Abstract
To clarify the function of DivIVA in Streptococcus pneumoniae, we localized this protein in exponentially growing cells by both immunofluorescence microscopy and immunoelectron microscopy and found that S. pneumoniae DivIVA (DivIVASPN) had a unique localization profile: it was present simultaneously both as a ring at the division septum and as dots at the cell poles. Double-immunofluorescence analysis suggested that DivIVA is recruited to the septum at a later stage than FtsZ and is retained at the poles after cell separation. All the other cell division proteins that we tested were localized in the divIVA null mutant, although the percentage of cells having constricted Z rings was significantly reduced. In agreement with its localization profile and consistent with its coiled-coil nature, DivIVA interacted with itself and with a number of known or putative S. pneumoniae cell division proteins. Finally, a missense divIVA mutant, obtained by allelic replacement, allowed us to correlate, at the molecular level, the specific interactions and some of the facets of the divIVA mutant phenotype. Taken together, the results suggest that although the possibility of a direct role in chromosome segregation cannot be ruled out, DivIVA in S. pneumoniae seems to be primarily involved in the formation and maturation of the cell poles. The localization and the interaction properties of DivIVASPN raise the intriguing possibility that a common, MinCD-independent function evolved differently in the various host backgrounds.
A number of cell division proteins have been identified in Streptococcus pneumoniae and have been shown to localize at midcell to form the septal machinery (the septosome or divisome), consistent with what is known about the best-characterized rod-shaped model organisms, Escherichia coli and Bacillus subtilis (for recent reviews, see references 12, 16, 49, and 50).
These proteins include the cell division initiator proteins FtsZ and FtsA, which are required at the early stages of the process (25, 29, 32), and some of the later proteins, DivIB/FtsQ, DivIC/FtsB, FtsL, FtsW, PBP 2X, and PBP 1A (29, 32, 33, 38), which are the septal markers for S. pneumoniae cells. Recent studies have confirmed that, overall, the major events in septation are conserved in S. pneumoniae. However, other aspects related to the division process, such as the associated morphological changes, the correct choice of the division site, and proper chromosome segregation, and the factors that regulate these aspects remain largely unknown.
We have described characterization of a chromosome region in S. pneumoniae, downstream of the ftsZ gene, that is well conserved among gram-positive bacteria and is physically and transcriptionally related to the division and cell wall (dcw) cluster. We showed that functional inactivation of each of the five genes in the region resulted in defects in cell morphology, chromosome segregation, and/or cell division (13), and the importance of these genes in other species has been confirmed (18, 23, 30). In S. pneumoniae, the most dramatic effect on division was observed after inactivation of a gene designated divIVA, after its closest homolog in B. subtilis (13, 29).
Clear orthologs of DivIVA are found in a wide range of gram-positive bacteria and in other phylogenetically distinct species, including Deinococcus radiodurans and cyanobacteria. Multiple-sequence alignment of the DivIVA proteins has shown that despite great variation in length, the N-terminal part of DivIVA is significantly conserved, while the C-terminal part is much more varied. However, the part C-terminal contains predicted repetitive coiled-coil regions, supporting the notion that DivIVA is a coiled-coil protein (11).
Despite the significant degree of sequence similarity of the DivIVA proteins, studies of the physiological role of DivIVA in cell division and related processes have not revealed a unified function. Indeed, in B. subtilis DivIVA (DivIVABS) was proposed to be the equivalent of the missing MinE determinant and was shown to be involved in division site selection, attracting the MinCD cell division inhibitors away from midcell (3, 10, 28). Other studies have shown that DivIVABS also has a second, quite distinct function during sporulation, in which it is involved in chromosome segregation, attracting another complex that consists of the chromosome origin and the DNA-binding protein RacA together with Spo0J and Soj (1, 48, 51). In Synechococcus elongatus, which has both MinE and DivIVA, disruption of minE resulted in filamentation, while disruption of divIVA resulted in cells that were two or three times longer than the wild-type cells and exhibited reduced frequency and misplacement of the Z ring (30).
Gene inactivation or depletion in other species that lack MinC, MinD, and MinE homologs has revealed a variety of other phenotypes. In Streptomyces coelicolor and Brevibacterium glutamicum, DivIVA has been shown to be essential for growth, and both protein depletion and overexpression studies have shown that its function is related to polar growth and morphogenesis (14, 44). In S. pneumoniae, disruption of divIVA resulted in the formation of chains of unseparated, morphologically altered cells with incomplete septa, often devoid of nucleoids. This complex phenotype suggested that DivIVA has a role in cell shape, septum assembly, and completion, as well as chromosome segregation through an unknown mechanism (13). A similar phenotype was observed for Enterococcus faecalis when DivIVA was depleted (43). Finally, in Staphylococcus aureus, a mutant in which the divIVA gene was inactivated did not have any distinctive phenotype related to morphology, growth, chromosome partitioning, and division, suggesting that in this species DivIVA is dispensable (42).
In agreement with the proposed function(s) in the different host backgrounds, localization of DivIVA by immunofluorescence and/or by green fluorescent protein (GFP)-DivIVA fusion revealed that the protein localizes at the cell division site and is retained or localizes directly at the cell poles (10, 11, 14, 44), except in S. aureus, where it localizes only at the cell center (42).
An additional characteristic of DivIVABS is its ability to form oligomers in vitro (34, 46), and E. faecalis DivIVA (DivIVAEF) can self-interact in vivo (43). Nothing is known about the interaction(s) of DivIVA with proteins other than itself.
In this paper, we report the cytological and interaction properties of DivIVASPN. We found that in addition to its unique pattern of localization simultaneously at the cell center and the cell poles, DivIVASPN interacts with a number of different proteins and that some of these interactions are crucial for its function.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The S. pneumoniae Rx1 strain, a nonencapsulated well-characterized laboratory strain (45), was used in this study. Rx1 was routinely grown in tryptone soya broth (TSB) (Oxoid) or on tryptone soya agar (Oxoid) supplemented with 5% defibrinated sheep blood and incubated at 37°C in an atmosphere containing 5% CO2. When appropriate, chloramphenicol was added to the growth medium at a concentration of 4.5 μg/ml.
E. coli strain DH5α was used as a host for cloning, and E. coli strain BL21(DE3)AI (Invitrogen) was used as a host to overexpress the DivIVASPN fusion protein. Luria-Bertani (LB) broth and agar supplemented with ampicillin (150 μg/ml), when required, were used for routine growth of E. coli at 37°C.
E. coli strain R721 was used as a host for the two-hybrid assay. LB broth and agar were used for bacterial cultures and plating, and suspension medium was used for dilution as described previously (7, 8). Ampicillin (50 μg/ml), chloramphenicol (34 μg/ml), and kanamycin (30 μg/ml) were added when necessary.
Plasmid construction.
For cloning and expression purposes, the divIVASPN gene was amplified by PCR from S. pneumoniae Rx1 chromosomal DNA (13), using the spnF_divIVA and spnR_divIVA primers (Table 1); the only difference was the incorporated restriction sites used for cloning, BamHI (forward primer) and EcoRI (reverse primer). The amplified 848-bp DNA fragments were purified, double digested with BamHI and EcoRI (Roche Molecular Biochemicals, Mannheim, Germany), repurified, and ligated to the BamHI-EcoRI-digested pRSETA vector (Invitrogen). The ligation mixture was transformed into E. coli DH5α competent cells (Invitrogen), and transformants were selected after overnight growth on LB medium plates with ampicillin. Clones containing recombinant plasmid pRSETA_divIVASPN were verified by restriction analysis and DNA sequencing of the inserts and were used as sources of plasmid DNA to transform E. coli strain BL21(DE3)AI (Invitrogen) to produce the corresponding protein fused to a His tag at the N-terminal end (see below).
TABLE 1.
