Abstract
CovR, the two-component response regulator of Streptococcus pyogenes (group A streptococcus [GAS]) directly or indirectly represses about 15% of the genome, including genes encoding many virulence factors and itself. Transcriptome analyses also showed that some genes are activated by CovR. We asked whether the regulation by CovR of one of these genes, dppA, the first gene in an operon encoding a dipeptide permease, is direct or indirect. Direct regulation by CovR was suggested by the presence of five CovR consensus binding sequences (CBs) near the putative promoter. In this study, we identified the 5′ end of the dppA transcript synthesized in vivo and showed that the start of dppA transcription in vitro is the same. We found that CovR binds specifically to the dppA promoter region (PdppA) in vitro with an affinity similar to that at which it binds to other CovR-regulated promoters. Disruption of any of the five CBs by a substitution of GG for TT inhibited CovR binding to that site in vitro, and binding at two of the CBs appeared cooperative. In vivo, CovR activation of transcription was not affected by individual mutations of any of the four CBs that we could study. This suggests that the binding sites are redundant in vivo. In vitro, CovR did not activate transcription from PdppA in experiments using purified GAS RNA polymerase and either linear or supercoiled DNA template. Therefore, we propose that in vivo, CovR may interfere with the binding of a repressor of PdppA.
Streptococcus pyogenes (group A streptococcus [GAS]) is an exclusively human pathogen that causes multiple diseases, both mild and severe. In some individuals, the serious poststreptococcal sequelae rheumatic fever and rheumatic heart disease or acute glomerulonephritis may occur. Mild GAS infections of the skin and throat include pharyngitis, impetigo, and scarlet fever, while more invasive life-threatening infections include necrotizing fasciitis and streptococcal toxic shock syndrome (7). Thus, GAS must be able to adapt to, and grow in, many different microenvironments within its human host. Since GAS encodes only one secondary sigma factor (σX) (43), and this is not expressed in laboratory culture (42), GAS must have alternate means to mount global changes in gene expression to adapt to environmental stresses.
Two-component signal transduction systems are often used by bacteria to sense and respond to alterations in environmental conditions, such as those encountered during infection by GAS (29, 50, 53). Typically, these systems consist of a membrane-bound sensor kinase and a cytoplasmic DNA-binding response regulator protein. The sensor kinase detects a change in the extracellular environment and responds by autophosphorylation. The phosphoryl group is then transferred by the kinase to the response regulator, resulting in enhanced DNA binding of the response regulator to target promoters for activation or repression of gene expression.
The two-component signal transduction system CovR/CovS (CsrR/CsrS) is a global regulator that controls the expression of about 15% of GAS genes (8, 20), including those encoding virulence factors that enable the organism to attach to host cells, tolerate stress conditions, and evade the immune system (8, 16, 27, 31). The importance of the CovR/CovS system in GAS pathogenesis is attested to by the findings that it is expressed in vivo in animal models of infection (5, 9, 55) and that its presence correlates with severe invasive disease in humans (54). Unlike most response regulators, CovR represses most of the genes that it controls, including its own expression (25). Regulation by CovR may be indirect, through a cascade involving an additional transcription factor (48), or direct, by binding at the regulated promoter. Direct regulation of transcription by CovR in vitro was demonstrated at the promoters for genes encoding has (capsule synthesis), sag (streptolysin S), covRS (18, 24, 25), and riv (48). Furthermore, in vivo, repression by CovR is reduced by substitution mutations in the CovR binding sites at the has and covRS promoters (17, 25).
The dppA promoter, PdppA, which is regulated positively by CovR, has five CovR consensus binding sequences (CBs) within 200 bp upstream of the first open reading frame, suggesting a direct interaction with CovR. Binding sites of transcriptional activators are usually located upstream of promoters to allow contact with RNA polymerase to increase the initiation of transcription (3). However, at PdppA, CovR binding sites are located both upstream and downstream of the putative promoter, which is similar to the location of CBs at promoters of genes repressed by CovR (18, 24, 25). Therefore, we wanted to investigate whether CovR binds PdppA directly to activate transcription and, if so, how CovR interacts differently at this promoter to increase, instead of decrease, transcription.
In this study, we confirmed that CovR increases dppA transcription in vivo. However, although CovR binds PdppA specifically in vitro at the predicted CBs, this binding alone does not stimulate transcription from the dppA promoter in vitro. It appears, therefore, that a factor is present in vivo that is absent from our in vitro transcription system. Thus, CovR activation at PdppA is unlike CovR repression at Phas, Pcov, Psag, and Priv, where no additional factor is required.
MATERIALS AND METHODS
Media and strains.
Escherichia coli K-12 strain DH5α was used for constructing all plasmids. All GAS strains are derivatives of the M6 serotype strain JRS4 (41). Escherichia coli strains were grown in LB broth (51), and GAS strains were grown at 37°C without agitation in Todd-Hewitt broth supplemented with 0.2% yeast extract.
Antibiotics were used at the following concentrations: ampicillin at 100 μg/ml for E. coli, kanamycin at 50 μg/ml for E. coli and 200 μg/ml for GAS, and spectinomycin at 100 μg/ml for both E. coli and GAS.
RPAs.
RNase protection assays (RPAs) were performed on late-log-phase RNA harvested from GAS as described previously (10). The dppA segment was amplified using primers dppA-M6-F and dppA-M6-R (all primers are listed in Table 1) to construct pEU7564. Labeling and preparation of an antisense RNA probe for dppA from the pEU7564 template were the same as those for the gusA probe (10).
TABLE 1.
