Abstract
The level of expression of conjugation genes in Enterococcus faecalis strains carrying the pheromone-responsive transferable plasmid pCF10 is determined by the ratio in the culture medium of two types of signaling peptides, a pheromone (cCF10) and an inhibitor (iCF10). Recent data have demonstrated that both peptides target the cytoplasmic receptor protein PrgX. However, the relative importance of the interaction of these peptides with the pCF10 protein PrgZ (which enhances import of cCF10) versus PrgX is not fully understood, and there is relatively little information about specific amino acid sequence determinants affecting the functional interactions of cCF10 with these proteins in vivo. To address these issues, we used a pheromone-inducible reporter gene system where various combinations of PrgX and PrgZ could be expressed in an isogenic host background to examine the biological activities of cCF10, iCF10, and variants of cCF10 isolated in a genetic screen. The results suggest that most of the amino acid sequence determinants of cCF10 pheromone activity affect interactions between the peptide and PrgX, although some sequence variants that affected peptide/PrgZ interactions were also identified. The results provide functional data to complement ongoing structural studies of PrgX and increase our understanding of the functional interactions of cCF10 and iCF10 with the pheromone-sensing machinery of pCF10.
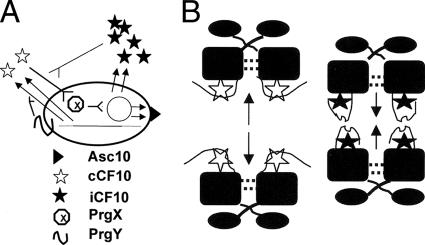
Enterococcus faecalis strains frequently harbor transmissible plasmids whose conjugation functions are regulated by extracellular signaling peptides (15, 17). In contrast to peptide signaling systems where a two-component signal transduction phosphorelay activates a cellular response upon peptide binding to the cell surface, the enterococcal pheromones must be imported into responder cells by the concerted action of a plasmid-encoded pheromone binding protein and the chromosomal oligopeptide permease system Opp (33). Initial studies of these plasmids suggested a unidirectional communication circuit where a mating pheromone produced by plasmid-free recipient cells was specifically sensed by the plasmid-containing donor cells (19, 21). This model is insufficient because pheromone production ability is encoded by the chromosome (13), implying that a plasmid-containing cell must avoid self-induction by its own endogenous pheromone while retaining the ability to sense the same pheromone from an exogenous source. Subsequent studies of several different pheromone plasmids suggested that, in addition to sensing exogenous pheromone, the inducibility of these systems also depends on two plasmid-encoded gene products, a membrane protein that partially shuts down or inactivates pheromone produced by donor cells (1, 8, 37) and an inhibitor peptide which reduces the activity of pheromone in solution (14, 16, 36, 38). The inhibitor appears to function in donor cells by neutralizing the endogenously produced pheromone that escapes the pheromone reduction activity of the membrane protein. Both gene products are required to prevent the donor cell from a wasteful constitutive self-induction of conjugation in the absence of recipients. In the case of plasmid pCF10, conjugation is positively regulated by the pheromone peptide cCF10 (35) and is negatively regulated by the inhibitor peptide iCF10 (38). The level of endogenous pheromone production by pCF10-containing donor cells is reduced by the PrgY membrane protein (8, 9). Ultimately, the induction state of a pCF10-containing donor cell depends on the molar ratio of the two peptides in the growth medium rather than the absolute pheromone concentration; the mechanism by which inhibitors inhibit pheromones is not completely clear. A model depicting the major peptide-mediated control circuits in the pCF10 system is shown in Fig. 1A.
FIG. 1.
Extracellular (A) and intracellular (B) peptide-mediated control circuits in the pCF10 system. (A) Pheromone cCF10 (LVTLVFV) is produced (double arrows) from the chromosome (thin line with broken end) by proteolytic processing of the chromosomal ccfA gene product (3), whereas inhibitor iCF10 (AITLIFI) is produced by processing of the polypeptide encoded by the prgQ gene (38) of pCF10 (thin circle); prgQ is actually the first gene in a long, pheromone-inducible conjugation operon. Either endogenously produced or exogenous cCF10 pheromone can induce expression of Asc10 and other pCF10-encoded conjugation gene products. Pheromone activity of cCF10 is inhibited by iCF10, but previous work has not fully elucidated whether this occurs inside the cell, outside the cell, or in both locations. In a plasmid-containing donor cell, the pCF10-encoded PrgY protein reduces the amount of cCF10 produced, and the cell produces enough iCF10 to neutralize the residual cCF10 that escapes PrgY control (9, 10, 25); this precise balance in opposing activities can be shifted by addition of exogenous cCF10 from recipient cells. (B) Working model for the function of PrgX as the intracellular switch controlling pheromone-inducible conjugation. The protein consists of an N-terminal DNA binding domain (oval) connected by a flexible linker to a central dimerization domain (filled box), which also contains a peptide binding pocket (39). When cCF10 pheromone occupies this pocket, the PrgX C-terminal regulatory domain (represented by a thin, curved line) undergoes a marked shift to allow for interactions of amino acids between positions 295 and 300 with the bound peptide; this displaces a loop in this domain (residues 287 to 294) that is important for PrgX tetramer formation (39). In complexes with iCF10, PrgX amino acids between residues 311 and 315 interact with the bound peptide; this actually stabilizes the position of the tetramer-promoting loop. Tetramers of PrgX are predicted to simultaneously bind two operator sites in pCF10 DNA more stably than a pair of unlinked dimers. One of these operators overlaps the prgQ promoter, such that conjugation is repressed when it is occupied by PrgX (6, 7).
