Abstract
A DNA-damaging agent, mitomycin C, derepresses the site-specific excision of two integrative and potentially conjugative elements from Streptococcus thermophilus, ICESt1 and ICESt3. The regulation pathway involves a repressor related to phage lambda cI repressor. It could also involve a putative regulator related to another type of phage repressors, the “cI-like” repressors.
Whereas in silico analyses revealed that numerous genomic islands could be integrative and conjugative elements (ICEs) or elements derived from ICEs (2, 4, 9, 13, 15, 19, 25, 26), only a few ICEs have been described. ICEs excise by site-specific recombination, transfer through conjugation, and integrate into a replicon of the recipient cell (8). Whereas the regulation of the excision of numerous prophages is well known, the regulation of site-specific excision of only a few ICEs, including Tn916, ICEs from Bacteroides, pSAM2, and clc, has been described (6, 11, 22, 23). These few regulation systems are very different from each other. Recently, DNA-damaging agents were found to derepress the excision and transfer of two other ICE types, ICEBs1 from Bacillus subtilis (1) and IncJ elements, including SXT from Vibrio cholerae (3) and SXT-related elements from enterobacteria (18). Such regulation systems are similar to the derepression of the site-specific excision of numerous prophages by DNA damage.
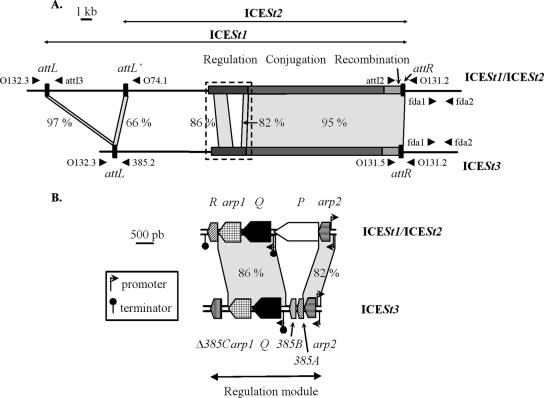
Two putative ICEs, ICESt1 and ICESt3, are integrated in the 3′ end of the fda locus of the lactic acid bacteria Streptococcus thermophilus CNRZ368 and CNRZ385, respectively (Fig. 1A). These ICEs harbor almost identical recombination and conjugation modules (20). The tyrosine integrase and the excisionase encoded by the ICESt1 recombination module catalyze its excision by recombination between the attL and attR flanking sites, leading to an excised circular ICE harboring an attI site and to a chromosomal attB site (10). Furthermore, ICESt1 carries an internal recombination site related to attL, attL′ (Fig. 1A) (20). Recombination between attL′ and attR leads to excision of the circular form of a shorter putative ICE, ICESt2, carrying an attI′site. A genomic island corresponding to the left part of ICESt1 and flanked by attL and attB′ sites remains integrated in fda.
FIG. 1.
Comparison of ICESt1/ICESt2 and ICESt3 maps. (A) Location of the recombination, conjugation, and regulation modules and the recombination sites on the ICESt1/ICESt2 and ICESt3 maps. The recombination sites are indicated by solid rectangles and are magnified. The schematic localizations and orientations of oligonucleotides used for PCRs are indicated by arrowheads. The following primer pairs were used for amplification of fragments containing the recombination sites resulting from ICE excision: primers O131.2 and O132.3 (attB from ICESt1 and ICESt3), primers attI2 and attI3 (attI from ICESt1), primers attI2 and O74.1 (attI′ from ICESt2), and primers O131.5 and 385.2 (attI from ICESt3). The fda1-fda2 primer pair was used for amplification of an fda internal region. The dotted rectangle indicates the area shown in panel B. (B) Map of regulation modules of ICESt1/ICESt2 and ICESt3. ORF designations beginning with “orf” are abbreviated with the corresponding letter or name. The locations and orientations of ORFs and truncated ORFs (Δ385C) belonging to the regulation modules are indicated by arrow boxes. The angled arrows and the lollipops indicate the putative promoters and rho-independent transcription terminators deduced from in silico analyses. The gray areas join closely related modules and att sites belonging to the ICEs (>65% nucleotide identity); the levels of identity are indicated. The close relationships between short sequences and insertion elements are not shown.