Primers used in this study
| Primer | Sequence (5′-3′)a | Use |
|---|---|---|
| PdppA S1 BamHI | cggcgcggatccTGTATTAATGGCTATCTGAAG | PdppA-gusA transcriptional fusions and construction of pEU7535 |
| PdppA A2 PstI | atagaaaactgcagTGATATTCCCTTGATTGTGG | Construction of pEU7535 |
| PdppA A1 XhoI | gggccgctcgagCCCAGTCAAAAATAACGTGA | PdppA-gusA transcriptional fusions |
| PdppA PE3 | TGATAATAGAGAAGTATTTTAGG | Primer extension |
| PdppA S mut CB-1 | TGATCAGTTTCCTTCTTTTTTTAATCTAAAAT | PdppA CB-1 mutagenesis |
| PdppA A mut CB-1 | AAAAAAAGAAGGAAACTGATCACTAAGAAATA | PdppA CB-1 mutagenesis |
| PdppA S mut CB-2 | TTCTTTTTTTCCTCTAAAATAAGTCATGAAAC | PdppA CB-2 mutagenesis |
| PdppA A mut CB-2 | TTATTTTAGAGGAAAAAAAGAATTAAACTGAT | PdppA CB-2 mutagenesis |
| PdppA S mut CB-3 | CTTTTGTTCCTACATCAACTTATAAAAAAGCC | PdppA CB-3 mutagenesis |
| PdppA A mut CB-3 | GTTGATGTAGGAACAAAAGATAAAGAGGTTTC | PdppA CB-3 mutagenesis |
| PdppA S mut CB-4 | CAAAAATTCTCCTTAACCAGTTAAACAATTGC | PdppA CB-4 mutagenesis |
| PdppA A mut CB-4 | AACTGGTTAAGGAGAATTTTTGTTATTTTCAG | PdppA CB-4 mutagenesis |
| PdppA S mut CB-5 | TATTTATTAAGGAGAAATAAGGAGATTTGATTG | PdppA CB-5 mutagenesis |
| PdppA A mut CB-5 | CCTTATTTCTCCTTAATAAATAATAATAGTAAC | PdppA CB-5 mutagenesis |
| codY S1 EcoRI | atccggaattcGTAAAATCACATCTATTTTGC | codY insertional inactivation |
| codY A1 SacI | gaatcgcgagctcATCTAACTCTCCAAGAATTGC | codY insertional inactivation |
| PdppA-M6-F | CCCTCCCAACAAGCTATTTTTGTTCGTAACC | RPA probe construction |
| PdppA-M6-R | GGGAGGGGTGACATCATTTCCTACAGGATAACC | RPA probe construction |
| Ptrc99a-EcoRI | gggaattccGTAGCGCCGATGGTAGTG | Construction of pEU7535 |
| Ptrc99a-BamHI | gggggatccGCTTCCTCGCTCACTGAC | Construction of pEU7535 |
Uppercase letters indicate sequences derived from the genes of interest, lowercase letters indicate sequences added for cloning purposes, and underlined sequences indicate restriction sites.
Primer extension.
An antisense primer to the dppA open reading frame (PdppA PE3) was radiolabeled with [γ-32P]ATP (38) and used in a reverse transcription reaction of late-log-phase RNA isolated from JRS4. Primer extension was performed using the protocol for reverse transcriptase Superscript Enzyme II (Invitrogen).
Construction of pEU7535.
The PdppA promoter region was cloned upstream of transcriptional terminators in pEU2146, a derivative of pTrc99a (1). To construct pEU2146, an EcoRI- and BamHI-digested Prns promoter fragment was cloned into a 2.1-kb fragment of pTrc99a using primers Ptrc99a-EcoRI and Ptrc99a-BamHI (G. P. Munson and J. R. Scott, unpublished data). The Prns promoter was excised using enzymes BamHI and EcoRI. Primers PdppA S1 BamHI and PdppA A2 PstI were used to PCR amplify the dppA promoter region (positions −233 to +102 with respect to the dppA transcriptional start) from the JRS4 chromosome. The resulting PCR product was cloned into the BamHI and EcoRI sites of pEU2146 to construct pEU7535.
Mutation of CBs.
CBs at PdppA were mutated by substituting guanine residues for thymine residues in the ATTARA sequence by site-directed mutagenesis using complementary primer pairs (Table 1). Each sense strand primer containing the desired mutation was used with PdppA XhoI and each antisense primer containing the desired mutation was used with PdppA BamHI to amplify a segment of PdppA from the JRS4 chromosome. These PCR products subsequently served as templates in an overlapping PCR with the outside primers PdppA BamHI and PdppA XhoI to produce the mutated PdppA fragments (positions −233 to +102) with flanking BamHI and XhoI sites.
Construction of reporter fusions.
All PdppA-gusA reporter fusions were cloned upstream of the gusA reporter gene in pJRS462 (25), a suicide vector containing regions of homology to the VIT locus of RTG229 (19) for single-copy introduction into the chromosome of GAS. To construct the PdppA-gusA wild-type transcriptional fusion, primers PdppA S1 BamHI and PdppA A1 XhoI were used to amplify a 335-bp segment of PdppA (positions −233 to +102) from the JRS4 chromosome. This segment was cloned upstream of the gusA reporter gene in pJRS462 between the BamHI and XhoI sites to construct pEU7526. To construct the PdppA-gusA CB mutant fusions (with TT-to-GG substitutions), PCR products containing the mutated PdppA regions with flanking BamHI and XhoI sites (see above) were cloned into pJRS462 to create the following plasmids with the indicated promoter mutations (Table 2): pEU7524 (CB-1*), pEU7527 (CB-2*), pEU7531 (CB-3*), pEU7525 (CB-4*), and pEU7528 (CB-5*). The resulting GAS strains are listed in Table 2.
TABLE 2.
PdppA-gusA transcriptional fusions in E. coli and GAS and locations of consensus sequence mutations
| PdppA-gusA fusion | Location of TT-to-GG mutationa | Plasmidb | GAS strainc
|
|
|---|---|---|---|---|
| CovR+ | CovR− | |||
| Wild type | pEU7526 | JRS675 | JRS679 | |
| CB-1* | −158, −157 | pEU7524 | JRS661 | JRS671 |
| CB-2* | −146, −145 | pEU7527 | JRS662 | JRS672 |
| CB-3* | −108, −107 | pEU7531 | JRS680 | JRS685 |
| CB-4* | −51, −52 | pEU7525 | JRS663 | JRS676 |
| CB-5* | +29, +30 | pEU7528 | JRS664 | JRS674 |
The location of thymine base pair substitutions is indicated with respect to the dppA transcriptional start.
PdppA wild-type and CB mutant promoter regions were cloned upstream of the gusA reporter in pJRS462 in E. coli.
PdppA-gusA fusions were integrated in a single copy into the chromosome of GAS strain RTG229 in CovR+ or CovR− backgrounds.
To construct the PdppA-gusA ΔCB-5 fusion, a region including the dppA promoter from positions −233 to +14 was amplified from the JRS4 chromosome using primers PdppA S1 BamHI and PdppA A3 XhoI. The resulting PCR product was cloned into the BamHI and XhoI sites of pJRS462 to construct pEU7540. The plasmid was linearized with XmnI, and the reporter fusion was introduced in a single copy into the chromosome as described above.
Deletion of the covR gene.
