Abstract
The FCT regions of Streptococcus pyogenes strains encode a variety of cell wall-anchored surface proteins that bind the extracellular matrix of the human host and/or give rise to pilus-like appendages. Strong linkage is evident between transcription-regulatory loci positioned within the FCT and emm regions and the emm pattern genotype marker for preferred infection of the throat or skin. These findings provide a basis for the hypothesis that FCT region gene products contribute to tissue-specific infection. In an initial series of steps to address this possibility, the FCT regions of 13 strains underwent comparative sequence analysis, the gene content of the FCT region was characterized for 113 strains via PCR, and genetic linkage was assessed. A history of extensive recombination within FCT regions was evident. The emm pattern D-defined skin specialist strains were highly homogenous in their FCT region gene contents, whereas the emm pattern A-C-defined throat specialist strains displayed a greater variety of forms. Most pattern A-C strains harbored prtF1 (75%) but lacked cpa (75%). In contrast, the majority of emm pattern D strains had cpa (92%) but lacked prtF1 (79%). Models based on FCT and emm region genotypes revealed the most parsimonious pathways of evolution. Using niche-determining candidate genes to infer phylogeny, emm pattern E strains—the so-called generalists, which lack a strong tissue site preference—occupied a transition zone separating most throat specialists from skin specialists. Overall, population genetic analysis supports the possibility that the FCT region gives rise to surface proteins that are largely necessary, but not always sufficient, to confer tissue site preference for infection.
Streptococcus pyogenes can cause a wide variety of diseases in its human host (13). However, most often it causes infection of the throat mucosal epithelium or skin epidermal layers. In most parts of the world, streptococcal pharyngitis and/or impetigo attains a high disease incidence (13, 15). Colonization, followed by a low level of bacterial growth, tends to lead to asymptomatic carriage. In contrast, infection is characterized by high numbers of organisms due to replication in situ and an accompanying host immune response. Since the throat and skin provide an easy exit for progeny, infection at superficial tissue sites provides a key source for new transmission.
Numerous epidemiological studies report differences in the strains of S. pyogenes recovered from oropharyngeal infection (pharyngitis) versus superficial skin infection (impetigo) (4, 13, 15, 16, 18, 34, 43, 49, 65). Markers used to identify distinct strains were often based on antigenic forms of the M surface protein (M serotypes). The epidemiologic observations led to the recognition of distinct tissue tropisms among members of the species. More recent analysis of the emm genes encoding M proteins led to the definition of three emm genotype patterns, based on the number of emm genes, their ancestral lineages, and their chromosomal arrangements (29, 30).
Multiple isolates of most emm types studied are restricted to a single emm pattern genotype. Among 495 isolates represented by 160 emm types, and including numerous isolates of the more common emm types, only two emm types were observed in association with more than one pattern group (44) despite the fact that a given emm type can often be recovered from strains with distant genetic backgrounds. Thus, the emm pattern can be deduced from the emm type with reasonable accuracy. Population-based surveillance in temperate regions (the United States and Italy), where streptococcal pharyngitis is far more prevalent than impetigo, shows that the vast majority of isolates recovered from cases of throat infection have emm types characteristic of the emm pattern A-C or E genotype, with fewer than 1% of the isolates having pattern D emm types (17, 55). Likewise, nearly all isolates recovered from cases of acute pharyngitis in Mexico (22), Spain (1), and Germany (14) are of emm types characteristic of the emm pattern A-C or E genotype. In a population-based study in tropical Australia, where impetigo is endemic and both pharyngitis and throat colonization are rare, the majority of impetigo isolates had the emm pattern D (46%) or E (40%) genotype (9). In a recent population-based survey in Nepal, 19% of the impetigo isolates were emm pattern A-C (53), higher than the 13% of impetigo isolates from tropical Australia that were pattern A-C (9). A recent study in Ethiopia showed that none of the >60 impetigo isolates had emm types associated with the emm pattern A-C genotype (63).
The findings from population-based collections of isolates are largely consistent with initial observations made from a worldwide collection (12). Taken together, these data provide strong support for the idea that the emm pattern genotype can serve as a reliable genetic marker for tissue site preferences among S. pyogenes causing infection. Strains with the emm pattern A-C genotype tend to show a strong preference for throat infection (throat specialists), emm pattern D strains have a predilection for causing impetigo (skin specialists), and emm pattern E strains readily infect both tissues (generalists).
The observed emm pattern correlations with tissue site preference are restricted to cases of infection and do not necessarily extend to asymptomatic carriage (53). It is also important to emphasize that the association of the emm pattern genotype with throat versus skin infection reflects a general trend, and there are exceptions. For example, in the Ethiopia study, 28% of tonsillitis isolates were of emm types characteristic of emm pattern D (63). Although the relative rates of tonsillitis versus impetigo in Ethiopia are not known, there appear to be ample levels of both forms of disease; a problem inherent in assigning a particular isolate to throat infection is a streptococcal carrier state that coincides with pharyngitis or tonsillitis due to another cause (e.g., virus). Unusual or exceptional clones may also contribute to deviations from the general trend. Nearly all of the pattern A-C impetigo isolates from the Nepal (53) and tropical Australia (9) studies were dominated by just one or two clones, and the pattern A-C clones of both studies were recovered more often from impetigo lesions than from throat carriage.
T antigens provide the basis for another serological typing scheme, in which T and M types share some degree of concordance (33). At least some T antigens are cell wall-anchored surface proteins encoded by genes positioned within the FCT region of the S. pyogenes genome (10, 42, 45, 54). Included among the FCT region surface proteins are several microbial surface cell recognition adhesion matrix molecules (MSCRAMMs) that bind human fibronectin or collagen (24, 38, 39). Recent findings show that at least some T proteins form pilus-like appendages on the bacterial cell surface (42, 45). Thus, FCT region gene products of S. pyogenes are good candidates to have key roles in pathogenesis by mediating bacterial adherence to host tissue. In fact, strong genetic linkage is observed between the emm pattern genotype and rofA versus nra lineage-specific alleles of the transcription-regulatory locus that lies within the FCT region and regulates the expression of at least some of the FCT surface protein genes (10, 11).
