Abstract
The expression of two Enterococcus faecalis virulence-related proteases, gelatinase (GelE) and serine protease (SprE), is positively regulated by a quorum-sensing system encoded by the fsr gene cluster. In this system, E. faecalis secretes an autoinducing peptide, gelatinase biosynthesis-activating pheromone (GBAP), which triggers the FsrC-FsrA two-component regulatory system controlling the expression of two transcripts, fsrBDC and gelE-sprE. In the present study, we screened actinomycete metabolites for inhibitors of fsr quorum sensing. E. faecalis was cultured with each actinomycete culture supernatant tested, and the production of gelatinase and the production of GBAP were examined using the first screening and the second screening, respectively. Culture supernatant of Streptomyces sp. strain Y33-1 had the most potent inhibitory effect on both gelatinase production and GBAP production without inhibiting E. faecalis cell growth. The inhibitor in the culture supernatant was identified as a known peptide antibiotic, siamycin I. Siamycin I inhibited both gelatinase production and GBAP production at submicromolar concentrations, and it inhibited E. faecalis cell growth at concentrations above micromolar concentrations. Quantitative analysis of fsrBDC and gelE-sprE transcripts revealed that siamycin I suppressed the expression of both transcripts at a sublethal concentration. Siamycin I attenuated gelatinase production even when an overdose of GBAP was exogenously added to the culture. These results suggested that siamycin I inhibited the GBAP signaling via the FsrC-FsrA two-component regulatory system in a noncompetitive manner. The sublethal concentrations of siamycin I also attenuated biofilm formation. Treatment with siamycin could be a novel means of treating enterococcal infections.
Enterococcus faecalis is a gram-positive intestinal commensal of humans and other animals, but it sometimes causes opportunistic infections, including urinary tract, bloodstream, and wound infections, endophtalmitis, and endocarditis (22). Notably, in the past two decades, nosocomial infections caused by multiple-antibiotic-resistant or vancomycin-resistant E. faecalis have become a serious clinical problem (6, 33, 36, 49).
Besides cytolysin, which is lethal by itself for a broad range of prokaryotic and eukaryotic cells (10), several virulence-related factors have been found in E. faecalis, including aggregation substance (Agg), enterococcal surface protein (Esp), and two extracellular proteases, gelatinase (GelE) and serine protease (SprE) (35, 59). These factors have been thought to act synergistically to enhance virulence by facilitating colonization, translocation, and biofilm formation (8, 16, 23, 28, 34, 35, 52, 60, 62, 65). GelE and SprE are encoded in an operon, gelE-sprE, whose expression is positively regulated by a quorum-sensing system encoded by the fsr locus (45, 46). Several in vivo studies using animal or nematode models have shown that the fsr system contributes to virulence (17, 19, 37, 46, 53).
The fsr locus is comprised of four genes, designated fsrA, fsrB, fsrC, and fsrD (38, 40, 45, 46). In this system, a cyclic peptide, gelatinase biosynthesis-activating pheromone (GBAP), acts as an autoinducer (38, 39). It has been proposed that the prepropeptide of GBAP is translated from fsrD and then processed and cyclized by FsrB, resulting in the mature form of GBAP (40). When the concentration of GBAP that accumulates outside cells reaches a threshold level that is around 1 nM, it triggers the two-component regulatory system consisting of a histidine kinase (FsrC) and a response regulator (FsrA). The activated FsrA induces expression of the fsrBDC transcript, which is involved in an autoregulatory circuit resulting in a boost of GBAP signaling, and eventually induces gelE-sprE transcription.
Quorum sensing has recently been proposed as a new target for antimicrobial drug therapy (42, 48, 56). A compound which attenuates virulence without bactericidal or bacteriostatic activity is called “antipathogenic.” For example, macrolides such as azithromycin, which inhibit N-acylhomoserine lactone-mediated quorum sensing but do not inhibit the growth of Pseudomonas aeruginosa, are known to efficiently decrease the symptoms of cystic fibrosis and diffuse panbronchiolitis (57, 58). Furthermore, a number of other studies have revealed inhibitors targeting N-acylhomoserine lactone-mediated quorum sensing of gram-negative bacteria (26, 43, 50, 54, 55). In the case of gram-positive pathogens, quorum-sensing inhibitors have been investigated with staphylococci, which have a well-known regulatory system designated agr (32, 41). The agr system is mediated by a cyclic peptide pheromone, like the enterococcal fsr system, and positively regulates expression of some virulence factors via a regulatory RNA molecule designated RNA-III. Lyon et al. attempted to rationally design a peptide antagonist of the agr pheromone and were successful (31, 32). An RNA-III-inhibiting peptide found in culture filtrates of some staphylococcal strains is also thought to be an antistaphylococcal agent (1, 4, 9, 13, 21, 63).
In the present study, we screened inhibitors of E. faecalis fsr quorum sensing from actinomycete culture supernatants, because actinomycetes are rich sources of biologically active compounds. To our knowledge, this is the first screening study to target natural compounds in order to obtain a quorum-sensing inhibitor of a gram-positive pathogen.
MATERIALS AND METHODS
E. faecalis strains, media, and culture conditions.
E. faecalis OG1RF was used as a standard gelatinase-positive strain in this study (15). E. faecalis OU510 was a clinical isolate with an fsrB mutation resulting in a lack of GBAP production and was used as an indicator strain for the GBAP assay because in this strain gelatinase production depends solely on exogenously added GBAP (40). E. faecalis OU510B was strain OU510 carrying fsrB translationally fused to pNZ8048 NcoI site (29). This strain was used to screen fsr quorum-sensing inhibitors because of its high gelatinase and GBAP activities. For all analyses except the liquid chromatography-mass spectrometry (LC/MS) experiment and the biofilm formation assay, an overnight culture of E. faecalis was inoculated into Todd-Hewitt broth (THB) (Oxoid, Basingstoke, Hampshire, United Kingdom) to an optical density at 660 nm (OD660) of 0.01 and was then cultivated at 37°C with gentle shaking. For the LC/MS experiment, E. faecalis was cultivated in a chemically defined medium (CDM) developed for Lactobacillus plantarum (27). An overnight CDM culture (0.5 ml) of E. faecalis OG1RF was inoculated into 10 ml of fresh CDM and grown at 37°C for 7 h with gentle shaking.
Isolation and culture of actinomycetes.
Soil samples were collected from 33 places in Kyushu, Japan. One hundred milligrams of a soil sample was suspended in 1 ml of phosphate-buffered saline by 10 s of vortexing, 30 s of sonication, and 15 min of gentle mixing at 37°C in an Eppendorf tube. Then 100 μl of the suspension was mixed with 900 μl of a sodium dodecyl sulfate-yeast extract solution (0.05% [wt/vol] sodium dodecyl sulfate, 6% [wt/vol] yeast extract, 5 mM phosphate-buffered saline) and incubated at 40°C for 20 min with gentle mixing. Serial dilutions of the suspension were spread onto humic acid-vitamin agar (24), Bennett agar (2% [wt/vol] peptone, 0.1% [wt/vol] yeast extract, 0.1% [wt/vol] meat extract, 1% [wt/vol] glucose, 50 mg/liter cycloheximide, 1.5% [wt/vol] agar; pH 7.2), and yeast extract-starch agar (0.2% [wt/vol] yeast extract, 1% [wt/vol] soluble starch, 1% [wt/vol] glucose, 50 mg/liter cycloheximide, 1.5% [wt/vol] agar; pH 7.2). The agar plates were incubated at 30°C for 1 to 2 weeks. As a result, 179 actinomycete strains were isolated. For preparation of the culture supernatants used for screening, actinomycete isolates were inoculated into three liquid media (5 ml), Bennett medium, yeast extract-starch medium, and humic acid-vitamin medium, and were grown aerobically at 30°C for 3 days, 7 days, or 14 days.