Cloning primers used in this studya
| Primer | Sequence | Product size (bp) |
|---|---|---|
| spnF_divIVA | ATGCCAATTACATCATTAGAA | 848 |
| spnR_divIVA | AGTTTCTGTAGCTGGTGTTGGAC | |
| spnF_ftsA | ATGGCTAGAGAAGGCTTTTTTACAGG | 1,442 |
| spnR_ftsA | CTTGAGCAGCAGCTGTATCAAATG | |
| spnF_ftsZ | ATGACATTTTCATTTGATACAGCTG | 1,360 |
| spnR_ftsZ | CCACTCTCTCGATGAGCACTCA | |
| spnF_zapA | GTGAATTTTATGGCAAATCTAAAT | 337 |
| spnR_zapA | AAGACCAATAGAAGAAGGAATGA | |
| spnF_ezrA | TTGTTTCAAGTAAAAAAGGAGTTT | 1,787 |
| spnR_ezrA | TGCTCCTCACACAATAAAATCTT | |
| spnF_ftsK | ATGGCAAACAAGAATACAAGTAC | 2,330 |
| spnR_ftsK | TCCAAACTTGGAAAGAAGCTATT | |
| spnF_ftsL | ATGGCAGAAAAAATGGAAAAAAC | 347 |
| spnR_ftsL | ACTCTTTTTGTCCACTTCATATC | |
| spnF_ftsB | ATGTCTAAAAATATTGTAC | 369 |
| spnR_ftsB | TCACCTTTGAATCAAGTC | |
| spnF_ftsQ | ATGTCAAAAGATAAGAAAAATGAG | 1,234 |
| spnR_ftsQ | TGGTCACTAAATAACTATAAGAGA | |
| spnF_ftsW | ATGAAGATTAGTAAGAGGCACTT | 1,260 |
| spnR_ftsW | TGAAGAGACATAAACTATCCTTT | |
| spnF_pbp2X | ATGAAGTGGACAAAAAGAGTAAT | 2,280 |
| spnR_ pbp2X | ATTCCAGCACTGATGGAAATAAA | |
| spnF_pbp1A | ATGAACAAACCAACGATTCTGCG | 2,189 |
| spnR_ pbp1A | ATCACCCAGAAAAATCTGGATGA | |
| spnF_pcsB | ATGAAGAAAAAAATCTTAGCGTCA | 1,206 |
| spnR_ pcsB | AGGGCTCTATTCGAGTCCCT | |
| spnF_spo0J | ATGGAAAAATTTGAAATGATT | 760 |
| spnR_ spo0J | TTATTTCAGGCTGTTGATAAT | |
| spnF_pfrA | ATGGTCAACTATCCACATA | 597 |
| spnR_ pfrA | TCATCTTGTTTTACCACC | |
| spnF_lytB | ATGAATTTAGGAGAATTTTGG | 2,166 |
| spnR_ lytB | CTAATCTTTGCCACCTAGC |
Additional bases, containing the appropriate restriction sites, were added to the sequences to allow cloning in the different plasmid vectors, as described in the text. The product size is total length, including linkers.
Similarly, recombinant plasmids pTTQ18(gfp_divIVA), pTTQ18(gfp_ftsZ), pTTQ18(gfp_spo0J), and pTTQ18(gfp_pfrA) were obtained by cloning the amplified DNA fragments of each corresponding gene in frame with the gfp gene from Aequorea victoria in the plasmid vector pTTQ18 (47), using the specific primers (Table 1) containing restriction sites for PstI and HindIII.
Finally, the recombinant plasmids used in the E. coli two-hybrid assays were constructed by cloning the genes of interest into the pcIP22 and pcI434 vectors (7). Each gene was amplified by PCR using the specific oligonucleotides shown in Table 1 carrying at the ends compatible restriction sites for SalI (or XhoI) (forward primer) and BamHI (reverse primer). The resulting recombinant plasmids were designated pcI434(x) and pcIP22(y), where x is the gene to be tested and y is divIVA or vice versa.
Purification of DivIVASPN-His6 and production of anti-DivIVASPN antibodies.
E. coli BL21(DE3)AI (Invitrogen) cells were used to overexpress the protein from plasmid pRSETA_divIVA. The pellet obtained from a 1.5-liter culture induced with 0.2% (wt/vol) arabinose for 3.5 h was disrupted as described above. Purification was performed with a Ni2+-charged column, using a His-Bind kit (Novagen) according to the manufacturer's instructions. The integrity and purity of the His-tagged DivIVA fusion were verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the fusion was quantified using the Bradford method (Bio-Rad).
Polyclonal antibodies against the purified DivIVASPN protein were obtained from Custom Hybridoma.com (PickCell Laboratories, Leiden, The Netherlands). No cross-reactions in any of the preimmune sera were observed with any S. pneumoniae or E. coli protein. The DivIVA protein was detected in streptococcal cells by Western blotting as previously described (25). S. pneumoniae cells were cultured in TSB, and the growth was monitored turbidimetrically until the optical density at 650 nm (OD650) was 0.45. Cultures (100 ml) were rapidly chilled in an ice bath, centrifuged with a Sorvall centrifuge (20,000 × g, 15 min, 4°C), washed once in 10 mM phosphate (pH 7.0), and resuspended in 1 ml of the same buffer. Cells were disrupted mechanically using glass beads and a Hybaid Ribolyser (Hybaid Ltd., Ashford, United Kingdom) according to the manufacturer's instructions. After cells were broken, they were mixed 1:1 with Laemmli buffer (Bio-Rad) and boiled for 5 min. Crude extracts were stored at −20°C until they were used. Proteins were separated on 12.5% Criterion precast gels (Bio-Rad) and transferred to a nitrocellulose membrane (Protran BA83; Schleicher & Schuell) at 100 V for 30 min with a Criterion Blotter apparatus (Bio-Rad). After transfer, the membranes were stained with Ponceau S (Sigma-Aldrich) and then incubated in 5% nonfat dry milk in PBST (phosphate-buffered saline [PBS] containing 0.05% Tween) overnight at 4°C. After blocking, membranes were incubated with an anti-DivIVA antibody solution (1:25,000) in PBST for 1 h at room temperature and, after washing, with horseradish peroxidase-conjugated antibodies (Bio-Rad). FtsA and FtsZ antibodies were used as previously described (25). Chemiluminescent bands were detected using an Immun-Star horseradish peroxidase chemiluminescence kit (Bio-Rad) and Kodak Biomax light film.
Cellular immunolocalization of the DivIVA protein.
For immunofluorescence, exponentially growing cells were fixed and treated as described by Lara et al. (25). Cells were then transferred onto poly-l-lysine-coated slides (Sigma). The slides were washed twice with PBS, air dried, dipped in methanol at −20°C for 10 min, and allowed to dry. After rehydration with PBS, the slides were blocked for 1 h at room temperature with 2% (wt/vol) bovine serum albumin (BSA) and 0.2% Triton X-100 (vol/vol) in PBS (BSA-PBST) and for 1 h with appropriate dilutions of anti-DivIVA or other antibodies in BSA-PBST. The slides were then washed five times with PBST and incubated for 30 min with a 1:500 dilution of anti-rabbit immunoglobulin (IgG) Alexa Fluor 488 (Invitrogen, Molecular Probes) or with a 1:500 dilution of anti-mouse IgG Alexa Fluor 488 or Alexa Fluor 594 in BSA-PBST. To check the specificity of the secondary antibodies, incubation with primary antibodies was omitted in negative controls. Preparations were finally stained with a fluorescence antifade solution containing propidium iodide (0.5 μg/ml) or 4′,6-diamidino-2-phenylindole (DAPI) (0.2 μg/ml) and 2% (wt/vol) 1,4-diazabicyclo[2.2.2] octane (DABCO), all obtained from Sigma. Slides were observed using a Zeiss Axioplan 2 equipped with a 100× Achroplan and fluorescence objective and standard filter sets (Zeiss no. 01, no. 09, no. 15, and no. 24). Photographs were taken with a Canon Powershot G6 digital camera, acquired with a Canon Zoom Browser, and processed with Adobe Photoshop 6.0.
A double-immunofluorescence analysis was performed as described by Morlot et al. (32), using polyclonal rabbit anti-DivIVA antibodies in combination with polyclonal mouse anti-FtsZ antibodies, kindly provided by T. Vernet.