The closely related regulation modules of ICESt1/ICESt2 and ICESt3 contain three shared open reading frames (ORFs) (arp1, orfQ, and arp2) (Fig. 1B). The ICESt3 recombination module also includes three specific ORFs or pseudogenes (orf385A, orf385B, and Δorf385C), and the ICESt1/ICESt2 recombination module contains two specific ORFs (orfP and orfR). The putative regulatory proteins encoded by arp1, arp2, and orf385A have a helix-turn-helix (HTH) DNA binding domain. The functions of the putative proteins encoded by the other ORFs are unknown.
The 5′ part of arp1 encodes an HTH domain, and its 3′ part encodes a region characteristic of the COG2932 protein family, including the cI repressor of phage λ (9, 20). This region has two functions, cI autoproteolysis and cI oligomerization. In the presence of damaged DNA, the RecA protein, activated by single-stranded DNA (RecA*), induces autoproteolysis of cI and related proteins. Cleaved proteins are not able to oligomerize and therefore are not able to repress their target genes (12). In silico analysis suggested that DNA damage could derepress excision of ICEs from S. thermophilus.
To test this hypothesis, strains harboring ICESt1/ICESt2 (CNRZ368) or ICESt3 (CNRZ385) were grown at 42°C in HJL medium (24). Exponentially growing cells (optical density at 600 nm for MIC/4 and MIC/2, 0.04; optical density at 600 nm for 2× MIC and 4× MIC, 0.4) were treated with mitomycin C (MC) concentrations close to the MIC for 2.5 h to induce DNA damage. Then the recombination sites resulting from excision, attI and attB (ICESt1 and ICESt3) or attI′ (ICESt2), were amplified by PCR using 1 μg genomic DNA for CNRZ368 and 1 ng genomic DNA for CNRZ385. PCR experiments were performed in 25-μl mixtures using 0.5 U of Taq DNA polymerase (Biolabs) according to the manufacturer's specifications and primers (melting temperatures, 49.6 to 53.1°C) described in the legend to Fig. 1. After an initial denaturation step consisting of 4 min at 95°C, PCR was performed for 30 cycles of denaturation for 30 s at 95°C, annealing for 30 s at 47.5°C, and extension for 30 s at 72°C, followed by final extension for 7 min at 72°C.
Treatment of CNRZ368 with MC induced an increase in the PCR signal intensity corresponding to the attB and attI sites, while this treatment did not induce an increase in the PCR signal intensity corresponding to the fda gene used as a control (Fig. 2). The most intense signal for both attB and attI sites was obtained when 0.1 μg/ml MC (MIC/2) was used. The effects of MC were also determined by PCR amplification of fragments harboring attI and attB, using 1 μg to 1 ng genomic CNRZ368 DNA. The minimum amounts of template DNA that resulted in positive PCR amplification of the attB and attI sites were 100 ng using CNRZ368 DNA in the absence of MC and 10 ng after treatment with MC (MIC/2), suggesting that DNA damage caused by MC induced at least a 10-fold increase in ICESt1 excision.
FIG. 2.
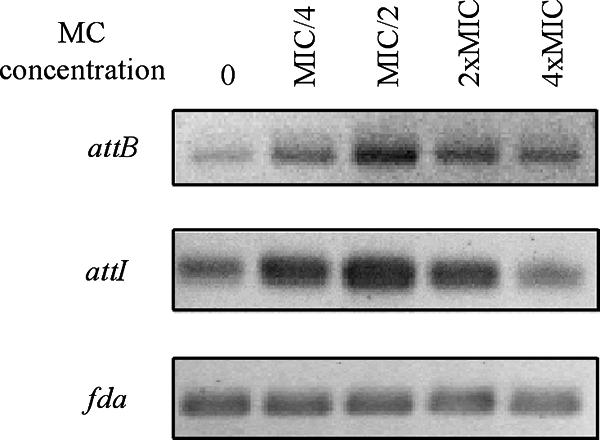
Effects of MC treatment of exponentially growing cells on the excision frequency of ICESt1. Fragments containing the attB (361 bp) or attI (400 bp) recombination site were amplified using 1 μg template DNA from MC-treated cells and primers described in the legend to Fig. 1. An internal fragment of the fda gene (339 bp) was amplified as a control using 1 ng template DNA.
