Abstract
Group A Streptococcus (GAS) is a leading human pathogen associated with a wide spectrum of mucosal and invasive infections. GAS expresses a large number of virulence determinants whose expression is under the control of several transcriptional regulatory networks. Here we performed the first mutational analysis of a genetic locus immediately upstream of the streptolysin S biosynthetic operon in several GAS genome sequences, including that of the M1T1 serotype, the leading isolates associated with serious invasive disease. The locus consists of a predicted RofA-like stand-alone transcriptional regulator (RALP3) and the largest open reading frame in the GAS genome, encoding a predicted LPXSG motif cell wall-anchored protein we have named LSA (for “large surface-anchored” protein). Comparative reverse transcription-PCR analysis of wild-type M1T1 GAS and an isogenic RALP3-deficient mutant identifies RALP3 as a global transcriptional regulator affecting expression of numerous virulence factor genes, including those for strong repression of the hyaluronic acid capsule and cysteine protease production. RALP3 contributed to GAS epithelial cell invasion and bloodstream survival. LSA was found to be under negative regulation by RALP3 and to influence GAS-epithelial cell interactions and GAS antimicrobial peptide sensitivity. Isogenic M1T1 GAS mutants lacking either RALP3 or LSA were attenuated in a murine model of systemic infection, indicating that this locus plays a role in the virulence potential of the organism.
Group A Streptococcus (GAS; Streptococcus pyogenes) is the causative agent of a wide array of human infections, ranging from simple pharyngitis to life-threatening necrotizing fasciitis (NF) and toxic shock syndrome (TSS). GAS is also the trigger for the postinfectious, immunologically mediated syndromes of rheumatic fever and glomerulonephritis. The prominence of GAS as a human pathogen and the diversity of its disease manifestations reflect a large array of genetically encoded virulence factors that promote attachment to and invasion of host epithelium, resistance to host immune clearance mechanisms, and direct or inflammatory response-mediated injury to host cells and tissues (6, 14, 44). The expression of GAS virulence factors is controlled at the transcriptional level by an array of two-component signal transduction systems and “stand-alone” response regulators (38) and is further modulated posttranslationally by the actions of the broad-spectrum GAS cysteine protease, SpeB (2, 13).
One well-studied virulence factor of GAS is the cytolytic toxin streptolysin S (SLS), which is responsible for producing the hallmark zone of β-hemolysis surrounding colonies of the organism grown on blood agar media. The nine-gene GAS sag operon has been found to be necessary and sufficient for production of the small SLS toxin, which exhibits characteristics of a bacteriocin-like peptide (48, 49). Targeted mutagenesis of SLS production yields nonhemolytic mutants with a reduced capacity to injure eukaryotic cells, resist phagocytic clearance, or establish infection in a murine model of GAS NF (4, 18, 21, 32, 45).
While GAS strains of many genotypes are capable of producing serious infections, strains representing one globally disseminated M1T1 clone have persisted for over 2 decades as the most prevalent invasive isolates (8, 11, 12, 46), including those collected at the nine surveillance centers of the United States Centers for Disease Control Emerging Infections Program Network each year from 1997 though 2005 (http://www.cdc.gov/ncidod/dbmd/abcs). Immediately upstream of the sagA-to-sagI operon for SLS biosynthesis in the published M1 GAS genome (23) lies a locus which contains two open reading frames (ORFs) which have interesting sequence features but whose potential role in GAS biology and pathogenesis has been unexplored. The first ORF (Spy0737) encodes over 2,000 amino acids, by far the largest putative protein in the M1 GAS genome, including a C-terminal LPASG cell wall anchor motif, leading us to name the candidate gene product LSA (for “large surface-anchored” protein). The second gene (Spy0735), divergently transcribed, is predicted to encode a protein resembling the stand-alone global transcriptional regulator RofA (3, 24) and was recently named RALP3 for inclusion in the RofA-like protein family (26).
In this study, we performed a targeted allelic replacement mutagenesis of ralp3 in an invasive M1T1 GAS isolate, initially to ascertain whether this gene affected expression of the nearby sag operon for SLS biosynthesis. We discovered that RALP3 impacts sagA expression and indeed functions in global control of several other GAS virulence phenotypes, including down-regulation of expression of genes encoding streptococcal inhibitor of complement (sic), hyaluronic acid capsule (hasA), and cysteine protease (speB). RALP3 also negatively regulates expression of the adjacent LSA, which through additional mutagenesis studies was found to influence GAS-epithelial cell interactions. Isogenic ΔRALP3 and ΔLSA GAS mutants show significant loss of virulence potential in vivo, indicating that this locus plays a role in the pathogenesis of invasive M1T1 GAS infection.
MATERIALS AND METHODS
Bacteria and growth conditions.
M1T1 GAS wild-type (WT) strain 5448 is an isolate from a patient with NF and TSS that is genetically representative of a globally disseminated clone associated with invasive GAS infections (8). GAS strains were grown in Todd-Hewitt broth (THB), pH 7.5, or on THB agar plates. For antibiotic selection, 2 μg/ml erythromycin (Em) or 2 μg/ml chloramphenicol was used. Escherichia coli strains were grown in Luria-Bertani broth; antibiotic selection employed 500 μg/ml Em. For functional assays, unless otherwise noted, bacteria were grown to early log phase, i.e., an optical density at 600 nm (OD600) of 0.4 (∼1 × 108 CFU per ml) in THB and resuspended in appropriate buffers, and concentrations were confirmed by plating dilutions and enumerating CFU.
Allelic exchange mutagenesis of the GAS ralp3 and lsa genes.