The strong associations observed between particular S. pyogenes strains and certain forms of disease raise the possibility that throat-tropic strains share genotypes that distinguish them from skin-tropic strains. Alternatively, there may be a multitude of molecular strategies that are used to achieve the same disease phenotype. In this report, the FCT region gene contents are defined for >100 strains representing a broad segment of the S. pyogenes population. Genetic linkage among various combinations of FCT region genes is assessed, and the relationships between FCT region genes and emm markers for preferred tissue sites for infection are delineated.
MATERIALS AND METHODS
Bacterial strains.
The sample set of 113 strains of S. pyogenes chosen for genotype analysis is listed in Table S1 in the supplemental material and matches a data set described previously (10), with the exclusion of one isolate (MGAS2140). The multilocus sequence type (MLST), emm type, and emm pattern genotype findings were previously reported (11); strain ACTH81 was reassigned emmst11014. Strains used for T-serotype determination and/or nucleotide sequence analysis were obtained as described previously (10) or purchased from the American Type Culture Collection. The T agglutination test was performed following trypsin treatment of whole bacteria (32).
Nucleotide sequence determination and analysis.
The nucleotide sequence of the FCT region was determined according to previously described methods (10), using long overlapping PCR fragments and primer walking. The sequencing strategy included strands in both directions and resulted in at least twofold coverage for all regions. Contigs were assembled and sequences were analyzed for open reading frames (ORFs) using Lasergene software (DNASTAR, Inc., Madison, WI). BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) was used for sequence alignments via the Clustal W algorithm and for calculation of sequence similarity matrices.
PCR amplification-based mapping.
PCR amplifications were performed using an annealing temperature of 55°C and extension times of at least 1 min per 0.8 kb of expected product. Two different sets of primers were used for four genes displaying high levels of nucleotide sequence heterogeneity (prtF1, prtF2, cpa, and fctA). For fctA, the second reaction used a forward primer specific for the 3′ end of sipA2 and a reverse primer targeting the 5′ end of srtC2 (designated a “bridge” reaction). All strains were tested two or more times for each reaction, except that the fctA bridge reaction was used only for strains that were negative for fctA but positive for sipA2 and srtC2. For prtF1, prtF2, cpa, and fctA, a strain was scored positive if at least one of the two reactions (each tested in duplicate) yielded a product.
The oligonucleotide primer pairs tested for 13 different PCR amplification reactions were as follows: reaction PrtF1-A (amplifying an ∼0.55-kb fragment of prtF1) forward, 5′-TGCGC GGGTT CTATC GGTTT TGGTC AAGTA-3′, and reverse, 5′-AATTA GTTTT YTCAR WGCYT CACGC ATTAA-3′; reaction PrtF1-B (amplifying an ∼1.2-kb fragment of prtF1) forward, same as for reaction PrtF1-A, and reverse, 5′-CTCCG TCTCA CCAGA CTCAC CCGCT AGAGG TGATT GGTC-3′; reaction Cpa-A (amplifying an ∼0.75-kb fragment of cpa) forward, 5′-GGATA TGAGA TTGCC GAACC TATTA CTTTT AAAG-3′, and reverse, 5′-GGAGC CTGTT TATCT TCCAT TCGAA TAATA TCCAC-3′; reaction Cpa-B (amplifying an ∼1.3-kb fragment of cpa) forward, 5′-GAAGG TGACT ACTCT AAACT TCTAG AGGGA GCAAC-3′, and reverse, 5′-CCAGT TGGTG GGACA AGATC TTTWC GG-3′; reaction PrtF2-A (amplifying an ∼0.60-kb fragment of prtF2) forward, 5′-GCTGG TGCAA CTATG GAGTT GCGTG ATTCA TCTGG T-3′, and reverse, 5′-CCAGT TGCTG GTAAA CTAGT ATTAC TCTTT GGC-3′; reaction PrtF2-B (amplifying an ∼1.2-kb fragment of prtF2) forward, same as for reaction PrtF1-B, and reverse, 5′-CCCTG GTTAT ACTGG TTGGA GTCCT TCTCT AG-3′; reaction SipA2 (amplifying an ∼0.46-kb fragment of sipA2) forward, 5′-GCTTT CATAC GGTTA GTACT TAAGA TTTCT ATTAT TGG-3′, and reverse, 5′-CCTCT CACTC TTAAT AGAGT TGAGA TTTTC CC-3′; reaction FctA (amplifying an ∼1.0-kb fragment of fctA) forward, 5′-AAATT ATTAC TTGCT ACTGC AATCT TAGCA ACTGC-3′, and reverse, 5′-CTCCA CCAAT AGCCA CAATG CTAAG AACTG CAAAT GGAGC-3′; SipA-SrtC bridge reaction (amplifying an ∼1.3-kb fragment that encompasses fctA) forward, 5′-GGGAA AATCT CAACT CTATT AAGAG TGAGA GG-3′, and reverse, 5′-GGCTT TATTG ATAAC CTGTA CAATT GTCAT C-3′; reaction SrtC2 (amplifying an ∼0.72-kb fragment of srtC2) forward, 5′-GATGA CAATT GTACA GGTTA TCAAT AAAGC C-3′, and reverse, 5′-CTTGA ATAGT ACCGA CAACG ATAAC ACGAT TGTCA G-3′; reaction FctB (amplifying an ∼0.53-kb fragment of fctA) forward, 5′-ATGTT ATTTT CTGTC GTAAT GATAT TAACC-3′, and reverse, 5′-CTAGT AACCC CAGTA ATACG ATACT TAAGA TACCC-3′; reaction SrtB (amplifying an ∼0.74-kb fragment of srtB) forward, 5′-CTAAA ATAAT AGCTA TAACC ACCCC GAAAG CAGCA C-3′, and reverse, 5′-CTAAA ATAAT AGCTA TAACC ACCCC GAAAG CAGCA C-3′; and reaction Sof (amplifying an ∼0.65-kb fragment of sof) forward, 5′-GTATA AACTT AGAAA GTTAT CTGTA GG-3′, and reverse, 5′-GGCCA TAACA TCGGC ACCTT CGTCA ATT-3′.
