Abstract
The response regulator CovR acts as a master regulator of virulence in Streptococcus pyogenes by repressing transcription of approximately 15% of the group A streptococcus genome directly or indirectly. We demonstrate that phosphorylated CovR represses transcription of rivR directly by binding to conserved sequences located downstream from the promoter to block procession of RNA polymerase. This establishes the first link in a regulatory network where CovR interacts directly with other proteins that modulate gene expression.
As bacteria evolve resistance to antimicrobial therapies, our understanding of their basic pathogenic strategies must evolve as well. The group A streptococcus (GAS) (Streptococcus pyogenes), an important human pathogen, is particularly challenging to understand because, despite its relatively small genome, some strains can cause a wide spectrum of diseases, which differ both in severity and in location within the human body. To cause such diverse diseases, GAS must sense cues from the host microenvironment and respond by expressing specific subsets of virulence factors. Many bacteria use secondary sigma factors to alter gene expression in response to external signals. However, because GAS lacks secondary sigma factors, it appears to rely instead on an intricate network of transcriptional regulators. Interplay among these regulators allows the organism to fine-tune its transcriptome so that it is able to survive and grow in a variety of sites within the human host.
The major global regulator of virulence in GAS is the two-component signal transduction system CovR/S (CsrR/S). In such systems, a sensor kinase, usually located in contact with the outside of the cell, responds to extracellular stimuli by phosphorylating a response regulator. This alters the affinity of the latter compound for specific sites on DNA to which it binds to regulate the level of expression of genes. CovR is a response regulator that acts primarily as a repressor, in contrast to most other response regulators. It is of central importance to GAS because it controls expression of at least 15% of the GAS genome, including many genes that code for virulence factors (3, 11). CovR expression is increased during the acute infection phase in pharyngitis in macaques, suggesting that it has a fundamental role in disease progression (20). Furthermore, there is a correlation between mutations in covS and/or covR and isolates of GAS that are invasive, emphasizing the importance of the CovR/S regulatory system in controlling virulence (19).
Included among the genes repressed by CovR are many genes that code for stress response proteins. The CovR/S system appears to be the primary regulator of stress response in GAS because ΔcovS mutants cannot grow under heat and acid stress conditions unless second-site mutations occur in CovR (4). Thus, under stress conditions, CovS inactivates CovR, presumably by dephosphorylation, so that CovR-repressed stress response proteins can be expressed (3, 4).
While the repression of several key virulence factors by CovR has been shown to be direct, it is likely that at additional promoters, repression by CovR is mediated indirectly through other transcriptional regulators. Comparison of the transcriptome of a ΔcovR strain to that of its wild-type parent showed that CovR represses expression of several proven and putative transcriptional regulators (3, 11). CovR also represses its own transcription directly (14). To begin an investigation of the CovR network, we have started to explore this regulatory cascade by focusing on CovR regulation of a potential transcriptional regulator gene found in microarray analyses to be repressed by CovR, SPy0216 (3, 11), which encodes Ralp4.
SPy0216 (which we have renamed RivR, where the first “R” stands for Ralp and “iv” represents the Roman numeral iv) is a member of the RofA-like protein family of transcriptional regulators, which is unique to, and conserved in, streptococci, including all strains of GAS (12, 16). In strains of some serotypes of GAS, RofA, the prototypical member of the family, binds to the promoter of prtF, which encodes a fibronectin-binding protein, to activate its transcription (8, 9). In addition, RofA binds to its own promoter to activate its own transcription (12). In strains of other serotypes of GAS, Nra, another member of the RofA-like protein family of transcriptional regulators, represses transcription of cpa, which encodes a collagen-binding protein (18). Because RivR exhibits 29% identity with RofA and 31% identity with Nra at the amino acid level, it is likely that RivR also plays a role in modulation of gene expression.