For production of siamycin I, Streptomyces sp. strain Y33-1 was precultured in 10 ml of liquid Bennett medium at 30°C for 1 week. One milliliter of the preculture was inoculated into 100 ml of the same medium in a flat-bottom flask. A total of 10 flasks were prepared for a 1-liter culture. The culture was grown aerobically at 30°C for 1 week.
Gelatinase and GBAP assays.
Gelatinase activity was measured by using azocoll (Calbiochem, San Diego, CA) as a substrate for gelatinase according to the protocol described previously (38). Briefly, 25 μl of E. faecalis culture supernatant was added to 0.5 ml of an azocoll suspension, incubated for 4 h with constant mixing (170 rpm), and centrifuged at 20,000 × g for 5 min, and then the OD540 of the supernatant was determined.
To screen fsr quorum-sensing inhibitors, 4 ml of fresh THB was mixed with 1 ml of culture supernatant of one of the actinomycetes tested and was inoculated into an overnight culture (30 μl) of E. faecalis OU510B. After 5 h of incubation at 37°C, the culture supernatant was collected by centrifugation at 6,500 × g for 5 min. For the first screening, to examine inhibition of gelatinase production, 25 μl of the OU510B culture supernatant was used for direct measurement of gelatinase activity. For the second screening, to examine inhibition of GBAP production, 2 ml of the OU510B culture supernatant was applied to a Sep-Pak Plus C18 cartridge column (100 mg; Waters, Milford, MA), washed with 2 ml of 20% (vol/vol) acetonitrile containing 0.1% (vol/vol) trifluoroacetic acid (TFA), and then eluted with 1 ml of 40% acetonitrile containing 0.1% TFA. The eluate was dried with a Speed-vac concentrator, redissolved in 5 ml of fresh medium, and then inoculated with 30 μl of an E. faecalis OU510 overnight culture. After 5 h of incubation at 37°C, the gelatinase activity was measured using 25 μl of the OU510 culture supernatant as described above, and the gelatinase activity was considered GBAP activity.
The GBAP activity was also determined by LC/MS. CDM culture supernatant (10 ml) was loaded onto a Sep-Pak Plus C18 cartridge column (360 mg; Waters), washed with 10 ml of 10% acetonitrile containing 0.1% TFA, and then eluted with 5 ml of 60% acetonitrile containing 0.1% TFA. The eluate was evaporated to dryness and then redissolved in 200 μl of 10% acetonitrile containing 0.1% TFA, and 80 μl of the resulting solution was injected into an LC/MS (Agilent HP1100 LC; Agilent Zorbax Eclipse XDB-C18 column [2.1 by 50 mm]; Accutof T100LC MS [JEOL, Tokyo, Japan]). The column was eluted at a flow rate of 0.2 ml/min at 30°C with a linear acetonitrile gradient (20 to 40% acetonitrile in 20 min after 5 min of 20% acetonitrile) in an aqueous 0.05% TFA solution. The eluates were directly loaded into the electrospray ionization-time of flight mass spectrometer. Mass analyses were performed under the following conditions: positive polarity; capillary temperature, 260°C; needle voltage, 2.0 kV; orifice voltage, 70 V; and ring lens voltage, 10 V. After scanning for molecular ions derived from column eluates in the m/z range from 100 to 2,000, extracted ion chromatograms were plotted with detector counts at m/z values ranging from 1,303.5 to 1,304.2, which covered a protonated molecular ion of GBAP. GBAP was detected at 19 min.
Purification of siamycin I.
Streptomyces sp. strain Y33-1 culture supernatant (850 ml) was collected by centrifugation at 6,520 × g and applied to an Amberlite XAD-7 column (50 ml; Sigma, St. Louis, MO). The column was eluted by using 300 ml of methanol. The eluate was evaporated to dryness, redissolved in 50 ml of Milli-Q water, and then applied to a Sep-Pak Vac C18 cartridge column (35 ml; 10 g; Waters). After washing with 150 ml of 35% acetonitrile containing 0.1% TFA, the column was eluted with 50 ml of 50% acetonitrile containing 0.1% TFA. The eluate was evaporated, lyophilized, and then redissolved in 50 ml of 10% acetonitrile containing 0.1% TFA. The solution was divided into five aliquots, and each aliquot was applied to a reverse-phase high-performance liquid chromatography column (Inertsil ODS-3; 20 by 150 mm; GL Sciences Inc., Tokyo, Japan). The column was developed by using a gradient of 10 to 80% acetonitrile in 0.1% TFA for 35 min at a rate of 10 ml/min. The active fractions were pooled, lyophilized, and then rechromatographed in the same column by using a gradient of 40 to 65% acetonitrile in 0.1% TFA for 35 min. Finally, 2.8 mg of siamycin I was purified.
NMR experiment.
Nuclear magnetic resonance (NMR) spectra were measured with a Varian Unity INOVA600 spectrometer at 10°C and 40°C. A 2 mM sample solution was prepared in 50% (vol/vol) CD3OD-50% (vol/vol) H2O. Chemical shifts were referenced to the water resonance (4.92 and 4.63 ppm at 10 and 40°C, respectively). Standard 1H-1H homonuclear NMR methods were used to obtain a series of two-dimensional spectra by double quantum filtered correlation spectroscopy, total correlation spectroscopy (mixing time, 45 ms), and nuclear Overhauser enhancement spectroscopy (mixing time, 300 ms).
Effect of siamycin I on the NisK-NisR two-component regulatory system.
The Escherichia coli lacZ gene was cloned into NcoI and SphI sites of pNZ8048 (29), and the resultant plasmid, designated pNZ8048lacZ, was introduced into E. faecalis OG1RF together with pNZ9530 carrying nisRK. The resultant strain, OG1RF(pNZ8048lacZ/pNZ9530), was cultured in THB containing 3 nM nisin A and various concentrations of siamycin I for 5 h at 37°C. The nisin-induced β-galactosidase activity was measured and expressed in Miller units using the method described at a website (http://rothlab.ucdavis.edu/protocols/beta-galactosidase-3.html).
Real-time quantitative RT-PCR.