Targeting vectors for precise, in-frame allelic replacement of ralp3 and lsa with the chloramphenicol acetyltransferase gene (cat) were constructed using our published method (39) along with sequence information from the GAS M1 serotype SF370 genome (23). Briefly, ∼500 bp of chromosomal DNA immediately upstream of each gene was amplified with the primer sets ralp3-upF (5′-ACACCCAGACTGACTGATGA-3′) plus ralp3-upR (5′-AACTGAACTCCTTTCATTTTTATA-3′) and lsa-upF (5′-ATGAAGTCGTGTAGGCTCAG-3′) plus lsa-upR (5′-TCTGTATTCCCCCTTATTTTTTCA-3′), where the two reverse primers were designed to possess a 25-bp 5′ extension corresponding to the negative strand of the 5′ end of cat. Likewise, ∼500 bp of chromosomal DNA immediately downstream of each gene was amplified with the primer sets ralp3-downF (5′-GCATCCCTGCGTCCACTACTAG-3′) plus ralp3-downR (5′-CAGGTATTGCCTATCTGCGA-3′) and lsa-downF (5′-GAACCTGGCTTTATCCAAATTGCT-3′) plus lsa-downR (5′-GTAGCACGAAAATTCGCTC-3′), where the two forward primers were engineered to possess a 25-bp 5′ extension corresponding to the positive strand of the 3′ end of cat. Fusion PCRs were performed with the ralp3 upstream amplicon plus a 658-bp amplicon of cat (from pACYC) plus the ralp3 downstream amplicon, or the corresponding series of DNA fragments for lsa. The resulting PCR products, containing an in-frame substitution of either ralp3 or lsa with cat in the genomic context, were subcloned into the temperature-sensitive vector pHY304. Subsequent steps in the transformation of GAS strain 5448, procedures for antibiotic and temperature selection for single- and double-crossover events, and final PCR confirmation of the allelic replacement mutants were performed as previously described (33).
Reverse transcription and real-time quantitative PCR.
WT and mutant GAS strains were grown to logarithmic phase (OD600 = 0.4) or stationary phase (18-h culture), and RNA was isolated using QIAGEN′s RNeasy minikit with in-column DNase digestion according to the manufacturer's protocol. First-strand synthesis and real-time PCR were performed as previously described (52) in TaqMan Universal MasterMix SYBR Green (Applied Biosystems, Foster City, CA) using primer sets for individual GAS genes, each time normalizing rates to the expression level of gyrase A. The specific primer sets utilized for reverse transcription-PCR (RT-PCR) detection of mRNA for the GAS genes ralp3, lsa, speB, hasA, sda1, mga, covR, sagA, spn, sic, grab, emm1, eno, scpC, ska, cpa, scpA, cfa, mac, endoS, smeZ, pulA, and gyrA are available on request.
Hyaluronic acid and cysteine protease quantifications.
Levels of hyaluronic acid on the GAS cell surface and released into the culture supernatant were measured using a hyaluronic acid enzyme-linked immunosorbent assay (ELISA) kit (Corgenix) per the manufacturer's instructions. GAS strains were grown to mid-log (OD600 = 0.4), late log (OD600 = 0.8), or stationary (18-h culture) phase for collection of supernatants, with the corresponding cell pellets equilibrated in PBS before detection. The mature cysteine functional proteolytic activity of SpeB was measured with a EnzCheck protease assay kit (Molecular Probes) as previously described (34). Two-dimensional gel analysis of secreted GAS proteins was performed using published methods (2).
Epithelial cell adherence and invasion.
GAS adherence and invasion assays with human A549 lung epithelial cells were performed as previously described (19), with some modifications. Briefly, newly confluent monolayers of ∼2 × 105 A549 cells/well in 24-well plates were washed three times with RPMI 1640. Early-log-phase GAS cells were pelleted, washed with PBS, resuspended in RPMI 1640 plus 10% FBS, and added to the wells at a multiplicity of infection of 1:1 (bacteria:epithelial cells). Plates were centrifuged at 200 × g for 5 min to place GAS cells on the monolayer surface and incubated at 37°C under 5% CO2. For adherence assays, the plates were incubated for 30 min, after which the wells were washed six times with PBS to remove nonadherent bacteria. Monolayers were disrupted with 100 μl of trypsin-EDTA and 400 μl of 0.025% Triton X-100, and dilutions were plated to enumerate total cell-associated CFU. For cellular invasion assays, after 2 h incubation under the above protocol, monolayers were washed three times with phosphate-buffered saline (PBS), and RPMI 1640 plus 10% fetal bovine serum, 100 μg/ml gentamicin, and 5 μg/ml penicillin was added to kill surface-associated, noninternalized bacteria. After subsequent incubation for 2 h, monolayers were washed three times with PBS and lysed as described above for enumeration of intracellular CFU. Assays were performed in quadruplicate and repeated four times.
Whole-blood and antimicrobial peptide killing assays.
Human venous blood was collected from healthy individuals and heparinized. GAS (200 CFU in 100 μl PBS) was added to 300 μl of blood and incubated at 37°C with rotation. At various intervals, 50-μl aliquots were removed and plated for quantification. The murine cathelicidin mCRAMP was synthesized and purified (>99%) by the Louisiana State University Protein Facility. In sterile 96-well microtiter plates, logarithmic-phase GAS cells were adjusted to 105 CFU/ml in 100 μl THB containing 28 μM mCRAMP and incubated at 37°C for 1 h prior to plating for enumeration of surviving CFU.
In vivo mouse infection model.
A murine model of GAS systemic infection was used to compare the virulence potential of WT and mutant strains (57). GAS cells were grown to mid-log phase in THB, washed, resuspended, and diluted to an appropriate concentration in PBS. An equal volume of gastric mucin was added for a final concentration of 5%. A total of 2 × 106 CFU were injected intraperitoneally in 10- to 12-week-old female CD1 mice. Blood samples were taken at 6 h from tail veins for quantification of bacterial CFU, and mice were monitored for mortality over a 13-day period.
Statistical analysis.
Data were analyzed in Excel using Student's t test; a P value of <0.05 was considered significant. The Kaplan-Meyer survival plot was analyzed using the log rank test in GraphPad Prism (GraphPad Software, San Diego, CA).
RESULTS
Real-time RT-PCR analysis of specific transcripts in WT versus ΔRALP3 mutant GAS identifies RALP3 as a global transcriptional regulator.