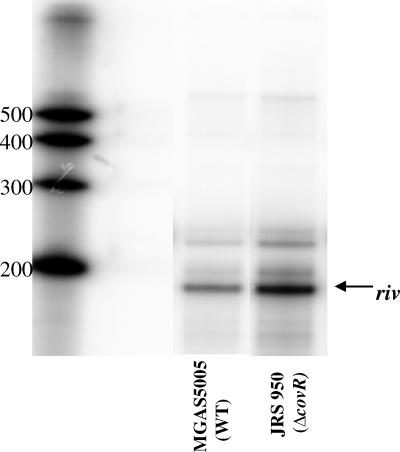
To verify that CovR represses rivR in vivo in the M1 strain MGAS5005, RNase protection assays (RPAs) were performed. Primers Ralp4-int-SG2 and Ralp4-int-AG1 were used to amplify an approximately 190-bp fragment internal to rivR from the chromosome of MGAS5005 for use as a probe template (4). T7 polymerase was used in an in vitro transcription reaction with the rivR probe template and [α-32P]UTP to create a labeled antisense RNA probe. RPAs (Fig. 1) were performed as previously described (4) using this probe and total late-exponential-phase RNA isolated from MGAS5005 or its ΔcovR derivative, JRS950 (6). The RPA results showed that CovR represses rivR about threefold. Furthermore, RPAs using late-exponential-phase RNA isolated from serotype M6 strain JRS4 or its ΔcovR derivative, JRS948 (6), showed that CovR represses rivR in this strain as well (data not shown). Therefore, repression of rivR by CovR does not appear to be strain specific.
FIG. 1.
CovR represses rivR. For the RNase protection assay, 100 μg of total RNA from strain MGAS5005 or JRS950 harvested at the late exponential phase was hybridized to an [α-32P]UTP-labeled rivR probe. The sizes (in nucleotides) of the RNA size markers are indicated on the left. The arrow indicates the band at the size expected for the rivR hybridization product. WT, wild type.
The ability of CovR to bind to promoter regions and the role of this binding have been characterized for several genes that are repressed by CovR (1, 7, 10, 14, 15, 17). At the has promoter (Phas), which directs transcription of genes required for synthesis of the hyaluronic acid capsule, CovR binds to five sites, each of which contains a single ATTARA motif (7). These sites surround the −10 and −35 promoter elements. Because changing the double thymine residues to double guanine residues in these ATTARA motifs prevents binding of CovR, this motif was suggested to be a consensus binding sequence (7). Transcription from Phas is repressed in vitro by CovR in the absence of other factors, showing that this repression is direct (14). Similar results were reported for the cov promoter (Pcov) (15). In contrast to Phas and Pcov, where CovR binds to discrete ATTARA motifs with little or no binding cooperativity, at the sagA promoter (Psag), which directs transcription of the streptolysin S toxin, direct repression by CovR requires cooperative binding to large segments of DNA. In these DNA regions, CovR appears to interact specifically with TTA repeats spaced with a periodicity of 10.9 bp (10). CovR also binds to AT-rich regions in the promoters of speB, ska, speMF/sda, and dppA, which code for cysteine protease, streptokinase, DNase, and dipeptide permease, respectively (5, 13, 17).
The AT-rich nature of the DNA upstream of rivR and the presence of three nearly consensus ATTARA motifs overlapping the putative ribosome-binding site of rivR suggested that CovR might bind to these sequences to repress transcription. To determine if this is correct, we first located the promoter. The 5′ end of the rivR transcript was identified by primer extension (data not shown). Two transcription start sites for rivR were identified using RNA isolated from a ΔcovR derivative of MGAS5005, JRS950; these sites were a major start site (P1) and a minor start site (P2) located approximately 84 bp and 60 bp, respectively, upstream from the start of translation (Fig. 2). The deduced locations of the corresponding promoters were confirmed by in vitro transcription experiments as described below.
FIG. 2.
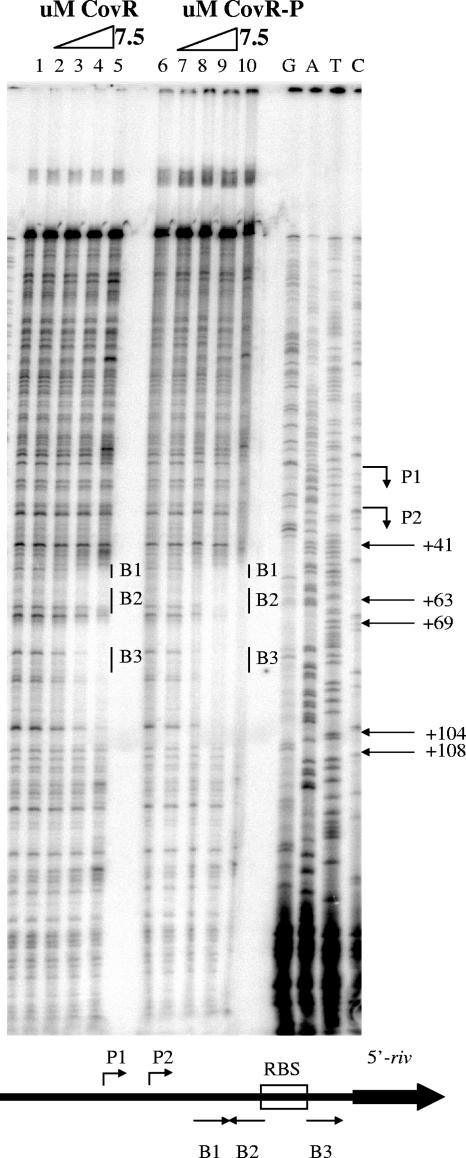
DNA sequence from position −51 to position 149 relative to the P1 start of rivR transcription. The starts of transcription, P1 and P2, are indicated by bent arrows. The nearly consensus ATTARA CovR-binding sites (B1, B2, and B3) are indicated by straight arrows whose directions indicate the orientations of the motif. The region of DNA protected by 7.5 μM CovR-P in a DNase I protection assay is enclosed in a box. The two regions of DNA protected by 7.5 μM unphosphorylated CovR are indicated by dotted lines. The ribosome-binding site is italicized, and the start of translation is indicated by an asterisk.