In the absence of MC, the following ratios of the excised circular form to the genome were found in the stationary phase of growth: 10−6 for ICESt1, <10−6 for ICESt2, and 9 × 10−3 for ICESt3 (20). However, treatment of the strains harboring ICESt2 or ICESt3 with MC (MIC/2) was also found to induce excision (data not shown). This suggests that DNA damage induced excision of the three ICEs.
In order to examine the arp1 function, two fragments containing the 5′ and 3′ ends of arp1 of ICESt1/ICESt2 were cloned in the thermosensitive vector pG+host9 (5, 17). The recombinant plasmid, carrying a very short deleted ORF, Δarp1, was introduced into S. thermophilus CNRZ368. A strain harboring the Δarp1 ORF instead of arp1 (CNRZ368 Δarp1) was obtained by two successive homologous recombination events.
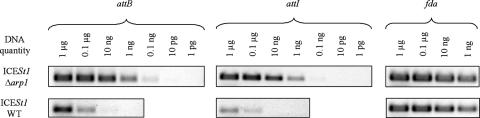
The effects of the arp1 deletion were determined by PCR amplification of fragments harboring attI and attB, using amounts of genomic DNA ranging from 1 μg to 1 ng for wild-type ICESt1 and from 1 μg to 1 pg for ICESt1 Δarp1 (Fig. 3). The minimum amounts of DNA that produced a positive result for the attB site were 10 ng of CNRZ368 template DNA and 10 pg of CNRZ368 Δarp1 template DNA. Using the same procedure, the minimum amounts of DNA that produced a positive PCR result for the attI site were 0.1 μg of CNRZ368 DNA and 0.1 ng of CNRZ368Δarp1 DNA. The arp1 deletion did not induce an increase in the PCR signal intensity when the fda gene was used as a control. Thus, deletion of the arp1 gene resulted in at least a 1,000-fold increase in the concentration of ICESt1 attB and attI sites. Furthermore, whereas ICESt1 and ICESt2 were excised at different frequencies, the concentrations of a fragment carrying the attI′ site from wild-type ICESt2 or ICESt2 Δarp1 (Fig. 1A) increased by a factor similar to that observed for ICESt1 (data not shown). Therefore, the arp1 gene repressed the excision of ICESt1 and ICESt2.
FIG. 3.
Effects of arp1 deletion on the excision frequency of ICESt1. Fragments containing the attB (361 bp) and attI (400 bp) recombination sites from the strains harboring wild-type ICESt1 (ICESt1 WT) or ICESt1 Δarp1 were amplified by PCR using primers described in the legend to Fig. 1 and amounts of template DNA ranging from 1 μg to 1 pg for CNRZ368 Δarp1 and from 1 μg to 1 ng for CNRZ368. An internal fragment of the fda gene (339 bp) was amplified as a control.
To validate the assumption that Arp1 autoproteolysis is involved in the induction of ICE excision by MC, exponentially growing cells of CNRZ368 Δarp1 were treated with MC at a concentration of 0.1 μg/ml (MIC/2). In three replicate experiments, the minimal amount of DNA that allowed detection of attB amplification (1 ng) was the same in the presence and in the absence of MC. Furthermore, in these three replicate experiments, the minimal amounts of DNA that allowed detection of attI amplification were 0.1 ng or 1 ng in the absence of MC and 0.1 ng after treatment with MC. Therefore, the inducibility of ICESt1 Δarp1 excision by MC was reduced or suppressed compared to the results for the same MC treatment in the wild-type strain. This suggests that MC treatment alleviates the repression of ICESt1 excision mediated by Arp1. The minimal amount of DNA that allowed detection of attI′ amplification (1 ng) was the same in the presence and in the absence of MC. This suggests that MC treatment also alleviates the repression of ICESt2 excision mediated by Arp1. In the same way, MC treatment derepressed the excision and transfer of another ICE, SXT from V. cholerae, probably by promoting the autocleavage of a cI homologue encoded by the element (3).