To address the potential problem of either false positives or false negatives, several quality controls were implemented. Controls for the template DNA, deoxynucleotide triphosphates, and Taq polymerase always included PCR amplification of a housekeeping gene (usually xpt or mutS) (21) performed in parallel. In addition, for a given primer pair set, all PCR amplifications were performed in parallel with a positive control template DNA and often with a known negative control DNA. All reactions were performed at least twice on separate days. To score PCRs as positive or negative, the PCR products were confirmed for their predicted sizes by agarose gel electrophoresis using molecular size markers and scored only if they were clearly negative or strongly positive based on the intensity of ethidium bromide staining. Discordant findings (e.g., two trials giving different results or bands that were present but weak) led to multiple rechecks of that strain using template DNA that was freshly prepared by boiling colony picks grown from the frozen stock culture; this helped to eliminate confounding results due to cross-contamination of the DNA template with DNAs from other strains. Together, the controls helped to ensure that both false positives (due to DNA cross-contamination) and false negatives (due to amplification of the wrong gene, partially evidenced by gel migration; bad reagents, template, or primers; or failure of the thermal-cycler machine) were minimized.
Additional controls to help rule out potential false positives among the PrtF1-A, Cpa-A, FctA, and SipA-SrtC bridge reaction products, each of which exhibited high levels of sequence heterogeneity among alleles, included nucleotide sequence determination and multiple-sequence alignment of a large selection of products; findings of well-aligned sequences signified that the PCR products were part of the same gene family (44, 51) (data not shown).
Statistical analyses.
Linkage disequilibrium between pairs of genes was calculated using 2-by-2 tests for independence; data are reported for χ2 analysis (two tailed) with Yates' correction (DnaSP 4.10). Unrooted phylogenetic trees were based on matrices of character states at several loci, defined as the presence/absence of a locus or a specified gene lineage, and were constructed by maximum parsimony (PAUP 4.0; Sinauer Associates). Diversity (D; Simpson's diversity index) among the S. pyogenes isolates was quantified according to methods that incorporate the number of distinct genotypes and their frequencies of occurrence (31), where a D value of 1.0 indicated that the genotype was able to discriminate between all isolates and a D value of 0.0 indicated that all isolates had identical genotypes. Confidence intervals (95%) were determined by the method of Grundmann et al. (26).
Nucleotide sequence accession numbers.
The complete nucleotide sequences of the FCT regions of strains ALAB49, 29487, and D633 have been assigned GenBank accession numbers DQ984656, EF025060, and EF025061.
RESULTS AND DISCUSSION
Comparative sequence analysis of FCT regions.
The nucleotide sequences of the complete FCT regions of S. pyogenes strains represented by 10 emm types have been reported; however, all 10 strains are either emm pattern genotype A-C or E. As an initial step toward understanding the relationship between FCT region genes and the so-called skin specialists (i.e., emm pattern D) and expanding our knowledge of the S. pyogenes population, the complete nucleotide sequences of the FCT regions for three isolates derived from impetigo skin lesions were determined. They were denoted ALAB49 (emm53), recovered in Alabama in 1987; 29487 (emm33), from the Czech Republic in 1986; and D633 (emmstD633), isolated in Trinidad in 1972.
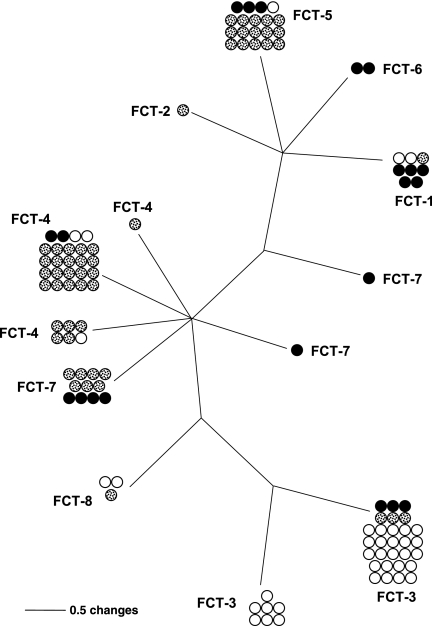
In an earlier report, the FCT regions of five S. pyogenes strains were subjected to a detailed sequence analysis (10). An update to that analysis is presented here and includes all 13 emm type-defined strains whose complete FCT region sequences are known. Levels of sequence identity established by BLASTN (3), combined with relative positions within the FCT region, were used to help distinguish between alleles belonging to the same locus and alleles derived from distinct genes. Based on gene content and order, each FCT region was assigned to one of six major forms (Fig. 1). Genotypes FCT-1 through FCT-4 were previously described (10, 46, 60). The recently reported genome sequences of an emm4 and an emm2 strain (8) indicate that each of the two strains represents a new FCT region genotype arrangement, designated FCT-5 and FCT-6, respectively. Only a single strain was identified for each of the FCT-1, -2, -5, and -6 genotypes. The three emm pattern D impetigo isolates that underwent nucleotide sequence determination for this study were found to share the previously recognized FCT-3 form with several emm pattern A-C and E strains. In all, 9 of the 13 strains displayed the FCT-3 or FCT-4 form.
FIG. 1.
