pathway, leading to a cascade of gene expression (37). This enables the cells to change their behavior and function as a group, like a multicellular organism. Quorum sensing is involved in the regulation of bioluminescence, genetic competence, mating, bacteriocin production, sporulation, stress responses, virulence expression, and biofilm formation (11-13, 23, 37). The ability of bacteria to use quorum-sensing mechanisms to communicate with each other and function as a group provides significant benefits to the bacteria during host colonization, defense against competitors, and adaptation to environments (11, 23, 37).
The gram-positive organism Streptococcus mutans, a leading cariogenic pathogen that causes dental caries worldwide, has a well-conserved, peptide-mediated quorum-sensing system that primarily regulates genetic competence (2, 11, 26). The full activity of this signaling system involves at least six gene products encoded by comCDE, comAB and comX (2, 3, 26). The comC gene encodes a signal peptide precursor, which is cleaved and exported to release a 21-amino-acid signaling peptide or competence-stimulating peptide (CSP) through a peptide-specific ABC transporter encoded by comAB (35). The comDE genes encode a two-component signal transduction system that specifically senses and responds to CSP. Another gene in the quorum-sensing cascade is comX, which encodes an alternative sigma factor that directs transcription of numerous genes required for uptake and integration of foreign DNA (3). When a critical CSP concentration is reached, the CSP interacts with the ComD histidine kinase receptor of neighboring cells and activates ComE via autophosphorylation. The phosphorylated ComE in turn activates its target genes, presumably comCDE, comAB, and comX, to trigger the signaling cascade for genetic competence, as well as formation of a positive feedback loop for quorum sensing (11, 26, 28), although multilevel regulation may be involved in competence development (1). This signaling system has also been found to play a regulatory role in biofilm formation (3, 11, 28), stress responses (27), and bacteriocin production (43), which are key virulence factors in S. mutans pathogenesis (11, 24).
The quorum-sensing circuit in S. mutans is a system in which chemical details of the signaling molecule are known. Only one type of signaling peptide has been identified so far in S. mutans wild-type strains (26). However, deletion of the three C-terminal amino acid residues from the peptide of S. mutans JH1005, a mutant that was constructed to produce a higher level of bacteriocin (20), resulted in an approximately 600-fold reduction in the transformation efficiency (26), suggesting that the C terminus might be important for activation of the quorum-sensing signaling pathway. To test this hypothesis, we used circular dichroism (CD) and nuclear magnetic resonance (NMR) spectroscopy to analyze and compare the three-dimensional structures of synthesized peptides from S. mutans wild-type strain UA159 (UA159sp) and mutant JH1005 (TPC3). Based on the structural characteristics of these peptides, we designed and synthesized a series of terminally truncated peptides and peptides with amino acid substitutions to analyze their structure-activity relationships. By analyzing these synthetic peptides, we investigated the following questions. (i) What is the major structural difference between wild-type signaling peptide UA159sp and C-terminally truncated peptide TPC3? (ii) Does TPC3 induce genetic competence in a comC null mutant that is unable to produce UA159sp but still responds to UA159sp? (iii) Does UA159sp complement the defect in genetic competence of mutant JH1005? (iv) How many amino acid residues have to be deleted from the C terminus to affect the quorum-sensing activity? (v) Does a modification to the amphipathic nature of the peptide affect the quorum-sensing activity? (vi) Does a deletion from the N terminus also affect the quorum-sensing activity? Here, we report the results of experiments in which we addressed these questions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and their characteristics are shown in Table 1. S. mutans wild-type strain UA159 was grown on Todd-Hewitt yeast extract (THYE) agar or broth (BBL, Becton Dickinson, Cockeysville, MD), whereas the mutants and lacZ reporter strains were maintained on THYE medium containing an appropriate antibiotic as indicated below. Escherichia coli host strains were grown in Luria-Bertani medium supplemented with an appropriate antibiotic when necessary.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| S. mutans strains | ||
| UA159 | Wild type, Ems Kms Specs | 2 |
| SMCC3 | UA159::pComC2-KO comC Specr | 3 |
| SMC-pYH2 | SMCC3 harboring pSL-PcomDE, comC Kanr Specr | This study |
| SMC-pSL | SMCC3 harboring pSL-lacZ, comC Kanr Specr | This study |
| SMEK-1 | UA159::pComE-KO, ComE− Ermr | This study |
| E. coli DH5α | Host competent cells | Invitrogen |
| Plasmids | ||
| pYH2 | pSL::PcomDE, Kanr | This study |
| pSL | pDL276 containing a 3.45-kb HindIII fragment of a promoterless lacZ gene from pALH122, Kanr | This study |
| pDL276 | Streptococcus-E. coli shuttle vector, Kanr | 14 |
| pALH122 | Streptococcus-E. coli shuttle vector harboring a promoterless lacZ gene, Ermr | 21 |
| pVA-gtfA | Streptococcal suicide vector, Ermr | 26 |
| pComE-KO | pVA8912 harboring a 462-bp comE fragment, Ermr | 26 |
Rational design and synthesis of peptides.
A series of truncated peptides and peptides with amino acid substitutions were synthesized to investigate the role of each peptide in inducing quorum sensing and its controlled genetic competence. The sequences of the peptides and their designations are shown in Table 2. All the peptides were chemically synthesized and purified by reversed-phase high-pressure liquid chromatography, and their identities were confirmed by mass spectrometry (GL Biochem Ltd., Shanghai, China). The peptides were stored in powder at −20°C until they were used. Each peptide was dissolved freshly in sterile distilled water at a concentration of 1.0 mM to obtain a stock solution, which was diluted to obtain the working concentrations required for individual experiments.
TABLE 2.
Amino acid sequences of chemically synthesized peptides
| Peptide | Amino acid sequence |
|---|---|
| UA159sp | SGSLSTFFRLFNRSFTQALGK |
| Truncated peptides | |
| TPC1 | SGSLSTFFRLFNRSFTQALG |
| TPC2 | SGSLSTFFRLFNRSFTQAL |
| TPC3 (JH1005sp) | SGTLSTFFRLFNRSFTQA |
| TPC4 | SGSLSTFFRLFNRSFTQ |
| TPN1 | -GSLSTFFRLFNRSFTQALGK |
| TPN2 | --SLSTFFRLFNRSFTQALGK |
| TPN3 | ---LSTFFRLFNRSFTQALGK |
| TPN4 | ----STFFRLFNRSFTQALGK |
| Peptides with substitutionsa | |
| F7Q | SGSLSTQFRLFNRSFTQALGK |
| F11Q | SGSLSTFFRLQNRSFTQALGK |
| F15Q | SGSLSTFFRLFNRSQTQALGK |
Amino acid substitutions are underlined.