An overnight culture (30 μl) of E. faecalis OG1RF was inoculated into 5 ml THB with or without 1 μM siamycin I and grown for 4 h. Cells were harvested by centrifugation at 8,500 × g for 5 min. Total RNA was extracted from the harvested cells by a method described previously (38) and then dissolved in 50 μl of diethyl pyrocarbonate-treated water to obtain a solution containing about 10 μg RNA/μl. Five microliters of this solution was treated with 1 μl of DNase (Nippon Gene, Tokyo, Japan) in a 50-μl (total volume) reaction mixture according to the manufacturer's instructions. After 15 min of incubation at 37°C, 5 μl of stop solution was added to the reaction solution, which was then incubated at 70°C for 10 min. The levels of transcription of 16S rRNA, fsrBDC, and gelE-sprE were determined by one-step real-time reverse transcription-PCR (RT-PCR). An RT-PCR was performed in duplicate for each RNA sample with a one-step SYBR RT-PCR kit (Takara, Kyoto, Japan). For quantification of fsrBDC and gelE-sprE, 1-μl portions of the DNase-treated RNA solution were used as templates in a 17-μl (total volume) reaction mixture containing 0.2 μM primer fsrBF1 (5′-TGGATCAGGAAGATCAATCAGG-3′) and 0.2 μM primer fsrBR1 (5′-GTACGACGTATACAATAAAGGTTTCG-3′) and in a 17-μl (total volume) reaction mixture containing 0.2 μM primer gelEF (5′-AGTGAACGCTACAGATGGAAC-3′) and 0.2 μM primer gelER (5′-CGTTCCGTGTAAAGCAATTCC-3′), respectively. For 16S rRNA, the DNase-treated RNA solution was diluted 100-fold, and 1 μl was used as a template in a 17-μl (total volume) reaction mixture containing 10 μM primer Enc-F-rt (5′-CCCTTATTGTTAGTTGCCATCATT-3′) and 10 μM primer Enc-FR-rt (5′-ACTCGTTGTACTTCCCATTGT-3′) (51). Real-time PCR monitoring of amplified products and comparative quantitation were carried out by using a real-time PCR system (Mx3000; Stratagene, La Jolla, CA) with the MxProTM software (version 3.00). The relative level of expression was calculated from the standard curve constructed with serial dilutions of the RNA sample prepared from the culture without siamycin I.
Biofilm formation assay.
Biofilm formation assays were performed by using the protocol of Seno et al. (52), with minor modifications. Briefly, E. faecalis OG1RF that had been grown overnight was diluted 1:100 in 200 μl of tryptic soy broth supplemented with 0.25% (wt/vol) glucose and inoculated into wells of a sterile flat-bottom 96-well polystyrene microtiter plate (Corning Inc., Corning, NY). Siamycin I was dissolved in methanol (10 μg/μl) and added to the medium at a final concentration of 0.125, 0.25, 0.5, or 1 μM. After 24 h of static incubation at 37°C, the plates were processed, stained with 0.3% crystal violet for 45 min, and rinsed with distilled water. The bound dye was solubilized in ethanol-acetic acid (95:5, vol/vol), and the optical density at 570 nm was determined using a microplate reader (model 680; Bio-Rad Japan, Tokyo, Japan). The effect of methanol was examined in the absence of siamycin I, and we confirmed that methanol did not have a significant effect on biofilm formation in the concentration range used in this experiment. Each assay was performed in quadruplicate on two occasions.
RESULTS
Screening of inhibitors targeting fsr quorum sensing from actinomycete culture supernatants.
A total of 179 actinomycete strains were isolated from several soil samples that were obtained in Kyushu, Japan, and subjected to screening. In the first screening, E. faecalis was grown with culture supernatant of each isolate, and gelatinase production was examined. Twenty-two strains had an inhibitory effect on gelatinase production without growth inhibition. In the second screening of these 22 strains, the inhibitory effect of culture supernatant on GBAP production was examined, and three strains significantly inhibited GBAP production. One of these strains, Streptomyces sp. strain Y33-1, which had the most potent inhibitory effect on GBAP production, was selected for further study.
The inhibitor in the Y33-1 culture supernatant was purified and subjected to structural analysis. The UV absorption spectrum of this compound was a typical profile of a peptidic compound containing a tryptophan residue, which produced a shoulder around 220 nm and a peak at 280 nm. Mass spectrometry suggested that the molecular mass of this compound was 2,164, which is identical to the molecular masses of siamycin I (14, 61), NP-06 (7), and RP-71955 (18). These compounds were previously screened for anti-human immunodeficiency virus (HIV) activity and are known to have antimicrobial activity against gram-positive bacteria as well (7, 18, 30, 61). They were found to be tricyclic peptides consisting of 21 amino acids. The same structure has been reported for siamycin I and NP-06. RP-71955 differs from the other compounds only at residues 4 and 17. A series of 1H-1H two-dimensional NMR, double quantum filtered correlation spectroscopy, total correlation spectroscopy, and nuclear Overhauser enhancement spectroscopy analyses allowed assignment of almost all protons in the inhibitor of Y33-1 (Table 1). Most of the chemical shifts indicated were identical to those of NP-06 (7), and the chemical shifts of the Val-4 and Ile-17 protons were clearly different from those of the Val-17 and Ile-4 protons of RP-71955 (18). These differences in chemical shifts were consistent with the data reported in a previous study in which NP-06 was compared with RP-71955 (7). As a result, the inhibitor of Y33-1 was identified as siamycin I and NP-06 (Fig. 1), and it is referred to below as siamycin I.
TABLE 1.
Observed proton chemical shifts of Y33-1 (siamycin I)a
| Residue | HN | Hαb | Hβ | Otherc |
|---|---|---|---|---|
| C1 | 8.83 | ND | 3.53, 2.54 | |
| L2 | 9.84 | 4.52 | 2.03, 1.57 | Hγ = 2.01, Mδ = 1.02, 0.89 |
| G3 | 8.97 | 4.17, 3.73 | ||
| V4 | 7.03 | 4.10 | 1.82 | Mγ = 0.74, 0.74 |
| G5 | 7.27 | 4.11, 3.56 | ||
| S6 | 7.98 | 4.57 | 3.91, 3.78 | |
| C7 | 8.16 | ND | 3.10, 3.40 | |
| N8 | 8.92 | ND | 2.41, 2.31 | |
| D9 | 7.98 | 4.40 | 2.87, 2.72 | |
| F10 | 8.59 | 4.30 | 2.90, 2.57 | Hδ = 7.20, Hɛ = 7.39, Hη = 7.78 |
| A11 | 8.83 | 3.89 | 1.23 | |
| G12 | 8.80 | 4.04, 3.58 | ||
| C13 | 8.34 | ND | 3.37, 3.10 | |
| G14 | 7.43 | 3.90, 3.14 | ||
| Y15 | 9.39 | 5.40 | 2.72, 1.99 | Hδ = 6.73, Hɛ = 6.58 |
| A16 | 8.40 | 4.68 | 1.19 | |
| I17 | 8.25 | 4.24 | 1.74 | Hγ1 = 1.47, 1.02, Mγ2 = 0.86, Mδ1 = 0.80 |
| V18 | 7.41 | 3.88 | 1.85 | Mγ = 0.82, 0.67 |
| C19 | 8.05 | 4.49 | 2.53, 2.30 | |
| F20 | 8.05 | 4.50 | 2.80, 2.57 | Hδ = 6.75, Hɛ = 7.02, Hη = 6.98 |
| W21 | 7.94 | 4.56 | 3.27, 3.09 | Hδ1 = 7.04, Hɛ1 = 10.16, Hζ2 = 7.03, Hη2 = 7.50, Hζ2 = 7.05, Hɛ3 = 7.33 |
The NMR data were obtained at 10°C.