The covR gene in each GAS transcriptional fusion strain was replaced with a nonpolar spectinomycin resistance cassette (32) using a temperature-sensitive plasmid, pJRS651 (25).
Purification and phosphorylation of CovR.
Native CovR protein was purified from E. coli and phosphorylated with acetyl phosphate as described previously (18). For in vitro transcriptions, following phosphorylation, acetyl phosphate was removed by purification through MicroBioSpin columns (Bio-Rad). The protein concentration was determined using the Micro BCA protein assay kit (Pierce) standardized against bovine serum albumin.
DNase I protection assay.
Primers PdppA S1 BamHI and PdppA A1 XhoI were used to amplify the 335-bp PdppA segment (positions −233 to +102) of the JRS4 chromosome. DNase I protection assays with purified CovR were performed as described previously (17).
Gus assays.
GAS cultures were grown to late log phase in Todd-Hewitt broth supplemented with 0.2% yeast extract and assayed for specific β-glucuronidase (GusA) activity (15).
Purification of GAS RNA polymerase and major sigma factor.
RNA polymerase from GAS was purified as described previously (43). The major sigma factor from GAS (RpoD) was expressed in E. coli and purified as described previously (25).
In vitro transcription.
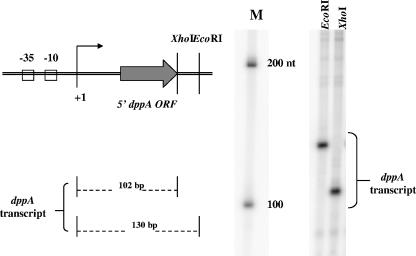
Transcription reactions were performed as described previously (18). To identify transcripts from the putative dppA promoter, pEU7526 (see reporter fusions) was digested separately at each of two sites (EcoRI and XhoI) downstream of the promoter so that transcript sizes could be predicted. Transcripts from the promoter were of the predicted size (∼130 bp and ∼102 bp for EcoRI and XhoI digests, respectively), indicating that the 5′ end of the transcript mapped by primer extension in vivo coincides with the dppA start of transcription in vitro.
Insertional inactivation of codY.
A 593-bp internal segment of the codY gene was amplified from the JRS4 chromosome using primers codY S1 EcoRI and codY A1 SacI and cloned between the EcoRI and SacI sites of the suicide vector pSK-Erm (42) to construct pEU7562. The plasmid was electroporated into JRS675 (PdppA-gusA reporter strain) and integrated into the chromosome by a single crossover. The presence of the integrated plasmid within the codY open reading frame was confirmed by PCR.
ISS1 mutagenesis.
Transposon mutagenesis using the temperature-sensitive plasmid pGhost9-ISS1 (33) was performed on JRS679 (Table 2), the isogenic covR derivative of the wild-type PdppA-gusA reporter strain. Survivors at the nonpermissive temperature were screened for increased transcription from the PdppA-gusA reporter by replica plating onto Thy plates spread with 5-bromo-4-chloro-3-indoxy-beta-d-glucuronide (X-glu; Gold BioTechnology, Inc.). Colonies with increased Gus activity (blue) were analyzed further.
RESULTS
CovR activates dppA expression in vivo.
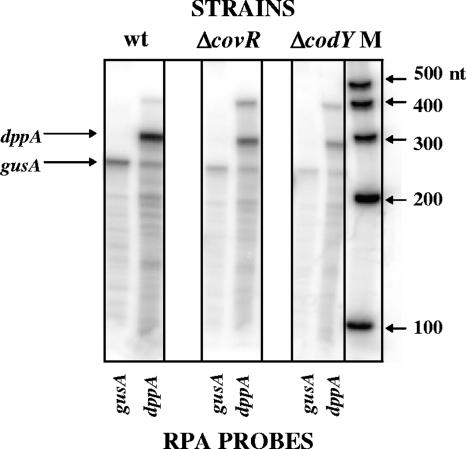
Microarray analyses showed that the amount of dppA transcript in a strain deleted for CovR is reduced about fivefold in late exponential phase in a serotype M1 GAS strain (20) and about fourfold in a serotype M6 strain (10). We confirmed these results with an RPA using radiolabeled antisense RNA probes to the dppA open reading frame and the gusA (β-glucuronidase) reporter gene fused to the dppA promoter region (Fig. 1). Controls for experiments described below using quantitative reverse transcription-PCR (data not shown) and reporter enzyme assays (see below) also corroborated these results.
FIG. 1.
CovR activates dppA expression in vivo. RPA was performed on RNA isolated from the PdppA-gusA transcriptional fusion parent strain (JRS675, designated wt) and derivative strains ΔcovR (JRS679) and ΔcodY (JRS692). RNA was isolated from late-log-phase cultures and probed with gusA and dppA antisense radiolabeled RNA. A ribonucleotide size marker (M) (Ambion) is shown to the right of the image. nt, nucleotides.
CovR binds PdppA in vitro.
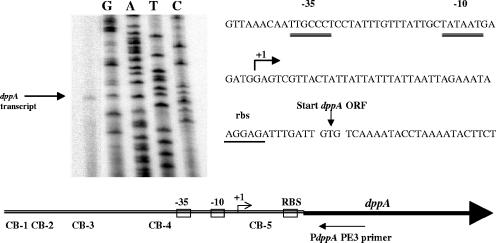
The 5′ end of the dppA transcript lies downstream of putative −35 and −10 promoter elements and 48 bp upstream of the dppA open reading frame, as shown by primer extension analysis of RNA isolated from JRS4 in late log phase (Fig. 2). The size of the transcript produced in vitro from PdppA using GAS RNA polymerase and GAS σ factor was consistent with this (Fig. 3).
FIG. 2.
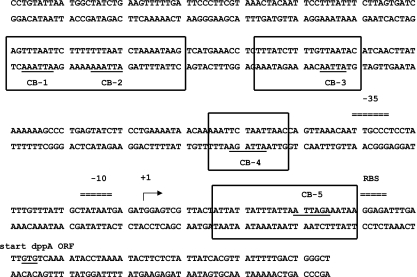
Primer extension to map the 5′ end of the dppA transcript. Antisense primer PdppA PE3 was annealed to RNA isolated from JRS4 in late log phase, and primer extension was performed using Superscript II enzyme (Invitrogen). The arrow (+1) indicates the 5′ end of the transcript. Predicted promoter elements (−35 and −10) and the ribosome binding site (rbs) are labeled. CovR consensus binding sequences (CB) are shown in relation to the start of dppA transcription. ORF, open reading frame.