An extensive series of genetic, biochemical and structural studies has shown that the 317-amino-acid protein PrgX is the cytoplasmic pheromone-responsive molecular switch of the pCF10 system (4-7, 29, 30, 39). PrgX controls expression of conjugation by negative regulation of the promoter of the prgQ operon, encoding the pCF10 conjugation machinery. It has been proposed that repression of prgQ transcription by PrgX is mediated by a mechanism involving PrgX occupancy of two relatively weak binding sites in pCF10 DNA (6), with a dimer bound to each site (39). Binding is predicted to be stabilized by protein/protein interactions between pairs of PrgX dimers and by formation of a DNA loop between the two PrgX binding sites (29, 39). Determination of high-resolution structures of PrgX and PrgX/cCF10 complexes indicated that a PrgX C-terminal loop (residues 287 to 294) comprising the major interface holding tetramers together in the apoprotein structure would be moved as a result of pheromone binding (39). The observed changes the crystal structure would likely destabilize tetramers in solution; our initial structural data also led to the prediction that iCF10 might bind the same pocket of PrgX but not cause the tetramer-destabilizing movement of the C-terminal loop. Figure 1B depicts the oligomeric structure of PrgX complexed with either iCF10 or cCF10, based on the data just presented and those described in the following paragraph.
Previously published literature suggested that the primary site of competition between the inhibitor and pheromone peptides might be the extracellular pheromone binding proteins (PrgZ in the pCF10 system and TraC in the case of plasmids pAD1 and pPD1) (17) that enhance pheromone uptake at low concentrations, but there was little direct evidence to support this contention. Very recently, we have shown that both cCF10 and iCF10 target PrgX (30). New structural data show that both proteins bind to the same region of PrgX but produce different conformational changes (30). As noted above, in the case of cCF10 binding (39), a subdomain of PrgX immediately adjacent to the loop encompassing residues 287 to 294 changes its structure dramatically. This is due to direct interactions between the bound pheromone and residues 296 to 299 of PrgX. Our recent data (30)show that in iCF10/PrgX complexes, PrgX residues 312 to 315 interact with the bound inhibitor, actually locking the loop encompassing residues 287 to 294 into a tetramer-promoting conformation. In light of these recent data, the extent to which direct interactions of cCF10 and iCF10 with PrgZ modulate the biological activities of these peptides is less clear. In addition, the means by which the specific amino acid sequences of these peptides determine their biological activities are not fully understood.
In the course of several studies of the pCF10 system (6, 7, 10, 29, 30), we have generated isogenic sets of strains containing a transcriptional reporter gene fused to the pheromone-inducible prgQ promoter and expressing various combinations of prgX, prgY, and prgZ in order to better understand the interactions between the products of these genes and the regulatory peptides. Here we report the use of similar reporter strains in a series of genetic and physiological experiments designed to complement recent structural studies (30, 39) by providing data on the functional interactions of cCF10 and iCF10 with the PrgX and PrgZ proteins. We also describe a genetic screen to identify peptide variants of cCF10 with altered biological activities, and we examine the extent to which PrgX and PrgZ affect these activities. The results suggest that the amino acid sequence determinants affecting interactions with PrgX are most important in the activities of both pheromone and inhibitor peptides. Comparative analysis of native and variant peptides also revealed amino acid residues of cCF10 that were important for efficient PrgZ-mediated import.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
E. faecalis was grown at 37°C either in Todd-Hewitt broth (Difco), in M9-YE glucose medium (20) (an M9-based medium supplemented with 0.3% yeast extract, 1% Casamino Acids, 0.1% glucose, 1 mM MgSO4, and 0.1 mM CaCl2), or on Todd-Hewitt broth plates containing 1.5% agar. Antibiotics were used at the following concentrations: tetracycline, 10 μg/ml; erythromycin, 10 μg/ml; chloramphenicol, 20 μg/ml; rifampin, 200 μg/ml; kanamycin, 1,000 μg/ml; and fuscidic acid, 25 μg/ml. Escherichia coli DH5α was used for cloning purposes and was grown in Luria-Bertani (LB) broth at 37°C with antibiotics at the following concentrations: erythromycin, 200 μg/ml; kanamycin, 50 μg/ml; and spectinomycin, 50 μg/ml. All bacterial strains and plasmids used in this study are summarized in Table 1. Plasmid pPCR-4, used for mutagenesis of the cCF10-coding sequence, contains the pCF10 prgQ gene cloned into the vector pDL278 with the 3′ sequence altered to encode cCF10 instead of the native iCF10 (8, 38). The plasmid pMSP6043-1 expresses PrgZ from a pCF10-derived carrying encoding prgN, -O, -P, -W, and -Z cloned into the vector pDL27 (9); pMSP6043-2, which does not express PrgZ, was made by ligating the EcoRI and PstI fragment from prgN through the middle of prgZ from pINY6023 into the EcoRI and PstI sites of pDL276 in the same orientation as that of pMSP6043-1. Kristich et al. (31) have recently described the integration of the pWV01 repA gene under control of a lactococcal promoter into the upp locus of the E. faecalis chromosome by homologous recombination using an integrative plasmid called pCJK20, where the cloned gene to be integrated is flanked by the 5′ and 3′ segments of upp to promote recombination. To construct an OG1S derivative with an expressed prgX gene integrated into the chromosome for this study, we used a vector plasmid, pCJK41, that is virtually identical to pCJK20 except that it contains a minor change in the ribosome binding site driving expression of the cloned gene (the sequence from the ribosome binding site to the prgX start codon of the construct used here is AGGAGGTATTATTACATG). The desired integration of prgX into the upp locus was confirmed by PCR and sequencing, and we confirmed expression of levels of PrgX protein comparable to those of cells carrying pCF10 by Western blot analysis (4).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) or description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | Cloning host | Invitrogen |
| E. faecalis | ||
| OG1RF | Rifr Far; derived from clinical strain OG1 | 19 |
| JRC104 | cCF10-deficient OG1RF derivative | 10 |
| OG1Sp | Spr; derived from clinical strain OG1 | 24 |
| 100-5 | Derived from OG1Sp, carries chromosomal copy of prgX | This study |
| Plasmids | ||
| pCF10 | cCF10-inducible conjugative plasmid | 18 |
| pPCR-4 | cCF10 expression construct derived from prgQ region cloned into pDL278 | 8, 38 |
| p043lacZ | pAT18 vector containing prgX-Q region of pCF10 with a pheromone-inducible lacZ reporter gene | 29 |
| p043lacZ dX | No PrgX protein produced, control strain | 30 |
| pMSP6043-1 | prgN, -O, -P, -W, and -Z in pDL276 | 9 |
| pMSP6043-2 | prgN, -O, -P, and -W in pDL276 | This study |
Oligonucleotide-directed random mutagenesis and screening.