ND, not detected.
M, methyl.
FIG. 1.
Structure of the inhibitor of Y33-1 (siamycin I [14, 61] or NP-06 [7]).
Effect of siamycin I on growth of and gelatinase production in E. faecalis.
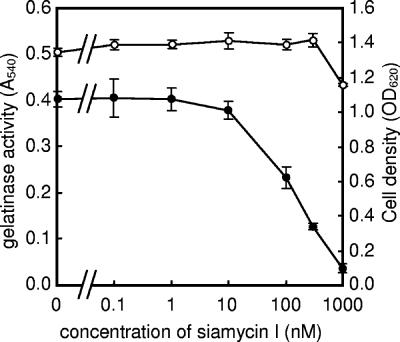
Siamycin I slightly inhibited the growth of E. faecalis at a concentration of 1 μM (80% growth 5 h after inoculation) and completely inhibited the growth at a concentration of 5 μM (no growth 12 h after inoculation). This growth-inhibiting activity is consistent with the data reported previously (61). Figure 2 shows the effects of various concentrations of siamycin I on gelatinase production in E. faecalis OG1RF. Siamycin I slightly inhibited gelatinase production at a concentration of 10 nM and strongly inhibited gelatinase production at a concentration of 1 μM, and the 50% inhibitory concentration was around 100 nM.
FIG. 2.
Effects of various concentrations of siamycin I on growth of and gelatinase production by E. faecalis OG1RF. E. faecalis OG1RF was grown for 5 h in the presence of different concentrations of siamycin I, and then the OD660 (○) and gelatinase activity at OD540 (•) in the culture supernatant were determined as described in Materials and Methods. The data are averages ± standard deviations of duplicate determinations.
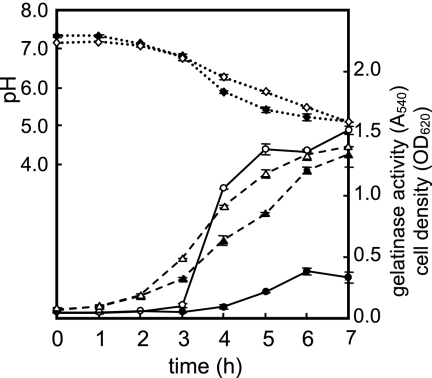
Figure 3 shows the time courses for cell growth, pH, and gelatinase activity of E. faecalis cultures with and without 1 μM siamycin I. The same level of inhibition of gelatinase production was observed until the end of the time course experiment (7 h, nearly the stationary phase). This indicated that siamycin I did not delay the onset of gelatinase production but constitutively inhibited this production. As shown in Fig. 3, the pH of the culture supernatant decreased during cell growth, which was due to lactate fermentation by E. faecalis cells. The decrease in the pH was slightly slower in the presence of siamycin I, as was cell growth, while gelatinase production was greatly reduced by addition of siamycin I. It is likely that the inhibition of gelatinase production by a sublethal concentration of siamycin I was due to more than the pleiotropic effect on general metabolism.
FIG. 3.
Time courses for cell growth, pH, and gelatinase activity of E. faecalis cultures with and without siamycin I (1 μM). E. faecalis OU510B was inoculated into fresh medium with (solid symbols) or without (open symbols) siamycin I (1 μM) and was then cultured. Culture supernatant was collected every hour, and the OD660 (triangles), pH (diamonds), and gelatinase activity at OD540 (circles) were determined. The data are averages ± standard deviations of duplicate determinations.
Inhibitory effect of siamycin I on GBAP production and GBAP response of E. faecalis.
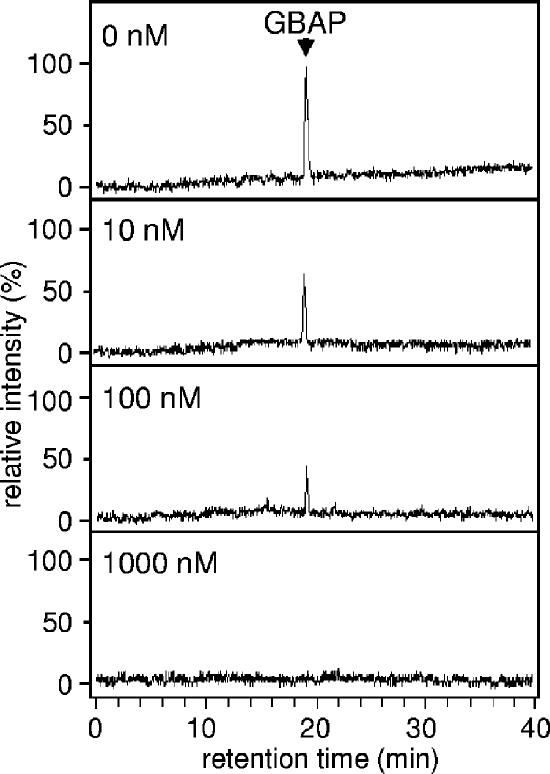
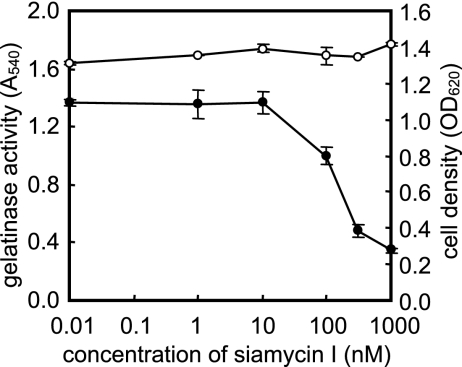
The effect of siamycin I on GBAP production was examined by LC/MS (Fig. 4). Siamycin I slightly inhibited GBAP production at a concentration of 10 nM and eliminated GBAP production at a concentration of 1 μM. Together, the data showed that siamyicin I attenuates GBAP production, as well as gelatinase production, at sublethal concentrations. These results suggested the following two possibilities: (i) siamycin I inhibits the FsrB function for GBAP biosynthesis and (ii) siamycin I inhibits GBAP signal transduction via the FsrC-FsrA two-component regulatory system. In order to examine these possibilities, the effect of siamycin I on the GBAP responsiveness of E. faecalis cells was examined by using E. faecalis OU510, which had an amber mutation of fsrB resulting in the loss of GBAP production. In this strain, induction by exogenous GBAP is necessary for gelatinase production. In this experiment, 100 nM GBAP (a dose 10-fold higher than the concentration in a late-log-phase culture) and various concentrations of siamycin I were added to early-log-phase cultures at the same time. The cultures were incubated for an additional 3 h, and then the gelatinase activities in the culture supernatants were measured. As shown in Fig. 5, siamycin I inhibited the GBAP-induced gelatinase production at concentrations higher than 100 nM. This suggested that the site of action of siamycin I is not GBAP biosynthesis but GBAP signal transduction.