FIG. 3.
Identification of the dppA transcript by in vitro transcription. In vitro transcription was performed using plasmid pEU7526 (containing the PdppA region from positions −233 to +102) linearized separately with two different enzymes (EcoRI and XhoI) at sites downstream of the 5′ end of the dppA transcript (+1). A ribonucleotide size marker (M) (Ambion) is shown to the right of the image. ORF, open reading frame. nt, nucleotides.
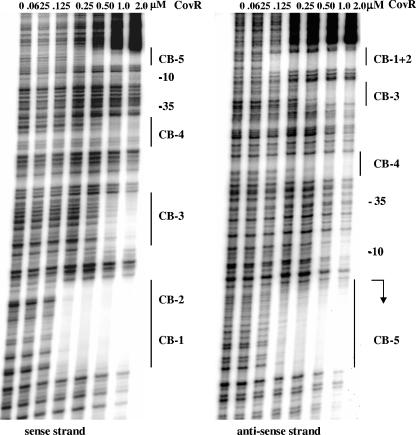
We found that CovR retarded the mobility of a 335-bp fragment (from positions −233 to +102) of the JRS4 chromosome that includes the dppA promoter (data not shown), demonstrating that CovR binds to this region. Binding of CovR was localized more precisely by DNase I protection assays. We found four regions of PdppA DNA, from positions −163 to +40, that were specifically protected by CovR (Fig. 4 and 5). Within each region of protected DNA, there is at least one ATTARA consensus or near-consensus binding sequence (CB). Binding at CB-3 and CB-4 requires a higher concentration of CovR than does binding to the other CBs, suggesting that the affinity of CovR for these sites is lower. The sequence ATTAAC, labeled CB-3, differs from the consensus by a single base, and at other promoters, Pcov and Psag, CovR was also shown to bind at near-consensus binding sequences (18, 25).
FIG. 4.
Protection by CovR at PdppA from DNase I. A 335-bp region including the dppA promoter was labeled and used as the DNA template. Regions of protection are indicated by solid lines, and CovR consensus binding sequences (CB) are labeled. The bent arrow indicates the start of dppA transcription.
FIG. 5.
CovR binding sites at PdppA. Regions of CovR protection at PdppA based on DNase I footprints shown in Fig. 4 are outlined. CovR consensus binding sequences (CB) are underlined. Promoter elements (−35 and −10) are indicated, and the bent arrow indicates the 5′ end of the dppA transcript.
In a DNase I protection assay of PdppA with phosphorylated CovR (incubated with acetyl phosphate), we found that phosphorylation increased the binding affinity of the protein to the CB-1 and CB-2 regions by twofold and the CB-5 region by at least threefold; however, the regions of DNA protected were the same (data not shown).
Mutation of CB sequences at PdppA disrupts binding of CovR in vitro.
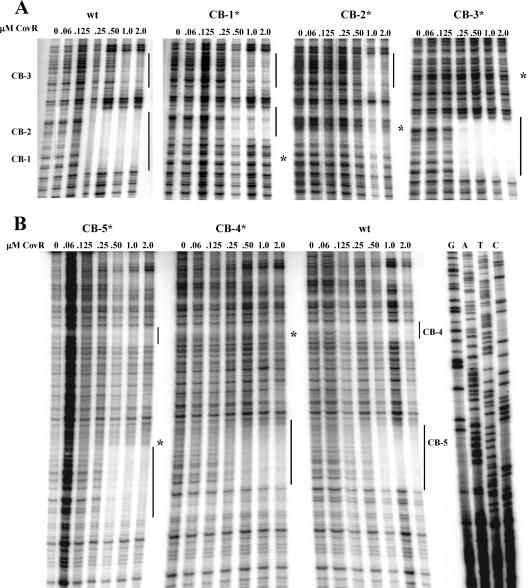
The thymine residues of CB sequence ATTARA are required for binding of CovR at Phas and Pcov (17, 25). To determine whether these residues are also required for CovR binding at PdppA, we substituted guanine residues for thymine residues at each CB by site-directed mutagenesis and used DNase I protection assays to evaluate the ability of CovR to bind to the mutated DNA. Within the region where the mutation was located, protection was reduced by the mutation of each CB sequence (Fig. 6), although the binding at the other protected sequences showed little if any effect of the mutation. The exception to this lack of cooperativity in binding occurred at CB-1 and CB-2, which are both located within the same protected region. In these cases, mutation of either CB prevented protection within the entire region (Fig. 6A). In addition to inhibiting binding in the protected region that included the mutation, the mutation at CB-5 also shifted the location of the protected region downstream from that protected on wild-type DNA (Fig. 6B). These results show that CovR binds specifically at PdppA in a manner similar to that at other promoters (17, 18, 25).
FIG. 6.
Binding of CovR to PdppA binding site mutant DNA. Shown is DNase I protection of wild-type PdppA (335-bp fragment) and each consensus binding site mutant DNA (CB-1* through CB-5*). Panel A shows the binding of CovR to binding site mutant DNA CB-1*, CB-2*, and CB-3* (sense strand), while panel B shows the binding of CovR to CB-4* and CB-5* DNA (antisense strand). Regions of protection are indicated by solid lines, and the location of each mutation (TT-to-GG substitution) is indicated to the right of each footprint by a large asterisk. The images are cropped to show the relevant data.
Effect of CB mutations on PdppA transcription in vivo.
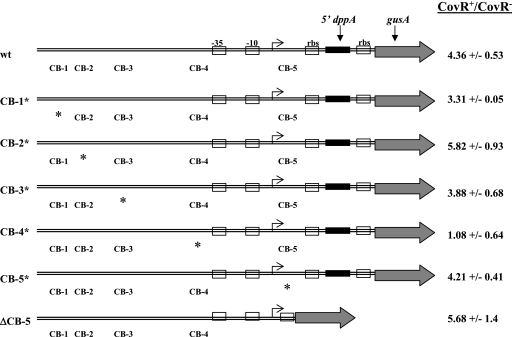
To determine whether CovR binding sites at PdppA are required for activation of the promoter in vivo, we constructed transcriptional fusions of the dppA promoter region (from positions −233 to +102 with respect to the start of transcription) to the gusA (β-glucuronidase) reporter gene. The wild-type promoter fusion (PdppA-gusA) as well as promoter fusions containing individual CB mutations (Table 2) were introduced in single a copy at an ectopic site into the GAS chromosome (see Materials and Methods), leaving the native dppA locus intact. Reporter activity (Gus activity) was measured in extracts of wild-type and isogenic covR strains harvested at late log phase.