Sample preparation for CD and NMR spectroscopy.
Synthetic peptides UA159sp and TPC3 were dissolved in either aqueous buffer/D2O/trifluoroethanol-d2 (TFE-d2) (95:5:0, 70:0:30, 30:0:70, or 0:0:100) or aqueous buffer-D2O (95:5) with 300 mM dodecylphosphocholine-d38 (DPC-d38) (98 atom% D). The aqueous buffer used for sample preparation contained 50 mM K2HPO4/KH2PO4 (pH 7.0). For NMR studies, the concentrations of peptides used for solutions containing 100% TFE-d2 or DPC-d38 were 2 and 5 mM, determined from the mass of the peptide. Neither UA159sp nor TPC3 dissolved completely in aqueous buffer at this pH, so the concentrations for the remainder of the samples were estimated, based on the signal-to-noise ratios of the NMR spectra, to be <0.5 mM. For CD studies, samples were prepared in a similar manner, except that the solvents were not deuterated and the concentrations of the peptides and DPC-d38 were 50 μM and 30 mM, respectively, concentrations at which both peptides completely dissolved.
CD and NMR spectroscopy.
CD measurements were obtained and analyzed to confirm the effects of TFE on the secondary structure of the peptides (see Fig. S1 in the supplemental material). Spectra were analyzed using the CD analysis software DICHROWEB (www.cryst.bbk.ac.uk/cdweb/html/home.html) (29). One-dimensional (1D) and two-dimensional (2D) 1H NMR data sets were collected with a Bruker AVANCE 500 spectrometer and processed using methods similar to previously described methods (4, 41) (see Fig. S2 to S6 in the supplemental material). Using diffusion ordered spectroscopy, the UA159sp, TPC3, and DPC-d38 diffusion constants were calculated to be identical (8 × 10−11 m2 s−1) within the experimental error. Both UA159sp and TPC3 diffused at the same rate as the DPC micelles and thus were strongly associated with the DPC micelles (see Fig. S2 in the supplemental material).
Structural determination.
For structural determination, spin systems were identified through chemical shifts and characteristic TOCSY (total correlation spectroscopy) cross-peak patterns (see Fig. S3 and S4 in the supplemental material). Sequence-specific assignments were determined by the methods described previously (41). Only i to i+1, i to i+3, and i to i+4 interresidue nuclear Overhauser effect spectroscopy (NOESY) cross-peaks were observed for residues between Leu4 and Gly20 of UA159sp and between Ser5 and Thr16 of TPC3 (see Fig. S5 and S6 in the supplemental material). Nevertheless, more than 320 (UA159sp) and 390 (TPC3) unique cross-peaks were assigned from the corresponding NOESY spectra (see Fig. S7 and S8 in the supplemental material).
Structure calculations.
For structure calculations, distance restraints determined from integration of the 250-ms NOESY cross-peaks were classified into four groups: strong, medium, weak, and very weak, corresponding to interproton distance ranges of <2.3, 2.0 to 3.5, 3.3 to 5.0, and 4.8 to 6.0 Å, respectively. Due to intramolecular motions, the 3JHNHα coupling values were averaged over a distribution of dihedral angles. Thus, dihedral angle constraints were conservatively assessed (see Table S2 in the supplemental material) (7). Couplings that were indicative of a random coil structure were not used for structure calculations. An important factor when a peptide structure is determined is the presence of hydrogen bonds that stabilize secondary and tertiary structures. An assessment of hydrogen bonding was carried out on the basis of Hαi-HNi+3 and Hαi-HNi+4 observed connectivities in the NOESY spectrum, dihedral angles, and chemical shift index values (44). Two series of structural calculations were performed with and without added hydrogen bonds to assess the effect of adding suspected hydrogen bonds on the structure calculation.
All structural calculations were based on previous studies (10, 34, 41), using the XPLOR 3.1 software package. A total of 33 and 21 UA159sp lowest-energy structures (with and without hydrogen bonds included in the simulation) and 43 and 36 TPC3 lowest-energy structures (with and without hydrogen bonds included in the simulation) were retained, which had no violations of NOESY constraints of >0.5 Å. The overall quality of these refined structures was examined with the program PROCHECK (25, 33). Except for random-coil sections, all backbone dihedral angles resided in the well-defined, acceptable regions of the Ramachandran plot.
Construction of a lacZ reporter strain.
To assay quorum-sensing activation by synthetic peptides, a lacZ transcriptional reporter strain was constructed to determine the promoter activity of comDE in response to the peptides. Briefly, a 450-bp fragment comprising the entire promoter region of comDE from S. mutans UA159 was amplified by PCR using primers PcomDE-F (5′-TTCTAGACAGGGATAGCGTCAATAAGTTG-3′; XbaI site underlined) and PcomDE-B (5′-AGGTACCAATAGCATAGGTGAGTCAAAGTG-3′; KpnI site underlined). The PCR in a 50-μl mixture was initiated using a Biometra T-personal thermocycler (Biometra, Goettingen, Germany) and the following conditions: initial heating at 94°C for 5 min, followed by 34 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min, followed by an additional 10-min extension at 72°C. The resulting PCR product was digested with KpnI and XbaI, gel purified, and cloned into the same restriction sites of pSL, which was previously constructed by cloning a 3.9-kb HindIII fragment carrying the promoterless lacZ gene from pALH122 (21) into the backbone of low-copy-number of Streptococcus-E. coli shuttle vector pDL276 (14). This vector allows virtually any promoter to insert into the location immediately upstream of the promoterless lacZ gene to generate a transcriptional lacZ fusion construct. The fusion construct was then cloned into the E. coli host DH5α (Invitrogen). The clones from Luria-Bertani medium plates containing kanamycin (50 μg/ml) were initially screened by restriction digestion analysis. One of the clones was selected for sequencing to confirm the fusion site using primer F-S1 (5′-ATCTGCCAGTTTGAGGGGAC-3′) or F-S2 (5′-CGAACATAATTTACAGCGGTTC-3′) at the Sequencing Center at Dalhousie University. The confirmed construct was designated pYH2 and transformed into a comC null mutant (SMCC3) to generate SMC-pYH2 (PcomDE::lacZ comC Specr Kanr). The vector pSL was also transformed into the comC mutant and used as a background control (SMC-pSL). These strains were then assessed to determine their lacZ reporter activities in response to addition of a synthetic peptide.
Peptide-dependent transformation assay.