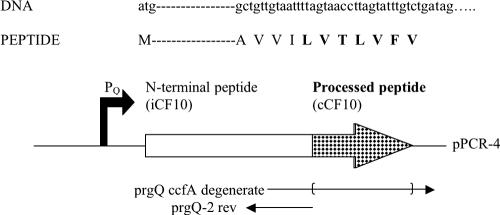
Oligonucleotide-directed random mutagenesis was carried out using Invitrogen's GeneTailor site-directed mutagenesis system. In this process, base substitutions in the cCF10 sequence were made using the mutagenic primer prgQ-ccfA degenerate, with the sequence 5′-TCA AGA TAT ATA GCT GTT GTA ATT TTA GTA ACC TTA GTA TTT GTC TGA TAG AAT TC-3′ (boldface indicates the degenerate portion), and the primer prgQ-2 rev, with the sequence 5′-TTA CAA CAG CTA TAT ATC TTG ATA G-3′. These mutagenic primers were synthesized using the five-bottle method described by Chiang (11) In this process, when the nucleotide targeted for mutagenesis is reached during primer synthesis, the wild-type nucleotide is mixed with a specific concentration of each of the non-wild-type nucleotides such that a certain percentage of misincorporation occurs (11). For this experiment, 2.5% misincorporation was used, meaning that each non-wild-type nucleotide had a 2.5% chance of being replaced at a given position. The coding sequences and locations of primers used for this oligonucleotide-directed random mutagenesis strategy are outlined in Fig. 2. The resulting PCR-generated mutated plasmids were then transformed into E. coli DH5α-T1R cells (Invitrogen) and screened for production of cCF10 with altered activity.
FIG. 2.
Diagram of pheromone-coding sequence (66 bp) and primers used for oligonucleotide-directed random mutagenesis of plasmid pPCR-4. The coding sequence is derived from the prgQ open reading frame in pCF10 and includes the prgQ promoter (PQ) and altered coding sequence so that cCF10 is expressed instead of the native iCF10 (indicated in checked region of the arrow); the complete DNA sequence for the start codon, the pheromone-coding region, and the four codons immediately upstream is shown. Primers used for random PCR mutagenesis are indicated below; the bracketed region of the prgQ-ccfA degenerate primer indicates the region directed for degenerate base incorporation (see Materials and Methods).
In E. coli, pPCR-4-produced cCF10 and the mutants are secreted into the surrounding medium. The peptides produced were then screened to identify those with altered activity by using an E. faecalis based-clumping assay (see below). E. coli clones whose supernatants showed altered activity compared with clones expressing wild-type cCF10 were sequenced to determine the amino acid substitutions giving the resulting activity. We determined the sequence of about 300 to 400 bp of plasmid DNA spanning the region from the upstream end of the prgQ promoter to about 200 bp downstream from the peptide-coding region. The only mutations detected were in the region where degenerate bases were incorporated into the mutagenic primers. The deduced peptides of selected plasmids from E. coli clones whose supernatants showed altered but not abolished activity were synthesized chemically (Mimotopes, Raleigh, NC) to further examine their activity.
Clumping assay.
The fact that pheromone induction can be detected by the formation of visible clumps in donor cell cultures exposed to the peptide (35) served as the basis for the biological assay used to screen mutants. E. coli strains harboring mutated plasmids were individually grown in LB medium for 15 h, and the cells were then centrifuged for 10 min at 11,000 × g and the pheromone activity in the supernatant was assayed. One hundred microliters of the supernatant was added to the first well of a microtiter plate, and twofold serial dilutions were made across a row of wells. Then 10 μl of an overnight culture of E. faecalis OGIRF(pCF10) was added to each well, and the plate was incubated at 37°C with shaking for 2 hours. Culture supernatant from E. coli DH5α-T1R(pPCR-4) was used as a positive control, and wells containing the indicator strain OGIRF(pCF10) and medium were used as a negative control. Pheromone activity was reported as the titer, which is the reciprocal of the highest twofold dilution showing a positive clumping reaction.
β-Galactosidase assay.
Derivatives of E. faecalis OG1RF carrying p043lacZ, containing a pheromone-inducible prgQ-lacZ transcriptional fusion (29) and in some cases a second plasmid (pMSP6043-1, described above) expressing prgZ, were grown overnight in M9-YE medium and then diluted 1:5 into fresh M9-YE medium and grown for an additional 1.5 h before harvesting for β-galactosidase assay. When indicated, strains were induced immediately after the 1:5 dilution with the indicated concentration of synthetic cCF10, iCF10, or mutated peptide. β-Galactosidase assays were performed as described previously (29). Results shown are representative of at least two independent experiments done in duplicate on different days.
RESULTS
PrgZ increases sensitivity to pheromone, while PrgX is required for the response to both pheromone and inhibitor.