FIG. 4.
Effects of various concentrations of siamycin I on GBAP production by E. faecalis OG1RF. E. faecalis OG1RF was grown for 7 h in CDM containing different concentrations of siamycin I. Then the GBAP in each culture supernatant was detected by LC/MS. Extracted ion chromatograms were plotted with detector counts at m/z values ranging from 1,303.5 to 1,304.2, which covered a protonated molecular ion of GBAP, and were normalized to the GBAP peak height in the absence of siamycin I (top chromatogram).
FIG. 5.
Effects of various concentrations of siamycin I on gelatinase production by E. faecalis OU510 induced by 100 nM GBAP. After 2 h of incubation, 100 nM synthetic GBAP and different amounts of siamycin I were added to cultures of E. faecalis OU510, and the cultures were incubated for another 3 h to induce gelatinase production. Then the OD660 (○) and gelatinase activity at OD540 (•) in the culture supernatants were determined. The data are averages ± standard deviations of duplicate determinations.
The inhibitory mode of siamycin I was examined by measuring the GBAP-induced gelatinase production in the presence of 1 μM of siamycin I and various concentrations of GBAP. As shown in Fig. 6, even in the presence of 1 mM GBAP (titer, ca. 1,000), 1 μM siamycin I inhibited the GBAP-induced gelatinase production. This suggested that siamycin I is not a competitive inhibitor of GBAP.
FIG. 6.
Effect of siamycin I (1 μM) on gelatinase production in E. faecalis OU510 induced by various concentrations of GBAP. E. faecalis OU510 was grown for 5 h in the presence of different concentrations of synthetic GBAP without (○) or with 1 μM siamycin I (•). Then the gelatinase activity in the culture supernatant was determined at OD540 in duplicate, and the average values were plotted.
Effect of siamycin I on NisK-NisR two-component regulatory system heterologously expressed in E. faecalis.
To examine the specificity of the inhibitory effect of siamycin I, the NisK-NisR two-component regulatory system from Lactococcus lactis (29) was heterologously expressed in E. faecalis, and the effect of siamycin I on NisK-NisR signal transduction was examined. In the reporter strain, the lacZ gene was cloned under the nisin-inducible nisA promoter controlled by the NisR-NisK two-component regulatory system. This reporter strain was cultured with 3 nM nisin A and various concentrations of siamycin I, and the nisin-induced β-galactosidase activity was measured. As shown in Fig. 7, the induction of β-galactosidase activity was partially reduced at a siamycin I concentration of 400 nM and almost eliminated at a siamycin I concentration of 600 nM. These result indicated that siamycin I inhibited the NisK-NisR two-component regulatory system, as well as the FsrC-FsrA system. However, siamycin I did not affect the induction of β-galactosidase activity at concentrations of 100 nM and 200 nM, whereas it clearly reduced gelatinase production at the same concentrations, suggesting that the NisK-NisR two-component regulatory system was less sensitive to siamycin I than the FsrC-FsrA system was.
FIG. 7.
Effects of various concentrations of siamycin I on β-galactosidase induction and gelatinase production in E. faecalis OG1RF(pNZ8048lacZ/pNZ9530). E. faecalis OG1RF(pNZ8048lacZ/pNZ9530) was cultured for 5 h in medium containing 3 nM nisin A and different concentrations of siamycin I. The cell growth (circo]), gelatinase activity (•), and β-galactosidase activity (⧫) of each culture were determined and expressed in OD660, OD540, and Miller units, respectively. The data are averages ± standard deviations of duplicate determinations.
Effect of siamycin I on transcription of fsrBDC and gelE-sprE.
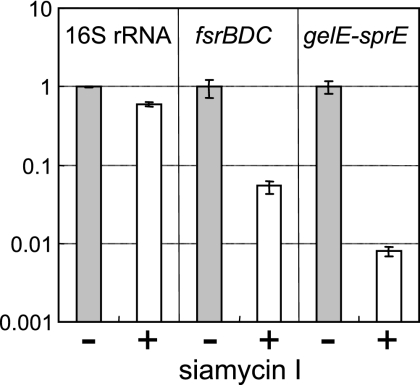
The effect of siamycin I on transcription of fsrBDC and gelE-sprE was examined by using real-time quantitative RT-PCR. As shown in Fig. 8, both transcription reactions were strongly suppressed. The transcription of gelE-sprE was more strongly suppressed than the transcription of fsrBDC. This result may have reflected the signal transduction pathway in which the fsr regulatory system positively regulates gelE-sprE expression.
FIG. 8.
Effects of siamycin I (1 μM) on transcription of 16S rRNA, fsrBDC, and gelE-sprE in E. faecalis OG1RF. E. faecalis OG1RF was grown for 4 h in the absence (control) (gray bars) or in the presence (open bars) of 1 μM siamycin I. Total RNA was prepared from the grown cells and used as a template for a one-step RT-PCR. The relative levels of expression determined by real-time PCR are expressed as the ratio of the value obtained with 1 μM siamycin I to the control value. For each RNA sample duplicate cultures were used and duplicate RT-PCR analyses were performed. The standard deviations of quadruplicate determinations are indicated by the error bars.
Effects of siamycin I on cell morphology of and biofilm formation by E. faecalis.
The effect of siamycin I on cell morphology was examined by microscopy. In the absence of siamycin I, the length of most E. faecalis cell chains was between two and four cells. On the other hand, in the presence of 0.5 or 1.0 μM siamycin I, the chain length clearly increased to around 10 cells (data not shown).
The effect on biofilm formation by E. faecalis was also examined. As shown in Fig. 9, siamycin I slightly inhibited biofilm formation at a concentration of 0.25 μM, and the inhibitory effect was marked at concentrations higher than 0.5 μM.
FIG. 9.
Effect of siamycin I on biofilm formation by E. faecalis OG1RF. Assays were performed by using tryptic soy broth supplemented with 0.25% glucose containing different concentrations of siamycin I and flat-bottom 96-well polystyrene microtiter plates. After 24 h of incubation at 37°C, biofilm formation was quantified by determining the OD570 of a crystal violet-stained biofilm. Each assay was performed in quadruplicate on two occasions; the bars indicate means, and the error bars indicate standard errors.
DISCUSSION
To target antipathogenic compounds against E. faecalis, we screened our actinomycete collection for inhibitors of GBAP-mediated quorum sensing. Inhibition of gelatinase production and inhibition of GBAP production were assayed in the first screening and second screening, respectively. The two-step screening procedure allowed efficient screening for inhibitors targeting GBAP-mediated quorum sensing. This screening system may detect two types of inhibitors; one type targets the FsrC-FsrA two-component regulatory system, and the other targets FsrB, which is the GBAP biosynthetic enzyme. These two types can be distinguished by adding physiological concentrations of GBAP at the same time that a test sample is added. Unlike the situation with siamycin I, if the inhibitor targets FsrB, gelatinase production can be recovered. Recently, it has been demonstrated that FsrB belongs to the AgrB protein family, which is predicted to have a cysteine protease-like function (40, 47). Inhibitors targeting FsrB should be effective against some other gram-positive bacteria, including staphylococci, which use cyclic peptide-mediated quorum sensing (32, 47).