To determine if a synthetic peptide activated quorum sensing for induction of genetic competence, we used an S. mutans comC null mutant (SMCC3) (3) that was unable to produce but still responded to CSP, enabling assays of peptide-dependent genetic transformation (3, 26). A streptococcal suicide vector, pVA-gtfA, which was constructed to harbor a 2.4-kb fragment of the S. mutans gtfA gene and an erythromycin resistance marker (26), was used as transforming DNA. Wild-type strain UA159 was used as a positive control, while mutants SMCC3 (comC) and SMEK-1 (comE) were used as negative controls. When a culture reached the early mid-log phase (optical density at 600 nm, ∼0.25), an aliquot of a test peptide was added to the culture at a final concentration of 50 nM, which was predetermined based on the half-maximal activation concentration (AC50) (∼25 nM) of UA159sp. After 10 min of incubation at 37°C, an aliquot of transforming DNA (1 μg/ml) was added, and the culture was incubated for an additional 2 h and then plated on THYE agar plates containing 10 μg/ml erythromycin. Prior to addition of the DNA, an aliquot of the cell suspension was plated on the same antibiotic-containing plates to monitor spontaneous mutation. The transformation frequency was expressed as the percentage of transformants (erythromycin-resistant colonies) based on the total number of viable recipient cells per milliliter of cell suspension.
lacZ reporter assay.
A lacZ reporter assay was performed to monitor quorum-sensing activation by synthetic peptides. SMC-pYH2 was used to assay β-galactosidase (β-Gal) activity in response to test peptides, while SMC-pSL was used as a background control. When a culture reached the early mid-log phase, it was supplemented with a test peptide at a final concentration of 50 nM. To assay β-Gal activity using a modification of the methods described previously (3, 21), aliquots of samples were taken at different times (0, 10, 20, and 30 min after addition of a test peptide). The cell suspensions were centrifuged at 10,000 × g at 4°C for 10 min, washed, and resuspended in 50 mM Tris-HCl buffer (pH 7.5) containing 0.27% (vol/vol) β-mercaptoethanol to obtain 1-ml (total volume) cultures. Cells were permeabilized with glass beads after addition of 50 μl chloroform and 20 μl of 0.1% sodium dodecyl sulfate. Following incubation at 37°C for 30 min, o-nitrophenyl-d-galactopyranoside (final concentration, 80 μM) was added as the substrate. The reaction mixtures were incubated for 1 h, and then the reactions were stopped by adding 500 μl of 1 M Na2CO3 and the results were quantified with a spectrophotometer at 420 nm. Protein concentrations of the supernatants were determined by a Bio-Rad protein assay. β-Gal specific activities (A420 per minute per milliliter per milligram of protein) were calculated for triplicate samples from two independent experiments. The data for β-Gal activity were then normalized using data for negative or background controls and plotted as percentages of maximal activation versus log peptide concentrations. The AC50 was determined from the sigmoidal dose-response curve using Prism 4 (GraphPad, San Diego, CA).
Competitive inhibitor experiment.
A competitive inhibitor experiment was performed to determine if any truncated peptide or peptide with an amino acid substitution acted as an antagonist to interfere with the quorum-sensing activity. The β-Gal activities were assayed to determine half-maximal inhibitory concentrations (IC50) of potential antagonists using increasing concentrations in the presence of an effective concentration (50 nM) of UA159sp. The IC50 were determined from an antagonist inhibition curve by plotting the data against various log concentrations of an antagonist in the presence of an effective concentration of UA159sp. All assays were performed in triplicate for two independent experiments.
Supplemental material.
The supplemental material consists of eight figures and two tables (Tables S1 and S2) and includes CD spectra, 2D diffusion ordered spectroscopy data for TPC3, 150-ms 2D 1H-1H TOCSY data, 500-ms 2D 1H-1H NOESY data for UA159sp and TPC3 in 100% TFE, and a 1H chemical shift table and derived structural constraints for UA159sp and TPC3 in 100% TFE. The structural coordinates for two peptides have been deposited in the RCSB Protein Data Bank (http://www.pdb.org/) under RCSB ID code rcsb039055 or PDB ID code 2I2J for UA159sp and RCSB ID code rcsb039053 or PDB ID code 212H for TPC3.
RESULTS
NMR structural analysis.
The initial NMR data for UA159sp and TPC3 were obtained with aqueous buffer to assess the structural nature of the peptides. Based on the narrow chemical shift distributions in the 1D spectrum, both of these peptides appeared to be in random coil conformations. Due to the limited solubility of the peptides in aqueous buffer and aqueous buffer-TFE solutions, the resultant 2D spectra were poor quality. Nevertheless, there were very few HN-to-HN connections, and the HNs were rapidly exchanged with water, confirming the assessment based on CD studies that these two peptides are most likely random coils in an aqueous buffer.
The 1H NMR spectra (Fig. 1) and the CD spectra (see Fig. S1 in the supplemental material) of the peptides in TFE and DPC-d38 were similar, with broader ranges of chemical shift distributions; therefore, a complete structural determination was performed in 100% TFE. Both UA159sp and TPC3 diffused at the same rate as the DPC-d38 micelles and thus were associated with the micelles. Based on the spectral similarity between UA159sp and TPC3 in TFE and DPC-d38 (in particular the aliphatic region, which is less susceptible to the effects of solvents), it is likely that UA159sp and TPC3 form similar structures in TFE and DPC-d38 (i.e., amphipathic α-helices). Sequence-specific assignment of backbone and side chain resonances were determined as described previously (7, 9, 15, 39) and briefly outlined in Table S1 in the supplemental material.
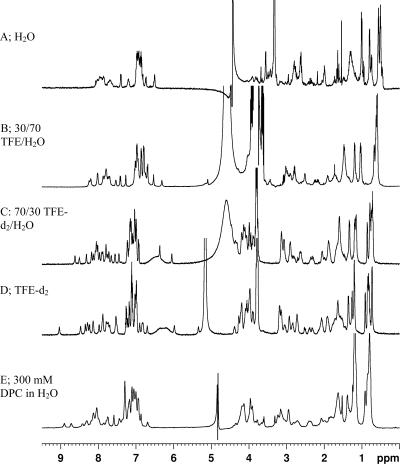
FIG. 1.
UA159sp in H2O is unstructured based on the chemical shifts from the NMR spectra (A). Increasing the TFE concentration (30% [B], 70% [C], and 100% [D]) resulted in a subsequent change in the secondary structure. The similarity between UA159sp in 100% TFE and in DPC (as determined by CD and 1D NMR spectra [E]) suggests that in DPC this peptide also adopts an amphipathic α-helical structure. Similar effects were observed for TPC3 (not shown).