For CovR binding analyses, a 505-bp DNA segment containing PrivR (from position −314 to position 191 with respect to P1) was amplified from the MGAS5005 chromosome using primers Ralp4-PEs4 and Ralp4-PEa2. DNase I protection assays were performed with unphosphorylated and phosphorylated CovR (CovR-P) as previously described (15). The antisense primer used in the PCR was radiolabeled with 32P. CovR was purified as described elsewhere following overexpression in Escherichia coli (10) and was phosphorylated by incubation with acetyl-phosphate for 2 h, and the protein concentration was determined (14).
The DNase I protection assay showed that CovR binds to two regions of PrivR DNA that include the predicted CovR-binding sites and the start of translation (Fig. 3) (position 41 to position 63 and position 69 to position 104 relative to P1). Furthermore, as observed for all other promoters tested except Psag, phosphorylation of CovR resulted in only a modest enhancement (about twofold) of the binding affinity. However, phosphorylation of CovR did extend the two regions of DNA which were protected to form one contiguous protected region (position 41 to position 108 relative to P1) that included the ribosome-binding site, suggesting that CovR-P polymerizes along the DNA template. CovR-P binding to the rivR ribosome-binding site might also suggest that CovR-P interferes with translation. At other promoters repressed by CovR (Phas, Pcov, and Psag), CovR or CovR-P binds to sequences overlapping the −10 and −35 elements. In contrast, all sequences protected by CovR or CovR-P at PrivR are at least 41 bases downstream of P1 and 17 bases downstream of P2 (Fig. 2). Additional DNase I protection assays revealed that no sequences further upstream were protected from DNase I cleavage by CovR (data not shown). Thus, the rivR promoter is unique among the promoters studied in that CovR-P does not bind to and protect sequences overlapping and surrounding the −10 and −35 elements.
FIG. 3.
CovR protects a region downstream of the P1 start of rivR transcription from DNase I cleavage. CovR or CovR-P was added to the radiolabeled antisense strand. Lanes 1 and 6, no CovR; lanes 2 to 5, twofold increases in the concentration of unphosphorylated CovR from 0.9 to 7.5 μM; lanes 7 to 10, twofold increases in the concentration of phosphorylated CovR from 0.9 to 7.5 μM. The positions of the three nearly consensus ATTARA CovR-binding sequences B1, B2, and B3 are indicated by vertical lines, the positions of the transcription start sites (P1 and P2) are indicated by bent arrows, and the positions of the coordinates relative to P1 are indicated by straight arrows. The ribosome-binding site is located between position 63 and position 69. Reaction mixtures were run next to sequencing ladders (G,A,T, and C). Below the DNase I footprint is a diagram indicating the positions of the CovR-binding sites in relation to the ribosome-binding site (RBS) and transcription start sites. The orientations of the predicted CovR-binding sites are indicated by the directions of the arrows.
The ability of CovR to bind specifically to PrivR in vitro supports the hypothesis that CovR or CovR-P represses transcription of rivR directly. However, CovR binding could mediate repression by another means (as it does at PdppA [13]), such as by preventing the binding of an activator protein. To determine whether CovR alone represses transcription from PrivR, in vitro transcription using GAS RNA polymerase and sigma factor was employed (14). To construct a PrivR template for in vitro transcription, the same 505-bp DNA segment used in the DNase I protection assay was amplified, digested with BamHI and XhoI, and cloned into the BamHI/XhoI sites of pJRS462 (2) to create pJRS1608. This plasmid was linearized with EcoRI, which cut at sites 215 bp and 191 bp downstream of the predicted P1 and P2 start sites for PrivR. The kanamycin resistance gene (aphA3) was used as an internal transcription control. For this, the plasmid was also digested with XcmI, which cut 510 bp downstream of the PaphA3 start site.