As a result of screening, a known secondary metabolite of actinomycetes, siamycin I, was found to be a potent inhibitor of GBAP-mediated quorum sensing. Three varieties of siamycin have been screened as anti-HIV compounds, and they have been found to be tricyclic peptides consisting of 21 amino acids. They differ from one another only at positions 4 (Val or Ile) and 17 (Val or Ile), and the differences are unlikely to make much difference in terms of conformational and functional properties (12). Due to their structural similarity, the other siamycins may inhibit GBAP-mediated quorum sensing.
The target molecule of siamycins in anti-HIV activity has been proposed to be HIV envelope glycoprotein gp41 because siamycins have a strong inhibitory effect on syncytium formation and exhibit sequence similarity to part of gp41 (7, 11, 18). However, unlike the sequence similarity between siamycin and gp41, there is no sequence similarity between siamycin I and GBAP. An overdose of GBAP could not outcompete the inhibition by siamycin I. These findings suggested that the inhibitory activity is not caused by specific binding of siamycin I to the GBAP-binding site on FsrC. It has been reported that siamycins also inhibit the growth of gram-positive bacteria at micromolar concentrations but do not inhibit the growth of gram-negative bacteria (61). Although no information about the antibacterial mode of action has been reported, the antibacterial activity specific for gram-positive bacteria is well known for bacteriocins, which are membrane-active peptides found in many gram-positive bacteria. Like bacteriocins, siamycins may also be integrated into the cell membrane and inhibit the growth of gram-positive bacteria (25). At sublethal concentrations, siamycins may disturb the receptor kinase function of FsrC by localizing in the cell membrane. It has been speculated that this mode of action is effective for the other membrane kinases. As expected, the NisK-NisR two-component regulatory system heterologously expressed in E. faecalis was also inhibited by sublethal concentrations of siamycin I. However, the FsrC-FsrA two-component regulatory system was very sensitive compared to the NisK-NisR system, suggesting that there is a specific interaction between siamycin I and FsrC.
At sublethal concentrations, siamycin I inhibited GBAP and gelatinase production by E. faecalis, while it did not have a great effect on cell growth and acid production. Transcriptional analysis also indicated that sublethal concentrations of siamycin I inhibited the expression of fsrBDC and gelE-sprE, while they slightly reduced the 16S rRNA expression. These results suggested that the inhibitory effect of a sublethal concentration of siamycin I on the fsr system is due to more than the pleiotropic effect on general metabolism. Understanding how siamycin I inhibits transcription would help in understanding the mode of action of siamycin. DNA microarray analysis is now available (5) and can provide information concerning genes suppressed by siamycin I. This kind of information would allow us to understand the precise mode of action of siamycin I, especially whether siamycin I targets only the fsr regulatory system or other genes in a different regulatory system.
The effects of siamycin I on cell morphology and biofilm formation were also examined, and it was found that siamycin I led to increased cell chain length and decreased biofilm formation at sublethal concentrations. It has been reported that knockout of gel or fsr led to the same changes (23, 44, 64). This coincidence suggests that the effects of siamycin which we observed may have been due to the inhibition of fsr quorum sensing associated with gelatinase induction. However, it should be noted that a number of factors other than gelatinase are involved in biofilm formation by E. faecalis (52), and siamycin I may also influence some of these factors. Since pathogenic bacteria are often capable of surviving antibiotic treatment through encapsulation in biofilms, these kinds of inhibitors are expected to efficiently eliminate biofilm- and quorum-sensing-associated infections and could be synergistic with other antibiotics (2, 3, 20, 21). Considering the fact that siamycin I has antibacterial activity at micromolar concentrations, it may have the potential to eliminate and inhibit biofilm-associated E. faecalis infections by itself.
This work was an initial screening study that targeted GBAP-mediated quorum sensing. Larger-scale screening of natural and/or synthetic compounds would provide more information concerning other compounds that are potentially antipathogenic against E. faecalis and perhaps other gram-positive pathogens.
Acknowledgments
This work was supported in part by grants-in-aid for scientific research from the Japan Society for the Promotion of Science (grants 15580065 and 17580068 to J. Nakayama and grant 15591688 to R. Kariyama), from the Ministry of Education, Culture, Sports and Technology of Japan (grant 16087203 to K. Nagata), and from the Kato Memorial Bioscience Foundation.
We thank Philip S. Stewart and Betsey Pitts of the Center for Biofilm Engineering, Montana State University, for critically reading the manuscript.
Footnotes
Published ahead of print on 27 October 2006.
REFERENCES
- 1.Balaban, N., L. V. Collins, J. S. Cullor, E. B. Hume, E. Medina-Acosta, O. Vieira da Motta, R. O'Callaghan, P. V. Rossitto, M. E. Shirtliff, L. Serafim da Silveira, A. Tarkowski, and J. V. Torres. 2000. Prevention of diseases caused by Staphylococcus aureus using the peptide RIP. Peptides 21:1301-1311. [DOI] [PubMed] [Google Scholar]
- 2.Balaban, N., A. Giacometti, O. Cirioni, Y. Gov, R. Ghiselli, F. Mocchegiani, C. Viticchi, M. S. Del Prete, V. Saba, G. Scalise, and G. Dell'Acqua. 2003. Use of the quorum-sensing inhibitor RNAIII-inhibiting peptide to prevent biofilm formation in vivo by drug-resistant Staphylococcus epidermidis. J. Infect. Dis. 187:625-630. [DOI] [PubMed] [Google Scholar]
- 3.Balaban, N., Y. Gov, A. Bitler, and J. R. Boelaert. 2003. Prevention of Staphylococcus aureus biofilm on dialysis catheters and adherence to human cells. Kidney Int. 63:340-345. [DOI] [PubMed] [Google Scholar]
- 4.Balaban, N., P. Stoodley, C. A. Fux, S. Wilson, J. W. Costerton, and G. Dell'Acqua. 2005. Prevention of staphylococcal biofilm-associated infections by the quorum sensing inhibitor RIP. Clin. Orthop. Relat. Res. 437:48-54. [DOI] [PubMed] [Google Scholar]
- 5.Bourgogne, A., S. G. Hilsenbeck, G. M. Dunny, and B. E. Murray. 2006. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J. Bacteriol. 188:2875-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chavers, L. S., S. A. Moser, W. H. Benjamin, S. E. Banks, J. R. Steinhauer, A. M. Smith, C. N. Johnson, E. Funkhouser, L. P. Chavers, A. M. Stamm, and K. B. Waites. 2003. Vancomycin-resistant enterococci: 15 years and counting. J. Hosp. Infect. 53:159-171. [DOI] [PubMed] [Google Scholar]