Based on the calculated structure, CD analysis, and α-helical wheel plot (Fig. 2), there is an amphipathic α-helical structure between residues Leu-4 and Gly-20 of UA159sp and between residues Ser-5 and Thr-16 of TPC3. No cross-peaks between distant residues in the linear sequence were observed, as would be expected for β-sheets. Furthermore, the intense HN-HN cross-peaks, the ratio of Hαi-HNi+1 to Hαi-HNi distances, the chemical shift index values (44), and the HN-Hα dihehdral angles (determined from the 3JHNHα coupling values) are indicative of α-helices. The root mean square deviations for structure calculations of the backbone atoms in the α-helical region with and without hydrogen bonding were very similar (0.33 and 0.55 Å for UA159sp and 0.23 and 0.40 Å for TPC3), indicating that although inclusion of hydrogen bonding did tighten up the structure, it was not necessary for defining the α-helical region.
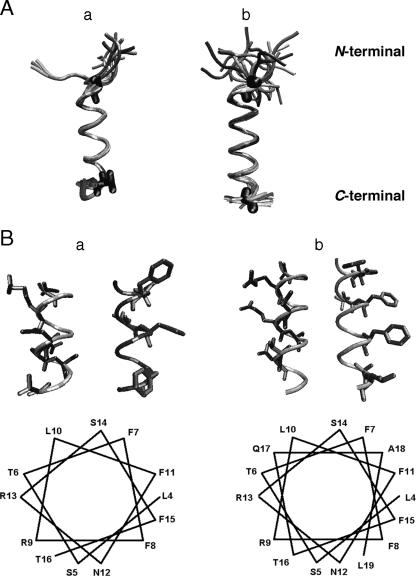
FIG. 2.
(A) Retained structures of TPC3 (a) and UA159sp (b) determined from NMR analysis and computer simulations. The structures that did not violate any structural constraints were retained. The N termini of both peptides are in randomly coiled conformations, but the core helical regions are comparable to each other (residues 4 and 20 for UA159sp and residues 5 and 16 for TPC3). (B) α-Helical wheel of TPC3 (a) and UA159sp (b). The hydrophobic residues are clustered around two-fifths of the α-helix, whereas the basic and hydrophilic residues comprise the remaining three-fifths. Only the α-helical portion of the peptides is shown. Note that TPC3 lacks the proposed C-terminal motif but still has the amphipathic α-helical structure.
Confirmation of lacZ reporter strain.
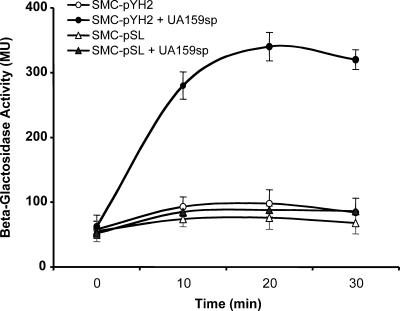
To assay quorum-sensing activation by UA159sp or other peptides, we constructed a lacZ transcriptional reporter strain to monitor the promoter activity of comDE in response to exogenous peptides, since the promoter is directly activated by the peptide-receptor interaction during quorum sensing (3, 26). The newly generated clones that presumably contained the lacZ fusion construct were initially confirmed by restriction digestion analysis. As expected, restriction digestion using KpnI and XbaI released a 450-bp fragment of the entire insert and an 11.3-kb fragment of the plasmid (data not shown). One of the clones was selected for sequencing the PcomDE-lacZ fusion site, which confirmed that the comDE promoter was fused correctly with the promoterless lacZ-containing vector. The confirmed construct was designated pYH2, and pYH2 was then transformed into an S. mutans comC mutant to generate SMC-pYH2 (PcomDE::lacZ comC Specr Kanr). pSL was also transformed into the same mutant to generate SMC-pSL (comC Specr Kanr) as a background control. Then the lacZ activity of these strains in response to exogenous UA159sp was assessed. Before the peptide was added, a basal level of β-Gal activity was detected in both strain SMC-pYH2 and strain SMC-pSL (Fig. 3). After addition of UA159sp, the β-Gal activity in SMC-pYH2 increased rapidly and then plateaued at 20 min, when the activity was nearly fivefold higher than that without the peptide. The activity began to decline after 20 min. In contrast to SMC-pYH2, in control strain SMC-pSL there was no significant difference in the β-Gal activities with and without the peptide. The results confirmed that the lacZ reporter strain constructed provided measurable reporter activity. Using both a lacZ reporter strain and a control strain to assay β-Gal activity provided a sensitive reporter system to assay peptide-dependent quorum-sensing activation or inhibition.
FIG. 3.
Constructed lacZ reporter strain SMC-pYH2 expresses predicted β-galactosidase activity in response to UA159sp, although a basal level of β-Gal activity was detected without UA159sp. The control strain, SMC-pSL, did not show the same response to UA159sp. MU, Miller units.
In vitro biological activity.
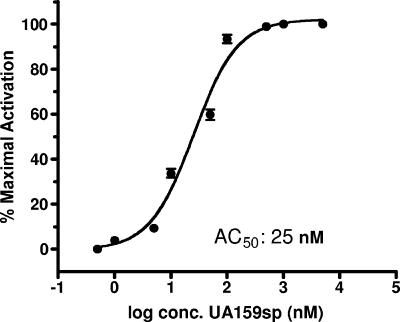
The synthetic 21-amino-acid signaling peptide (UA159sp) derived from S. mutans wild-type strain UA159 was first assessed to determine its in vitro ability to activate quorum sensing for genetic competence. To avoid interference by endogenous CSP, an S. mutans comC null mutant (SMCC3) that was unable to produce, but still responded to, the CSP was used to assay peptide-dependent induction of genetic competence. The results showed that synthetic UA159sp at a concentration as low as 5.0 nM was active in induction of genetic competence. A dose-dependent relationship was observed between the quorum-sensing activity and the concentration of UA159sp, and the AC50 was ∼25 nM (Fig. 4). Concentrations of the peptide higher than 1,000 nM did not significantly increase the activity or transformation frequency.
FIG. 4.
Dose-response assay for β-Gal activity of the reporter strain, SMC-pYH2, in response to synthetic UA159sp. There was a dose-dependent relationship between the quorum-sensing activity and the UA159sp concentration, and the AC50 was ∼25 nM. The half-maximal activation value was calculated using triplicate samples from two independent experiments.
Effects of C-terminal truncations on peptide activity.