Structural diversity of FCT regions. The FCT region structures are depicted for strains representing 13 emm types whose complete FCT region sequences are known. Regions FCT-1 through FCT-4 were assigned using nomenclature described previously (60); genotypes FCT-5 and FCT-6 were newly assigned. The lengths of the FCT regions of S. pyogenes range from ∼10 to 16 kb. Genes are designated, according to nucleotide sequence alignments with all other genes in this data set, as ranging from highly conserved to unique. The FCT regions are bounded by highly conserved ORFs encoding a putative chaperonin on the left flank, which is closest to the origin of replication, and a hypothetical protein on the right flank (10). Five of the shared loci encode (putative) cell wall-anchored surface proteins: cpa (collagen-binding protein), fctA (also known as orf100 in M type 3 strains and orf80 in M type 5 strains and eftLSLA in M type 12 strains), fctB (also known as orf102 in M type 3 strains and orf82 in M type 5 strains and orf2 in M type 49 strains), prtF1 (fibronectin-binding protein; also known as sfbI), and prtF2 (fibronectin-binding protein; also known as pfbpI and fbaB). FCT region sequences were based on the following strains and GenBank accession numbers: FCT-1, MGAS10394 (M6; CP000003) and D471 (M6) (10); FCT-2, SF370 (M1; AE04092) and MGAS5005 (M1; CP000017); FCT-3, MGA215 (M3; AE014074), Manfredo (M5 [http://www.sanger.ac.uk]), MGAS8232 (M18; AE009949), 29487 (M33), B737 and CS101 (M49; U49397 and AY049089), ALAB49 (M53), and D633 (stD633); FCT-4, A735 (M12; AF447492) and MGAS6180 (M28; CP000056); FCT-5, MGAS10750 (M4; CP000262); and FCT-6, MGAS10270 (M2; CP000260). The fctX product of the FCT-1 strain (emm6) has been referred to as cpa (45); however, when FctX is aligned with any of the Cpa family proteins, the amino acid identity does not exceed 10% (Table 1 and data not shown); thus, fctX appears to be unique among emm6 isolates and structurally unrelated to cpa.
Several genes are shared among multiple FCT region forms. Using the predicted ORFs of strain ALAB49 (FCT-3) for comparison, amino acid sequence identities based on Clustal W alignments were calculated (Table 1) . Positioned at the left end of all six FCT region forms is a transcriptional regulatory gene corresponding to either the nra or rofA lineage (Fig. 1). The predicted amino acid sequence identities to Nra of strain ALAB49 are ∼99 and 60%, respectively, for products of the nra and rofA lineage alleles (Table 1) (11). There are also several highly conserved gene products, exhibiting more than 95% amino acid sequence identity, that are shared among the nine strains of the FCT-3 or FCT-4 genotype. These include SipA2 (a putative signal peptidase), SrtC2 (a specialized sortase) (5), and MsmR (a transcriptional regulator) (46). Several distinct (putative) sortase genes were identified, and all six FCT region forms had at least one type of sortase gene (data not shown); however, aside from srtC2, only srtB is present in more than one FCT region form (FCT-1, -2, and -4) (Fig. 1). In contrast to sortase, putative signal peptidase genes are restricted to three FCT regions.
TABLE 1.
Amino acid sequence identities of FCT-region genes, based on Clustal W alignments with strain ALAB49
| Strain | emm type | emm pattern genotype | T serotype | Sequence identity
|
FCT-region genotype designation | STc | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nraa | cpa | sipA | fctA | srtC | fctB | msmR | prtF2 | ||||||
| ALAB49 | 53 | D | T3/13/B | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 (fba-like) | FCT-3 | 11 |
| 29487 | 33 | D | T3/13/B | 1.000 | 1.000 | 1.000 | 1.000 | 0.995 | 0.989 | 0.997 | 1.000 (fba-like) | FCT-3 | 3 |
| D633 | stD633 | D | T3/13/B | 0.998 | 0.783 | 0.976 | 0.631 | 0.995 | 0.962 | 0.992 | 0.987 (fba-like) | FCT-3 | 48 |
| MGAS8232 | 18 | A-C | TNTf | Frameshift | 0.777 | 0.982 | 0.637 | 0.991 | 0.994 | 0.995 | 0.942 (fba-like) | FCT-3 | 42 |
| Manfredo | 5 | A-C | T5/27/44 | 0.996 | 0.579 | 0.965 | 0.692 | 0.991 | 0.994 | 0.995 | 0.942 (fba-like) | FCT-3 | 99 |
| MGAS315 | 3 | A-C | TNT | 0.996 | 0.536 | 0.953 | 0.675 (orf100) | 0.983 | 0.978 (orf102) | 0.995 | 0.938 (fba-like) | FCT-3 | 15d |
| CS101; B737e | 49 | E | TNT | 0.996 | 0.463 | 0.953 (lepAL) | 0.652b | 1.000b | 0.978 (orf2) | 0.995 | 0.373 (pfbpI-like) | FCT-3 | 30 |
| A735 | 12 | A-C | T12 | 0.608 (rofA) | 0.395 | 0.971 | 0.628 (eftLSLA) | 0.995 (eftLSLB) | 0.978 | 0.995 | 0.372 (pfbpI-like) | FCT-4 | 36 |
| MGAS6180 | 28 | E | T28 | 0.604 (rofA) | 0.378 | 0.965 | 0.631 | 0.991 | 0.978 | 0.992 | 0.373 (pfbpI-like) | FCT-4 | 52 |
| MGAS5005; SF370 | 1 | A-C | T1 | 0.600 (rofA) | 0.354 | 0.393 (SPy0127) | 0.352 (SPy0128) | 0.473 (SPy0129) | 0.279 (SPy0130) | NA | NA | FCT-2 | 28 |
| MGAS10394; D471 | 6 | A-C | T6 | 0.604 (rofA) | 0.072 (fctX) | NA | NA | NA | NA | NA | NA | FCT-1 | 37, 382 |
| MGAS10750 | 4 | E | NDh | 0.605 (rofA) | NAg | NA | NA | NA | NA | NA | NA | FCT-5 | 39 |
| MGAS10270 | 2 | E | ND | 0.609 (rofA) | NA | NA | NA | NA | NA | NA | NA | FCT-6 | 55 |
Alternative gene names are given in parentheses.