For immunoelectron microscopy, S. pneumoniae Rx1 cells were cultured in TSB and growth was monitored turbidimetrically until the OD650 was 0.6. Cultures (4 ml) were rapidly chilled in an ice bath, centrifuged (10,000 × g, 15 min, 4°C), washed in 10 mM phosphate (pH 7.0), and fixed in 1 ml of a solution containing 2% paraformaldehyde and 0.05% glutaraldehyde in 1× PBS for 4 h at 4°C. The fixed cells were washed with PBS and stored at 4°C until they were used. Small pellets of fixed cells were cryoprotected with glycerol, applied to small pieces of filter paper, and quickly frozen in liquid ethane. Frozen specimens were transferred to a Reichert-Jung AFS freeze-substitution unit (Leica, Vienna, Austria) for 48 h at −90°C in a mixture of methanol and 0.5% (wt/vol) uranyl acetate for complete substitution of the water in the sample. After freeze-substitution, samples were infiltrated using Lowicryl K4M (EML Laboratories, Berkshire, United Kingdom) at 30°C and polymerized with UV light. Ultrathin sections of the samples were collected on 200-mesh gold grids covered with Formvar and carbon and processed for immunogold labeling as follows. After 40 min of blocking with Tris buffer-gelatin (TBG) (30 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% bovine serum albumin, 1% gelatin), sections were incubated for 1 h in TBG containing anti-DivIVA (1:750) and then washed with PBS. Next, grids were floated on three drops of TBG and incubated for 10 min on the last drop before a 45-min incubation with 10 nM gold-labeled goat anti-rabbit IgG (1:40) in TBG. The grids were then washed in PBS and distilled water before they were stained with saturated uranyl acetate for 30 min, followed by lead citrate for 1 min. Images were collected with a JEOL 1200-EXII electron microscope (Zeiss) operating at 100 kV. Electron micrographs were scanned using an Epson Stylus PHOTO 890 scanner and the Picture Publisher 8 software.
Bacterial two-hybrid assay.
All the different combinations of recombinant plasmids coding for the chimeric repressors, obtained as described above, were cotransformed into the recipient E. coli strain R721, and a β-galactosidase assay was performed as described previously (8). Bacterial cultures were grown at 34°C in Luria-Bertani medium supplemented with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to an OD600 of 0.5, and the residual β-galactosidase activity was evaluated for each strain. E. coli R721 without plasmids and E. coli R721 with plasmids pcI434434 and pcIP22434 were used as negative and positive controls, respectively. Plasmids pcIP22(divIVA)/pcI434434 and pcI434(divIVA)/pcIP22434 were used as additional negative controls to exclude self-interactions. Residual β-galactosidase activity that was less than 50% indicated that there was repression and hence a protein-protein interaction, whereas when the activity was greater than 50%, the interaction was uncertain or null. The use of these values has been discussed previously (8).
Coimmunoprecipitation.
Cultures of the E. coli DH5α strain containing the pairs of recombinant plasmids to be tested [pTTQ18(gfp) and p434(cI) derivatives] were grown overnight in LB medium supplemented with the appropriate antibiotic and then diluted 100-fold in fresh prewarmed medium. To induce expression, 0.1 mM (final concentration) IPTG was added to a culture, and the culture was incubated for 4 h. After this, the cells were centrifuged, washed and resuspended 1/300 (vol/vol) in lysis buffer (1 mM EDTA [pH 8], 25 mM HEPES [pH 7.6], 0.1 mg/ml lysozyme), and cooled on ice for 30 min. Membrane fractions were then prepared as described by Buddelmeijer and Beckwith (2). Soluble fractions were prepared by centrifuging a sample at 50,000 × g for 45 min following lysis and recovering the supernatant. Proteins concentrations were determined by the Bradford method.
The immunoprecipitation experiments were similar to those described by Duong and Wickner (9). The assays were performed using commercial antibodies, whose specificity was determined. The beads (protein A-Sepharose) were divided into 50- to 100-μl aliquots in microcentrifuge tubes, and 10 μl of anti-GFP mouse monoclonal antibodies (1:100; Santa Cruz Biotechnology, Inc.) was added to each tube and incubated for 15 to 60 min at room temperature with gentle mixing on a shaker. After centrifugation, samples were washed with 1 ml washing buffer (20 mM HEPES buffer [pH 7.5], 150 mM NaCl, 0.1% Triton X-100, 10% glycerol). Then 0.1 to 1 ml of cell lysate (corresponding to about 1 mg of protein) was added to each tube. Samples were incubated for 90 min to overnight at 4°C with gentle mixing on a shaker. Immunoprecipitated complexes were collected by centrifugation at 3,000 × g for 2 min at 4°C. Electrophoresis and Western blotting were performed as described above. The nitrocellulose membranes were probed with anti-cI rabbit polyclonal antibodies (1:6,000) and detected with an ECF Western blotting kit (Amersham Biosciences).
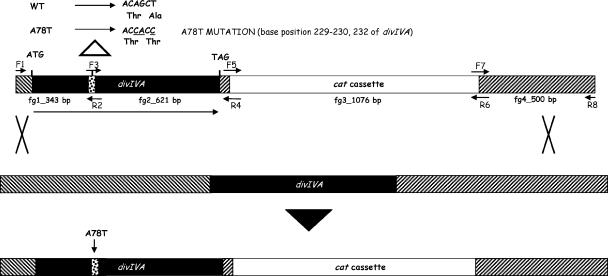
Allelic replacement mutagenesis.
S. pneumoniae genomic DNA isolated from strain Rx1 as previously described (13) was used as a template for PCR experiments. Constructs for allelic replacement were obtained by a two-step PCR method similar to the method described previously for gene insertion/deletion mutagenesis (13, 25). However, in these experiments four sets of primers (PF1-PR2, PF3-PR4, PF5-PR6, and PF7-PR8 [sequences available on request]) were used for each construct. The desired T→C mutation at nucleotide position 232, which resulted in an A78T substitution in the recombinant protein, was introduced into the PF3 primer. Two additional substitutions (A→ C and G→A at positions 229 and 230, respectively), resulting in silent changes, were also introduced in order to saturate and inhibit the mismatch repair system (4). PCR-derived fragments were purified using a QIAquick PCR purification kit (QIAGEN), mixed at a ratio of 1:1:1:1, and reamplified in a second PCR, using the external primers. Constructs for gene mutagenesis, obtained as described above, were used to transform S. pneumoniae Rx1 competent cells (13). Transformants were selected after 24 h of growth on tryptone soya blood agar plates containing chloramphenicol. The presence of the mutation was confirmed by PCR using specific primers, by a restriction digestion profile analysis of the PCR products using the PvuII restriction enzyme, and finally by sequence analysis of the entire constructs. As a control, a wild-type transformant containing the cat cassette inserted at same position was constructed by using the same procedure.
Growth studies, viability, and microscopy.
For physiological studies, bacterial cultures were inoculated from glycerol stocks into prewarmed TSB and incubated at 37°C in an atmosphere containing 5% CO2. Growth was monitored turbidimetrically at 650 nm every 30 min for 9 h with an Ultrospec 3100 (Amersham Pharmacia Biotech). Viable counts were determined every 90 min by serial dilution of the cultures and spot plating 20 μl on tryptone soya agar plates supplemented with 5% defibrinated sheep blood. At selected times during the exponential phase of growth, samples (100 μl) were transferred into Eppendorf tubes and fixed with 1% formaldehyde for 15 min at room temperature. Aliquots (10 μl) were transferred to poly-l-lysine-coated slides to which 2 μl of DAPI (0.5 μg/ml) was added. Samples were incubated for 5 to 10 min at room temperature in the dark before microscopic examination using a Zeiss Axioskop HBO 50 equipped with 100× phase-contrast and fluorescence objectives. To determine the presence of dead cells in the various strains, exponentially growing unfixed cells were washed and resuspended in 10 mM phosphate (pH 7.0) and then treated with propidium iodide (3 μg/ml) and DAPI (0.5 μg/ml). Cells were then incubated at room temperature for 15 min in the dark, transferred to poly-l-lysine-coated slides, and examined with the microscope as described above. Photographs were taken with a Zeiss MC100 Spot camera.
For scanning electron microscopy S. pneumoniae cells were cultured in TSB and growth was monitored turbidimetrically until the OD650 was 0.45. Cultures were rapidly chilled in an ice bath, centrifuged (10,000 × g, 15 min, 4°C), washed in 10 mM phosphate (pH 7.0), and fixed in 1% paraformaldehyde and 1.25% glutaraldehyde in 0.15 M sodium cacodylate buffer for 4 h at room temperature. Fixed cells were washed with PBS, transferred onto a circular cover glass, and dehydrated using increasing concentrations of acetone; this was followed by critical-point drying using CO2. Samples were then coated with platinum using an Emitech 575 turbo sputtering apparatus and examined with an FE Hitachi S4000 scanning electron microscope operating at 15 to 20 kV.