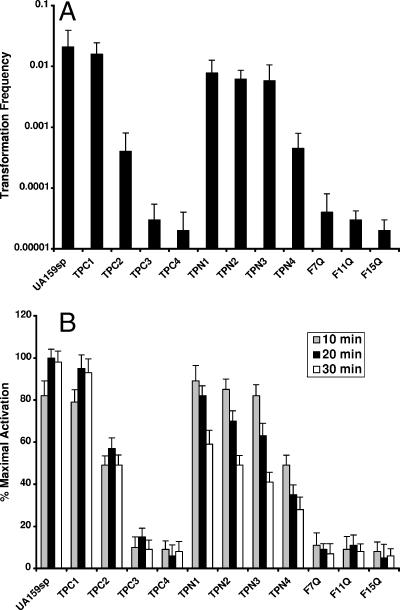
A previous study showed that the C-terminally truncated peptide from mutant JH1005 did not activate quorum sensing for genetic competence (26). However, this finding did not rule out the possibility that the defect might result from the inability of the cells to respond to a CSP or from the Ser-3 mutation at the N terminus of the JH1005 peptide. To answer this question, we first induced genetic competence in mutant JH1005 by adding UA159sp. We found that addition of UA159sp to a JH1005 culture almost completely restored genetic competence in this strain (Fig. 5), suggesting that mutant JH1005 could respond normally to the CSP. We then used the truncated JH1005 peptide (TPC3) to induce genetic competence in a comC mutant (SMCC3) defective in production of CSP. The results showed that TPC3 did not induce genetic competence of mutant SMCC3, although this strain responded to UA159sp normally. We also tested three more C-terminally truncated peptides for induction of genetic competence (Fig. 6A). Deletion of the C-terminal lysine (TPC1) did not affect the genetic competence. However, deletion of the C-terminal glycine and lysine (TPC2) resulted in a significant decrease in the transformation frequency, although a low level of activity remained. The peptides in which three (TPC3) or four (TPC4) C-terminal amino acid residues were deleted did not induce genetic competence. Consistent with these results, the lacZ reporter assay confirmed that both TPC3 and TPC4 did not activate the quorum-sensing activity, as shown by significantly lower levels of β-Gal activity at multiple times (Fig. 6B). These results imply that the C-terminal residues are required to activate the quorum-sensing activity.
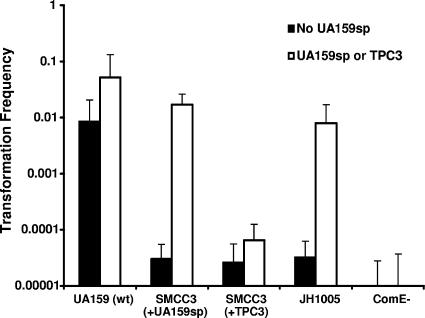
FIG. 5.
Induction of genetic competence of S. mutans strains in response to UA159sp or TPC3. The strains used included SMCC3 (comC), which was unable to produce but still respond to UA159sp; SMEK-1 (comE), which did not respond to UA159sp at all; and JH1005, which had a peptide (TPC3) of the C-terminal truncation. S. mutans UA159 was used as a positive control, while mutant SMEK-1 (comE) was used as a negative control.
FIG. 6.
Effects of truncated peptides or peptides with amino acid substitutions on the activation of quorum sensing for genetic competence. (A) Peptide-dependent induction of genetic competence. (B) Percentages of maximal activation by synthetic peptides. The β-Gal activities were assayed in triplicate in two independent experiments.
Since the C-terminally truncated peptides did not induce genetic competence, we examined whether the defect resulted from an inability of the peptides to activate the signal transduction pathway or from an inability to bind to the receptor. To answer this question, a competitive inhibitor experiment was performed to assay the CSP-dependent quorum-sensing activity in the presence of increasing concentrations of a potential antagonist. The results showed that at least two C-terminally truncated peptides, TPC3 (IC50, ∼368 nM) and TPC4 (IC50, ∼292 nM), at higher concentrations competitively inhibited the quorum-sensing activity induced by UA159sp (Table 3). The results suggested that the TPC3 and TPC4 truncated peptides appeared to bind to the receptor but did not activate the signal transduction pathway. It is interesting that these truncated peptides did not completely inhibit the quorum-sensing activity in the presence of UA159sp, suggesting that wild-type CSP interacts more effectively with the receptor.
TABLE 3.
Activation or inhibition of quorum sensing by peptide analogs
| Peptide | AC50 (nM)a | IC50 (nM)b |
|---|---|---|
| UA159sp | 25 | NAc |
| TPC1 | 29 | NA |
| TPC2 | 132 | NA |
| TPC3 | NA | 368 |
| TPC4 | NA | 292 |
| TPN1 | 29 | NA |
| TPN2 | 33 | NA |
| TPN3 | 35 | NA |
| TPN4 | 143 | NA |
| F7Q | 322 | 10,300 |
| F11Q | 328 | 11,500 |
| F15Q | 334 | 11,800 |
The AC50 were calculated from a dose-response curve using GraphPad Prism 4.
The IC50 were determined from an antagonist inhibition curve by plotting the data against various log concentrations of an antagonist in the presence of an effective concentration (50 nM) of UA159sp.
NA, not assayed.
Effects of substitution of a hydrophilic residue for Phe-7, Phe-11, or Phe-15 on peptide activity.
Our NMR analysis showed that the core structure of UA159sp was an amphipathic α-helix with a well-defined hydrophobic face characterized by a row of four highly hydrophobic phenylalanines. To test the hypothesis that the hydrophobic face of the CSP might be crucial for receptor binding, three peptides, designated F7Q, F11Q, and F15Q, were synthesized, and their abilities to activate quorum sensing were assessed. The results showed that none of the peptides activated quorum sensing for genetic competence (Fig. 6A). In contrast to TPC3 or TPC4, these peptides with amino acid substitutions did not competitively inhibit quorum sensing induced by UA159sp (Table 3). These data suggest that replacing the phenylalanines in the hydrophobic face appears to reduce peptide binding to the receptor.
Effects of N-terminal truncations on peptide activity.
We also tested four N-terminally truncated peptides to determine their activities in induction of genetic competence. The synthetic peptides with up to three amino acid residues deleted from the N terminus still activated quorum sensing for genetic competence. The transformation efficiencies of these peptides were as high as the transformation efficiency of UA159sp (Fig. 6A). These data suggest that the three N-terminal amino acid residues appear to be neither crucial for activation of the signal transduction pathway nor important for receptor binding. The data also rule out the possibility that the defect in genetic competence in mutant JH1005 was a result of the Thr-3 substitution. Based on the β-Gal activity assay (Fig. 6B), it is interesting that these three N-terminally truncated peptides appeared to be normal for activation of the quorum-sensing activity in the first 10 min following addition to the culture supernatants. However, the levels of β-Gal activity induced by these peptides began to decline at 20 min and dropped to basal levels after 30 min, in contrast to the effects of C-terminally truncated peptides. These results suggest that the N-terminally truncated peptides activate quorum sensing but lose their activity more rapidly than CSP. However, removal of the fourth residue at the N terminus significantly affected the quorum-sensing activity, and the resulting peptide did not induce wild-type levels of genetic transformation.