The eftLSL sequence is published as readthrough fctA and srtC2; the calculated values in the table assume a stop codon at nucleotides 1033 to 1035 of eftLSL.
From Table S1 in the supplemental material or http://www.mlst.net or determined in silico from whole genome sequences (GenBank accession numbers are listed in the legend to Fig. 1).
Single-locus variant of ST16, the MLST genotype of the strain used for FCT region mapping (see Table S1 in the supplemental material).
CS101 (Minnesota) is a laboratory derivative of B737 (Lancefield) (10); the right half of the FCT region sequence was determined using the CS101 culture, whereas for the right half, the B737 culture was used.
TNT, T nontypeable.
NA, not applicable.
ND, not determined.
The combined FCT regions of the 13 sequenced strains contain at least 15 distinct gene types encoding (putative) cell wall-anchored surface proteins (Fig. 1 and data not shown). However, only five surface protein genes are present in multiple FCT region forms: prtF1, cpa, fctA, fctB, and prtF2. Sequence identities, based on alignment with the predicted amino acid sequences of the surface protein genes present in strain ALAB49, were ascertained for all other strains that also had that gene. The most highly conserved gene encoded FctB, with amino acid sequence identities exceeding 95% (Table 1). Clustal W alignment of PrtF2 gene products revealed two discrete gene lineages, designated fbaB and pfbpI (50). The products of fbaB- versus pfbpI-like lineage alleles display only ∼ 35% overall amino acid sequence identity (data not shown), although within a lineage, the gene products are highly similar in sequence (Table 1). The fbaB and pfbpI lineage alleles correspond to the FCT-3 and FCT-4 genotypes, respectively, with the exception of the emm49 strain, which has an FCT-3 genotype and a pfbpI lineage allele (Table 1). The latter finding is indicative of a history of genetic recombination within the FCT region following interstrain horizontal gene transfer events.
The cell wall-anchored surface protein families of Cpa and FctA each display extensive amino acid heterogeneity. The magnitude of sequence similarity among Cpa proteins tends to be discordant with that found among FctA proteins (Tables 2 and 3). For example, the FctA proteins of strains D633 and MGAS6180 are 81% identical, whereas their Cpa proteins display only 37% identity; even their prtF2 genes belong to different lineages. Conversely, strains D633 and MGAS8232 have highly similar Cpa and PrtF2 proteins (98 and 94% identity, respectively), but their FctA sequences diverge by more than 35%. The finding on Cpa-FctA discordance may have biological implications for the direct molecular interactions that are observed between Cpa and FctA (42, 45). Furthermore, the Cpa-FctA discordance provides additional evidence for genetic recombination within the FCT region leading to the generation of mosaic-like structures.
TABLE 2.
Sequence similarity matrix for Cpa following amino acid sequence alignment using the Clustal W algorithm
| emm type | FCT region form | Strain | Sequence similarity
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALAB49 | 29487 | D633 | MGAS8232 | Manfredo | MGAS315 | CS101 | A735 | MGAS6180 | SF370 | MGAS5005 | |||
| 53 | 3 | ALAB49 | IDa | 1.000 | 0.783 | 0.777 | 0.579 | 0.536 | 0.463 | 0.395 | 0.378 | 0.354 | 0.354 |
| 33 | 3 | 29487 | 1.000 | ID | 0.783 | 0.777 | 0.579 | 0.536 | 0.463 | 0.395 | 0.378 | 0.354 | 0.354 |
| stD633 | 3 | D633 | 0.783 | 0.783 | ID | 0.984 | 0.566 | 0.544 | 0.449 | 0.397 | 0.372 | 0.334 | 0.334 |
| 18 | 3 | MGAS8232 | 0.777 | 0.777 | 0.984 | ID | 0.568 | 0.540 | 0.448 | 0.397 | 0.372 | 0.333 | 0.333 |
| 5 | 3 | Manfredo | 0.579 | 0.579 | 0.566 | 0.568 | ID | 0.421 | 0.548 | 0.528 | 0.639 | 0.378 | 0.378 |
| 3 | 3 | MGAS315 | 0.536 | 0.536 | 0.544 | 0.540 | 0.421 | ID | 0.702 | 0.534 | 0.534 | 0.470 | 0.470 |
| 49 | 3 | CS101 | 0.463 | 0.463 | 0.449 | 0.448 | 0.548 | 0.702 | ID | 0.713 | 0.707 | 0.507 | 0.507 |
| 12 | 4 | A735 | 0.395 | 0.395 | 0.397 | 0.397 | 0.528 | 0.534 | 0.713 | ID | 0.704 | 0.516 | 0.516 |
| 28 | 4 | MGAS6180 | 0.378 | 0.378 | 0.372 | 0.372 | 0.639 | 0.534 | 0.707 | 0.704 | ID | 0.552 | 0.551 |
| 1 | 2 | SF370 | 0.354 | 0.354 | 0.334 | 0.333 | 0.378 | 0.470 | 0.507 | 0.516 | 0.552 | ID | 0.998 |
| 1 | 2 | MGAS5005 | 0.354 | 0.354 | 0.334 | 0.333 | 0.378 | 0.470 | 0.507 | 0.516 | 0.551 | 0.998 | ID |
ID, identical.