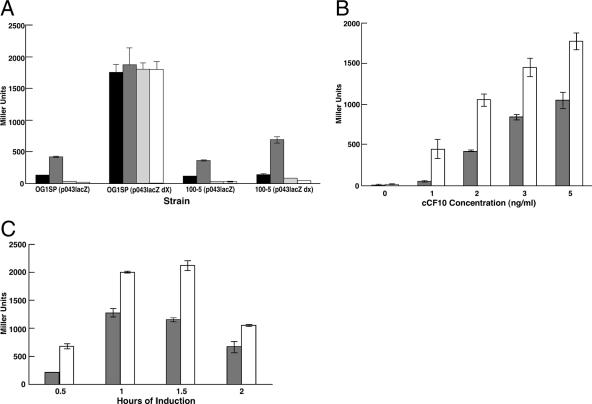
Several previous studies (7, 29, 30) have employed plasmid p043lacZ, containing a pheromone-inducible prgQ-lacZ transcriptional reporter gene fusion as a “minimal” genetic system for detailed analysis of the pheromone-sensitive repression of the prgQ promoter by PrgX in E. faecalis. This system has proven to be very useful in genetic studies and in quantitative analysis of the effects of pheromone induction on PrgX-regulated prgQ transcription in vivo. However, the high copy number of the vector plasmid and the resulting high levels of expression of the negative regulatory elements PrgX and iCF10 make it necessary to use higher levels of pheromone to obtain induction (7, 29, 30), relative to the amounts required to induce cells containing pCF10. We wished to employ this system for the analysis of cCF10 variants, as described below. Therefore, we carried out experiments to confirm the critical role of PrgX in interaction with both cCF10 and iCF10 in this system and to determine the feasibility of using this system to examine the effects of PrgZ on pheromone. We recently reported (30) that a strain carrying p043lacZdX (containing a prgX deletion removing 67% of the coding sequence from the 3′ end) showed high-level constitutive expression of β-galactosidase and was unresponsive to cCF10 or iCF10 (Fig. 3A), indicating that PrgX is essential for response to both peptides. Due to a limited number of compatible cloning vectors and instability of many chimeric plasmids carrying cloned prgX, we have had limited success in carrying out genetic complementation of prgX mutations (7). To overcome this problem, prgX was cloned into the chromosome where it is expressed constitutively in strain 100-5 (see Materials and Methods), and this strain was used to test whether the p043lacZdX plasmid phenotype could be complemented. The results demonstrated that expression of a functional prgX in trans in a strain carrying p043lacZdX fully restored (i) repression of prgQ in the absence of exogenously added peptides, (ii) induction with exogenous cCF10, and (iii) iCF10 inhibition of cCF10 induction (Fig. 3A).
FIG. 3.
Importance of PrgZ and PrgX in cCF10 response. (A) PrgX requirement for cCF10 response as shown by measuring β-galactosidase induction of plasmid p043lacZ carrying prgX (see text) or a an isogenic plasmid with a deletion in prgX, p043lacZ dX (dX), harbored by either a wild-type strain (OG1Sp) or an isogenic strain with a chromosomal copy of prgX (100-5). Strains grown overnight in M9 medium were diluted 1:5 with no peptides added (black bars), or peptides were added as indicated. cCF10 was added at 10 ng/ml (dark gray bars), and in some cases iCF10 was also added at 10 times (100 ng/ml, light gray bars) or 100 times (1,000 ng/ml, white bars) the cCF10 concentration. (B and C) Response to exogenously added cCF10 in the absence (vector control, gray bars) or presence (pMSP6043-1, white bars) of PrgZ. Cultures grown overnight in M9 medium were diluted 1:5, and cCF10 was added in increasing amounts and harvested at 90 min (B) or added at 5 ng/ml and harvested at the indicated time points (C). Data shown are representative of at least two independent experiments done in duplicate; error bars represent one standard deviation of the mean.
In cells where the pheromone response genes of pCF10 are carried on a low-copy replicon, PrgZ is required for an active response to cCF10 in concentration ranges (10−11 to 10−12 M) comparable to those found in recipient cell culture supernatants (33, 35). The defects in pheromone response associated with deletion of prgZ could be overcome by addition of higher concentrations of peptide, and it was concluded that the host OppA peptide binding protein in conjunction with the rest of the oligopeptide permease system could mediate cCF10 import at higher peptide concentrations. Because induction of (PrgZ−) cells carrying only p043lacZ requires substantially higher levels of pheromone, we wanted to determine whether PrgZ could enhance the pheromone response at these higher peptide levels. E. faecalis carrying p043lacZ along with either pMSP6043-1 (conferring PrgZ expression) or pMSP6043-2 (vector control) was used in time course and dose-response analyses. As shown in Fig. 3B and C, strains expressing PrgZ had a higher response to cCF10 than strains expressing the control at several different concentrations of cCF10 as well as at different time points. While the relative increase in response that was attributable to PrgZ was highest at the lowest pheromone level, there was a significant difference at all concentrations tested; furthermore, the increase in induction observed in the presence of PrgZ in the range of 3 to 5 ng/ml was comparable to the previously observed increase of about twofold in our study where the reporter genes were expressed from a lower-copy replicon (33). In the time course studies we saw the largest effect of PrgZ during a 90-min induction period. We therefore used a 90-min induction period with 5 ng/ml peptide to analyze the cCF10 variant peptides described in the following section; we chose not to use a lower peptide concentration because the significantly reduced activities of several of these peptides would not generate a detectable response at lower concentrations.
Construction of random cCF10 mutants and determination of biological activity.
To explore the cCF10 amino acid sequence determinants critical in the functional interactions with PrgZ and PrgX, oligonucleotide-directed random mutagenesis of a chimeric open reading frame encoding the cCF10 peptide (Fig. 2) (see Materials and Methods) in the shuttle plasmid pPCR-4 was performed. Both E. faecalis and E. coli cells carrying this plasmid excrete substantial quantities of cCF10 into the culture medium. To screen these peptides for biological activity, mutated pPCR-4 plasmids were transformed into E. coli, and supernatant preparations from transformants were screened for their ability to cause clumping of a wild-type strain harboring pCF10 (see Materials and Methods). Preliminary analysis indicated that a high percentage of clones expressing no clumping-inducing activity contained stop codons and other severe mutations abolishing peptide production that were uninteresting for our purposes. Therefore, we focused our attention on those clones expressing altered but not abolished activities indicative of production of a variant peptide with amino acid substitutions.