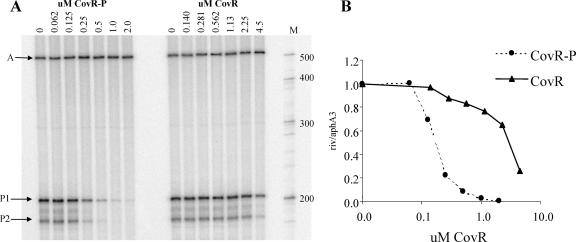
Incubation of the linear template with increasing concentrations of CovR-P led to decreased synthesis of the rivR transcript, while the levels of the aphA3 transcript were unaffected (Fig. 4). Thus, purified CovR-P is sufficient to repress transcription from PrivR, and this repression is specific. At PrivR, repression in vitro by CovR-P occurred at a concentration similar to that observed for previously studied promoters. However, it was surprising that although unphosphorylated CovR bound to PrivR with an affinity similar to that of CovR-P in DNase I protection assays (Fig. 2), phosphorylation enhanced repression of PrivR approximately 20-fold (3.4 μM CovR was required to repress PrivR transcription by 50%, while only 0. 17 μM CovR-P was needed to achieve 50% repression) (Fig. 4). At Phas, a discrepancy between phosphorylation-dependent enhancement of binding and enhancement of repression by CovR also occurred (2-fold versus 6.5-fold) (14), but this discrepancy is less severe than that observed for PrivR. At Phas, the effect of phosphorylation on repression was enhanced because RNA polymerase recruited phosphorylated, but not unphosphorylated, CovR to bind to the DNA. At PrivR, however, RNA polymerase had no effect on the ability of CovR-P to bind (data not shown). It appears, therefore, that phosphorylation of CovR may alter the protein-DNA interaction at PrivR so that it is more resistant to displacement by RNA polymerase. Several possible mechanisms can be proposed for this. Because phosphorylation causes dimerization of CovR (14), phosphorylated CovR binds as dimers while unphosphorylated CovR binds as monomers. Furthermore, phosphorylation of CovR extends protection from DNase I to a contiguous region. This suggests that the bound dimeric CovR-P may interact with additional CovR-P protein. Thus, at PrivR, it is possible that progression of RNA polymerase can sequentially displace discrete CovR monomers in a stepwise fashion, while a contiguous tract of phosphorylated CovR dimers may present a significant energetic barrier to polymerase progression. Additionally, polymerization of CovR-P along PrivR may lead to DNA bending that favors dissociation of the RNA polymerase.
FIG. 4.
(A) In vitro transcription from the aphA3 promoter (arrow A) and from the rivR promoter (start sites P1 and P2 [arrows P1 and P2]) with increasing amounts of CovR or phosphorylated CovR. Lane M contained an RNA size marker, and the sizes (in nucleotides) are indicated on the right. (B) Densitometric analysis (ImageQuant) was performed, and the P1rivR/PaphA3 ratio was normalized to 1. 0 in the absence of CovR. The same curves were obtained for both P1 and P2.
In summary, this work defines several new aspects of CovR regulation. First, PrivR is the first promoter investigated at which unphosphorylated CovR is essentially ineffective at repression in vitro. This suggests that GAS requires expression of RivR for growth in situations where CovR is present but dephosphorylated. This occurs under general stress conditions. Second, while at other promoters CovR binding occludes the promoter elements to retard initiation of transcription, at PrivR CovR-P acts as a roadblock, preventing elongation of the transcript. This is a new mode of regulation by CovR. The differences in the locations of CovR-binding sites and the consequent different mechanisms by which CovR interferes with transcription at different promoters suggest ways in which a single global regulator provides a large repertoire of growth-specific responses to control differential gene expression. This work is also the first demonstration that CovR directly represses a transcriptional regulator other than itself, and it thus demonstrates the ability of CovR to interact with other regulatory systems and to fine-tune the response of GAS to additional environmental signals.
Acknowledgments
This work was supported by NIH grant AI20723. S.A.R. was supported in part by NIH training grant T32-AI07470 and in part by an award from the American Heart Association.
We thank Virginia Stringer and JinXin Gao for technical assistance.
Footnotes
Published ahead of print on 8 September 2006.