RESULTS
DivIVA localizes both at midcell and at the cell poles in S. pneumoniae.
To determine the DivIVA localization, we used immunostaining techniques that have been successfully employed to study cell division in S. pneumoniae (25, 32, 33). For this purpose, the divIVASPN gene was first cloned in the pRSETA vector (Invitrogen), and the corresponding protein was expressed in E. coli, purified as a His-tagged fusion protein, and used to generate polyclonal anti-DivIVASPN antibodies in rabbits. The antibodies were tested by Western blotting with crude extracts of the Rx1 wild-type strain and the isogenic divIVA null mutant (13) and were shown to be specific for DivIVA, which was detected as a single band in the crude extracts of the Rx1 wild-type strain but was completely absent in the corresponding extracts of the divIVA null mutant (data not shown).
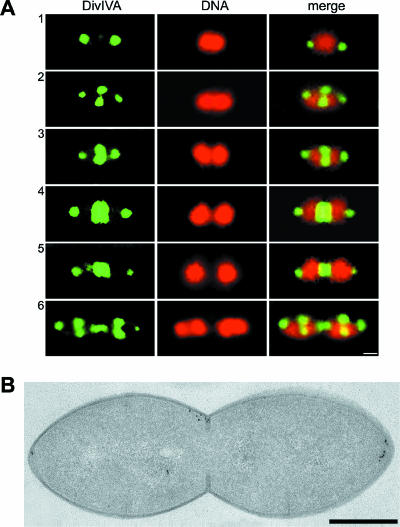
The subcellular localization of DivIVA in S. pneumoniae Rx1 cells was then examined by both immunofluorescence microscopy and immunoelectron microscopy using exponentially growing cells that were fixed and processed as previously described (25). Using both techniques, DivIVA was observed in more than 80% of the cells at the cell division septum and the cell poles simultaneously (Fig. 1).
FIG. 1.
Localization of the DivIVA protein in S. pneumoniae Rx1 cells. (A) Fluorescence micrographs showing the localization of DivIVA in representative cells at different stages of division, designated 1 to 6. Cells were stained to visualize DivIVA (green) and DNA (red); the merge images show the localization of DivIVA and DNA simultaneously. Scale bar = 0.25 μm. (B) Immunogold labeling of DivIVA. Exponentially growing cells were fixed and freeze-substituted. Thin sections were immunolabeled with specific antibodies and a gold-labeled secondary antibody and analyzed by electron microscopy. Dark grains show the localization of DivIVA. Scale bar = 0.25 μm.
The dynamics of the distribution of DivIVA was then monitored through the six main stages described for S. pneumoniae cells as they progress through the cell cycle (25, 32). Figure S1 in the supplemental material shows a representative immunofluorescence field, and cells in different stages (designated 1 to 6) are enlarged and arranged sequentially in Fig. 1A. In the 13% (49/372) of the cells in the first stage, representing newly generated single cells with a centrally located replicated chromosome that has just begun to segregate, DivIVA was observed predominantly as dots at both poles. In some cells at this stage, faint fluorescence with no clear pattern of localization was observed in the central region. In stages 2 to 5, as chromosome separation progressed, DivIVA was present at the center as an open or closed ring (stages 2 and 3, accounting for 29% [108/372] and 19% [71/372] of the cells, respectively) or as a band or disk (stages 4 and 5, accounting for 21% [77/372] and 12% [46/372] of the cells, respectively) and concurrently at the poles as dots that precisely marked the polar caps. Finally, in cells at stage 6, which had completed division but had not yet separated (6% [21/372] of the population), DivIVA was observed as dots at both old and newly formed poles and as an incipient ring at the new equatorial division sites of the future daughter cells. Interestingly, the DivIVA central band often had a doublet appearance, similar to the appearance of the B. subtilis band (19, 27). Moreover, as the septum closed and the central band progressively narrowed, the doublet appearance was clearly distinguishable as two dots, one for each separating newborn cell (Fig. 1A, stage 6).
Immunoelectron microscopy confirmed the pattern of localization observed by immunofluorescence microscopy. A representative dividing cell is shown in Fig. 1B. The distribution of the gold beads in the 360 cells examined showed that around 50% of the beads were at midcell and 24% were at the poles, primarily localized near the membrane. About 10% of the beads were outside the cells, and the remaining beads (around 16%) were observed in the cytoplasm, occasionally in a central position.
Differential localization and timing of DivIVA with respect to FtsZ during septation.
In dividing S. pneumoniae cells, FtsZ is targeted to the septum at early stages of cell division and remains there throughout the process (25, 32). Some cell division proteins, such as FtsA, exhibit the same pattern of localization (25; unpublished results), while others, such as PBP 2X, PBP 1A, FtsW, and DivIC, are targeted to the division site after FtsZ or, as is the case for DivIB/FtsQ and FtsL, are present transiently only during septation (32, 33, 38).
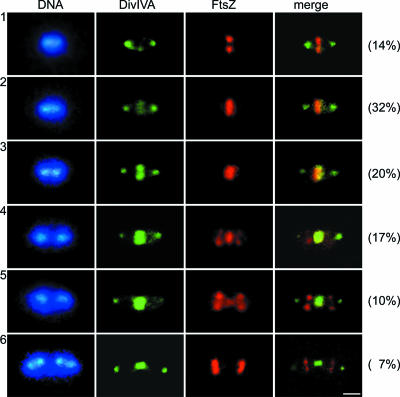
To determine the relative timing of assembly of DivIVASPN with respect to FtsZ during the cell cycle, we used double immunofluorescence with rabbit anti-DivIVA and mouse anti-FtsZ polyclonal antibodies, combined with DAPI nucleoid staining. Representative cells immunostained for FtsZ, DivIVA, FtsZ/DivIVA, and nucleoids are shown in Fig. 2. Six distinguishable patterns, in agreement with the patterns obtained previously for single localization, were observed. Immunocolocalization experiments showed that DivIVA is recruited to the septum at a later stage than FtsZ. Using the reconstruction of Morlot et al. (32) and assuming that all the cells were growing and dividing, the percentage in each stage was related to the duration of the phase. For cells growing with a generation time of 32 min, a delay of about 5 min was observed between FtsZ localization and DivIVA localization. Consistently, when FtsZ was already localized to the septum or relocalized to the equator of the daughter cells, DivIVA was still visible at the center and retained at the poles, and it was subsequently recruited to the future division sites only after the Z ring was assembled (Fig. 2).
FIG. 2.
Fluorescence micrographs showing the double localization of DivIVA and FtsZ in S. pneumoniae Rx1 cells at different stages of division, designated 1 to 6. Cells were stained to visualize DNA (blue), DivIVA (green), and FtsZ (red); the merge images show the localization of DivIVA and FtsZ simultaneously. The fluorescence micrographs are arranged to show the progression through the cell cycle. The percentages of cells in the different stages are indicated on the right. Scale bar = 0.5 μm.
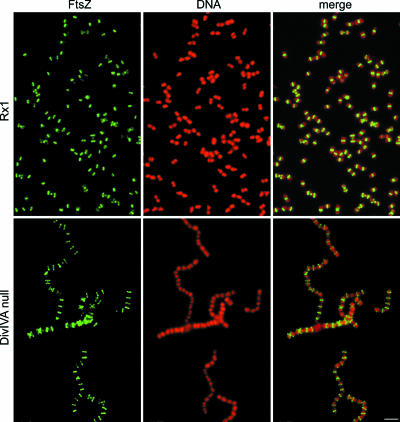
If, as indicated by the colocalization results, DivIVASPN is targeted to the septum at a later stage, FtsZ and FtsA and possibly also other cell division proteins should be localized in the absence of a functional DivIVA protein. To test this hypothesis, we analyzed the localization of FtsZ, FtsA, DivIC/FtsB, DivIB/FtsQ, FtsL, PBP 2X, and PBP 1A in the divIVA null mutant by immunofluorescence using the appropriate antibodies. The results for representative immunofluorescence fields for FtsZ in wild-type strain Rx1 and the divIVA null mutant are shown in Fig. 3, while the results for other proteins are shown in Fig. S2 and S3 in the supplemental material.
FIG. 3.
Fluorescence micrographs showing the localization of the FtsZ cell division protein in wild-type strain Rx1 and in the divIVA null mutant. Cells were stained to visualize FtsZ (green) and DNA (red); the merge images show the localization of DNA and proteins simultaneously. Scale bar = 1.5 μm.