We found that Gus activity from the wild-type promoter was about fourfold higher in the presence of CovR than in its absence (Fig. 7), consistent with previous results measuring dppA RNA (Fig. 1). CovR increased activity three- to fivefold from promoters mutated at CB-1, -2, -3, and -5, which is comparable to the amount of activation of the wild-type promoter (Fig. 7). Therefore, mutations at these CBs had little or no effect on the CovR activation of PdppA. Although no activation by CovR was detected from the CB-4 mutant promoter, the activity of this promoter approached the background level of detection for the assay (data not shown). Therefore, we are unable to rule out the possibility that the CB-4 mutation prevents transcription from PdppA, for example, by preventing binding of RNA polymerase.
FIG. 7.
CovR activation of wild-type and mutant PdppA-gusA fusions. Extracts of late-log-phase GAS cultures were harvested, and the specific activity of β-glucuronidase was measured in wild-type and isogenic CovR+ and CovR− derivatives of each strain. The ratio of the activity in a CovR+ strain divided by its activity in a CovR− strain is shown. The location of each binding site mutation (CB*) is indicated by an asterisk, the start of transcription is indicated by an arrow, and the −10 and −35 elements of the promoter are indicated by boxes.
Since the CB-5 mutation at PdppA did not fully disrupt the binding of CovR to this site in vitro (Fig. 6B) and was therefore unlikely to fully disrupt CovR binding in vivo, it remained possible that this binding site was required for PdppA activation. To evaluate this, we constructed a truncated PdppA-gusA fusion (from positions −233 to +14) that was deleted for the CB-5 sequence. This fusion, designated PdppA-gusA ΔCB-5, was introduced in a single copy into the GAS chromosome, as was done for the other fusions. Activity from the PdppA-gusA ΔCB-5 reporter in both wild-type and covR derivative strains was about twofold higher than the original PdppA-gusA fusion (data not shown). This may be due to the change in proximity of the promoter to the gusA ribosome binding site or to the deletion of a negative regulatory element located within the region omitted from the fusion. Despite the increase in reporter expression, activity from the PdppA-gusA ΔCB-5 fusion was about fivefold higher in the presence of CovR (Fig. 7), suggesting that CB-5 is not needed for CovR-dependent activation of PdppA.
CovR does not activate PdppA in vitro.
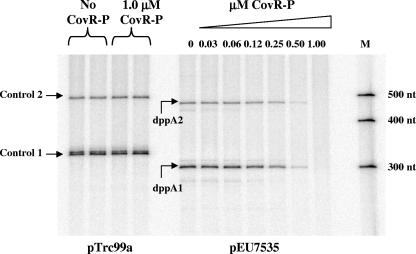
To determine whether phosphorylated CovR activates transcription from PdppA directly, we used an in vitro transcription system with purified GAS RNA polymerase (25) and a supercoiled DNA template. The 335-bp region including PdppA used in the DNA binding and reporter assays was cloned upstream of transcriptional terminators to produce pEU7535. As a control that should be unaffected by CovR, the Ptrc promoter in pTrc99a (1), which is also located upstream of transcriptional terminators, was used. The two transcripts produced from each promoter (PdppA and Ptrc) (Fig. 8) probably result from the inefficiency of termination of transcription by the first terminator, since they differ in size by the distance between the two terminators (175 bases). Unexpectedly, neither CovR (data not shown) nor CovR-P activated transcription from PdppA. Instead, at a concentration of CovR-P that had no effect on Ptrc, it repressed PdppA. Similarly, when PdppA was present on a linear template, CovR-P did not activate but repressed transcription (data not shown), although 50% repression of PdppA required twice the CovR-P concentration necessary to achieve 50% repression of Phas and Pcov on linear DNA templates (data not shown). Therefore, in the absence of other factors, CovR-P appears to repress (at high concentrations), not activate, transcription from PdppA in vitro using either a linear or supercoiled DNA template.
FIG. 8.
Effect of CovR-P on dppA expression in vitro. Transcripts from Ptrc (labeled Control 1 and Control 2) and PdppA (labeled dppA1 and dppA2) preincubated with or without CovR-P prior to the addition of RNA polymerase are shown. The DNA templates used are listed below the figure, and a ribonucleotide size marker (M) (Ambion) in nucleotides (nt) is shown to the right of the image.
CodY does not repress dppA expression in GAS.
The difference in the effect of CovR on PdppA in vivo and in vitro suggests that the in vitro system is missing a factor important for PdppA activation. Since CovR binds specifically to but does not activate PdppA in vitro, two simple explanations for the activation of PdppA in vivo by CovR are that a coactivator is required or that CovR interferes with the binding of a repressor at this promoter. In other bacteria, similar operons producing enzymes used to scavenge peptides from the environment are repressed by the pleiotropic repressor CodY. Both the dpp operon in Bacillus subtilis (37, 52) and the opp operon (oligopeptide transport system) in Lactococcus lactis (22) are repressed by CodY. Because GAS has a codY homolog in its genome, it seemed possible that CovR might interfere with CodY binding to the dppA promoter, thereby increasing dppA transcript levels.
To determine whether CodY represses dppA expression in GAS, we insertionally inactivated codY in the PdppA-gusA reporter strain (JRS675) and measured dppA and gusA transcript levels by RPA (Fig. 1). Unexpectedly, we found that in the absence of CodY, dppA and gusA expression levels were reduced eight- and fivefold, respectively, compared to that of the wild-type strain (Fig. 1). A Gus assay comparing activity in the wild type and codY isogenic strains confirmed the RPA result: PdppA activity in the codY strain was reduced fivefold (data not shown). This suggests that CodY does not repress PdppA in GAS but rather is required for wild-type expression from this promoter. Because the CodY mutation is polar, it is possible that the activation of PdppA might be due to a gene downstream of codY. However, the nearest downstream gene is a putative pyrazinamidase/nicotinamidase (SPy1776), which is unlikely to have this effect. After this work was completed (23), CodY activation of PdppA was reported in a different strain of GAS (34).
Isolation of mutants in a putative repressor or coactivator of dppA.