The FCT-1, -2, -5, and -6 region forms are strikingly different in both gene content and arrangement from the FCT-3 and FCT-4 region forms and from each other (Fig. 1 and Table 1). Their shared FCT region genes are limited to rofA. The FCT-5 strain has prtF1, whereas the FCT-2 strain has a cpa locus, although the amino acid sequence identity of the latter is less than 60% compared to all Cpa proteins derived from FCT-3 and FCT-4 strains (Table 2). The arrangement of the FCT-2 region genes strongly parallels a large segment shared by the FCT-3 and FCT-4 genotypes, namely, the series of cpa-sipA2-fctA-srtC2-fctB loci. However, the sipA2, srtC2, and fctB gene products of ALAB49, which display more than 95% identity with matching products from all other FCT-3 and FCT-4 strains, share only 39, 47, and 28% amino acid sequence identity, respectively, with their FCT-2 strain counterparts (Table 1). Thus, it seems likely that this set of contiguous genes in FCT-2, compared to FCT-3 and FCT-4, has an orthologous origin and comprises a discrete lineage.
T type and FCT region forms.
Historically, T typing has been widely used as a serological typing scheme (32). It was recently shown that at least some T antigens are cell wall-anchored surface proteins encoded by FCT region genes (10, 42, 45, 54). The extensive amino acid heterogeneity observed for Cpa and FctA (Tables 2 and 3) is suggestive of strong diversifying selection acting on cpa and fctA, such as that imposed by the host immune response. The T serotypes were determined for isolates having FCT-1 through FCT-4 genotypes via trypsin treatment, followed by agglutination in the presence of T-serotype-specific antiserum. Multiple T types were evident among strains assigned to either the FCT-3 or FCT-4 genotype (Table 1). One T type (T3/13/B) was present among multiple strains (all FCT-3), in which Cpa and FctA proteins diverged in amino acid sequence by more than 20 and 35%, respectively. The magnitude of amino acid homology that defines each T type and the extent to which trypsin removes antigenic epitopes remain to be established through more extensive strain sampling.
TABLE 3.
Sequence similarity matrix for FctA following amino acid sequence alignment using the Clustal W algorithm
| emm type | FCT region form | Strain | Sequence similarity
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ALAB49 | 29487 | D633 | MGAS8232 | Manfredo | MGAS315 | A735 | MGAS6180 | |||
| 53 | 3 | ALAB49 | IDa | 1.000 | 0.631 | 0.637 | 0.692 | 0.675 | 0.628 | 0.631 |
| 33 | 3 | 29487 | 1.000 | ID | 0.631 | 0.637 | 0.692 | 0.675 | 0.628 | 0.631 |
| stD633 | 3 | D633 | 0.631 | 0.631 | ID | 0.638 | 0.635 | 0.662 | 0.605 | 0.806 |
| 18 | 3 | MGAS8232 | 0.637 | 0.637 | 0.638 | ID | 0.601 | 0.669 | 0.608 | 0.660 |
| 5 | 3 | Manfredo | 0.692 | 0.692 | 0.635 | 0.601 | ID | 0.638 | 0.643 | 0.616 |
| 3 | 3 | MGAS315 | 0.675 | 0.675 | 0.662 | 0.669 | 0.638 | ID | 0.6 | 0.715 |
| 12 | 4 | A735 | 0.628 | 0.628 | 0.605 | 0.608 | 0.643 | 0.600 | ID | 0.576 |
| 28 | 4 | MGAS6180 | 0.631 | 0.631 | 0.806 | 0.660 | 0.616 | 0.715 | 0.576 | ID |
ID, identical.
FCT region structures of the global S. pyogenes population.
A major aim of this study was to define the gene contents of FCT regions within the global S. pyogenes population in order to understand the extent of FCT region diversity, perform linkage analysis, and devise evolutionary models. A genetically diverse sample set was assembled for this purpose. The 113 isolates of S. pyogenes chosen included 112 emm types (11) (see Table S1 in the supplemental material). By including ∼70% of the ∼160 known emm types (http://www.cdc.gov/ncidod/biotech/strep/strepindex.htm), the near-complete range of FCT region genotypes within the species is likely to be represented. MLST based on seven housekeeping loci yielded 113 distinct sequence types (STs) and 113 unique combinations of emm type and ST. Each of the 113 isolates also differed from all others in the set at two or more of the seven housekeeping alleles, and 107 of the isolates differed from all others at three or more housekeeping alleles. Thus, the 113 isolates under evaluation are genetically distant from one another, and each can be regarded as a distinct strain.
The FCT-3 and FCT-4 region genotypes were chosen as a starting point for comparison, since they accounted for most of the strains whose complete FCT region sequences are known (Fig. 1). Nine of the 10 FCT region genes were targeted for PCR amplification in order to ascertain their presence or absence among the set of 113 diverse strains. Since alleles of the prtF1, prtF2, cpa, and fctA genes display extensive sequence heterogeneity (10, 44, 64) (Tables 1, 2, and 3), two primer pairs were used for PCR-based detection of each gene (see Materials and Methods). All oligonucleotide primer pairs displaying BLASTN hits with an FCT region whose complete nucleotide sequence is known also yielded a PCR amplification product of the expected size using template DNA derived from the sequenced isolate or a related strain sharing the same emm type and ST (Table 1; see Table S1 in the supplemental material). Similarly, FCT regions in which one or both primers of a pair did not align via BLASTN also failed to yield a PCR product. The observed correspondence between in silico analysis and experimental findings provided further validation of the approach.
Detailed analytic PCR findings on 113 genetically distinct strains are presented in Table S1 in the supplemental material. Although the relative order of the FCT region genes was not established by this approach, correspondence to the FCT region genotypes outlined in Fig. 1 was assumed based on the presence or absence of individual genes. This assumption seems reasonably well justified, because there were no examples of gene order reversal within the FCT regions whose complete nucleotide sequences are known (Fig. 1).