- 7.Chokekijchai, S., E. Kojima, S. Anderson, M. Nomizu, M. Tanaka, M. Machida, T. Date, K. Toyota, S. Ishida, K. Watanabe, H. Yoshioka, P. P. Roller, K. Murakami, and H. Mitsuya. 1995. NP-06: a novel anti-human immunodeficiency virus polypeptide produced by a Streptomyces species. Antimicrob. Agents Chemother. 39:2345-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow, J. W., L. A. Thal, M. B. Perri, J. A. Vazquez, S. M. Donabedian, D. B. Clewell, and M. J. Zervos. 1993. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob. Agents Chemother. 37:2474-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirioni, O., A. Giacometti, R. Ghiselli, G. Dell'acqua, F. Orlando, F. Mocchegiani, C. Silvestri, A. Licci, V. Saba, G. Scalise, and N. Balaban. 2006. RNAIII-inhibiting peptide significantly reduces bacterial load and enhances the effect of antibiotics in the treatment of central venous catheter-associated Staphylococcus aureus infections. J. Infect. Dis. 193:180-186. [DOI] [PubMed] [Google Scholar]
- 10.Coburn, P. S., and M. S. Gilmore. 2003. The Enterococcus faecalis cytolysin: a novel toxin active against eukaryotic and prokaryotic cells. Cell. Microbiol. 5:661-669. [DOI] [PubMed] [Google Scholar]
- 11.Constantine, K. L., M. S. Friedrichs, D. Detlefsen, M. Nishio, M. Tsunakawa, T. Furumai, H. Ohkuma, T. Oki, S. Hill, R. E. Bruccoleri, P.-F. Lin, and L. Mueller. 1995. High-resolution solution structure of siamycin II: novel amphipathic character of a 21-residue peptide that inhibits HIV fusion. J. Biomol. NMR 5:271-286. [DOI] [PubMed] [Google Scholar]
- 12.De Clercq, E. 2000. Current lead natural products for the chemotherapy of human immunodeficiency virus (HIV) infection. Med. Res. Rev. 20:323-349. [DOI] [PubMed] [Google Scholar]
- 13.Dell'Acqua, G., A. Giacometti, O. Cirioni, R. Ghiselli, V. Saba, G. Scalise, Y. Gov, and N. Balaban. 2004. Suppression of drug-resistant staphylococcal infections by the quorum-sensing inhibitor RNAIII-inhibiting peptide. J. Infect. Dis. 190:318-320. [DOI] [PubMed] [Google Scholar]
- 14.Detlefsen, D. J., S. E. Hill, K. J. Volk, S. E. Klohr, M. Tsunakawa, T. Furumai, P. F. Lin, M. Nishio, K. Kawano, T. Oki, and M. S. Lee. 1995. Siamycins I and II, new anti-HIV-1 peptides. II. Sequence analysis and structure determination of siamycin I. J. Antibiot. 48:1515-1517. [DOI] [PubMed] [Google Scholar]
- 15.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 75:3470-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupont, H., P. Montravers, J. Mohler, and C. Carbon. 1998. Disparate findings on the role of virulence factors of Enterococcus faecalis in mouse and rat models of peritonitis. Infect. Immun. 66:2570-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelbert, M., E. Mylonakis, F. M. Ausubel, S. B. Calderwood, and M. S. Gilmore. 2004. Contribution of gelatinase, serine protease, and fsr to the pathogenesis of Enterococcus faecalis endophthalmitis. Infect. Immun. 72:3628-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frechet, D., J. D. Guitton, F. Herman, D. Faucher, G. Helynck, B. Monegier du Sorbier, J. P. Ridoux, E. James-Surcouf, and M. Vuilhorgne. 1994. Solution structure of RP 71955, a new 21 amino acid tricyclic peptide active against HIV-1 virus. Biochemistry 33:42-50. [DOI] [PubMed] [Google Scholar]
- 19.Garsin, D. A., C. D. Sifri, E. Mylonakis, X. Qin, K. V. Singh, B. E. Murray, S. B. Calderwood, and F. M. Ausubel. 2001. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 98:10892-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giacometti, A., O. Cirioni, R. Ghiselli, G. Dell'Acqua, F. Orlando, G. D'Amato, F. Mocchegiani, C. Silvestri, M. S. Del Prete, M. Rocchi, N. Balaban, V. Saba, and G. Scalise. 2005. RNAIII-inhibiting peptide improves efficacy of clinically used antibiotics in a murine model of staphylococcal sepsis. Peptides 26:169-175. [DOI] [PubMed] [Google Scholar]
- 21.Giacometti, A., O. Cirioni, Y. Gov, R. Ghiselli, M. S. Del Prete, F. Mocchegiani, V. Saba, F. Orlando, G. Scalise, N. Balaban, and G. Dell'Acqua. 2003. RNA III inhibiting peptide inhibits in vivo biofilm formation by drug-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:1979-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock, L. E., and M. S. Gilmore. 1999. Pathogenicity of enterococci, p. 251-258. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, DC.
- 23.Hancock, L. E., and M. Perego. 2004. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 186:5629-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayakawa, M., and H. Nonomura. 1987. Humic acid-vitamin agar, a new medium for the selective isolation of soil acinomycetes. J. Ferment. Technol. 65:501-509. [Google Scholar]
- 25.Hechard, Y., and H. G. Sahl. 2002. Mode of action of modified and unmodified bacteriocins from Gram-positive bacteria. Biochimie 84:545-557. [DOI] [PubMed] [Google Scholar]
- 26.Hentzer, M., and M. Givskov. 2003. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J. Clin. Investig. 112:1300-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kets, E. P. W., E. A. Galinski, and J. A. M. de Bont. 1994. Carnitine: a novel compatible solute in Lactobacillus plantarum. Arch. Microbiol. 162:243-248. [Google Scholar]
- 28.Kristich, C. J., Y. H. Li, D. G. Cvitkovitch, and G. M. Dunny. 2004. Esp-independent biofilm formation by Enterococcus faecalis. J. Bacteriol. 186:154-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuipers, O. P., P. G. G. A. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 30.Lin, P. F., H. Samanta, C. M. Bechtold, C. A. Deminie, A. K. Patick, M. Alam, K. Riccardi, R. E. Rose, R. J. White, and R. J. Colonno. 1996. Characterization of siamycin I, a human immunodeficiency virus fusion inhibitor. Antimicrob. Agents Chemother. 40:133-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyon, G. J., P. Mayville, T. W. Muir, and R. P. Novick. 2000. Rational design of a global inhibitor of the virulence response in Staphylococcus aureus, based in part on localization of the site of inhibition to the receptor-histidine kinase, AgrC. Proc. Natl. Acad. Sci. USA 97:13330-13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyon, G. J., and R. P. Novick. 2004. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 25:1389-1403. [DOI] [PubMed] [Google Scholar]
- 33.Marothi, Y. A., H. Agnihotri, and D. Dubey. 2005. Enterococcal resistance—an overview. Indian J. Med. Microbiol. 23:214-219. [PubMed] [Google Scholar]