In an attempt to identify a putative regulator of PdppA, the covR deletion strain containing the PdppA-gusA transcriptional fusion at an ectopic location (JRS679) was subjected to ISS1 transposon mutagenesis (33). Of about 15,000 colonies containing the inserted transposon in 12 independent mutagenesis experiments, three independent mutants were found to have increased GusA activity. This is an unexpectedly low number for GAS, which has only about 2,000 genes. Quantitative reverse transcription-PCR demonstrated that there was at least a fivefold increase in transcription from PdppA for two of these mutants compared with that of the parental strain (data not shown). Sequence analysis of both Pdpp regions of each of the two mutants showed no changes from the wild-type Pdpp sequence. Because there was an increase in gusA activity from PdppA-gusA as well as an increase in transcription from PdppA at its native location in these mutants (data not shown), it is likely that both of these independently derived mutations affect a regulator of dppA expression. Unfortunately, we were unable to map the location of these mutations because the ISS1 element was not present in the chromosome of these strains, indicating that the phenotype resulted from a spontaneous mutation. The isolation of spontaneous mutations with increased activity of PdppA, accompanied by the failure to isolate insertion mutations with a similar phenotype, suggests that the regulator of PdppA in GAS is essential for growth.
DISCUSSION
Because they interact differently with RNA polymerase, most transcriptional activators bind to promoter regions of DNA at different locations from those to which transcriptional repressors bind (3). To recruit RNA polymerase to target promoters, most activators bind upstream of the core −35 and −10 promoter elements. For example, class I activators, which interact with the C-terminal domain of the α subunit of RNA polymerase, bind promoter DNA upstream of the −35 sequence (14). The precise location of these activator binding sites can vary due to flexibility in the linker region connecting the α C-terminal domain and α N-terminal domain subunit of RNA polymerase. Class II activators contact the σ70 subunit of the RNA polymerase holoenzyme by binding upstream of promoters at or near position −41.5 (13, 40). A third class of activators, which includes members of the MerR family, binds the spacer region between the −35 and −10 elements to activate transcription. These activators change the conformation of DNA at the promoter to shorten the distance between the −35 and −10 elements, thus promoting binding of the RNA polymerase holoenzyme (2, 28).
In contrast to transcriptional activators, whose binding sites are generally located upstream of promoter DNA, binding sites for transcriptional repressors usually overlap the −35 and −10 promoter elements and also may occur downstream of the start of transcription (3, 49). Repressors may inhibit transcription by sterically interfering with RNA polymerase binding to a promoter or by inhibiting later steps in the transcription process, including the transition of the RNA polymerase-promoter complex from closed to open, promoter clearance, and transcriptional elongation (49). Repressors may also bind at sites flanking the −35 and −10 promoter elements to create a DNA loop that prevents access of RNA polymerase (6).
The presence of CovR CBs at PdppA suggested that the activation of this promoter might be direct. In agreement with this, we found that CovR binds specifically to PdppA in vitro (Fig. 4), and, as at other CovR-regulated promoters, the thymine residues of the ATTARA consensus sequences are required for the CovR-PdppA interaction (Fig. 6). Consistent with binding of an activator protein, four CovR binding sites at PdppA are upstream of the −35 sequence, and CovR binding does not protect the −35 and −10 elements from DNase I digestion. However, CovR also protected a region (CB-5) downstream of the promoter in DNase footprints (Fig. 4). Usually, transcription factors that bind downstream of promoters act to repress transcription, although in E. coli, activation by the AraC family member Rns requires a site downstream of the start of transcription at Prns as well as upstream of the −35 element (39).
To determine the role of CB-5 and the other CBs at PdppA in vivo, we examined transcription from each mutated promoter at an ectopic chromosomal location. Individual mutations at CB-1, CB-2, CB-3, and CB-5 had no effect in vivo on the activation of PdppA by CovR. The CB-4 mutation, which is centered 18 bp upstream of the −35 sequence, reduced promoter activity to the background level, probably because it disrupts the binding of the α subunit of RNA polymerase. Therefore, we cannot determine whether binding at this site is required for CovR activation. That single CB mutations did not reduce CovR activation of PdppA in vivo suggests that their function is redundant. This differs from the binding of CovR at promoters that it represses, since at these promoters, binding to multiple sites is required to achieve full repression in vivo (17, 25).
Direct repression by CovR at Phas, Pcov, Psag, and Priv was demonstrated in vitro using a purified GAS transcription system (18, 24, 25, 48). Using the same transcription system, we were surprised to find that the addition of CovR to either a linear or supercoiled PdppA template did not result in activation (Fig. 8). Instead, transcription from PdppA was repressed in vitro by CovR-P (Fig. 8). The lack of direct activation of PdppA by purified CovR-P suggests that although it can bind to this promoter, CovR alone is unable to activate it. It appears that the in vitro system lacks a factor required for activation by CovR.
Although it is always possible that the salt conditions in the in vitro transcription system preclude the detection of CovR activation, there are several other possible explanations. One is that to activate transcription, CovR requires a cofactor or small molecule to alter its conformation so that it can interact with PdppA or RNA polymerase to facilitate, instead of inhibit, transcription. The best-known example of a transcriptional regulator that changes from a repressor to an activator on binding a small molecule is probably MerR. MerR binds between the −10 and −35 elements of the merTPCAD promoter to sequester the RNA polymerase holoenzyme and repress transcription. However, when HgII binds to MerR, the resulting conformation change in MerR brings these DNA regions into a position that facilitates RNA polymerase binding and leads to the activation of transcription (30). We do not favor this explanation, however, because CovR is a response regulator in the OmpR family. Although OmpR family proteins may require metal ions (as well as phosphorylation) to achieve the conformation optimal for binding to DNA, the lack of such ligands reduces their affinity for DNA but does not alter the contacts made between the protein and the DNA (26, 35). Therefore, if ligand binding by CovR converts it from a repressor to an activator, this would be unique among this well-studied class of response regulator proteins.
Our ability to isolate mutants in a covR deletion strain that showed increased expression of both the PdppA transcriptional reporter as well as the native dppA locus suggests that GAS produces a trans-acting protein that affects the transcription of PdppA. The simplest explanation for this is that CovR functions as an antirepressor by competing with a repressor for binding to sites at PdppA or by interfering with the interaction of the repressor with RNA polymerase. In support of the existence of a repressor for PdppA, we found very low expression in vivo of GusA from PdppA (about 25 units) compared to its expression from Pska (about 70 units) or Pcov (200 units) (data not shown), although the amount of transcript produced in vitro from linear templates was at least as great for PdppA as for each of these promoters (data not shown). The putative repressor of PdppA in GAS is not CodY (Fig. 1), although the CodY homolog in B. subtilis represses dppA expression in that organism (37, 52). It appears instead that the putative repressor is an essential protein, since we were not able to obtain transposon-mediated insertional mutations in the gene encoding it. It is possible that the presumed PdppA repressor might be a histone-like DNA-binding protein similar to H-NS of gram-negative bacteria, which binds promoters that it regulates as a multimer to alter the conformation of DNA and repress transcription (12, 47).