The most prevalent FCT region genotypes uncovered by the PCR-based assessment were FCT-3 and FCT-4, representing 32 and 27% of the 113 strains, respectively (Table 4). The FCT-5 form ranked next in prevalence (17%), although correspondence to FCT-5 was characterized by an absence of many of the genes found in FCT-3 and FCT-4, and thus, probes designed to specifically target FCT-5 genes might reveal additional FCT region genotypes. A derivative of the FCT-4 genotype, in which only the cpa gene went undetected, was found among 12% of the strains; this combination of genes is designated the FCT-7 genotype (Table 4). Another derivative of the FCT-4 genotype, in which nra replaces rofA, is designated FCT-8, although this combination of genes was limited to just three strains. The FCT-1 genotype corresponded to eight strains. Both the FCT-6 and FCT-2 forms were rare among the sample set, found in association with only one and two strains, respectively.
TABLE 4.
FCT region genotypes for 113 diverse strains
| FCT region genotype | FCT region locus
|
No. of isolates (%) | |||||
|---|---|---|---|---|---|---|---|
| rofA/nra | prtF1 | srtB | cpa | sipA-(fctA)-srtC-fctB | prtF2 | ||
| FCT-1 | rofA | Present | Present | Absent | Absent | Absent | 8 |
| FCT-2 | rofA | Absent | Present | Absenta | Absent | Absent | 2 |
| FCT-3 | nra | Absent | Absent | Present | Present | Present | 36 (32) |
| FCT-4 | rofA | Present | Present | Present | Present | Present | 31 (27) |
| FCT-5 | rofA | Present | Absent | Absent | Absent | Absent | 19 (17) |
| FCT-6 | rofA | Absent | Absent | Absent | Absent | Absent | 1 |
| FCT-7 | rofA | Present | Present | Absent | Present | Present | 13 (12) |
| FCT-8 | nra | Present | Present | Present | Present | Present | 3 |
No PCR product was obtained with either the Cpa-A or Cpa-B reactions for the two FCT-2 isolates, although amino acid alignment (Table 2) provides evidence for a divergent cpa-like gene in two emm1-type strains.
Genetic linkage within the FCT region.
S. pyogenes ranks high among bacterial species in terms of its rate of homologous recombination arising from horizontal gene transfer relative to mutation (23, 27, 44). For highly recombinogenic bacteria, genetic linkage analysis can provide a foundation leading to the formulation of testable hypotheses, which in turn may yield new insights into key biological interactions (58, 59).
In general terms, the probability of coinheritance increases as the length of the intergenic region available for crossover decreases. Although recombination can disrupt physical connections, strong linkage among genes within the FCT region may be the consequence of their very close proximity. All pairwise combinations of FCT region genes were assessed for linkage using two-by-two tests for independence, in which the presence or absence of one gene was compared to the presence or absence of the other gene. Pairwise comparisons showing highly significant, nonrandom associations between genes were taken as evidence for strong linkage disequilibrium. As expected for closely positioned loci, nearly all pairwise comparisons of FCT region genes (42 of 45; 93%) displayed evidence for strong linkage (P < 0.05) (Fig. 2). Applying the Bonferroni correction, which factors in the total number of pairwise comparisons, reduced the number of gene combinations displaying strong linkage to 32 (71% of the total).
FIG. 2.
Linkages between pairs of genes within FCT regions. The statistical significances of linkages between pairs of genes within the FCT regions of 113 S. pyogenes strains are shown. The P values are based on the two-by-two χ2 test (two tailed) with Yates' correction. When the values are Bonferroni corrected, based on 45 pairwise comparisons in which the fctA and fctA bridge reactions are considered to be variants and are counted together only once, the corrected P value for significant differences shifts from 0.05 to 0.0011 and the corrected P value for highly significant differences shifts from 0.01 to 0.0002. Values that shift to nonsignificance following Bonferroni correction are indicated in italics. Examples of strong negative and positive linkages (defined in text) are also indicated. n/a, not applicable.
The relationship between pairs of loci displaying linkage can sometimes be further classified as either strong negative linkage, where few or no strains have both genes, or strong positive linkage, where the genes, if present at all, usually cooccur. Positive linkage may signify a codependent gene interaction that underlies a biologically important phenotype. Negative linkage may signify an antagonistic interaction between the two loci that leads to decreased fitness of the bacterium. There are three occurrences of strong negative linkage within this data set of 113 strains, and all involve nra (Fig. 2). As reported previously, either nra or rofA, but not both alleles, is detected in each of the 113 S. pyogenes strains via the PCR-based method (11). nra and prtF are also mutually exclusive, but strong positive linkage is evident for rofA and prtF1, a finding which may indicate that only RofA, and not Nra, is capable of regulating prtF1 expression (24). Strong positive linkage is also observed for prtF1 and srtB; the latter encodes the alternative sortase B. However, SrtB does not anchor PrtF1 to the cell wall in the one strain tested; instead, that is achieved by sortase A (6). The pairing of nra and srtB is not observed among any of the 113 strains, but strong positive linkage is observed for rofA and srtB. Taken together, it stands to reason that the observed patterns of linkage are probably the consequence of RofA being required for transcriptional regulation of both the prtF1 and srtB genes. This hypothesis can be experimentally tested.
Other examples of strong positive linkage among the 113 strains involve pairings of sipA2, srtC2, fctB, and prtF2 (Fig. 2). The fctA “bridge” PCR amplification (see Table S1 in the supplemental material) is designed to capture the most highly divergent fctA alleles; when the fctA bridge-positive strains were combined with the fctA-positive strains, additional gene pairs exhibiting strong positive linkage were identified. The sipA2, fctA, srtC2, fctB, and prtF2 loci are contiguous in the FCT-3 and FCT-4 genotypes (Fig. 1). In an emm49 strain (FCT-3), cpa plus four downstream loci—sipA2, fctA, srtC2, and fctB—are transcribed as a single polycistronic transcript (48). A general property of operons is that genes are transcribed in a coordinated manner in order to fulfill a biological need, and thus, the operon itself can become the unit of selection (41). The operon structure could explain the strong positive linkage observed for the pairwise comparisons made with sipA2, fctA, srtC2, and fctB (Fig. 2). It may be that cpa is not included among this group because it has divergent forms that were not detected by the PCR-based method employed. In fact, exclusion of the FCT-7 genotype strains lacking cpa (Table 4) from the above-mentioned calculations yielded strong positive linkage between cpa and the downstream genes of the operon (data not shown).