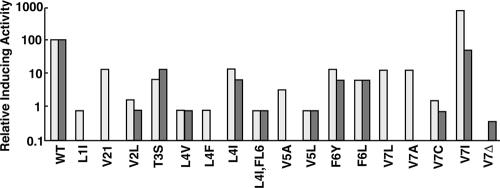
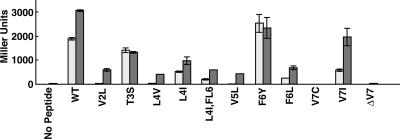
Of 600 clones tested from three independent transformations, 14 unique mutations resulting in culture supernatants yielding reduced activity were identified, and 1 unique mutation (V7I) resulted in increased supernatant activity (Fig. 4). Predicted amino acid substitutions resulting in decreased activity were identified at each position along the cCF10 peptide. Synthetic peptides were made for 10 representative peptides and tested using the same clumping assay to ensure that the observed effects were due to the inherent activity of the peptide as opposed to differences in expression from the E. coli host (Fig. 4). With the exception of V7I, the synthetic peptide activity was similar to the E. coli-produced peptide activity. E. coli supernatants from the strain encoding V7I had increased activity compared with wild-type cCF10, whereas synthetic V7I exhibited activity comparable to that of cCF10. The elevated activity of E. coli-produced V7I may have been due to increased synthesis or more efficient processing of this peptide, but this was not investigated further. All of the peptide variants identified in the genetic screen contained single amino acid substitutions, with the exception of L4I,F6L. For this variant, we also identified each of the corresponding single substitutions in separate clones, and the activity of the double variant was approximately what would be predicted from independent additive effects of the single substitutions (Fig. 4). It was considered possible that some of the peptides with very low pheromone activity might actually act as cCF10 inhibitors, but we were unable to detect any inhibitory activity when responder cells were treated with mixtures of cCF10 and excess amounts of the test peptides (data not shown).
FIG. 4.
Activity of cCF10 and variant peptides to induce clumping of OG1RF(pCF10) cultures. Activity was measured as the reciprocal of the highest dilution of E. coli-produced or synthetically derived cCF10 or mutant derivatives to aggregate an OG1RF(pCF10) indicator strain. The relative amounts of pheromone activity are representative of at least two independent experiments. The peptide LVTLVF- (V7Δ) was not found in the original assay but was synthesized and tested in the clumping assay. Supernatants from E. coli strains expressing wild-type (WT) cCF10 (LVTLVFV) from pPCR-4 or each of the mutant peptides were assayed for activity (light bars) following overnight growth with aeration in LB medium. Activities of synthesized peptides (dark bars) were assayed from twofold serial dilutions of a 50-ng/ml stock solution.
PrgZ alters sensitivity of pheromone response to mutant cCF10 peptides.
A second test of peptide activity levels was undertaken, which made use of plasmid p043lacZ. This plasmid was used as a reporter plasmid in a cCF10-negative host strain JRC104 (to remove any confounding effects of endogenous pheromone production by the responder strain), and the ability of each peptide to induce β-galactosidase expression was determined. In the case of cCF10, the presence of PrgZ increased the induction level by about 1.6-fold; most of the variant peptides had significantly lower absolute inducing activities, but the increase in activity associated with expression of PrgZ was similar to that observed with cCF10 (Fig. 5). In addition, the relative inducing activities of most of the variants in the p043lacZ system were similar to those observed in the clumping assays using pCF10-containing responders (Fig. 4 and 5). Three notable exceptions to this pattern were T3S, F6Y, and V7I. In the absence of PrgZ, F6Y actually had a higher inducing activity in the p043lacZ system than in the pCF10 system (Fig. 5), whereas in the presence of PrgZ, cCF10 was more active, similar to the result obtained with pCF10 responder cells (Fig. 4). T3S showed a similar pattern, with a higher activity relative to cCF10 in the absence of PrgZ. In contrast, V7I had about two-thirds of the inducing activity of cCF10 in the presence of PrgZ but much lower activity in the absence of PrgZ (Fig. 4 and 5).
FIG. 5.
PrgZ effects on p043lacZ induction ability of peptides. Synthetic cCF10 variant peptides were added at 5 ng/ml to cCF10-deficient strain JRC104 harboring p043lacZ with a second plasmid expressing PrgZ from pMSP6043-1 (Z+, dark bars) or a vector control, pMSP6043-2 (Z−, light bars). Peptides were added immediately after a 1:5 dilution from stationary-phase cultures, and β-galactosidase induction levels were measured after 90 min incubations. Data shown are representative of at least two independent experiments done in duplicate; error bars represent one standard deviation of the mean.
These results indicate that peptides T3S and F6Y may interact with PrgX as well as cCF10 (or even better in the case of F6Y), whereas the functional interaction of these peptides with PrgZ leading to their import appears to be impaired. In contrast, the activities of peptide V7I suggest that the substitution of isoleucine for valine at the cCF10 C terminus resulted in a less optimal interaction with PrgX, but this effect was partially abrogated by a more efficient import of the peptide by PrgZ. The importance of the C-terminal residue is also shown by the fact that peptide V7Δ (this peptide was synthesized and tested, though it was not identified in the genetic screen), comprised of the first six residues of cCF10, exhibited a nearly undetectable pheromone activity (Fig. 4). From these data, we conclude that productive interactions with both PrgX and PrgZ have driven the evolution of the biological activity of the native cCF10 sequence and that the specific peptide sequence determinants for productive interaction with each protein do not overlap completely.