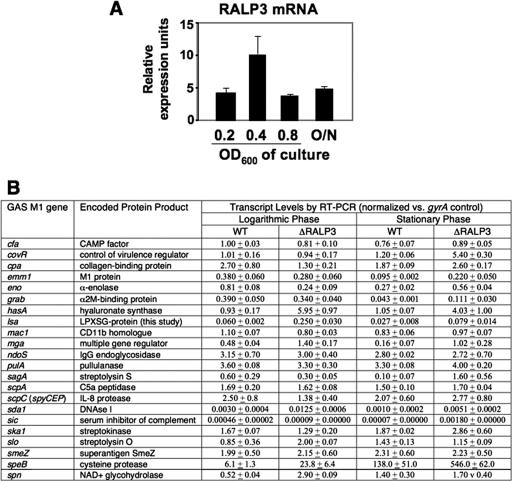
Isogenic ΔRALP3 and ΔLSA mutants were generated in WT M1T1 strain 5448 by precise, in-frame allelic replacement (Fig. 1). Each mutant exhibited growth characteristics identical to those of the parent GAS strain in THB medium (data not shown). Due to its homology to the RofA-like family of stand-alone response regulators, we predicted that RALP3 could be involved in transcriptional regulation of GAS genes relevant to disease pathogenesis. Quantitative real-time RT-PCR analysis of 23 proven or hypothesized virulence factors of M1 was performed to compare the transcriptional profiles of the WT GAS M1 strain and the isogenic ΔRALP3 mutant at logarithmic and stationary phases of growth. We collected RNA samples from logarithmic-phase (OD600 = 0.4) and stationary-phase (18-h growth) cultures of each strain and performed real-time RT-PCR analysis of selected virulence factor genes using specific primer sets with transcript amounts normalized to an internal gyrA control. Transcription of mRNA for the ralp3 gene was evident in all phases of growth (Fig. 2A), with the greatest level at the mid-logarithmic phase.
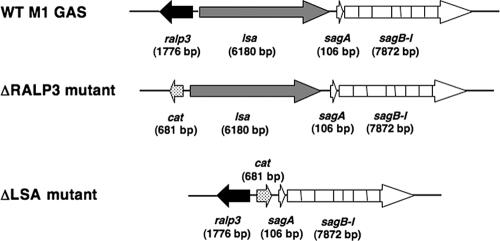
FIG. 1.
The ralp3/lsa locus. The ralp3/lsa locus is located upstream of the streptolysin S biosynthetic (sag) operon in the M1T1 GAS genome. Mutagenesis of ralp3 and that of lsa were each achieved through precise, in-frame allelic replacement with the chloramphenicol acetyltransferase (cat) gene.
FIG. 2.
Real-time RT-PCR analysis of selected GAS virulence genes. A. Real-time RT-PCR (TaqMan) analysis of expression of the ralp3 gene at various phases of growth. B. RNA was prepared from WT M1 GAS and isogenic ΔRALP3 mutant cultures at mid-logarithmic (OD600 = 0.4) and stationary (18-h culture) phases and analyzed with quantitative real-time RT-PCR. Transcript levels after normalization to the gyrA housekeeping gene control are shown.
The results of the RT-PCR studies are reported in Fig. 2B as a primary data set of expression levels of each transcript normalized to the gyrA housekeeping gene control and in Fig. 3 as the ratios of specific transcript expression levels in the ΔRALP3 mutant and WT parent strains. Using twofold change as a cutoff, RALP3 was identified as having either negative or positive regulatory effects on several genes. Five genes were upregulated in the ΔRALP3 mutant strain during both mid-log and stationary phases, namely, mga, sdaI, speB, hasA, and lsa. Expression of the multigene regulator Mga, itself involved in transcriptional control of multiple GAS genes important in colonization and immune evasion, was increased 2.9-fold at logarithmic phase and 6.4-fold at stationary phase (43). The mRNA levels for phage-encoded DNase Sda1, a proven virulence factor promoting M1 GAS escape from neutrophil extracellular traps (7, 55), were also increased (4.2- to 5.1-fold) in the ΔRALP3 mutant. The transcript levels for the hyaluronan synthase gene (hasA) were increased in the ΔRALP3 mutant in both logarithmic (6.4-fold) and stationary (3.8-fold) phases. GAS hyaluronic acid capsule expression is known to play important roles in virulence by promoting phagocyte resistance and CD44-mediated epithelial cell invasion (15, 58, 59). The cysteine protease gene speB showed a 3.9-fold increase at both phases, which is notable since significant SpeB expression by the WT M1T1 parent strain normally occurs only in late logarithmic and stationary phases (35). Finally, the lsa gene, adjacent to and divergently transcribed directly upstream of ralp3, appeared to be under negative control of the candidate regulator, with 4.2-fold-greater logarithmic-phase and 3.0-fold-greater stationary-phase transcript levels being found in the ΔRALP3 mutant.
FIG. 3.
Comparison of transcript levels in ΔRALP3 mutant versus WT M1 GAS (graphic depiction of RT-PCR data derived from Fig. 2B). Bars extending to the left indicate transcript levels higher in the ΔRALP3 mutant; bars extending to the right indicate transcript levels more highly expressed in the WT strain. Dotted lines demarcate a cutoff ratio of 2.0.
Three additional genes, slo, spn, and cpa, were upregulated in the ΔRALP3 mutant strain compared to the WT only during logarithmic-phase growth. The gene for streptolysin O (slo), which plays a role in GAS avoidance of lysosomal killing and animal virulence (29, 40), was upregulated 2.4-fold, and that for SPN, a NAD-glycohydrolase exerting toxic effects upon induction into host cells (41), was upregulated 5.5-fold. The gene encoding collagen type I-binding surface protein, cpa, was also negatively regulated twofold in the ΔRALP3 mutant during logarithmic growth.
Four more genes were found to be upregulated only in the ΔRALP3 mutant in the 18-h culture analysis, potentially reflecting transcript accumulated in late logarithmic and stationary phases. Strikingly, the structural gene for the SLS peptide, sagA, adjacent on the chromosome to the RALP3/LSA locus, was upregulated 15.6-fold in the ΔRALP3 mutant. Stationary-phase levels of transcript for the CovR response regulator, involved in global control of several GAS virulence genes (22), were increased 4.5-fold in the ΔRALP3 mutant. Finally, levels of transcript for GRAB and M protein (emm1) were upregulated in the ΔRALP3 mutant 2.5- and 2.3-fold, respectively.