Although the mechanism of CovR activation at PdppA remains unclear, it is clear that in GAS, the dpp operon is regulated by more than one transcriptional regulator in response to more than one signal. Since GAS is auxotrophic for 13 of the 20 amino acids (11), GAS requires peptide permeases to use small peptides in the environment as a source of these essential amino acids. In GAS, unlike other lactic acid bacteria, dipeptides are taken up exclusively by the DppA-E system encoded in this operon, which suggests that this operon is of critical importance for GAS growth (44). At least three global regulatory proteins are involved in the control of transcription from PdppA: CovR (8, 20), Mga (44), and CodY (34) (Fig. 1). Each of these regulators responds to a different set of environmental signals. CovR responds to stress signals including changes in pH, temperature, and osmolarity (10) and may respond to extracellular Mg2+ concentrations (21); Mga responds to elevated CO2, temperature, and iron levels (4, 36, 45); and CodY responds to changes in nutritional status (34). With all of these signals feeding into dpp regulation, it is even possible that several regulatory proteins are present at the promoter simultaneously to control expression. The multiple regulatory circuits controlling PdppA suggest that the ability to use dipeptides in the environment is very important for growth of GAS under many different stress conditions.
This study increases our appreciation of the complexity of the CovR regulatory network. Previous work defined promoters that are directly repressed by CovR and require no other factors for this repression. We showed above that one promoter activated by CovR requires some additional factor whose availability is presumably subject to additional regulatory systems. Thus, the mechanism of activation at PdppA represents an additional way for CovR to provide regulatory input and to interact with other regulons. Several other GAS promoters are activated by CovR, and it will be interesting to learn whether this activation is direct or indirect and whether these promoters require additional regulatory factors that are the same or different from that required for the activation of PdppA. The complexity of these interacting regulatory networks in GAS has probably evolved to facilitate the growth of these bacteria under many different environmental conditions in the absence of alternative sigma factors.
Acknowledgments
We thank Jinxin Gao and Gordon Churchward for supplying purified CovR used in some of the experiments.
This work was funded by NIH grant AI20723. A.A.G. was supported in part by NIH training grant T32-AI07470 and a UNCF-Merck Graduate Science Research Dissertation Fellowship.
Footnotes
Published ahead of print on 22 September 2006.
REFERENCES
- 1.Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. [DOI] [PubMed] [Google Scholar]
- 2.Brown, N. L., J. V. Stoyanov, S. P. Kidd, and J. L. Hobman. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27:145-163. [DOI] [PubMed] [Google Scholar]
- 3.Browning, D. F., and S. J. Busby. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:57-65. [DOI] [PubMed] [Google Scholar]
- 4.Caparon, M. G., R. T. Geist, J. Perez-Casal, and J. R. Scott. 1992. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J. Bacteriol. 174:5693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho, K. H., and M. G. Caparon. 2005. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol. Microbiol. 57:1545-1556. [DOI] [PubMed] [Google Scholar]
- 6.Choy, H., and S. Adhya. 1996. Negative control, p. 1287-1299. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 7.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalton, T. L., J. T. Collins, T. C. Barnett, and J. R. Scott. 2006. RscA, a member of the MDR1 family of transporters, is repressed by CovR and required for growth of Streptococcus pyogenes under heat stress. J. Bacteriol. 188:77-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalton, T. L., R. I. Hobb, and J. R. Scott. 2006. Analysis of the role of CovR and CovS in the dissemination of Streptococcus pyogenes in invasive skin disease. Microb. Pathog. 40:221-227. [DOI] [PubMed] [Google Scholar]
- 10.Dalton, T. L., and J. R. Scott. 2004. CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes. J. Bacteriol. 186:3928-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, H. C., F. Karush, and J. H. Rudd. 1965. Effect of amino acids on steady-state growth of a group A hemolytic streptococcus. J. Bacteriol. 89:421-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorman, C. J. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391-400. [DOI] [PubMed] [Google Scholar]
- 13.Dove, S. L., S. A. Darst, and A. Hochschild. 2003. Region 4 of sigma as a target for transcription regulation. Mol. Microbiol. 48:863-874. [DOI] [PubMed] [Google Scholar]
- 14.Ebright, R. H. 1993. Transcription activation at class I CAP-dependent promoters. Mol. Microbiol. 8:797-802. [DOI] [PubMed] [Google Scholar]
- 15.Eichenbaum, Z., M. J. Federle, D. Marra, W. M. de Vos, O. P. Kuipers, M. Kleerebezem, and J. R. Scott. 1998. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl. Environ Microbiol. 64:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Federle, M. J., K. S. McIver, and J. R. Scott. 1999. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J. Bacteriol. 181:3649-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Federle, M. J., and J. R. Scott. 2002. Identification of binding sites for the group A streptococcal global regulator CovR. Mol. Microbiol. 43:1161-1172. [DOI] [PubMed] [Google Scholar]
- 18.Gao, J., A. A. Gusa, J. R. Scott, and G. Churchward. 2005. Binding of the global response regulator protein CovR to the sag promoter of Streptococcus pyogenes reveals a new mode of CovR-DNA interaction. J. Biol. Chem. 280:38948-38956. [DOI] [PubMed] [Google Scholar]
- 19.Geist, R. T., N. Okada, and M. G. Caparon. 1993. Analysis of Streptococcus pyogenes promoters by using novel Tn916-based shuttle vectors for the construction of transcriptional fusions to chloramphenicol acetyltransferase. J. Bacteriol. 175:7561-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham, M. R., L. M. Smoot, C. A. Migliaccio, K. Virtaneva, D. E. Sturdevant, S. F. Porcella, M. J. Federle, G. J. Adams, J. R. Scott, and J. M. Musser. 2002. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. USA 99:13855-13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gryllos, I., J. C. Levin, and M. R. Wessels. 2003. The CsrR/CsrS two-component system of group A Streptococcus responds to environmental Mg2+. Proc. Natl. Acad. Sci. USA 100:4227-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guedon, E., P. Serror, S. D. Ehrlich, P. Renault, and C. Delorme. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 40:1227-1239. [DOI] [PubMed] [Google Scholar]
- 23.Gusa, A. A. 2005. Ph.D. thesis. Emory University School of Medicine, Atlanta, GA.