DISCUSSION
Recent genetic, biochemical, and structural studies elucidating the molecular interactions between PrgX and both iCF10 and cCF10 (7, 29, 30, 39) have allowed the formulation of detailed molecular models describing how the changes in PrgX structure caused by peptide binding could alter the PrgX oligomerization state in vivo and in turn affect PrgX-mediated repression of the prgQ operon (7). While it is possible to use molecular modeling to predict effects of specific amino acid substitutions on the structure of the corresponding PrgX/peptide complex, making accurate predictions of how these changes might affect biological activity is more complicated because the analysis of iCF10- and cCF10-containing complexes has already shown that residues in different segments of the PrgX C-terminal region can interact with different peptides occupying the same binding pocket. Thus, one can use modeling to test whether a given amino acid substitution would disrupt interactions with PrgX C-terminal residues that interact with cCF10, but it might be difficult to predict whether a different set of C-terminal residues could interact with the variant peptide. In addition, the biological activity of a given peptide depends both on its interaction with PrgX and on the PrgZ-mediated import of the peptide (which determines its intracellular concentration). Therefore, we decided to take a genetic approach to identify a collection of cCF10 variants with altered biological activity, which could then be used in molecular and structural studies to provide a more complete picture of the pheromone-sensitive molecular switch controlling E. faecalis conjugation. Since cCF10 pheromone activity is dependent on interactions with both PrgZ and PrgX, we developed the use of the p043lac reporter plasmid for pheromone induction. This facilitated expression of PrgZ in trans such that effects of this protein on pheromone activity could be distinguished from effects on PrgX. In the course of experiments (Fig. 3) to validate the system for the peptide mutagenesis study, we confirmed recent evidence showing that PrgX is the biologically significant target for iCF10 inhibition of cCF10 activity (31). While it had been suggested that the inhibitor peptides might function by blocking pheromone import (22, 23), making inhibitor interactions with PrgX irrelevant, our cumulative results indicate that PrgX/iCF10 interactions are highly significant. As shown previously and in Fig. 3A, PrgX is sufficient and necessary for iCF10 to function as an inhibitor of cCF10, at least at the peptide levels used in these experiments. We have been unable to obtain data to indicate that the presence of PrgZ in responder cells enhances iCF10 activity to a significant extent. Some of our data suggest that PrgZ expression actually decreased the ability of exogenous iCF10 to inhibit cCF10 (data not shown), suggesting that PrgZ might enhance import of cCF10 but not iCF10; however, we cannot rule out the possibility that the peptides could compete for PrgZ under some conditions. It could be argued that these concentrations are higher than those found in natural mating mixtures, but it should be pointed out that PrgZ does significantly enhance the response to cCF10 under the same conditions (Fig. 3). While our cumulative data indicate that PrgX may be the most important site of direct competition between iCF10 and cCF10, the results shown in Fig. 3B and C confirm that the net biological activity of cCF10 as a mating pheromone depends on interactions with both PrgX and PrgZ.
The genetic screen carried out in this study identified 15 variants of cCF10 with amino acid substitutions. With a single exception (V7I), determination of the relative pheromone activities of all the synthetic cCF10 variants gave the same results as those obtained in the assays of E. coli supernatants. Although we limited our screen to testing about 600 clones from a total of three independent transformations, we did identify mutations resulting in the same amino acid sequence change in multiple independent transformants. This suggests that, given the phenotypic constraints used, we may have approached saturation in this screen.
Relative to cCF10, all of the synthetic peptides had reduced pheromone activity when tested against responder cells expressing PrgZ (Fig. 4 and 5). In most cases, the presence of PrgZ increased the absolute inducing activities of the variants in a fashion similar to that observed with cCF10. For the two peptides (T3S and F6Y) whose activity was not enhanced by PrgZ, it was concluded that the substitutions probably had no deleterious effects on the peptide interactions with PrgX (and even enhanced this interaction slightly in the case of F6Y). However, these variants had a reduced productive interaction with PrgZ, with the overall effect being a slight reduction of pheromone activity when the responder cells expressed both PrgZ and PrgX. Peptide V7I had a much higher increase in inducing activity when assayed against p043lacZ-containing responder cells containing PrgZ than all the other peptides, including native cCF10. In this case the mutation probably enhanced a productive interaction with PrgZ, resulting in increased import into the responder cell, but at the same time the interaction with PrgX was impaired slightly, reducing the overall activity. The molecular nature of “productive” peptide/PrgZ interaction that was affected in peptides T3S, F6Y, and V7I is essentially unknown. One formal possibility is that the optimal interactions could increase the binding affinity of the peptide for PrgZ. However kinetic studies of oligopeptide permease-mediated peptide import in Lactococcus lactis by the Poolman group (32) suggested the possibility that release of previously bound peptide from the OppA binding protein (PrgZ is an OppA homolog specific for cCF10) into the transmembrane channel formed by the other Opp components could be the rate-limiting step in peptide import. Comparative biochemical and structural studies of PrgZ interactions with cCF10 and the three variant peptides discussed above could help distinguish between these two possibilities. It is also formally possible that certain amino acid substitutions could affect efficiency of import by the chromosomal OppA binding protein, but we lack the tools to examine this possibility at present.
The majority of amino acid substitutions identified in this study appeared to affect peptide/PrgX interactions. Since all of these variants retained some pheromone activity but caused no detectable inhibition of cCF10, it is likely that they all produced structural changes in PrgX similar to those observed with cCF10 but that the binding affinities were reduced or the rotation of the C-terminal arm of PrgX that leads to destabilization of tetramers occurred to a lesser extent. Previous structural studies of PrgX suggested that nearly all of the hydrogen bonding interactions between the peptide and residues in the C-terminal domain of PrgX involve the peptide backbone, and this is also the case for many of the interactions between the peptide and the PrgX residues lining the binding pocket. This is consistent with the fact that there are almost no charged side groups in any of the residues of cCF10 or iCF10. The hydrophobic side chains of these peptides likely affect PrgX interactions either by providing spatial constraints or by affecting the binding affinity. The available structural data suggest some explanations for the results presented here. For example, the L4V substitution, which reduced the inducing activity by >95% in responder cells lacking PrgZ (Fig. 5), replaced a large hydrophobic side chain with a smaller one. The three-carbon valine side chain of L4V would easily fit within the space occupied by four-carbon leucine side chain of cCF10, and the formation of an important hydrogen bond between the peptide backbone and PrgX residue Y298 (39) should not be impaired in the variant. However, the smaller side chain would probably confer weaker hydrophobic interactions with the binding pocket, reducing affinity of the peptide for PrgX. Similar lines of reasoning can be used to suggest structural interactions to account for the activities of the other variant peptides, including F6Y, where the extra hydroxyl group on the aromatic side chain would not appear to cause any spatial problems and might actually provide an additional hydrogen bond with PrgX to stabilize the complex, accounting for its increased inducing activity (in cells lacking PrgZ). As noted above, these kind of predictions need to be verified experimentally. Our efforts to obtain quantitative solution binding affinity data have been hampered by the tendency for the peptides and protein to precipitate from aqueous solutions and to stick nonspecifically to dialysis membranes and other surfaces, but further structural and biochemical studies on these peptides are in progress.