Two genes in our analysis appeared to be differentially regulated by RALP3 in logarithmic versus stationary phase. Of these, sic (streptococcal inhibitor of complement) was the most significantly changed, with a 25-fold increase compared to the WT in stationary phase but a 5-fold decrease in logarithmic phase. These data should be interpreted with caution due to the very low levels of transcript detected for sic; however, similar results were found repeatedly and thus can be considered at least qualitatively. An alpha-enolase gene situated immediately downstream of RALP3 in the GAS chromosome was down-regulated during logarithmic growth 3.4-fold but upregulated 2.0-fold in stationary phase.
Eight other genes that play important roles in GAS pathogenesis did not show marked (>2-fold) differences in expression levels detected by RT-PCR between the ΔRALP3 mutant and the WT parent strain in either logarithmic or stationary growth phase (Fig. 3). These included the genes encoding the strongly immunoactive superantigen SmeZ, the pore-forming cytotoxin CAMP factor, streptokinase, pullulanase, EndoS, Mac, C5a peptidase, and interleukin 8 protease (ScpC).
Hyaluronic acid capsule and cysteine protease expression are increased in the ΔRALP3 mutant.
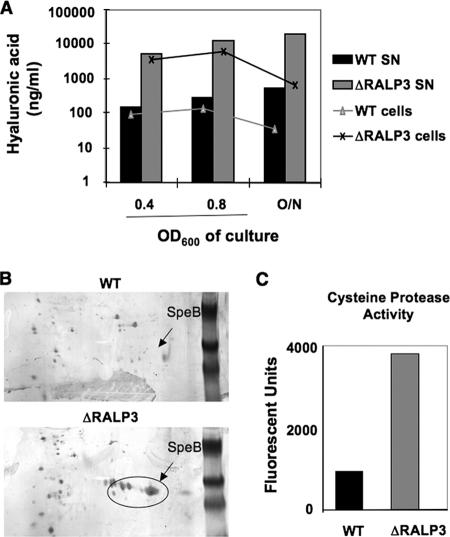
Real-time PCR analysis showed that transcription of the genes hasA, encoding hyaluronan synthase, and speB, encoding streptococcal cysteine protease, were increased in the ΔRALP3 mutant compared to the WT parent strain in both logarithmic and stationary growth phases. To corroborate these changes at the phenotypic level, we characterized the levels of hyaluronic acid in the supernatant and on the surface of wild-type and Δralp3 mutant bacteria by ELISA (Fig. 4A). The levels of hyaluronic acid released into the supernatant were 36, 47, and 36 times higher in the ΔRALP3 mutant bacteria at mid-log phase, late log phase, and 18 h of growth, respectively. Levels of hyaluronic acid maintained on the cell surface were nearly equal to that released at mid-log- and late-log-phase growth but reduced 50% by 18 h of growth. The ratios of cell surface versus released hyaluronic acid remained the same, indicating that increased hyaluronic acid in the ΔRALP3 supernatant likely mirrored increased biosynthesis. To compare the expression of cysteine proteinase SpeB, two-dimensional gel electrophoresis was performed on the supernatants of WT and ΔRALP3 mutant cultures grown in the presence of E64, an inhibitor of cysteine protease activity. At both early log phase (Fig. 4B) and stationary phase (data not shown), the Δralp3 mutant expressed much more SpeB protein than WT bacteria. Activity of SpeB in the culture supernatant was next assayed using a casein substrate, showing that the ΔRALP3 mutant had a fourfold-greater level of protease activity than WT GAS (Fig. 4C).
FIG. 4.
RALP3 negatively regulates the GAS hyaluronic acid capsule and cysteine protease. A. Hyaluronic acid levels found in the supernatant and on the surface of bacteria measured by ELISA. B. Two-dimensional gel electrophoresis of proteins in the supernatant of WT M1 GAS and the ΔRALP3 mutant. C. Cysteine protease activity in supernatants from WT M1 GAS and the ΔRALP3 mutant as measured using a fluorescent casein substrate assay.
Adherence and invasion of human epithelial cells are modulated by LSA and RALP3.
GAS adherence to and invasion of host epithelial cells are considered an essential early event in invasive infection (14). Because of its cell wall anchor domain and large size, suggesting that it could reach far outward from the bacterial surface, we hypothesized that the LSA protein could influence GAS interactions with host epithelial cells. The portion of the LSA ORF corresponding to the N-terminal domain possesses multiple repeat sequences with sequence homology (22 to 26% identity) to that of emb, a 14-kb gene encoding the major extracellular matrix-binding protein of Abiotrophia defectiva (42), and 13% identity to ebh, the gene encoding a 1.1-MDa fibronectin-binding protein of Staphylococcus aureus also hypothesized to play a role in cellular adhesion (10).
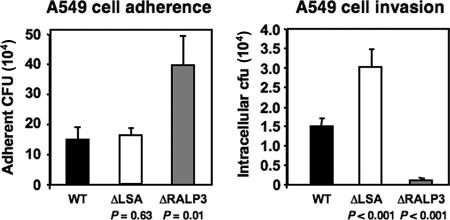
The relative abilities of the isogenic GAS ΔLSA mutant and the WT M1 GAS parent strain to adhere to and invade human A549 lung epithelial cells were compared in tissue culture assays. As shown in Fig. 5, the ΔLSA mutant adhered to the epithelial cells similarly to the WT strain but was recovered intracellularly in twice the numbers (P < 0.001). No differences in susceptibility of the WT and ΔLSA mutant to the antibiotics (penicillin or gentamicin) or detergent (Triton X-100) used in the invasion assay were identified (data not shown). The results indicate that LSA does not play a role in promoting GAS lung epithelial cell adherence but that expression of LSA decreased recovery of GAS from infected host cells.
FIG. 5.
LSA and RALP3 modulate GAS-epithelial cell interactions. The rates of (A) adherence and (B) intracellular uptake of WT M1 GAS and the isogenic ΔLSA and ΔRALP3 mutants to cultured monolayers of A549 human lung epithelial cells are shown.
Because RALP3 was found to negatively regulate GAS lsa expression (Fig. 2), we hypothesized that the effects of eliminating RALP3 on GAS epithelial cell adherence and invasion might run opposite to those observed in the ΔLSA mutant. Conversely, the GAS ΔRALP3 mutant demonstrated a 3-fold increase in adherence to A549 cells (P = 0.01) yet was 12-fold less invasive in the parallel in vitro invasion assay (P < 0.001) (Fig. 5).