DISCUSSION
The discovery of bacterial cell-cell communication or quorum-sensing mechanisms that regulate virulence and pathogenicity has provided a new opportunity to combat bacterial infections by designing drugs that block bacterial quorum-sensing and associated pathogenic activities (8, 18, 31, 38, 40). One approach is to discover natural or synthetic compounds that are structurally similar to quorum-sensing signal molecules. These structural analogs may competitively bind to the receptors and inhibit quorum-sensing-controlled pathogenicity (8, 30-32). Knowing the molecular details of quorum-sensing signal molecules should facilitate the development of inhibitors. In several recent studies workers have described the use of quorum-sensing antagonists to achieve inhibition of the quorum-sensing circuits in bacteria (8, 19, 30-32, 45). These studies have generated substantial information concerning structure-function relationships of quorum-sensing signal molecules and their receptors, which has great value for searching quorum-sensing inhibitors.
In this study, we analyzed and compared the three-dimensional structures of S. mutans wild-type signaling peptide UA159sp and the C-terminally truncated peptide TPC3 from mutant JH1005 defective in genetic competence in an effort to understand the structural characteristics of these two peptides. In the presence of water, neither UA159sp nor TPC3 was soluble enough for extensive NMR studies. From the profiles for 1D NMR and CD spectra, however, it is likely that these peptides are in a random coil conformation, which was observed for a related CSP of Streptococcus pneumoniae (22) during this study. In TFE, both peptides formed well-defined α-helices (Fig. 2) from residues Leu-4 to Gly-20 (UA159sp) and from residues Ser-5 to Thr-16 (TPC3), and the residues near the N termini were disordered. The last few residues at the C termini have slightly more motional freedom than the core of the helix. We dissolved the peptides in the widely used solvent TFE, because it improves the structural stability of peptides and is considered to be biologically relevant (7, 41). TFE is also thought to mimic a protein receptor environment by stabilizing helices in regions with intrinsic α-helical propensity that are likely to form helices when they bind to their protein partner (36, 44). An important feature observed from the NMR-determined structure is that both peptides form amphipathic α-helices with a well-defined row of phenylalanine residues along the hydrophobic side that comprises about 40% of the surface of the helix. Hydrophilic and charged residues comprise the remainder of the surface. Based on the experimentally determined structures of other similar binding peptides (9, 22), a well-defined row of hydrophobic residues on an amphipathic α-helix is common and is necessary for binding to the receptor protein. The UA159sp and TPC3 peptides fall into the FXXFF motif family; the FXXFF motif is a general protein-protein interaction motif in which the interaction occurs through a hydrophobic patch formed by the hydrophobic residues packing into a hydrophobic pocket of its receptor (33, 34, 45). The fact that the amphipathic helical structure is reflected in the binding and induction of the quorum-sensing response suggests that the peptide may be helical within the active site. The membrane environment may help form the helical structure, thus increasing the rate of binding to the receptor, but this does not imply that the structures of the peptides in TFE and DPC and the active site are identical; nevertheless, the structures that we have determined are similar.
The results of the activity analysis show that the quorum-sensing signaling peptide from S. mutans wild-type strains has at least two functional domains. The C-terminal structural motif appears to be crucial for activation of the quorum-sensing signal transduction pathway, while the core amphipathic α-helical structure with the hydrophobic domain is required for receptor binding, as demonstrated by the competitive inhibition studies. Our work revealed that deletion of even two amino acid residues from the C terminus significantly affects the activity of the CSP in induction of genetic competence. Deletion of three or four amino acid residues from the C terminus, which completely removes the C-terminal structural motif, eliminates the activity of the CSP in activation of quorum sensing for genetic competence. However, truncation of the C-terminal motif does not seem to affect peptide binding, because the C-terminally truncated peptides, at higher concentrations, competitively inhibit genetic competence, suggesting that the truncated peptides are still able to bind to the receptor but do not activate the signal transduction pathway. This suggestion is supported by the NMR structural analysis of TPC3 since C-terminal deletion of three amino acid residues does not seem to affect the α-helical structure. Since they competitively inhibit the quorum-sensing activity, these C-terminally truncated peptides may be valuable for the development of quorum-sensing inhibitors. We are currently developing a peptide receptor binding assay and an NMR analysis assay with bicelles to further study this question.
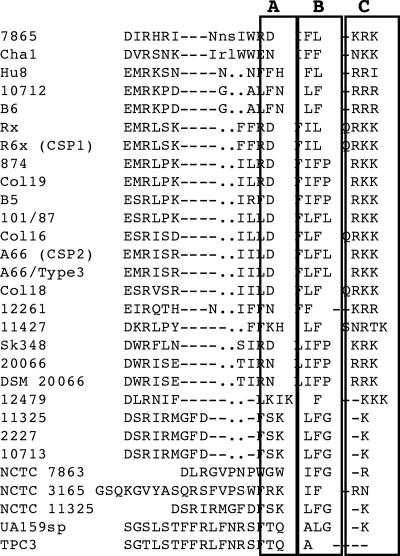
Since all C-terminally truncated peptides are shorter than UA159sp, there is a potential argument that the shorter sequences may be a reason why the truncated peptides lose activity. To address this question, we assessed four N-terminally truncated peptides. Deletion of one, two, or three amino acid residues did not affect the activity of the peptide, while removal of the fourth residue had a significant impact on the activity. The evidence clearly shows that the intact C-terminal structural motif rather than the shorter sequence is crucial for activation of the signal transduction pathway. Thus, the results of this study confirm that mutant JH1005 is defective in genetic competence likely because of the C-terminal truncation that disrupted the structural motif crucial for activation of the quorum-sensing signal transduction pathway. However, none of the truncated peptides completely inhibited genetic competence in the presence of UA159sp, suggesting that the wild-type CSP is more effective in interacting with the receptor. To the best of our knowledge, this is the first report showing that the C-terminal structural motif of streptococcal CSPs plays an important role in activating quorum sensing for genetic competence. A sequence alignment of CSPs from various strains of Streptococcus (16, 17, 38) showed that most CSPs that have been identified appear to have a charge or polar hydrophobic charged structural motif at the C terminus (Fig. 7). The peptide from S. mutans mutant JH1005 is the only example where the C-terminal structural motif is truncated with a significant defect in genetic competence, further supporting the argument that the C-terminal motif is important for activation of the signal transduction pathway. It is, therefore, likely that the C-terminal motifs of the CSPs from other streptococci have a similar role in activation of quorum-sensing signal transduction pathways.
FIG. 7.
Sequence alignment of CSPs from different streptococcal strains. The amino acid residues are identical or similar in at least 50% of the sequences compared. All CSPs likely form amphipathic α-helical structures with a C-terminal structural motif consisting of polar (box A), hydrophobic (box B), and charged residues (box C). The region enclosed in a boxes is separated to better indicate the structural motifs.