As shown in Fig. 3, FtsZ was localized in the divIVA::cat mutant; however, the percentage of cells of the mutant having constricted Z rings was significantly reduced (P < 0.001) compared with the percentage observed for the wild-type (75/500 and 190/500, respectively), indicating that the cells were arrested at the rate-limiting step and supporting our previous finding that DivIVA is somehow needed for correct septum assembly and closure (13). The same could be said for the other cell division proteins, and it was even more striking for the proteins, such as FtsQ/DivIB and FtsL, that were previously found to be present at the septum only for a short time during the division process (38) (see Fig. S2 and S3 in the supplemental material).
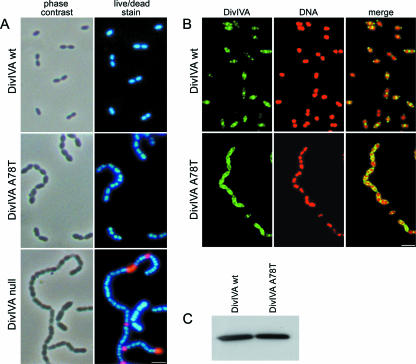
Unexpectedly, we observed no structurally intact anucleate cells in the immunofluorescence images of the divIVA null mutant, which we anticipated from a previous characterization of this mutant (13), suggesting that such cells could have been lost during the immunofluorescence procedure. This hypothesis was tested by performing a live/dead assay with unfixed, exponentially growing cells, which revealed that the anucleate cells observed in the divIVA null mutant (corresponding to 15% to 20% of the population) were indeed dead cells in the process of lysing (see below).
S. pneumoniae DivIVA protein interacts with itself and with other proteins of the septation machinery.
The septal localization of DivIVASPN suggested that the protein could interact with other components of the streptococcal division machinery; however, polar localization did not exclude the possibility that there were interactions with other proteins involved in chromosome segregation. To determine the interaction partners of DivIVASPN, we used a bacterial two-hybrid assay that is a powerful system for analyzing the protein-protein network of the E. coli components of the divisome (8).
Among the most prominent candidates, we tested the cell division proteins that are known or are thought to participate in divisome formation in S. pneumoniae, like FtsZ, FtsA, FtsK, FtsL, FtsQ/DivIVB, FtsB/DivIC, FtsW, PBP 2X, and PBP 1A (25, 32, 33, 38), as well as homologs of proteins that have been shown to have a role in division in other bacteria, like ZapA (17) and EzrA (26). In addition, we tested interactions with two other proteins, PcsB and LytB; PcsB is an essential putative murein hydrolase, whose precise function remains unclear (35, 36, 37), while LytB is a endo-β-N-acetylglucosaminidase involved in the separation of daughter cells (6). Finally, we examined the possible interactions with the homolog of Spo0J (15) and PfrA (SP1020/Spr0924), whose orthologs are involved in chromosome segregation in B. subtilis (22, 39).
The divIVASPN gene was fused in frame with the segment encoding the N-terminal portion of the phage 434 cI repressor (cI434-divIVA), while the various genes tested were fused in frame with the segment encoding the N-terminal portion of the phage P22 cI repressor (cIP22-y), as described in Materials and Methods. Pairs of the resulting recombinant plasmids, expressing the chimeric repressors, were then cotransformed into the reporter strain E. coli R721 carrying the lacZ gene under control of the hybrid 434-P22 promoter/operator, and β-galactosidase activity was measured as described previously (8).
As shown in Table 2, DivIVASPN was able to interact with itself. Moreover, high levels of interaction were observed with the cell division initiator proteins FtsZ, FtsA, and ZapA and also with FtsK and FtsL, as well as with the chromosome segregation protein Spo0J and the putative morphogenic determinant PcsB. Lower, but still significant, levels of interaction were detected with EzrA, DivIC/FtsB, DivIB/FtsQ, FtsW, and LytB. Finally, no interactions between DivIVA and PBP 2X, PBP 1A, or PrfA were detected.
TABLE 2.
Interactions of the DivIVASPN and DivIVASPN A78T proteins
| pcI22 protein | % of residual β-galactosidase activitya
|
|
|---|---|---|
| pcI434 DivIVA | pcI434 DivIVA A78T | |
| Itself | 7 | 10 |
| FtsZ | 15 | 5 |
| FtsA | 23 | 30 |
| ZapA | 7 | 8 |
| EzrA | 37 | 20 |
| FtsK | 27 | 55 |
| FtsL | 23 | 57 |
| FtsB/DivIC | 38 | 54 |
| FtsQ/DivIB | 30 | 64 |
| FtsW | 45 | 54 |
| PBP 2X | 58 | 60 |
| PBP 1A | 73 | 60 |
| PcsB | 14 | 6 |
| LytB | 34 | 67 |
| Spo0J | 16 | 25 |
| PrfA | 100 | 100 |
The assay was performed as described in Materials and Methods. The data are the averages of at least three independent determinations. The residual β-galactosidase activities of the positive and negative controls were 5% and 100%, respectively. A value less than 50% indicates that there was an interaction between the two proteins investigated. No interactions (95% of the residual β-galactosidase activity) were detected between pcI22_divIVA and an empty pcI434 vector. Similar results were obtained using the genes cloned in the reciprocal vectors (not shown).
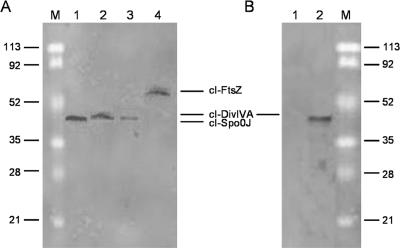
While localization data together with two-hybrid results should be sufficient to indicate that direct or indirect protein-protein interactions occur, in the absence of biochemical data, coimmunoprecipitation of putative interactive proteins has recently been performed to further corroborate the conclusion that the interactions observed are real. For this purpose, we selected two representative DivIVA-interacting proteins, FtsZ and Spo0J, and PfrA as a representative noninteractive protein and examined their abilities to coprecipitate in the presence of specific antibodies. The assay was performed by using the division proteins fused with the λ repressor (cI) or GFP, cloned and expressed in E. coli, and using commercial antibodies. Figure 4 shows the results of the coimmunoprecipitation experiments, which confirmed that there were interactions between FtsZ and DivIVA and between Spo0J and DivIVA (Fig. 4A) and that there were not interactions between DivIVA and PfrA (Fig. 4B), as detected by the two-hybrid assay. No cI-tagged proteins were detected in the pellets of control strains lacking GFP-tagged proteins (data not shown).
FIG. 4.
Immunoprecipitation to determine the specificity of DivIVA interactions. (A) Immunoprecipitation of GFP-DivIVA-cI-Spo0J complex (lane 1), GFP-Spo0J-cI-DivIVA complex (lane 2), GFP-FtsZ-cI-DivIVA complex (lane 3), and GFP-DivIVA-cI-FtsZ complex (lane 4). (B) Immunoprecipitation of cI-DivIVA-GFP-PrfA complex (lane 1). Protein extracts were immunoprecipitated with anti-GFP antibodies and were detected with anti-cI rabbit polyclonal antibodies by Western blotting as described in Materials and Methods. Lane M contained molecular weight markers. In panel B lane 2 contained crude extract of E. coli strain DH5α expressing cI-DivIVA, detected with anti-cI antibodies.
S. pneumoniae mutant carrying the A78T mutation is impaired in cell division but not in chromosome segregation.
Studies with B. subtilis showed that DivIVABS carrying an A78T allele had a phenotype similar to the null phenotype, affecting the site selection function of DivIVA (3, 10, 48). This residue, located in the hydrophobic core of the first predicted coiled-coil structure, is identical in the alignment of DivIVA proteins (11, 40), suggesting that it may also be important in S. pneumoniae and closely related bacteria. To test this hypothesis, we replaced, by allelic replacement, the divIVA wild-type gene with a gene containing the desired mutation, and constructs for gene mutagenesis were produced and transformed into S. pneumoniae competent cells, as shown in Fig. 5. Of 100 transformants, 4 that had a reduced growth rate and an altered phenotype as determined by phase-contrast microscopy and exhibited amplification by PCR using primers specific for the mutation were subjected to further analysis. The presence of the mutation in these mutants was confirmed by examining the restriction digestion profiles of the PCR products obtained using the PvuII restriction enzyme, which cut in the wild-type gene but not in the mutated gene, and by sequencing the entire construct. One of these mutants was chosen for further study. As a control, a wild-type transformant containing the cat cassette insertion at the same position but not the mutation was constructed using the same procedure (Fig. 5). Because the phenotype of this strain was indistinguishable from that of wild-type strain Rx1, we could effectively rule out the possibility that the cat cassette insertion had a polar effect.