Mutation of RALP3 promotes GAS survival in human whole blood and serum.
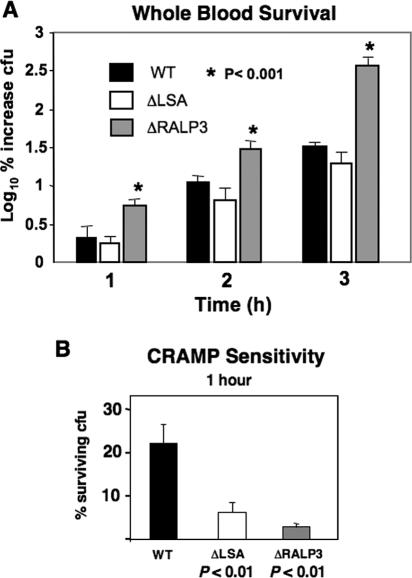
The GAS phenotype most strongly up-regulated upon elimination of RALP3 was hyaluronic acid capsule expression, which has a known role in resistance to phagocytic clearance (59). Compared to the WT strain in a fresh-human-blood killing assay, the ΔRALP3 mutant exhibited a ∼100-fold increase in survival after 3.0 h of incubation (Fig. 6A). Resistance to opsonization per se is not recognized as an attribute of the GAS hyaluronic acid capsule (16), and therefore the increased resistance to phagocytosis could also reflect in part increased expression of complement-impairing factors such as SIC (1, 5) in the ΔRALP3 mutant. The isogenic ΔLSA mutant did not differ significantly from the WT parent strain in the blood survival assays (Fig. 6A). Cathelicidin antimicrobial peptides are an important component of the innate immune killing capacity of phagocytic cells, and mice deficient in the sole murine cathelicidin mCRAMP are highly susceptible to invasive GAS infection (50). We found that both the ΔLSA and ΔRALP3 mutants demonstrate significantly increased sensitivity to mCRAMP killing (Fig. 6B).
FIG. 6.
Effects of LSA and RALP mutation on resistance to whole blood and antimicrobial peptide killing. (A) Survival of WT M1 GAS and isogenic ΔLSA and ΔRALP3 mutants in fresh human whole blood; (B) killing of WT M1 GAS and isogenic ΔLSA and ΔRALP3 mutants by the murine cathelicidin antimicrobial peptide mCRAMP (28 μM).
RALP3 and LSA contribute to GAS virulence in a mouse infection model.
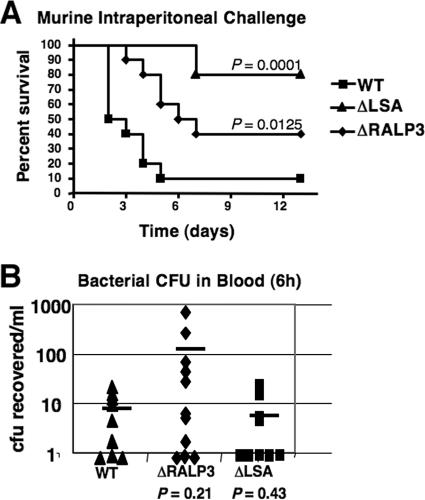
A murine intraperitoneal challenge model (57) was used to test the virulence potential of the ΔRALP3 and ΔLSA mutants compared to the WT M1T1 GAS strain. Mice were injected with 2 × 106 CFU of logarithmic-phase GAS and monitored for mortality over a 13-day period. Mortality caused by the WT strain was 50% by day 2 and 90% by day 5 (Fig. 7A). In contrast, the first death among the mice infected with the ΔRALP3 mutant occurred on day 3, and only 60% mortality was recorded by the end of the observation period (P < 0.0125). Attenuation of the ΔLSA mutant was more pronounced, with no deaths until 7 days after challenge and only 20% total mortality during the monitoring period (P < 0.0001). Thus, both RALP3 and LSA are required for the full virulence potential of M1T1 GAS in this in vivo model. Although not reaching statistical significance, a tendency toward higher numbers of CFU of the ΔRALP3 mutant than the WT strain in blood samples 6 h after intraperitoneal challenge was observed (Fig. 7B). While this effect of RALP3 deletion could be seen to correspond to the observed increases in blood survival of the ΔRALP3 mutant (Fig. 6A) and its upregulation of hyaluronic acid capsule expression (Fig. 4), it did not translate into increased mouse lethality during the ultimate infection process.
FIG. 7.
RALP3 and LSA contribute to animal virulence. WT M1 GAS and the isogenic ΔLSA and ΔRALP3 mutants were injected intraperitoneally into CD1 mice (10 per group). A. Kaplan-Meyer survival curve. B. Bacterial CFU in the blood 6 h after inoculation.
DISCUSSION
Immediately upstream of the sagA-to-sagI operon for SLS biosynthesis in the M1T1 GAS genome lies the a gene locus containing the RofA-like protein gene ralp3 and lsa, the largest ORF in the GAS genome, encoding a protein with a characteristic C-terminal LPXSG cell wall anchor motif. In this study, we use targeted mutagenesis to demonstrate that RALP3 is involved in the global transcriptional regulation of multiple GAS virulence genes, including prominently the negative control of hyaluronic acid capsule and cysteine protease expression. Further, we show that LSA plays a role in impeding intracellular uptake of the bacterium into host cells. RALP3 and LSA each contribute to the virulence potential of M1T1 GAS in an animal infection model.
Several mechanisms of transcriptional regulation have been characterized in GAS, including two-component signal transduction systems (22, 31, 37) and stand-alone response activators and repressors (9, 25, 26, 47, 51). RALP3 belongs to a major family of stand-alone response regulators with homology to RofA, originally discovered as a positive regulator of prtF, which encodes a fibronectin-binding adhesin in an M6 serotype strain of GAS (25). In an M49 serotype GAS strain, the negative regulator Nra was found to share 62% sequence identity with RofA and seen to suppress expression of prtF2 and a collagen-binding protein gene, cpa (53). In related bacterial species, at least two additional RALP family members have been discovered and preliminarily characterized. Group B Streptococcus expresses RogB, which is involved regulating the expression of surface pili (20, 28). The Streptococcus pneumoniae transcriptional regulator RlrA is required for colonization of the nasopharynx but is dispensable during systemic infection processes (30). Overall, RALPs are similar in size and in having an average of 30% of their predicted amino acid sequences shared with RofA. However, the predicted helix-turn-helix DNA-binding region of RofA is not well conserved among the group of RALP proteins, suggesting that the DNA targets of each stand-alone regulator may vary significantly (26, 28).