- 24.Gusa, A. A., J. Gao, V. Stringer, G. Churchward, and J. R. Scott. 2006. Phosphorylation of the group A streptococcal CovR response regulator causes dimerization and promoter-specific recruitment by RNA polymerase. J. Bacteriol. 188:4620-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gusa, A. A., and J. R. Scott. 2005. The CovR response regulator of group A streptococcus (GAS) acts directly to repress its own promoter. Mol. Microbiol. 56:1195-1207. [DOI] [PubMed] [Google Scholar]
- 26.Head, C. G., A. Tardy, and L. J. Kenney. 1998. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J. Mol. Biol. 281:857-870. [DOI] [PubMed] [Google Scholar]
- 27.Heath, A., V. J. DiRita, N. L. Barg, and N. C. Engleberg. 1999. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect. Immun. 67:5298-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heldwein, E. E., and R. G. Brennan. 2001. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature 409:378-382. [DOI] [PubMed] [Google Scholar]
- 29.Hoch, J. A., and K. I. Varughese. 2001. Keeping signals straight in phosphorelay signal transduction. J. Bacteriol. 183:4941-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulkarni, R. D., and A. O. Summers. 1999. MerR cross-links to the alpha, beta, and sigma 70 subunits of RNA polymerase in the preinitiation complex at the merTPCAD promoter. Biochemistry 38:3362-3368. [DOI] [PubMed] [Google Scholar]
- 31.Levin, J. C., and M. R. Wessels. 1998. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol. Microbiol. 30:209-219. [DOI] [PubMed] [Google Scholar]
- 32.Lukomski, S., N. P. Hoe, I. Abdi, J. Rurangirwa, P. Kordari, M. Liu, S. J. Dou, G. G. Adams, and J. M. Musser. 2000. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect. Immun. 68:535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malke, H., K. Steiner, W. M. McShan, and J. J. Ferretti. 2006. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int. J. Med. Microbiol. 296:259-275. [DOI] [PubMed] [Google Scholar]
- 35.McCleary, W. R. 1996. The activation of PhoB by acetylphosphate. Mol. Microbiol. 20:1155-1163. [DOI] [PubMed] [Google Scholar]
- 36.McIver, K. S., A. S. Heath, and J. R. Scott. 1995. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation on emm transcription. Infect. Immun. 63:4540-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molle, V., Y. Nakaura, R. P. Shivers, H. Yamaguchi, R. Losick, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munson, G. P., and J. R. Scott. 1999. Binding site recognition by Rns, a virulence regulator in the AraC family. J. Bacteriol. 181:2110-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munson, G. P., and J. R. Scott. 2000. Rns, a virulence regulator within the AraC family, requires binding sites upstream and downstream of its own promoter to function as an activator. Mol. Microbiol. 36:1391-1402. [DOI] [PubMed] [Google Scholar]
- 40.Nickels, B. E., C. W. Roberts, H. Sun, J. W. Roberts, and A. Hochschild. 2002. The sigma(70) subunit of RNA polymerase is contacted by the (lambda)Q antiterminator during early elongation. Mol. Cell 10:611-622. [DOI] [PubMed] [Google Scholar]
- 41.Norgren, M., M. G. Caparon, and J. R. Scott. 1989. A method for allelic replacement that uses the conjugative transposon Tn916: deletion of the emm6.1 allele in Streptococcus pyogenes JRS4. Infect. Immun. 57:3846-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Opdyke, J. A., J. R. Scott, and C. P. Moran, Jr. 2003. Expression of the secondary sigma factor σX in Streptococcus pyogenes is restricted at two levels. J. Bacteriol. 185:4291-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Opdyke, J. A., J. R. Scott, and C. P. Moran, Jr. 2001. A secondary RNA polymerase sigma factor from Streptococcus pyogenes. Mol. Microbiol. 42:495-502. [DOI] [PubMed] [Google Scholar]
- 44.Podbielski, A., and B. A. Leonard. 1998. The group A streptococcal dipeptide permease (Dpp) is involved in the uptake of essential amino acids and affects the expression of cysteine protease. Mol. Microbiol. 28:1323-1334. [DOI] [PubMed] [Google Scholar]
- 45.Podbielski, A., J. A. Peterson, and P. Cleary. 1992. Surface protein-CAT reporter fusions demonstrate differential gene expression in the vir regulon of Streptococcus pyogenes. Mol. Microbiol. 6:2253-2265. [DOI] [PubMed] [Google Scholar]
- 46.Reference deleted.
- 47.Rimsky, S. 2004. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 7:109-114. [DOI] [PubMed] [Google Scholar]
- 48.Roberts, S. A., G. Churchward, and J. R. Scott. 2007. Unraveling the regulatory network in Streptococcus pyogenes: the global response regulator CovR represses rivR directly. J. Bacteriol. 189:1459-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rojo, F. 2001. Mechanisms of transcriptional repression. Curr. Opin. Microbiol. 4:145-151. [DOI] [PubMed] [Google Scholar]
- 50.Russo, F. D., and T. J. Silhavy. 1993. The essential tension: opposed reactions in bacterial two-component regulatory systems. Trends Microbiol. 1:306-310. [DOI] [PubMed] [Google Scholar]
- 51.Scott, J. R. 1972. A new gene controlling lysogeny in phage P1. Virology 48:282-283. [DOI] [PubMed] [Google Scholar]
- 52.Slack, F. J., P. Serror, E. Joyce, and A. L. Sonenshein. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol. Microbiol. 15:689-702. [DOI] [PubMed] [Google Scholar]
- 53.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 54.Sumby, P., A. R. Whitney, E. A. Graviss, F. R. DeLeo, and J. M. Musser. 2006. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Virtaneva, K., S. F. Porcella, M. R. Graham, R. M. Ireland, C. A. Johnson, S. M. Ricklefs, I. Babar, L. D. Parkins, R. A. Romero, G. J. Corn, D. J. Gardner, J. R. Bailey, M. J. Parnell, and J. M. Musser. 2005. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc. Natl. Acad. Sci. USA 102:9014-9019. [DOI] [PMC free article] [PubMed] [Google Scholar]