FIG. 5.
Schematic diagram of the strategy used for allelic replacement mutagenesis and of the events that occurred after recombination via a double crossover. Four sets of primers were used to generate the final construct by a two-step PCR procedure as described in Materials and Methods. The resulting product was used to transform S. pneumoniae Rx1 competent cells, with selection for chloramphenicol resistance. As a result of the recombination event, in the transformants the cat cassette was inserted 40 bp after the end of divIVA. The correct gene replacement in the transformants was verified by PCR. Similarly, a transformant carrying a wild-type divIVA gene and the cat cassette inserted in same position was constructed as a control. WT, wild type.
The mutant Rx1 DivIVA A78T strain was characterized phenotypically with respect to growth, morphology, and nucleoid distribution, as well as for localization of the DivIVA protein in comparison with the controls, the wild-type Rx1 DivIVA and divIVA::cat strains. The results are shown in Fig. 6. The DivIVA A78T mutant had an intermediate phenotype compared to the DivIVA wild-type and divIVA null mutant strains; while the morphology of the cells was at least partially recovered, as observed by phase-contrast microscopy, and no anucleate or dead cells were detected (Fig. 6A), the chainy characteristics of the strain lacking a functional DivIVA were retained. The localization of DivIVA in the A78T mutant was altered compared with the localization in the wild type (Fig. 1 and 6B), and although it was visible at both the septum and the poles, bright diffuse fluorescence around the contour of the cells was clear. Western blot analysis showed that similar amounts of protein were present in the wild type and the A78T mutant, indicating that the altered localization in the latter was not due to a quantitative difference in the amount of the DivIVA protein (Fig. 6C).
FIG. 6.
Phenotypic characterization and localization of DivIVA in the wild-type DivIVA strain and the DivIVA A78T mutant. (A) Phase-contrast and live/dead stain fluorescence micrographs. (B) Fluorescence micrographs showing the localization of DivIVA. Cells were stained to visualize DNA (red) and DivIVA (green). The merge images show the localization of DNA and DivIVA simultaneously. Scale bar = 1.5 μm. (C) Western blot analysis of the DivIVA protein. WT, wild type.
Protein-protein interactions determined using the DivIVA A78T protein (Table 2) revealed that although the mutated protein was able to directly or indirectly interact with itself, with the early components of the divisome, like FtsZ, FtsA, EzrA, and ZapA, with PcsB, and with the putative chromosome segregation protein Spo0J, the interactions with FtsK and the so-called divisome late players, such as, FtsL, FtsQ, FtsB, FtsW, and LytB, appeared to be significantly reduced or lost.
DISCUSSION
The presence of DivIVA in dividing S. pneumoniae cells concurrently at the forming septa and the cell poles in a high percentage of the cells indicates that DivIVA is targeted to the septum during divisome assembly and is required there throughout cell septation and separation and that it is a marker for both the center and the poles. This localization differs from that of any other protein in S. pneumoniae described so far. In fact, in contrast to other streptococcal division proteins, which were shown to leave the center to initiate the next division cycle (25, 32) or to be only transiently visualized at one or both cell poles (33, 38) at the time that the septum initiated constriction, DivIVA appeared to split into symmetrically located disks that shrank to dots that were stably maintained at the newly formed poles upon completion of cell division and cell separation (Fig. 1A). In agreement with this, centrally located DivIVA bands with a doublet appearance were seen in the immunofluorescence fields. Similar doublets were reported previously for both vegetative B. subtilis cells and outgrowing spores when immunostaining with anti-DivIVABS antibodies was used but not when GFP-DivIVABS fusions were used and were interpreted to be a result of the lysozyme treatment (19, 27). However, streptococcal cells are much less sensitive to lysozyme than B. subtilis cells, and we obtained the same profile for cells prepared with and without lysozyme, suggesting that DivIVA localizes precisely on each side of the cytoplasmic membrane before septum completion and splitting.
Double-immunofluorescence analysis using anti-FtsZSPN antibodies and anti-DivIVASPN antibodies showed that DivIVA is recruited to the septum about 5 min (i.e., between one-sixth and one-seventh of the generation time) after FtsZ is recruited (Fig. 2). All the other cell division proteins that we tested were localized in the divIVA null mutant, indicating that they do not require DivIVA for localization and that, on the contrary, DivIVA may be dependent on them. However, proper placement of FtsZ and DivIVA in other cell division null mutants of S. pneumoniae, like Rx1 ftsK::cat and ftsQ::cat, which are viable (O. Massidda, unpublished results), suggests either that the formation of the Z ring alone is sufficient to target DivIVA to the septum and then to the new forming poles or that DivIVA is recruited to these sites independently. The presence of fluorescence at the division septa and, in particular, at the poles of E. coli cells expressing DivIVASPN-GFP (O. Massidda and P. Ghelardini, unpublished observations), as seen previously for DivIVBS-GFP (11), may be consistent with divisome-independent polar targeting, although the possibility of heterologous interactions with E. coli septal components cannot be excluded at this time.
While the localization profile observed for DivIVASPN was consistent with what was observed for the protein in B. subtilis, it was even more puzzling given the absence of both the MinC/MinD system and sporulation. On the one hand, septal localization strongly suggested that the protein could be necessary at the septum, raising the possibility that it interacts with other components of the streptococcal division machinery. On the other hand, polar localization did not allow us to exclude other functions, like chromosome segregation.
The ability of DivIVABS to oligomerize in vitro, forming elongated particles, the so-called “doggy-bone” structures, has been described previously (34, 46). Moreover, DivIVAEF was recently found to self-interact in vivo (43). In addition, it has been proposed that DivIVA could also interact with one or more components of the cell division machinery, as well as with other proteins (1, 11, 13, 34, 43, 51). Very recently, some interactive partners for DivIVABS have been determined (41). Therefore, as a step to further characterize DivIVASPN, we tested a number of different proteins using a two-hybrid assay.
As anticipated, DivIVASPN interacted with itself, indicating that it should at least dimerize efficiently. Moreover, in accordance with its localization profile and coiled-coil nature, DivIVASPN interacted directly or indirectly with a number of different proteins (Table 2). Although some interactions could be expected, we were surprised by the large number of them.
DivIVASPN interactions were detected with most components required at the early or late stage of septosome assembly but not with the septal peptidoglycan biosynthetic enzymes, PBP 2X and PBP 1A. However, DivIVASPN interacted with PcsB and LytB (6, 35, 36, 37), which were included because of their likely role in streptococcal cell division and because of the divIVA null phenotype. Inactivation of LytB resulted in chains of morphologically normal cells, consistent with the polar localization of this protein and its proposed function in daughter cell separation (6). In S. pneumoniae pcsB is essential, but reduced expression also results in chains of cells with abnormal morphology (36) that resemble the divIVA null cells remarkably. PcsB is a putative peptidoglycan hydrolase, conserved among streptococci, that is believed to be involved in cell growth and morphology, although an enzymatic activity on peptidoglycan has not been demonstrated yet (35, 36). Interestingly, LytB and PcsB are thought to cooperate in maintaining membrane and cell wall integrity and are positively regulated by the essential YycFG/VicRK two-component system (31, 35, 36, 37). We also found that DivIVASPN interacts with Spo0J, the only ortholog of the Spo0J/Soj/RacA complex of B. subtilis that could be identified in S. pneumoniae. Interactions between DivIVA and Spo0J were somewhat expected, given the chromosome segregation defects of both S. pneumoniae and E. faecalis that lacked divIVA or in which divIVA was depleted. Additionally, this interaction was postulated to occur in B. subtilis, certainly for the two proteins during sporulation and possibly also during vegetative growth (40, 51). Interestingly, Perry and Edwards have demonstrated that in B. subtilis DivIVABS coimmunoprecipitates with FtsZ and MinD and that a mutant allele, DivIVABSR18C, also coimmunoprecipitates with Spo0J (41).