Among the 12 published complete GAS genomes, 7, representing four serotypes (M1, M4, M12, and M28) in the published literature and 1 (M49) available in the public databases, contain ralp3 homologues. In every case, the lsa gene is colocalized with ralp3 and the ralp3/lsa locus is situated upstream of the sag operon for SLS biosynthesis. However, each sequenced strain of the different serotypes is characterized by an lsa gene of a different length (e.g., 6,180 bp in M1, 5,040 bp in M12, etc.).
Because of the large size of LSA, we speculate that the increased recovery of ΔLSA mutant bacteria versus WT bacteria from infected host epithelial cells could reflect LSA steric interference or cloaking of key domains in shorter surface-anchored proteins (e.g., Sfb1, M protein) capable of mediating cellular uptake. Correspondingly, the significantly decreased intracellular uptake seen in the ΔRALP3 mutant could reflect increased LSA levels on the cell surface.
Two of the most prominent phenotypes under transcriptional control by RALP3 appear to be hyaluronic acid capsule and cysteine protease expression, each of which is markedly increased in the ΔRALP3 mutant. When invasive GAS strains are passaged in vivo, hyaluronic acid capsule expression is increased but cysteine protease expression is decreased (2, 27, 58). These phase shifts are consistent with clinical data correlating mucoid strains of GAS with increased pathogenic potential and the inverse correlation of cysteine protease expression with severity of invasive GAS infection (34, 36). The in vivo advantage associated with increased capsule expression likely reflects increased resistance to phagocytosis (59), while downregulation of cysteine protease expression preserves the expression of several known GAS virulence factors such as M protein, DNase, and streptokinase (2, 13, 54). Recently, the in vivo phenotypic switch to upregulation of capsule gene expression and downregulation of cysteine protease expression has been attributed to selection for specific mutations in the sequence of either component gene of the global transcriptional regulator CovR/S (17, 56).
In contrast to CovR, which negatively regulates hasA expression but positively regulates speB expression, we found that RALP3 negatively regulates both genes. As RALP3 downregulated covR transcription (Fig. 2B and 3), the effect of RALP3 of downregulating speB expression could in part be attributable to reduced covR transcription; in contrast, RALP3 downregulation of hasA expression could be mitigated by the same pathway. The in vivo challenge with the ΔRALP3 mutant dissociates the observed in vivo-selected phenotypes of enhanced capsule expression and decreased cysteine protease expression. The finding of decreased virulence of the ΔRALP3 strain compared to the WT suggests that overexpression of the SpeB cysteine protease and its attendant effects of degrading GAS virulence factors can effectively negate the expected survival advantage conferred by increased hyaluronic acid capsule expression.
In sum, we have performed the first characterization of a two-gene locus situated upstream of the sag operon in invasive M1T1 GAS and found that both genes contribute to the virulence potential of the pathogen. The RofA family stand-alone regulator, RALP3, functions in global transcriptional control of several phenotypes relevant to disease progression and can be added to the complex network of virulence gene regulators in this leading human pathogen (38). The mechanism by which LSA enhances GAS virulence in vivo remains to be elucidated, although a role in antimicrobial peptide resistance may be a contributing factor. Because of its large size and predicted surface localization, LSA may represent an attractive target to elicit protective immunity against M1T1 GAS and other strains in which it is expressed.
Footnotes
Published ahead of print on 17 November 2006.
REFERENCES
- 1.Akesson, P., A. G. Sjoholm, and L. Bjorck. 1996. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J. Biol. Chem. 271:1081-1088. [DOI] [PubMed] [Google Scholar]
- 2.Aziz, R. K., M. J. Pabst, A. Jeng, R. Kansal, D. E. Low, V. Nizet, and M. Kotb. 2004. Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol. Microbiol. 51:123-134. [DOI] [PubMed] [Google Scholar]
- 3.Beckert, S., B. Kreikemeyer, and A. Podbielski. 2001. Group A streptococcal rofA gene is involved in the control of several virulence genes and eukaryotic cell attachment and internalization. Infect. Immun. 69:534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betschel, S. D., S. M. Borgia, N. L. Barg, D. E. Low, and J. C. De Azavedo. 1998. Reduced virulence of group A streptococcal Tn916 mutants that do not produce streptolysin S. Infect. Immun. 66:1671-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binks, M. J., B. A. Fernie-King, D. J. Seilly, P. J. Lachmann, and K. S. Sriprakash. 2005. Attribution of the various inhibitory actions of the streptococcal inhibitor of complement (SIC) to regions within the molecule. J. Biol. Chem. 280:20120-20125. [DOI] [PubMed] [Google Scholar]
- 6.Bisno, A. L., M. O. Brito, and C. M. Collins. 2003. Molecular basis of group A streptococcal virulence. Lancet Infect. Dis. 3:191-200. [DOI] [PubMed] [Google Scholar]
- 7.Buchanan, J. T., A. J. Simpson, R. K. Aziz, G. Y. Liu, S. A. Kristian, M. Kotb, J. Feramisco, and V. Nizet. 2006. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol. 16:396-400. [DOI] [PubMed] [Google Scholar]