- 34.Mohamed, J. A., W. Huang, S. R. Nallapareddy, F. Teng, and B. E. Murray. 2004. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect. Immun. 72:3658-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mundy, L. M., D. F. Sahm, and M. S. Gilmore. 2000. Relationships between enterococcal virulence and antimicrobial resistance. Clin. Microbiol. Rev. 13:513-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray, B. E. 2000. Vancomycin-resistant enterococcal infections. N. Engl. J. Med. 342:710-721. [DOI] [PubMed] [Google Scholar]
- 37.Mylonakis, E., M. Engelbert, X. Qin, C. D. Sifri, B. E. Murray, F. M. Ausubel, M. S. Gilmore, and S. B. Calderwood. 2002. The Enterococcus faecalis fsrB gene, a key component of the fsr quorum-sensing system, is associated with virulence in the rabbit endophthalmitis model. Infect. Immun. 70:4678-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakayama, J., Y. Cao, T. Horii, S. Sakuda, A. D. Akkermans, W. M. de Vos, and H. Nagasawa. 2001. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41:145-154. [DOI] [PubMed] [Google Scholar]
- 39.Nakayama, J., Y. Cao, T. Horii, S. Sakuda, and H. Nagasawa. 2001. Chemical synthesis and biological activity of the gelatinase biosynthesis-activating pheromone of Enterococcus faecalis and its analogs. Biosci. Biotechnol. Biochem. 65:2322-2325. [DOI] [PubMed] [Google Scholar]
- 40.Nakayama, J., S. Chen, N. Oyama, K. Nishiguchi, E. A. Azab, E. Tanaka, R. Kariyama, and K. Sonomoto. 2006. Revised model for Enterococcus faecalis fsr quorum-sensing system: small open reading frame, fsrD, encodes GBAP propeptide corresponding to staphylococcal AgrD. J. Bacteriol. 188:8321-8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 42.Otto, M. 2004. Quorum-sensing control in staphylococci—a target for antimicrobial drug therapy? FEMS Microbiol. Lett. 241:135-141. [DOI] [PubMed] [Google Scholar]
- 43.Persson, T., M. Givskov, and J. Nielsen. 2005. Quorum sensing inhibition: targeting chemical communication in gram-negative bacteria. Curr. Med. Chem. 12:3103-3115. [DOI] [PubMed] [Google Scholar]
- 44.Pillai, S. K., G. Sakoulas, G. M. Eliopoulos, R. C. Moellering, Jr., B. E. Murray, and R. T. Inouye. 2004. Effects of glucose on fsr-mediated biofilm formation in Enterococcus faecalis. J. Infect. Dis. 190:967-970. [DOI] [PubMed] [Google Scholar]
- 45.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183:3372-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68:2579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu, R., W. Pei, L. Zhang, J. Lin, and G. Ji. 2005. Identification of the putative staphylococcal AgrB catalytic residues involving the proteolytic cleavage of AgrD to generate autoinducing peptide. J. Biol. Chem. 280:16695-16704. [DOI] [PubMed] [Google Scholar]
- 48.Raffa, R. B., J. R. Iannuzzo, D. R. Levine, K. K. Saeid, R. C. Schwartz, N. T. Sucic, O. D. Terleckyj, and J. M. Young. 2005. Bacterial communication (“quorum sensing”) via ligands and receptors: a novel pharmacologic target for the design of antibiotic drugs. J. Pharmacol. Exp. Ther. 312:417-423. [DOI] [PubMed] [Google Scholar]
- 49.Rice, L. B. 2001. Emergence of vancomycin-resistant enterococci. Emerg. Infect. Dis. 7:183-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rice, S. A., D. McDougald, N. Kumar, and S. Kjelleberg. 2005. The use of quorum-sensing blockers as therapeutic agents for the control of biofilm-associated infections. Curr. Opin. Investig. Drugs 6:178-184. [PubMed] [Google Scholar]
- 51.Rinttila, T., A. Kassinen, E. Malinen, L. Krogius, and A. Palva. 2004. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 97:1166-1177. [DOI] [PubMed] [Google Scholar]
- 52.Seno, Y., R. Kariyama, R. Mitsuhata, K. Monden, and H. Kumon. 2005. Clinical implications of biofilm formation by Enterococcus faecalis in the urinary tract. Acta Med. Okayama 59:79-87. [DOI] [PubMed] [Google Scholar]
- 53.Sifri, C. D., E. Mylonakis, K. V. Singh, X. Qin, D. A. Garsin, B. E. Murray, F. M. Ausubel, and S. B. Calderwood. 2002. Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect. Immun. 70:5647-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith, K. M., Y. Bu, and H. Suga. 2003. Induction and inhibition of Pseudomonas aeruginosa quorum sensing by synthetic autoinducer analogs. Chem. Biol. 10:81-89. [DOI] [PubMed] [Google Scholar]
- 55.Smith, K. M., Y. Bu, and H. Suga. 2003. Library screening for synthetic agonists and antagonists of a Pseudomonas aeruginosa autoinducer. Chem. Biol. 10:563-571. [DOI] [PubMed] [Google Scholar]
- 56.Smith, R. S., and B. H. Iglewski. 2003. Pseudomonas aeruginosa quorum sensing as a potential antimicrobial target. J. Clin. Investig. 112:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tateda, K., R. Comte, J. C. Pechere, T. Kohler, K. Yamaguchi, and C. Van Delden. 2001. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 45:1930-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tateda, K., T. J. Standiford, J. C. Pechere, and K. Yamaguchi. 2004. Regulatory effects of macrolides on bacterial virulence: potential role as quorum-sensing inhibitors. Curr. Pharm. Des. 10:3055-3065. [DOI] [PubMed] [Google Scholar]
- 59.Tendolkar, P. M., A. S. Baghdayan, and N. Shankar. 2003. Pathogenic enterococci: new developments in the 21st century. Cell. Mol. Life Sci. 60:2622-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Toledo-Arana, A., J. Valle, C. Solano, M. J. Arrizubieta, C. Cucarella, M. Lamata, B. Amorena, J. Leiva, J. R. Penades, and I. Lasa. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67:4538-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsunakawa, M., S. L. Hu, Y. Hoshino, D. J. Detlefson, S. E. Hill, T. Furumai, R. J. White, M. Nishio, K. Kawano, S. Yamamoto, Y. Fukagawa, and T. Oki. 1995. Siamycins I and II, new anti-HIV peptides. I. Fermentation, isolation, biological activity and initial characterization. J. Antibiot. 48:433-434. [DOI] [PubMed] [Google Scholar]
- 62.Vergis, E. N., N. Shankar, J. W. Chow, M. K. Hayden, D. R. Snydman, M. J. Zervos, P. K. Linden, M. M. Wagener, and R. R. Muder. 2002. Association between the presence of enterococcal virulence factors gelatinase, hemolysin, and enterococcal surface protein and mortality among patients with bacteremia due to Enterococcus faecalis. Clin. Infect. Dis. 35:570-575. [DOI] [PubMed] [Google Scholar]
- 63.Vieira-da-Motta, O., P. D. Ribeiro, W. Dias da Silva, and E. Medina-Acosta. 2001. RNAIII inhibiting peptide (RIP) inhibits agr-regulated toxin production. Peptides 22:1621-1627. [DOI] [PubMed] [Google Scholar]
- 64.Waters, C. M., M. H. Antiporta, B. E. Murray, and G. M. Dunny. 2003. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J. Bacteriol. 185:3613-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng, J., F. Teng, and B. E. Murray. 2005. Gelatinase is important for translocation of Enterococcus faecalis across polarized human enterocyte-like T84 cells. Infect. Immun. 73:1606-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]