To understand the biological meaning of the DivIVASPN interactions, we generated a missense isogenic S. pneumoniae DivIVA A78T mutant and compared it with the isogenic DivIVA wild-type strain in terms of growth, morphology, cellular localization, and interaction profile. The DivIVA A78T mutant was of particular interest since it has been shown that in B. subtilis this amino acid substitution correlates with cell division defects, possibly because it is located in the hydrophobic core of the first predicted coiled-coil domain (3, 10, 34). The DivIVA A78T mutant had an intermediate phenotype compared with the phenotypes of the DivIVA wild-type strain and the null mutant (Fig. 6), suggesting that DivIVA A78T is at least partially functional. Immunofluorescence revealed an altered localization profile, and although the protein was still visible at the septum and the poles, the majority was located diffusely around the cell. The two-hybrid assay showed that DivIVASPN A78T was able to self-interact, consistent with the finding of Muchova et al. (34) that DivIVABS A78T oligomerizes, and this suggests that oligomerization per se is not sufficient for the function of the protein.
Some of the interactions were also observed with the A78T protein, while others were significantly reduced or absent (Table 2), suggesting that they are crucial for proper function. The interaction profile of DivIVASPN A78T correlates well with the S. pneumoniae DivIVA A78T mutant phenotype. The DivIVA-PcsB and DivIVA-Spo0J interactions, both of which occurred in the A78T mutant, appear to be sufficient to reduce and eliminate, respectively, the cell morphology and nucleoid segregation defects evident in the null mutant. On the other hand, the DivIVA-LytB interactions, which did not occur with the mutated protein, could explain the chainy phenotype. Finally, while the interactions with the early cell division proteins may correlate with some localization of DivIVA A78T seen at the expected sites, the complete absence of interactions with the other division components that follows may be responsible for the abnormal localization of this mutant, although we cannot exclude other possible explanations.
In agreement with the original phenotype of the S. pneumoniae divIVA null mutant (13), these results indicate that DivIVA has a multifaceted role in controlling cell morphology and completion of cell division and separation, as well as chromosome segregation, through a complex interaction web. We speculate that DivIVA is a cytoplasmic cytoskeleton-like element that acts like a scaffold to direct cross wall formation and ultimately the formation of mature poles. Additionally, since this protein is stably present at the poles, it is not surprising that it may also cooperate with a putative segregation system that includes Spo0J to ensure proper chromosome partitioning in each daughter cell. Interestingly, a similar scaffolding role was postulated for DivIVA in S. coelicolor (14).
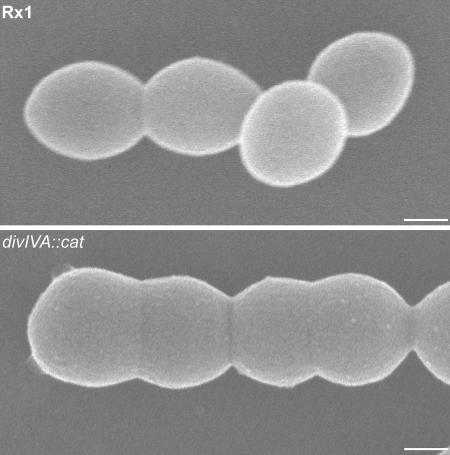
One possible model is that DivIVASPN arrives at the septum at some point during septation and assembles with the rest of the cell division proteins. Once there, DivIVA then ensures correct division through positioning the peptidoglycan hydrolytic enzymes necessary for septum splitting and possibly the late cell division proteins, determining the formation and the maturation of the distinctive pointed streptococcal poles. In the absence of DivIVA, this organization is lost, resulting in cells whose shape and diameter are altered, in which septum assembly and closure are delayed and (in some cells) chromosome segregation is impaired. In support of this is the fact that streptococci have rigid metabolically inert poles that are necessary to maintain cell shape (5) and the fact that the divIVA null cells form clearly defective poles (Fig. 7) (13). This view agrees well with the model for streptococcal growth and division of Higgins and Shockman (20) that predicts that in dividing cells, after initial inward growth of the cross wall, centripetal penetration remains relatively constant until the two newly synthesized internal hemispheres have reached the size of the external hemispheres. Moreover, in a subsequent study, the area of a streptococcal pole was found to be larger than the area expected for a pole formed from a flat, although complete, septum, suggesting that stretching alone is not sufficient to form a complete pole (21). This led to Koch's proposal that to form a prolate pole, like that in streptococci, new cell wall material should be added, in addition to stretching, in accordance with a “split and splay” model rather than the “split and stretch” model proposed for bacilli (24).
FIG. 7.
Scanning electron micrographs showing the morphological differences between wild-type Rx1 and Rx1 divIVA::cat cells. Note that the poles of the divIVA null mutant have an oblate rather than prolate shape, which correlated with the approximately 20% difference in cell diameter reported previously (13). Scale bar = 0.25 μm.
The proposed role of DivIVASPN is also consistent with its cellular localization and the role in polar growth proposed for DivIVA in phylogenetically distant bacteria that also lack the Min system (14, 44), raising the intriguing possibility of a common, MinCD-independent function that eventually evolved differently in the various host backgrounds. While other studies to clarify the complexity of the DivIVA function are in progress, this work provides new evidence about how this protein may work in streptococci and possibly also in a group of closely related bacteria.
Supplementary Material
Acknowledgments
This work was financed in part by EC project QLK3-CT-2000-00079 (SANITAS) within the Fifth Framework Program. Work in the laboratory of O.M. was also supported by ex-60% funds from MURST. A fellowship for Antonella Santona was funded by “Fondazione Banco di Sardegna.”
We thank Loredana Onidi for small-scale purification of the DivIVASPN protein and Stefania Collu for assistance with immunofluorescence. We thank A. Riva, Dipartimento di Citomorfologia, Università di Cagliari, for his help with the electron microscopy and Miguel Vicente for helpful comments. We thank Luciano Paolozzi for his constant support and advice. Finally, we thank Thierry Vernet for the generous gift of anti-FtsZ and other polyclonal antibodies and for lively discussions.