- 8.Chatellier, S., N. Ihendyane, R. G. Kansal, F. Khambaty, H. Basma, A. Norrby-Teglund, D. E. Low, A. McGeer, and M. Kotb. 2000. Genetic relatedness and superantigen expression in group A streptococcus serotype M1 isolates from patients with severe and nonsevere invasive diseases. Infect. Immun. 68:3523-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaussee, M. S., G. L. Sylva, D. E. Sturdevant, L. M. Smoot, M. R. Graham, R. O. Watson, and J. M. Musser. 2002. Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. Infect. Immun. 70:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke, S. R., L. G. Harris, R. G. Richards, and S. J. Foster. 2002. Analysis of Ebh, a 1.1-megadalton cell wall-associated fibronectin-binding protein of Staphylococcus aureus. Infect. Immun. 70:6680-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleary, P. P., D. LaPenta, R. Vessela, H. Lam, and D. Cue. 1998. A globally disseminated M1 subclone of group A streptococci differs from other subclones by 70 kilobases of prophage DNA and capacity for high-frequency intracellular invasion. Infect. Immun. 66:5592-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cockerill, F. R., III, K. L. MacDonald, R. L. Thompson, F. Roberson, P. C. Kohner, J. Besser-Wiek, J. M. Manahan, J. M. Musser, P. M. Schlievert, J. Talbot, B. Frankfort, J. M. Steckelberg, W. R. Wilson, and M. T. Osterholm. 1997. An outbreak of invasive group A streptococcal disease associated with high carriage rates of the invasive clone among school-aged children. JAMA 277:38-43. [PubMed] [Google Scholar]
- 13.Cole, J. N., J. D. McArthur, F. C. McKay, M. L. Sanderson-Smith, A. J. Cork, M. Ranson, M. Rohde, A. Itzek, H. Sun, D. Ginsburg, M. Kotb, V. Nizet, G. S. Chhatwal, and M. J. Walker. 2006. Trigger for group A streptococcal M1T1 invasive disease. FASEB J. 20:1745-1747. [DOI] [PubMed] [Google Scholar]
- 14.Courtney, H. S., D. L. Hasty, and J. B. Dale. 2002. Molecular mechanisms of adhesion, colonization, and invasion of group A streptococci. Ann. Med. 34:77-87. [DOI] [PubMed] [Google Scholar]
- 15.Cywes, C., and M. R. Wessels. 2001. Group A Streptococcus tissue invasion by CD44-mediated cell signalling. Nature 414:648-652. [DOI] [PubMed] [Google Scholar]
- 16.Dale, J. B., R. G. Washburn, M. B. Marques, and M. R. Wessels. 1996. Hyaluronate capsule and surface M protein in resistance to opsonization of group A streptococci. Infect. Immun. 64:1495-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalton, T. L., R. I. Hobb, and J. R. Scott. 2006. Analysis of the role of CovR and CovS in the dissemination of Streptococcus pyogenes in invasive skin disease. Microb. Pathog. 40:221-227. [DOI] [PubMed] [Google Scholar]
- 18.Datta, V., S. M. Myskowski, L. A. Kwinn, D. N. Chiem, N. Varki, R. G. Kansal, M. Kotb, and V. Nizet. 2005. Mutational analysis of the group A streptococcal operon encoding streptolysin S and its virulence role in invasive infection. Mol. Microbiol. 56:681-695. [DOI] [PubMed] [Google Scholar]
- 19.Doran, K. S., J. C. Chang, V. M. Benoit, L. Eckmann, and V. Nizet. 2002. Group B streptococcal beta-hemolysin/cytolysin promotes invasion of human lung epithelial cells and the release of interleukin-8. J. Infect. Dis. 185:196-203. [DOI] [PubMed] [Google Scholar]
- 20.Dramsi, S., E. Caliot, I. Bonne, S. Guadagnini, M. C. Prevost, M. Kojadinovic, L. Lalioui, C. Poyart, and P. Trieu-Cuot. 2006. Assembly and role of pili in group B streptococci. Mol. Microbiol. 60:1401-1413. [DOI] [PubMed] [Google Scholar]
- 21.Engleberg, N. C., A. Heath, K. Vardaman, and V. J. DiRita. 2004. Contribution of CsrR-regulated virulence factors to the progress and outcome of murine skin infections by Streptococcus pyogenes. Infect. Immun. 72:623-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Federle, M. J., K. S. McIver, and J. R. Scott. 1999. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J. Bacteriol. 181:3649-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fogg, G. C., and M. G. Caparon. 1997. Constitutive expression of fibronectin binding in Streptococcus pyogenes as a result of anaerobic activation of rofA. J. Bacteriol. 179:6172-6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fogg, G. C., C. M. Gibson, and M. G. Caparon. 1994. The identification of rofA, a positive-acting regulatory component of prtF expression: use of an m gamma delta-based shuttle mutagenesis strategy in Streptococcus pyogenes. Mol. Microbiol. 11:671-684. [DOI] [PubMed] [Google Scholar]
- 26.Granok, A. B., D. Parsonage, R. P. Ross, and M. G. Caparon. 2000. The RofA binding site in Streptococcus pyogenes is utilized in multiple transcriptional pathways. J. Bacteriol. 182:1529-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gryllos, I., C. Cywes, M. H. Shearer, M. Cary, R. C. Kennedy, and M. R. Wessels. 2001. Regulation of capsule gene expression by group A Streptococcus during pharyngeal colonization and invasive infection. Mol. Microbiol. 42:61-74. [DOI] [PubMed] [Google Scholar]
- 28.Gutekunst, H., B. J. Eikmanns, and D. J. Reinscheid. 2003. Analysis of RogB-controlled virulence mechanisms and gene repression in Streptococcus agalactiae. Infect. Immun. 71:5056-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Håkansson, A., C. C. Bentley, E. A. Shakhnovic, and M. R. Wessels. 2005. Cytolysin-dependent evasion of lysosomal killing. Proc. Natl. Acad. Sci. USA 102:5192-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hava, D. L., C. J. Hemsley, and A. Camilli. 2003. Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA. J. Bacteriol. 185:413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heath, A., V. J. DiRita, N. L. Barg, and N. C. Engleberg. 1999. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect. Immun. 67:5298-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Humar, D., V. Datta, D. J. Bast, B. Beall, J. C. De Azavedo, and V. Nizet. 2002. Streptolysin S and necrotising infections produced by group G streptococcus. Lancet 359:124-129. [DOI] [PubMed] [Google Scholar]