The functions of the other genes harbored by the ICESt1/ICESt2 and ICESt3 regulation modules (Fig. 1B) remain unknown. Nevertheless, the regulation modules of numerous Firmicutes prophages, such as TP901-1 from Lactococcus lactis (16), and of another ICE, ICEBs1 from B. subtilis (1), encode an OrfQ homologue and a “cI-like” repressor (i.e., a repressor related to Arp2), while none of these elements encode a genuine homologue of the cI repressor. OrfQ and the OrfQ homologues have a DUF955 domain that has a conserved H-E-X-X-H motif, suggesting that these proteins could be Zn2+ metalloproteinases (21). However, the activity of these proteins has not been demonstrated. Whereas the “cI-like” proteins encoded by prophages repress their lytic growth, these proteins are shorter than genuine cI homologues. Indeed, the cI-like repressors contain an HTH DNA binding domain but lack a region harboring autoproteolytic and oligomerization functions related to the genuine cI. However, the TP901-1 “cI-like” repressor binds to operators as dimers and higher multimers (14). Furthermore, the excision of prophages with cI-like genes and orfQ homologues, such as φSfi21 from S. thermophilus (7) or TP901-1 (16), is inducible by MC, and the prophage TP901-1 cI-like gene is involved in the lytic phase induction pathway. Moreover, the induction of the prophage TP901-1 (16) and of ICEBs1 by MC (1) is RecA dependent. Thus, arp2 and orfQ might also be involved in the derepression pathway of the excision of the S. thermophilus ICEs.
To our knowledge, ICESt1/ICESt2 and ICESt3 are the only integrative elements (i.e., phages, ICEs, or related elements) that encode a genuine cI homologue and might encode a cI-like homologue. Since MC induces the conjugative transfer of SXT, an ICE from V. cholerae coding for a genuine cI (3), and of ICEBs1, an ICE from B. subtilis coding for a cI-like repressor (1), DNA damage could regulate not only the excision but also the conjugative transfer of the ICEs from S. thermophilus.
Footnotes
Published ahead of print on 17 November 2006.
REFERENCES
- 1.Auchtung, J. M., C. A. Lee, R. E. Monson, A. P. Lehman, and A. D. Grossman. 2005. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc. Natl. Acad. Sci. USA 102:12554-12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacic, M., A. C. Parker, J. Stagg, H. P. Whitley, W. G. Wells, L. A. Jacob, and C. J. Smith. 2005. Genetic and structural analysis of the Bacteroides conjugative transposon CTn341. J. Bacteriol. 187:2858-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72-74. [DOI] [PubMed] [Google Scholar]
- 4.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 5.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonheyo, G. T., B. D. Hund, N. B. Shoemaker, and A. A. Salyers. 2001. Transfer region of a Bacteroides conjugative transposon contains regulatory as well as structural genes. Plasmid 46:202-209. [DOI] [PubMed] [Google Scholar]
- 7.Brüssow, H., and A. Bruttin. 1995. Characterization of a temperate Streptococcus thermophilus bacteriophage and its genetic relationship with lytic phages. Virology 212:632-640. [DOI] [PubMed] [Google Scholar]
- 8.Burrus, V., G. Pavlovic, B. Decaris, and G. Guédon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 9.Burrus, V., G. Pavlovic, B. Decaris, and G. Guédon. 2002. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 48:77. [DOI] [PubMed] [Google Scholar]
- 10.Burrus, V., Y. Roussel, B. Decaris, and G. Guédon. 2000. Characterization of a novel integrative element, ICESt1, in the lactic acid bacterium Streptococcus thermophilus. Appl. Environ. Microbiol. 66:1749-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clewell, D. B., D. D. Jaworski, S. E. Flannagan, L. A. Zitzow, and Y. A. Su. 1995. The conjugative transposon Tn916 of Enterococcus faecalis: structural analysis and some key factors involved in movement. Dev. Biol. Stand. 85:11-17. [PubMed] [Google Scholar]