Footnotes
Published ahead of print on 10 November 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ben-Yehuda, S., D. Z. Rudner, and R. Losick. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299:532-536. [DOI] [PubMed] [Google Scholar]
- 2.Buddelmeijer, N., and J. Beckwith. 2004. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol. Microbiol. 52:1315-1327. [DOI] [PubMed] [Google Scholar]
- 3.Cha, J. H., and G. C. Stewart. 1997. The divIVA minicell locus of Bacillus subtilis. J. Bacteriol. 179:1671-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claverys, J. P., M. Prudhomme, I. Mortier-Barriere, and B. Martin. 2000. Adaptation to the environment: Streptococcus pneumoniae, a paradigm for recombination-mediated genetic plasticity? Mol. Microbiol. 35:251-259. [DOI] [PubMed] [Google Scholar]
- 5.Cole, R. M., and J. J. Hahn. 1962. Cell wall replication in Streptococcus pyogenes. Science 135:722-724. [DOI] [PubMed] [Google Scholar]
- 6.De Las Rivas, B., J. L. Garcia, R. Lopez, and P. Garcia. 2002. Purification and polar localization of pneumococcal LytB, a putative endo-beta-N-acetylglucosaminidase: the chain-dispersing murein hydrolase. J. Bacteriol. 184:4988-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Lallo, G., L. Castagnoli, P. Ghelardini, and L. Paolozzi. 2001. A two-hybrid system based on chimeric operator recognition for studying protein homo/heterodimerization in Escherichia coli. Microbiology 147:1651-1656. [DOI] [PubMed] [Google Scholar]
- 8.Di Lallo, G., M. Fagioli, D. Barionovi, P. Ghelardini, and L. Paolozzi. 2003. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 149:3353-3359. [DOI] [PubMed] [Google Scholar]
- 9.Duong, F., and W. Wickner. 1997. Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 16:2756-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards, D. H., and J. Errington. 1997. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol. 24:905-915. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, D. H., H. B. Thomaides, and J. Errington. 2000. Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J. 19:2719-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Errington, J., R. A. Daniel, and D.-J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fadda, D., C. Pischedda, F. Caldara, M. B. Whalen, D. Anderluzzi, E. Domenici, and O. Massidda. 2003. Characterization of divIVA and other genes located in the chromosomal region downstream of the dcw cluster in Streptococcus pneumoniae. J. Bacteriol. 185:6209-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flärdh, K. 2003. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol. Microbiol. 49:1523-1536. [DOI] [PubMed] [Google Scholar]
- 15.Gasc, A. M., P. Giammarinaro, B. Ton-Hoang, P. Geslin, M. van der Giezen, and M. Sicard. 1997. Structural organization of the Streptococcus pneumoniae chromosome and relatedness of penicillin-sensitive and -resistant strains in type 9V. Microb. Drug Resist. 3:65-72. [DOI] [PubMed] [Google Scholar]
- 16.Goehring, N. W., and J. Beckwith. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15:R514—R526. [DOI] [PubMed] [Google Scholar]
- 17.Gueiros-Filho, F. J., and R. Losick. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamoen, L. W., J. C. Meile, W. de Jong, P. Noirot, and J. Errington. 2006. SepF, a novel FtsZ-interacting protein required for a late step in cell division. Mol. Microbiol. 59:989-999. [DOI] [PubMed] [Google Scholar]
- 19.Harry, E. J., and P. J. Lewis. 2003. Early targeting of Min proteins to the cell poles in germinated spores of Bacillus subtilis: evidence for division apparatus-independent recruitment of Min proteins to the division site. Mol. Microbiol. 47:37-48. [DOI] [PubMed] [Google Scholar]
- 20.Higgins, M. L., and G. D. Shockman. 1970. Model for cell wall growth of Streptococcus faecalis. J. Bacteriol. 101:643-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgins, M. L., and G. D. Shockman. 1976. Study of cycle of cell wall assembly in Streptococcus faecalis by three-dimensional reconstructions of thin sections of cells. J. Bacteriol. 127:1346-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ireton, K., N. W. Gunther IV, and A. D. Grossman. 1994. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 176:5320-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishikawa, S., Y. Kawai, K. Hiramatsu, M. Kuwano, and N. Ogasawara. 2006. A new FtsZ-interacting protein, YlmF, complements the activity of FtsA during progression of cell division in Bacillus subtilis. Mol. Microbiol. 60:1364-1380. [DOI] [PubMed] [Google Scholar]
- 24.Koch, A. L. 1992. Differences in the formation of poles of Enterococcus and Bacillus. J. Theor. Biol. 154:205-217. [DOI] [PubMed] [Google Scholar]
- 25.Lara, B., A. I. Rico, S. Petruzzelli, A. Santona, J. Dumas, J. Biton, M. Vicente, J. Mingorance, and O. Massidda. 2005. Cell division in cocci: localization and properties of the Streptococcus pneumoniae FtsA protein. Mol. Microbiol. 55:699-711. [DOI] [PubMed] [Google Scholar]
- 26.Levin, P. A., I. G. Kurtser, and A. D. Grossman. 1999. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 96:9642-9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marston, A. L., H. B. Thomaides, D. H. Edwards, M. E. Sharpe, and J. Errington. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role of the mid-cell division site. Genes Dev. 12:3419-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marston, A. L., and J. Errington. 1999. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol. Microbiol. 33:84-96. [DOI] [PubMed] [Google Scholar]
- 29.Massidda, O., D. Anderluzzi, L. Friedli, and G. Feger. 1998. Unconventional organization of the division and cell wall gene cluster of Streptococcus pneumoniae. Microbiology 144:3069-3078. [DOI] [PubMed] [Google Scholar]
- 30.Miyagishima, S. Y., C. P. Wolk, and K. W. Osteryoung. 2005. Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol. Microbiol. 56:126-143. [DOI] [PubMed] [Google Scholar]
- 31.Mohedano, M. L., K. Overweg, A. de la Fuente, M. Reuter, S. Altabe, F. Mulholland, D. de Mendoza, P. Lopez, and J. M. Wells. 2005. Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane composition. J. Bacteriol. 187:2357-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morlot, C., A. Zapun, O. Dideberg, and T. Vernet. 2003. Growth and division of Streptococcus pneumoniae: localization of the high molecular weight penicillin-binding proteins during the cell cycle. Mol. Microbiol. 50:845-855. [DOI] [PubMed] [Google Scholar]
- 33.Morlot, C., M. Noirclerc-Savoye, A. Zapun, O. Dideberg, and T. Vernet. 2004. The D,D-carboxypeptidase PBP3 organizes the division process of Streptococcus pneumoniae. Mol. Microbiol. 51:1641-1648. [DOI] [PubMed] [Google Scholar]
- 34.Muchova, K., E. Kutejova, D. J. Scott, J. A. Brannigan, R. J. Lewis, A. J. Wilkinson, and I. Barak. 2002. Oligomerization of the Bacillus subtilis division protein DivIVA. Microbiology 148:807-813. [DOI] [PubMed] [Google Scholar]
- 35.Ng, W. L., G. T. Robertson, K. M. Kazmierczak, J. Zhao, R. Gilmour, and M. E. Winkler. 2003. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol. Microbiol. 50:1647-1663. [DOI] [PubMed] [Google Scholar]
- 36.Ng, W. L., K. M. Kazmierczak, and M. E. Winkler. 2004. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol. Microbiol. 53:1161-1175. [DOI] [PubMed] [Google Scholar]
- 37.Ng, W. L., H. C. Tsui, and M. E. Winkler. 2005. Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae. J. Bacteriol. 187:7444-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noirclerc-Savoye, M., A. Le Gouellec, C. Morlot, O. Dideberg, T. Vernet, and A. Zapun. 2005. In vitro reconstitution of a trimeric complex of DivIB, DivIC and FtsL, and their transient co-localization at the division site in Streptococcus pneumoniae. Mol. Microbiol. 55:413-424. [DOI] [PubMed] [Google Scholar]
- 39.Pedersen, L. B., and P. Setlow. 2000. Penicillin-binding protein-related factor A is required for proper chromosome segregation in Bacillus subtilis. J. Bacteriol. 182:1650-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perry, S. E., and D. H. Edwards. 2004. Identification of a polar targeting determinant for Bacillus subtilis DivIVA. Mol. Microbiol. 54:1237-1249. [DOI] [PubMed] [Google Scholar]
- 41.Perry, S. E., and D. H. Edwards. 2006. The Bacillus subtilis DivIVA protein has a sporulation-specific proximity to Spo0J. J. Bacteriol. 188:6039-6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinho, M. G., and J. A. Errington. 2004. A divIVA null mutant of Staphylococcus aureus undergoes normal cell division. FEMS Microbiol. Lett. 240:145-149. [DOI] [PubMed] [Google Scholar]
- 43.Ramirez-Arcos, S., M. Liao, S. Marthaler, M. Rigden, and J. A. Dillon. 2005. Enterococcus faecalis divIVA: an essential gene involved in cell division, cell growth and chromosome segregation. Microbiology 151:1381-1393. [DOI] [PubMed] [Google Scholar]
- 44.Ramos, A., M. P. Honrubia, N. Valbuena, J. Vaquera, L. M. Mateos, and J. A. Gil. 2003. Involvement of DivIVA in the morphology of the rod-shaped actinomycete Brevibacterium lactofermentum. Microbiology 149:3531-3542. [DOI] [PubMed] [Google Scholar]
- 45.Ravin, A. W. 1959. Reciprocal capsular transformations of pneumococci. J. Bacteriol. 77:296-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stahlberg, H., E. Kutejova, K. Muchova, M. Gregorini, A. Lustig, S. A. Muller, V. Olivieri, A. Engel, A. J. Wilkinson, and I. Barak. 2004. Oligomeric structure of the Bacillus subtilis cell division protein DivIVA determined by transmission electron microscopy. Mol. Microbiol. 52:1281-1290. [DOI] [PubMed] [Google Scholar]
- 47.Stark, M. J. 1987. Multicopy expression vectors carrying the lac repressor gene for regulated high-level expression of genes in Escherichia coli. Gene 51:255-267. [DOI] [PubMed] [Google Scholar]
- 48.Thomaides, H. B., M. Freeman, M. El Karoui, and J. Errington. 2001. Division site selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation. Genes Dev. 15:1662-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vicente, M., A. I. Rico, R. Martinez-Arteaga, and J. Mingorance. 2006. Septum enlightenment: assembly of bacterial division proteins. J. Bacteriol. 188:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiss, D. S. 2004. Bacterial cell division and the septal ring. Mol. Microbiol. 54:588-597. [DOI] [PubMed] [Google Scholar]
- 51.Wu, L. J., and J. Errington. 2003. RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol. Microbiol. 49:1463-1475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.