Genetic analyses of signaling peptide specificity have been carried out in a few other systems, generally those involving membrane histidine kinase peptide receptors (12, 26, 27, 34). For the pneumococcal competence-stimulating peptide, genetic analyses have been coupled with solution structural analyses using spectroscopic techniques (27). In the case of the enterococcal pheromone peptides, the biological activities of synthetic peptides selected based on visual inspection of native pheromone sequences have been tested (28), but this was done before the identities of the sex pheromone receptors were actually proven. In the pAD1 system, An and Clewell synthesized cAD1 variants containing single alanine substitutions at each nonalanine residue and found that all of these except V5A dramatically reduced pheromone activity (2). The present study provides peptide reagents with a range of biological activities for a comprehensive high-resolution structure/function analysis of the pCF10 cell-cell signaling system.
Acknowledgments
This work was supported by PHS grant GM 49530 and by a grant-in aid from the University of Minnesota Graduate School to G.M.D. K.R.F. was a recipient of an undergraduate research fellowship from the American Society for Microbiology. B.K.K. was supported by NIH training grant T32 GM08347. J.R.C. was supported by NIH training grant T32 DE07288.
We thank Ke Shi, Douglas Ohlendorf, and Cathleen Earhart for helpful discussions about this work.
Footnotes
Published ahead of print on 10 November 2006.
REFERENCES
- 1.An, F. Y., and D. B. Clewell. 1994. Characterization of the determinant (traB) encoding sex pheromone shutdown by the hemolysin/bacteriocin plasmid, pAD1 in Enterococcus faecalis. Plasmid 31:215-221. [DOI] [PubMed] [Google Scholar]
- 2.An, F. Y., and D. B. Clewell. 2002. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J. Bacteriol. 184:1880-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antiporta, M. H., and G. M. Dunny. 2002. ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J. Bacteriol. 184:1155-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae, T., S. Clerc-Bardin, and G. M. Dunny. 2000. Analysis of expression of prgX, a key negative regulator of the transfer of the Enterococcus faecalis pheromone-inducible plasmid pCF10. J. Mol. Biol. 297:861-875. [DOI] [PubMed] [Google Scholar]
- 5.Bae, T., and G. M. Dunny. 2001. Dominant negative mutants of prgX: evidence for a role of PrgX dimerization in negative regulation of pheromone-inducible conjugation. Mol. Microbiol. 39:1307-1320. [DOI] [PubMed] [Google Scholar]
- 6.Bae, T., B. Kozlowicz, and G. M. Dunny. 2002. Two targets in pCF10 DNA for PrgX binding: their role in production of Qa and prgX mRNA and in regulation of pheromone-inducible conjugation. J. Mol. Biol. 315:995-1007. [DOI] [PubMed] [Google Scholar]
- 7.Bae, T., B. K. Kozlowicz, and G. M. Dunny. 2004. Characterization of cis-acting prgQ mutants: evidence for two distinct repression mechanisms by Qa RNA and PrgX protein in pheromone-inducible enterococcal plasmid pCF10. Mol. Microbiol. 51:271-281. [DOI] [PubMed] [Google Scholar]
- 8.Buttaro, B. A., M. H. Antiporta, and G. M. Dunny. 2000. Cell-associated pheromone peptide (cCF10) production and pheromone inhibition in Enterococcus faecalis. J. Bacteriol. 182:4926-4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandler, J. R., A. R. Flynn, E. M. Bryan, and G. M. Dunny. 2005. Specific control of endogenous cCF10 pheromone by a conserved domain of the pCF10-encoded regulatory protein PrgY in Enterococcus faecalis. J. Bacteriol. 187:4830-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandler, J. R., H. Hirt, and G. M. Dunny. 2005. A paracrine peptide sex pheromone also acts as an autocrine signal to induce plasmid transfer and virulence factor expression in vivo. Proc. Natl. Acad. Sci. USA 102:15617-15622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang, L. W. 1996. Saturation mutagenesis by mutagenic oligonucleotide-directed PCR amplification (Mod-PCR). Methods Mol. Biol. 57:311-321. [DOI] [PubMed] [Google Scholar]
- 12.Claverys, J. P., and L. S. Havarstein. 2002. Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front. Biosci. 7:1798-1814. [DOI] [PubMed] [Google Scholar]
- 13.Clewell, D. B., F. Y. An, S. E. Flannagan, M. Antiporta, and G. M. Dunny. 2000. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol. Microbiol. 35:246-248. [DOI] [PubMed] [Google Scholar]
- 14.Clewell, D. B., F. Y. An, M. Mori, Y. Ike, and A. Suzuki. 1987. Streptococcus faecalis pheromone cAD1 response: evidence that the peptide inhibitor exccreted by pAD1-containing cells may be plasmid determined. Plasmid 17:65-68. [DOI] [PubMed] [Google Scholar]
- 15.Clewell, D. B., M. V. Francia, S. E. Flannagan, and F. Y. An. 2002. Enterococcal plasmid transfer: sex pheromones, transfer origins, relaxases, and the Staphylococcus aureus issue. Plasmid 48:193-201. [DOI] [PubMed] [Google Scholar]
- 16.Clewell, D. B., L. T. Pontius, F. Y. An, Y. Ike, A. Suzuki, and J. Nakayama. 1990. Nucleotide sequence of the sex pheromone inhibitor (iAD1) determinant of Enterococcus faecalis conjugative plasmid pAD1. Plasmid 24:156-161. [DOI] [PubMed] [Google Scholar]
- 17.Clewell, D. B., and G. M. Dunny. 2002. Conjugation and genetic exchange in enterococci., p. 265-300. In M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. B. Rice (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC.