- 12.Dodd, I. B., K. E. Shearwin, A. J. Perkins, T. Burr, A. Hochschild, and J. B. Egan. 2004. Cooperativity in long-range gene regulation by the lambda CI repressor. Genes Dev. 18:344-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaillard, M., T. Vallaeys, F. J. Vorholter, M. Minoia, C. Werlen, V. Sentchilo, A. Puhler, and J. R. van der Meer. 2006. The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J. Bacteriol. 188:1999-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansen, A. H., L. Brondsted, and K. Hammer. 2003. Identification of operator sites of the CI repressor of phage TP901-1: evolutionary link to other phages. Virology 311:144-156. [DOI] [PubMed] [Google Scholar]
- 15.Larbig, K. D., A. Christmann, A. Johann, J. Klockgether, T. Hartsch, R. Merkl, L. Wiehlmann, H. J. Fritz, and B. Tummler. 2002. Gene islands integrated into tRNAGly genes confer genome diversity on a Pseudomonas aeruginosa clone. J. Bacteriol. 184:6665-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madsen, P. L., A. H. Johansen, K. Hammer, and L. Brondsted. 1999. The genetic switch regulating activity of early promoters of the temperate lactococcal bacteriophage TP901-1. J. Bacteriol. 181:7430-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maguin, E., H. Prévost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGrath, B. M., J. A. O'Halloran, and J. T. Pembroke. 2005. Pre-exposure to UV irradiation increases the transfer frequency of the IncJ conjugative transposon-like elements R391, R392, R705, R706, R997 and pMERPH and is recA+ dependent. FEMS Microbiol. Lett. 243:461-465. [DOI] [PubMed] [Google Scholar]
- 19.Mohd-Zain, Z., S. L. Turner, A. M. Cerdeno-Tarraga, A. K. Lilley, T. J. Inzana, A. J. Duncan, R. M. Harding, D. W. Hood, T. E. Peto, and D. W. Crook. 2004. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J. Bacteriol. 186:8114-8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pavlovic, G., V. Burrus, B. Gintz, B. Decaris, and G. Guédon. 2004. Evolution of genomic islands by deletion and tandem accretion by site-specific recombination: ICESt1-related elements from Streptococcus thermophilus. Microbiology 150:759-774. [DOI] [PubMed] [Google Scholar]
- 21.Rawlings, N. D., and A. J. Barrett. 1995. Evolutionary families of metallopeptidases. Methods Enzymol. 248:183-228. [DOI] [PubMed] [Google Scholar]
- 22.Sentchilo, V., R. Ravatn, C. Werlen, A. J. Zehnder, and J. R. van der Meer. 2003. Unusual integrase gene expression on the clc genomic island in Pseudomonas sp. strain B13. J. Bacteriol. 185:4530-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sezonov, G., C. Possoz, A. Friedmann, J. L. Pernodet, and M. Guérineau. 2000. KorSA from the Streptomyces integrative element pSAM2 is a central transcriptional repressor: target genes and binding sites. J. Bacteriol. 182:1243-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stingele, F., and B. Mollet. 1996. Disruption of the gene encoding penicillin-binding protein 2b (pbp2b) causes altered cell morphology and cease in exopolysaccharide production in Streptococcus thermophilus Sfi6. Mol. Microbiol. 22:357-366. [DOI] [PubMed] [Google Scholar]
- 25.Toussaint, A., C. Merlin, S. Monchy, M. A. Benotmane, R. Leplae, M. Mergeay, and D. Springael. 2003. The biphenyl- and 4-chlorobiphenyl-catabolic transposon Tn4371, a member of a new family of genomic islands related to IncP and Ti plasmids. Appl. Environ. Microbiol. 69:4837-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Meer, J. R., and V. Sentchilo. 2003. Genomic islands and the evolution of catabolic pathways in bacteria. Curr. Opin. Biotechnol. 14:1-7. [DOI] [PubMed] [Google Scholar]