REFERENCES
- 1.Bernish, B., and I. van de Rijn. 1999. Characterization of a two-component system in Streptococcus pyogenes which is involved in regulation of hyaluronic acid production. J. Biol. Chem. 274:4786-4793. [DOI] [PubMed] [Google Scholar]
- 2.Biswas, I., and J. R. Scott. 2003. Identification of rocA, a positive regulator of covR expression in the group A streptococcus. J. Bacteriol. 185:3081-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalton, T. L., J. T. Collins, T. C. Barnett, and J. R. Scott. 2006. RscA, a member of the MDR1 family of transporters, is repressed by CovR and required for growth of Streptococcus pyogenes under heat stress. J. Bacteriol. 188:77-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalton, T. L., and J. R. Scott. 2004. CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes. J. Bacteriol. 186:3928-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Federle, M. J. 2002. Ph.D. thesis. Emory University School of Medicine, Atlanta, GA.
- 6.Federle, M. J., K. S. McIver, and J. R. Scott. 1999. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J. Bacteriol. 181:3649-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Federle, M. J., and J. R. Scott. 2002. Identification of binding sites for the group A streptococcal global regulator CovR. Mol. Microbiol. 43:1161-1172. [DOI] [PubMed] [Google Scholar]
- 8.Fogg, G. C., and M. G. Caparon. 1997. Constitutive expression of fibronectin binding in Streptococcus pyogenes as a result of anaerobic activation of rofA. J. Bacteriol. 179:6172-6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fogg, G. C., C. M. Gibson, and M. G. Caparon. 1994. The identification of rofA, a positive-acting regulatory component of prtF expression: use of an m gamma delta-based shuttle mutagenesis strategy in Streptococcus pyogenes. Mol. Microbiol. 11:671-684. [DOI] [PubMed] [Google Scholar]
- 10.Gao, J., A. A. Gusa, J. R. Scott, and G. Churchward. 2005. Binding of the global response regulator protein CovR to the sag promoter of Streptococcus pyogenes reveals a new mode of CovR-DNA interaction. J. Biol. Chem. 280:38948-38956. [DOI] [PubMed] [Google Scholar]
- 11.Graham, M. R., L. M. Smoot, C. A. Migliaccio, K. Virtaneva, D. E. Sturdevant, S. F. Porcella, M. J. Federle, G. J. Adams, J. R. Scott, and J. M. Musser. 2002. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. USA 99:13855-13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Granok, A. B., D. Parsonage, R. P. Ross, and M. G. Caparon. 2000. The RofA binding site in Streptococcus pyogenes is utilized in multiple transcriptional pathways. J. Bacteriol. 182:1529-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gusa, A. A., B. Froehlich, D. Desai, V. Stringer, and J. R. Scott. 22. September 2006. CovR activation of the dipeptide permease promoter (PdppA) in group A streptococcus. J. Bacteriol. doi: 10.1128/JB.01036-06. [DOI] [PMC free article] [PubMed]
- 14.Gusa, A. A., J. Gao, V. Stringer, G. Churchward, and J. R. Scott. 2006. Phosphorylation of the group A streptococcal CovR response regulator causes dimerization and promoter-specific recruitment by RNA polymerase. J. Bacteriol. 188:4620-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gusa, A. A., and J. R. Scott. 2005. The CovR response regulator of group A streptococcus (GAS) acts directly to repress its own promoter. Mol. Microbiol. 56:1195-1207. [DOI] [PubMed] [Google Scholar]
- 16.Gutekunst, H., B. J. Eikmanns, and D. J. Reinscheid. 2003. Analysis of RogB-controlled virulence mechanisms and gene repression in Streptococcus agalactiae. Infect. Immun. 71:5056-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, A. A., N. C. Engleberg, and V. J. DiRita. 2001. Repression of virulence genes by phosphorylation-dependent oligomerization of CsrR at target promoters in S. pyogenes. Mol. Microbiol. 40:976-990. [DOI] [PubMed] [Google Scholar]
- 18.Podbielski, A., M. Woischnik, B. A. Leonard, and K. H. Schmidt. 1999. Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31:1051-1064. [DOI] [PubMed] [Google Scholar]
- 19.Sumby, P., A. R. Whitney, E. A. Graviss, F. R. DeLeo, and J. M. Musser. 2006. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Virtaneva, K., S. F. Porcella, M. R. Graham, R. M. Ireland, C. A. Johnson, S. M. Ricklefs, I. Babar, L. D. Parkins, R. A. Romero, G. J. Corn, D. J. Gardner, J. R. Bailey, M. J. Parnell, and J. M. Musser. 2005. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc. Natl. Acad. Sci. USA 102:9014-9019. [DOI] [PMC free article] [PubMed] [Google Scholar]