- 18.Dunny, G., C. Funk, and J. Adsit. 1981. Direct stimulation of the transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid 6:270-278. [DOI] [PubMed] [Google Scholar]
- 19.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunny, G. M., and D. B. Clewell. 1975. Transmissible toxin (hemolysin) plasmid in Streptococcus faecalis and its mobilization of a noninfectious drug resistance plasmid. J. Bacteriol. 124:784-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunny, G. M., R. A. Craig, R. L. Carron, and D. B. Clewell. 1979. Plasmid transfer in Streptococcus faecalis: production of multiple pheromones by recipients. Plasmid 2:454-465. [DOI] [PubMed] [Google Scholar]
- 22.Dunny, G. M., and B. A. B. Leonard. 1997. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 51:527-564. [DOI] [PubMed] [Google Scholar]
- 23.Fujimoto, S., and D. B. Clewell. 1998. Regulation of the pAD1 sex pheromone response of Enterococcus faecalis by direct interaction between the cAD1 peptide mating signal and the negatively regulating, DNA-binding TraA protein. Proc. Natl. Acad. Sci. USA 95:6430-6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gold, O., H. V. Jordan, and J. van Houte. 1975. The prevalence of enterococci in the human mouth and their pathogenicity in animal models. Archs. Oral Biol. 20:473-477. [DOI] [PubMed] [Google Scholar]
- 25.Hirt, H., P. M. Schlievert, and G. M. Dunny. 2002. In vivo induction of virulence and antibiotic resistance transfer in Enterococcus faecalis mediated by the sex pheromone-sensing system of pCF10. Infect. Immun. 70:716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji, G. Y., R. C. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 27.Johnsborg, O., P. E. Kristiansen, T. Blomqvist, and L. S. Havarstein. 2006. A hydrophobic patch in the competence-stimulating peptide, a pneumococcal competence pheromone, is essential for specificity and biological activity. J. Bacteriol. 188:1744-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitada, C., M. Fujino, M. Mori, Y. Sakagami, A. Isogai, A. Suzuki, D. Clewell, and R. Craig. 1985. Synthesis and structure/activity relationships of Streptococcus faecalis sex pheromones cPD1 and cAD1, p. 43-48. In N. Izumiya (ed.), Peptide chemistry 1984. Protein Research Foundation, Osaka, Japan.
- 29.Kozlowicz, B. K., T. Bae, and G. M. Dunny. 2004. Enterococcus faecalis pheromone-responsive protein PrgX: genetic separation of positive autoregulatory functions from those involved in negative regulation of conjugative plasmid transfer. Mol. Microbiol. 54:520-532. [DOI] [PubMed] [Google Scholar]
- 30.Kozlowicz, B. K., K. Shi, Z.-Y. Gu, D. H. Ohlendorf, C. A. Earhart, and G. M. Dunny. 2006. Molecular basis for the control of conjugation by bacterial pheromone and inhibitor peptides. Mol. Microbiol. 62:958-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kristich, C. J., J. R. Chandler, and G. M. Dunny. Development of a host genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis. Plasmid, in press. [DOI] [PMC free article] [PubMed]
- 32.Lanfermeijer, F. C., A. Picon, W. N. Konings, and B. Poolman. 1999. Kinetics and consequences of binding of nona- and dodecapeptides to the oligopeptide binding protein (OppA) of Lactococcus lactis. Biochemistry 38:14440-14450. [DOI] [PubMed] [Google Scholar]
- 33.Leonard, B. A., A. Podbielski, P. J. Hedberg, and G. M. Dunny. 1996. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl. Acad. Sci. USA 93:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayville, P., G. Ji, R. Beavis, H. Yang, M. Goger, R. P. Novick, and T. W. Muir. 1999. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. USA 96:1218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mori, M., Y. Sakagami, Y. Ishii, A. Isogai, C. Kitada, M. Fujino, J. C. Adsit, G. M. Dunny, and A. Suzuki. 1988. Structure of cCF10, a peptide sex pheromone which induces conjugative transfer of the Streptococcus faecalis tetracycline resistance plasmid, pCF10. J. Biol. Chem. 263:14574-14578. [PubMed] [Google Scholar]
- 36.Mori, M., H. Tanaka, Y. Sakagami, A. Isogai, M. Fujino, C. Kitada, D. B. Clewell, and A. Suzuki. 1987. Isolation and structure of the sex pheromone inhibitor, iPD1, excreted by Streptococcus faecalis donor strains harboring plasmid pPD1. J. Bacteriol. 169:1747-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakayama, J., A. Isogai, D. B. Clewell, and A. Suzuki. 1995. Molecular and genetic analysis of a region of Enterococcus faecalis plasmid pPD1 containing sex pheromone sensitivity (traC), pheromone shutdown (traB), and pheromone inhibitor (ipd) genes. J. Bacteriol. 177:5567-5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakayama, J., R. E. Ruhfel, G. M. Dunny, A. Isogai, and A. Suzuki. 1994. The prgQ gene of the Enterococcus faecalis tetracycline resistance plasmid pCF10 encodes a peptide inhibitor, iCF10. J. Bacteriol. 176:7405-7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi, K., C. K. Brown, Z. Y. Gu, B. K. Kozlowicz, G. M. Dunny, D. H. Ohlendorf, and C. A. Earhart. 2005. Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proc. Natl. Acad. Sci. USA 102:18596-18601. [DOI] [PMC free article] [PubMed] [Google Scholar]