The second functional domain of the signal peptide is the core α-helical structure that forms a typical amphipathic helix with a well-defined row of hydrophobic residues lining one side and hydrophilic residues lining another side. The hydrophobic residues form a potential binding site for the signal peptide to interact with the receptor (22, 30, 45). The data obtained in this study show that substitution of a hydrophilic residue, glutamine, for Phe-7, Phe-11, or Phe-15 significantly affected the activity of the signal peptide in activation of quorum sensing. It is important that unlike the C-terminally truncated peptides, the peptides with a glutamine substitution did not competitively inhibit genetic competence induced by UA159sp. The evidence suggests that disruption of the hydrophobic domain results in an inability of the signal peptide to bind to the receptor. We propose that the core amphipathic helix and hydrophobic face of the signaling peptide are important for receptor binding. During this study, Håvarstein and colleagues reported similar findings that support this hypothesis. They found that a hydrophobic patch of CSP1 from S. pneumoniae strains contributed mainly to receptor recognition and CSP specificity (22). Interestingly, the hydrophobic patch in S. pneumoniae CSP1 also consists of three highly hydrophobic phenylalanines at positions 7, 8, and 11 and an isoleucine at position 12 (22).
The synthetic peptides with one, two, or three amino acid residues deleted from the N terminus are still active in induction of genetic competence. The transformation efficiencies with these peptides are not significantly different from the transformation efficiency with UA159sp. These truncated peptides appear to be normal for activation of the quorum-sensing activity for induction of genetic competence in the first 10 min following addition to the culture supernatants. Clearly, the three N-terminal amino acid residues are essential neither for activation of the signal transduction pathway nor for peptide binding to the receptor. Since CSPs from S. pneumoniae strains have a conserved N-terminal sequence, the N termini of CSPs from this organism have been suggested to play a role in receptor activation (22). However, the N-terminal sequence of the signal peptides from S. mutans strains differs significantly from the N-terminal sequence of the S. pneumoniae CSPs (16, 22, 26). Sequence divergence at the N termini of CSPs from other species of Streptococcus, including Streptococcus gordonii, Streptococcus sanguis, Streptococcus cristatus, Streptococcus anginosus, and Streptococcus salivarius (Fig. 7), has also been observed (16, 17, 38). The evidence obtained in this study suggests that the N-terminally truncated peptides appear to be active for a shorter time than UA159sp. This raises the question of whether the N terminus may play a role in the stability of the signal peptide. It is known that the N terminus of a protein or peptide frequently plays a rule in its in vivo life span, a phenomenon called the N end role, in which N-terminal amino acid residues determine the life span of a protein (5, 42). Whether signal peptides secreted from bacteria follow the N end rule is currently unknown. Further study is necessary to analyze individual CSPs to determine the role of the N termini in the CSP activity and the fate of the peptides after binding to the receptor.
Based on the results presented in this paper and the findings of other workers for S. pneumoniae (22) during this study, we propose the following model for quorum-sensing activation by CSP. In aqueous environments, the CSP has no significant structure but is likely randomly coiled, soluble, and diffusible, which may facilitate diffusion of the protein in the environment. Upon recognition of the membrane-bound ComD receptor, the randomly coiled CSP is subject to folding or conformational change that forms an amphipathic α-helix with a hydrophobic face. This allows the CSP to interact with the binding pocket of the receptor via a hydrophobic interaction, enabling the CSP to bind to the receptor. However, the receptor is activated only by the presence of the C-terminal motif, which likely causes additional interactions within the binding pocket of the receptor. These interactions then initiate a signaling transmission through autophosphorylation and activate the ComE response regulator, in turn triggering the cascade of quorum sensing for genetic competence.
In conclusion, this study is the first study to recognize the importance of the signaling peptide C-terminal residues in streptococcal quorum sensing, and our results provide novel insights into the structure-activity relationship of quorum-sensing signal peptides in streptococci.
Supplementary Material
Acknowledgments
We thank John Walter and Ian Burton, Institute for Marine Biosciences, National Research Council of Canada, for their assistance with, and access to, an NMR spectrometer and Alison Thompson, Department of Chemistry, Dalhousie University, for access to a CD instrument. We also thank Dennis Cvitkovitch of the University of Toronto for providing bacterial strains and plasmids.
This work was supported in part by grant MOP-74487 from the Canadian Institute for Health Research, by grant 10313 from the Canadian Foundation for Innovation, by the Nova Scotia Health Research Foundation, and by research funds from the Faculty of Dentistry at Dalhousie University. R.T.S. was a recipient of an Izaak Walton Killam Memorial Post-Doctoral Fellowship from the Killam Trust Foundation and a Cancer Research Training Program fellowship.
Footnotes
Published ahead of print on 25 August 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ahn, S. J., Z. T. Wen, and R. A. Burne. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect. Immun. 74:1631-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aspiras, M. B., R. P. Ellen, and D. G. Cvitkovitch. 2004. ComX activity of Streptococcus mutans growing in biofilms. FEMS Microbiol. Lett. 238:167-174. [DOI] [PubMed] [Google Scholar]
- 4.Aue, W. P., E. Bartholdi, and R. R. Ernst. 1976. Two-dimensional spectroscopy. Application to nuclear magnetic resonance. J. Chem. Phys. 64:2229-2246. [Google Scholar]
- 5.Bachmair, A., and A. Varshavsky. 1989. The degradation signal in a short-lived protein. Cell 56:1019-1032. [DOI] [PubMed] [Google Scholar]
- 6.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 7.Cavanagh, J., W. J. Fairbrother, A. G. Palmer III, and N. J. Skelton. 1996. Protein NMR spectroscopy: principles and practice. Academic Press, New York, NY.