- 33.Jeng, A., V. Sakota, Z. Li, V. Datta, B. Beall, and V. Nizet. 2003. Molecular genetic analysis of a group A Streptococcus operon encoding serum opacity factor and a novel fibronectin-binding protein, SfbX. J. Bacteriol. 185:1208-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kansal, R. G., A. McGeer, D. E. Low, A. Norrby-Teglund, and M. Kotb. 2000. Inverse relation between disease severity and expression of the streptococcal cysteine protease, SpeB, among clonal M1T1 isolates recovered from invasive group A streptococcal infection cases. Infect. Immun. 68:6362-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kansal, R. G., V. Nizet, A. Jeng, W. J. Chuang, and M. Kotb. 2003. Selective modulation of superantigen-induced responses by streptococcal cysteine protease. J. Infect. Dis. 187:398-407. [DOI] [PubMed] [Google Scholar]
- 36.Kass, E. H., and C. V. Seastone. 1944. The role of the mucoid polysaccharide (hyaluronic acid) in the virulence of group A hemolytic streptococci. J. Exp. Med. 79:319-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kreikemeyer, B., M. D. Boyle, B. A. Buttaro, M. Heinemann, and A. Podbielski. 2001. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol. Microbiol. 39:392-406. [DOI] [PubMed] [Google Scholar]
- 38.Kreikemeyer, B., K. S. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224-232. [DOI] [PubMed] [Google Scholar]
- 39.Kristian, S. A., V. Datta, C. Weidenmaier, R. Kansal, I. Fedtke, A. Peschel, R. L. Gallo, and V. Nizet. 2005. d-Alanylation of teichoic acids promotes group a streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J. Bacteriol. 187:6719-6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Limbago, B., V. Penumalli, B. Weinrick, and J. R. Scott. 2000. Role of streptolysin O in a mouse model of invasive group A streptococcal disease. Infect. Immun. 68:6384-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madden, J. C., N. Ruiz, and M. Caparon. 2001. Cytolysin-mediated translocation (CMT): a functional equivalent of type III secretion in gram-positive bacteria. Cell 104:143-152. [DOI] [PubMed] [Google Scholar]
- 42.Manganelli, R., and I. van de Rijn. 1999. Characterization of emb, a gene encoding the major adhesin of Streptococcus defectivus. Infect. Immun. 67:50-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McIver, K. S., and R. L. Myles. 2002. Two DNA-binding domains of Mga are required for virulence gene activation in the group A streptococcus. Mol. Microbiol. 43:1591-1601. [DOI] [PubMed] [Google Scholar]
- 44.Mitchell, T. J. 2003. The pathogenesis of streptococcal infections: from tooth decay to meningitis. Nat. Rev. Microbiol. 1:219-230. [DOI] [PubMed] [Google Scholar]
- 45.Miyoshi-Akiyama, T., D. Takamatsu, M. Koyanagi, J. Zhao, K. Imanishi, and T. Uchiyama. 2005. Cytocidal effect of Streptococcus pyogenes on mouse neutrophils in vivo and the critical role of streptolysin S. J. Infect. Dis. 192:107-116. [DOI] [PubMed] [Google Scholar]
- 46.Murono, K., K. Fujita, M. Saijo, Y. Hirano, J. Zhang, and T. Murai. 1999. Emergence and spread of a new clone of M type 1 group A Streptococcus coincident with the increase in invasive diseases in Japan. Pediatr. Infect. Dis. J. 18:254-257. [DOI] [PubMed] [Google Scholar]
- 47.Nakata, M., A. Podbielski, and B. Kreikemeyer. 2005. MsmR, a specific positive regulator of the Streptococcus pyogenes FCT pathogenicity region and cytolysin-mediated translocation system genes. Mol. Microbiol. 57:786-803. [DOI] [PubMed] [Google Scholar]
- 48.Nizet, V. 2002. Streptococcal beta-hemolysins: genetics and role in disease pathogenesis. Trends Microbiol. 10:575-580. [DOI] [PubMed] [Google Scholar]
- 49.Nizet, V., B. Beall, D. J. Bast, V. Datta, L. Kilburn, D. E. Low, and J. C. De Azavedo. 2000. Genetic locus for streptolysin S production by group A streptococcus. Infect. Immun. 68:4245-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nizet, V., T. Ohtake, X. Lauth, J. Trowbridge, J. Rudisill, R. A. Dorschner, V. Pestonjamasp, J. Piraino, K. Huttner, and R. L. Gallo. 2001. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414:454-457. [DOI] [PubMed] [Google Scholar]
- 51.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peyssonnaux, C., V. Datta, T. Cramer, A. Doedens, E. A. Theodorakis, R. L. Gallo, N. Hurtado-Ziola, V. Nizet, and R. S. Johnson. 2005. HIF-1α expression regulates the bactericidal capacity of phagocytes. J. Clin. Invest. 115:1806-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Podbielski, A., M. Woischnik, B. A. Leonard, and K. H. Schmidt. 1999. Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31:1051-1064. [DOI] [PubMed] [Google Scholar]
- 54.Raeder, R., M. Woischnik, A. Podbielski, and M. D. Boyle. 1998. A secreted streptococcal cysteine protease can cleave a surface-expressed M1 protein and alter the immunoglobulin binding properties. Res. Microbiol. 149:539-548. [DOI] [PubMed] [Google Scholar]
- 55.Sumby, P., K. D. Barbian, D. J. Gardner, A. R. Whitney, D. M. Welty, R. D. Long, J. R. Bailey, M. J. Parnell, N. P. Hoe, G. G. Adams, F. R. Deleo, and J. M. Musser. 2005. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc. Natl. Acad. Sci. USA 102:1679-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sumby, P., A. R. Whitney, E. A. Graviss, F. R. DeLeo, and J. M. Musser. 2006. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Timmer, A. M., S. A. Kristian, V. Datta, A. Jeng, C. M. Gillen, M. J. Walker, B. Beall, and V. Nizet. 2006. Serum opacity factor promotes group A streptococcal epithelial cell invasion and virulence. Mol. Microbiol. 62:15-25. [DOI] [PubMed] [Google Scholar]
- 58.Wessels, M. R., and M. S. Bronze. 1994. Critical role of the group A streptococcal capsule in pharyngeal colonization and infection in mice. Proc. Natl. Acad. Sci. USA 91:12238-12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wessels, M. R., A. E. Moses, J. B. Goldberg, and T. J. DiCesare. 1991. Hyaluronic acid capsule is a virulence factor for mucoid group A streptococci. Proc. Natl. Acad. Sci. USA 88:8317-8321. [DOI] [PMC free article] [PubMed] [Google Scholar]