- 8.Chan, W. C., B. J. Coyle, and P. Williams. 2004. Virulence regulation and quorum sensing in staphylococcal infections: competitive AgrC antagonists as quorum sensing inhibitors. J. Med. Chem. 47:4633-4641. [DOI] [PubMed] [Google Scholar]
- 9.Compton, L. A., and W. C. Johnson, Jr. 1986. Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal. Biochem. 155:155-167. [DOI] [PubMed] [Google Scholar]
- 10.Corcoran, J. A., R. Syvitski, D. Top, R. M. Epand, R. F. Epand, D. Jakeman, and R. Duncan. 2004. Myristoylation, a protruding loop, and structural plasticity are essential features of a nonenveloped virus fusion peptide motif. J. Biol. Chem. 279:51386-51394. [DOI] [PubMed] [Google Scholar]
- 11.Cvitkovitch, D. G., Y. H. Li, and R. P. Ellen. 2003. Quorum sensing and biofilm formation in streptococcal infections. J. Clin. Investig. 112:1626-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunny, G. M., and B. A. Leonard. 1997. Cell-cell communication in Gram-positive bacteria. Annu. Rev. Microbiol. 51:527-564. [DOI] [PubMed] [Google Scholar]
- 14.Dunny, G. M., L. N. Lee, and D. J. LeBlanc. 1991. Improved electroporation and cloning vector system for gram-positive bacteria. Appl. Environ. Microbiol. 57:1194-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griesinger, C., G. Otting, K. Wuthrich, and R. R. Ernst. 1988. Clean tocsy for H-1 spin system-identification in macromolecules. J. Am. Chem. Soc. 110:7870-7872. [Google Scholar]
- 16.Håvarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Håvarstein, L. S., R. Hakenbeck, and P. Gaustad. 1997. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J. Bacteriol. 179:6589-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hentzer, M., and M. Givskov. 2003. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J. Clin. Investig. 112:1300-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hentzer, M., H. Wu, J. B. Andersen, K. Riedel, T. B. Rasmussen, N. Bagge, N. Kumar, M. A. Schembri, Z. Song, P. Kristoffersen, M. Manefield, J. W. Costerton, S. Molin, L. Eberl, P. Steinberg, S. Kjelleberg, N. Hoiby, and M. Givskov. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22:3803-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hillman, J. D., A. L. Dzuback, and S. W. Andrews. 1987. Colonization of the human oral cavity by a Streptococcus mutans mutant producing increased bacteriocin. J. Dent. Res. 66:1092-1094. [DOI] [PubMed] [Google Scholar]
- 21.Honeyman, A. L., C. K. Cote, and R. Curtiss III. 2002. Construction of transcriptional and translational lacZ gene reporter plasmids for use in Streptococcus mutans. J. Microbiol. Methods 49:163-171. [DOI] [PubMed] [Google Scholar]
- 22.Johnsborg, O., P. E. Kristiansen, T. Blomqvist, and L. S. Håvarstein. 2006. A hydrophobic patch in the competence-stimulating peptide, a pneumococcal competence pheromone, is essential for specificity and biological activity. J. Bacteriol. 188:1744-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Parmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuramitsu, H. K. 1993. Virulence factors of mutans streptococci: role of molecular genetics. Crit. Rev. Oral Biol. Med. 4:159-176. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski, R. A., J. A. C. Rullmann, M. W. MacArthur, R. Kaptein, and J. M. Thornton. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8:477-486. [DOI] [PubMed] [Google Scholar]
- 26.Li, Y. H., P. C. Y. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Y. H., M. N. Hanna, G. Svensäter, R. P. Ellen, and D. G. Cvitkovitch. 2001. Cell density modulates acid adaptation in Streptococcus mutans: implication for survival in biofilms. J. Bacteriol. 183:6875-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Y. H., N. Tang, P. C. Y. Lau, M. B. Aspiras, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobley, A., and B. A. Wallace. 2001. Dichroweb: a website for the analysis of protein secondary structure from circular dichroism spectra. Biophys. J. 80:373A. [DOI] [PubMed] [Google Scholar]
- 30.Lyon, G. J., P. Mayville, T. W. Muir, and R. P. Novick. 2000. Rational design of a global inhibitor of the virulence response in Staphylococcus aureus, based in part on localization of the site of inhibition to the receptor-histidine kinase, AgrC. Proc. Natl. Acad. Sci. USA 97:13330-13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayville, P., G. Ji, R. Beavis, H. Yang, M. Goger, R. P. Novick, and T. W. Muir. 1999. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. USA 96:1218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDowell, P., Z. Affas, C. Reynolds, M. T. G. Holden, S. J. Wood, S. Saint, A. Cockayne, P. J. Hill, C. E. R. Dodd, B. W. Bycroft, W. C. Chan, and P. Williams. 2001. Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus. Mol. Microbiol. 41:503-512. [DOI] [PubMed] [Google Scholar]
- 33.Morris, A. L., M. W. MacArthur, E. G. Hutchinson, and J. M. Thornton. 1992. Stereochemical quality of protein structure coordinates. Proteins 12:345-364. [DOI] [PubMed] [Google Scholar]
- 34.Nilges, M., J. Kuszewski, and A. T. Brunger. 1991. Computational aspects of the study of biological macromolecules by NMR. Plenum Press, New York, NY.
- 35.Petersen, F. C., and A. A. Scheie. 2000. Genetic transformation in Streptococcus mutans requires a peptide secretion-like apparatus. Oral Microbiol. Immunol. 15:329-334. [DOI] [PubMed] [Google Scholar]
- 36.Roccatano, D., G. Colombo, M. Fioroni, and A. Mark. 2002. Mechanism by which 2,2,2-trifluoroethanol/water mixtures stabilize secondary-structure formation in peptides: a molecular dynamics study. Proc. Natl. Acad. Sci. USA 99:12179-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schauder, S., and B. L. Bassler. 2001. The languages of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 38.Scheie, A. A., and F. C. Petersen. 2004. The biofilm concept: consequences for future prophylaxix of oral diseases? Crit. Rev. Oral Biol. Med. 15:4-12. [DOI] [PubMed] [Google Scholar]
- 39.Smith, L. J., K. A. Bolin, H. Schwalbe, M. W. MacArthur, J. M. Thornton, and C. M. Dobson. 1996. Analysis of main chain torsion angles in proteins: prediction of NMR coupling constants for native and random coil conformations. J. Mol. Biol. 255:494-506. [DOI] [PubMed] [Google Scholar]
- 40.Smith, R. S., and B. H. Iglewski. 2003. Pseudomonas aeruginosa quorum sensing as a potential antimicrobial target. J. Clin. Investig. 112:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Syvitski, R. T., I. Burton, N. R. Mattatall, S. E. Douglas, and D. L. Jakeman. 2005. Structural characterization of the antimicrobial peptide pleurocidin from winter flounder. Biochemistry 44:7282-7293. [DOI] [PubMed] [Google Scholar]
- 42.Tobias, J. W., T. E. Shrader, G. Rocap, and A. Varshavsky. 1991. The N-end rule in bacteria. Science 254:1374-1377. [DOI] [PubMed] [Google Scholar]
- 43.van der Ploeg, J. R. 2005. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J. Bacteriol. 187:3980-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wishart, D. S., B. D. Sykes, and F. M. Richards. 1992. The chemical-shift index—a fast and simple method for the assignment of protein secondary structure through NMR-spectroscopy. Biochemistry 31:1647-1651. [DOI] [PubMed] [Google Scholar]
- 45.Wright, J. S., III, G. J. Lyon, E. A. George, T. W. Muir, and R. P. Novick. 2004. Hydrophobic interactions drive ligand-receptor recognition for activation and inhibition of staphylococcal quorum sensing. Proc. Natl. Acad. Sci. USA 101:16168-16173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.