The tight association of prtF2 with genes of the adjacent operon could be the result of either physical proximity on the chromosome (Fig. 1) or positive selection that arises from interactions among the gene products. In support of the latter possibility, Cpa, FctA, and PrtF2 are each required for the formation of the T-antigen complex (42). prtF2 and fctB comprise the only gene pair that exhibits complete positive linkage, where none of the 113 strains has one locus in the absence of another (Fig. 2). However, the pfbpI lineage prtF2 allele present in the FCT-3 region of an emm49 strain (Table 1), in place of the fbaB lineage allele that is more typical of sequenced FCT-3 forms, provides at least one example showing that recombinational replacements involving prtF2 alleles have occurred.
Of the many possible pairwise comparisons between FCT region genes (Fig. 2), 12 involve gene pairs for which nearly all 113 strains harbor at least one of the two genes. Of special interest are the four pairs involving genes encoding cell wall-anchored surface proteins. Nearly every strain has either prtF1 or at least one of four other surface proteins (cpa, fctA, fctB, or prtF2) (Table 4 and data not shown). Only three strains are devoid of all five of these surface protein genes, although they have other genes corresponding to pilus-like structures. Specifically, FCT-2 has orthologs of the cpa through fctB series of genes, whereas FCT-6 has strong partial homology to Streptococcus agalactiae streptococcal pilus genes (19, 40, 52). Although the cell wall-anchored surface protein genes are considered to be accessory loci due to their differential presence among strains, the data presented here suggest that possession of either prtF1 or one of the other four surface protein genes is critical to the long-term survival of most S. pyogenes organisms in their natural environment.
Diversity in FCT region forms among emm pattern-defined strains.
The distribution of the eight FCT region genotypes within each emm pattern-defined subset of strains is summarized in Table 5. Statistical methods were used to compare the extents of diversity in FCT region genotypes among the emm pattern-defined subsets of isolates. The highest D value was observed for the emm pattern A-C strains, indicating that as a group, the so-called throat specialists display the most diversity in FCT region genotypes. The 20 emm pattern A-C strains are represented by six of the FCT region genotypes, with no single FCT region form corresponding to more than 25% of the strains. The lowest D value was found for the 38 emm pattern D strains, indicating that the so-called skin specialists are the most homogenous in their FCT region gene contents. The emm pattern D strains are distributed among five FCT region genotypes; however, the majority (79%) belong to the FCT-3 genotype. The 55 emm pattern E strains have an intermediate D value and correspond to seven of the FCT region genotypes, but most (75%) are restricted to two FCT region forms (FCT-4 and FCT-5). The 95% confidence intervals for the three emm pattern-defined groups indicate that each is significantly different from the other two groups in terms of diversity. If gene products of the entire FCT region are critical for infection at specific tissue sites, then the diversity measures support the notion that emm pattern A-C strains draw upon a variety of strategies to cause infection in the throat, whereas emm pattern D strains may be more dependent on a singular mechanism.
TABLE 5.
Distribution of FCT-region genotypes among emm pattern-defined strains
| FCT region genotype | No. of isolates according to emm pattern genotype
|
|||
|---|---|---|---|---|
| A-Ca | Db | Ec | Totald | |
| FCT-1 | 5 | 2 | 1 | 8 |
| FCT-2 | 2 | 0 | 0 | 2 |
| FCT-3 | 3 | 30 | 3 | 36 |
| FCT-4 | 2 | 3 | 26 | 31 |
| FCT-5 | 3 | 1 | 15 | 19 |
| FCT-6 | 0 | 0 | 1 | 1 |
| FCT-7 | 5 | 0 | 8 | 13 |
| FCT-8 | 0 | 2 | 1 | 3 |
| Total | 20 | 38 | 55 | 113 |
D, 0.853; 95% confidence interval, 0.797, 0.908.
D, 0.374; 95% confidence interval, 0.181, 0.567.
D, 0.690; 95% confidence interval, 0.605, 0.775.
D, 0.783; 95% confidence interval, 0.746, 0.819.
Linkage between emm and FCT region genes.
An alternative role for the FCT region in tissue-specific infection is that only some of its genes are essential. If this is correct, then those genes should be present in most isolates recovered in association with infection of that tissue. However, the 113 strains under evaluation were selected by maximizing their genetic distances, as defined by emm type and MLST, and not by their actual tissue site of isolation. Only 33 of the 113 isolates were known to be recovered from impetigo lesions, and even fewer (n = 6) isolates were recovered from known cases of pharyngitis or tonsillitis (see Table S1 in the supplemental material). Since the sample sizes for isolates recovered from infections of the skin or throat are rather small and large-scale analysis of population-based collections of isolates was beyond the scope of this study, the emm pattern was used as a surrogate marker for tissue site preference.
Five of the FCT region genes assessed by PCR encode cell wall-anchored surface proteins. Cpa, FctA, and FctB are associated with pilus-like appendages (42, 45). Cpa binds human type I collagen (38), and PrtF1 and PrtF2 each bind human fibronectin (28, 39). The strongest measure of positive linkage between the emm pattern-defined skin versus throat specialist strains and FCT region surface protein genes lies with cpa, which is present in nearly all (92%) emm pattern D strains and absent from 75% of emm pattern A-C strains (Table 6). However, the presence of cpa in several pattern A-C strains, particularly those of the highly prevalent classical throat types emm3, emm5, and emm18 (55), makes it unlikely that Cpa by itself directs S. pyogenes to the skin. Cpa may be necessary for most skin infections, but it does not appear to be sufficient. Otherwise, emm3, emm5, and emm18 isolates should be recovered from impetigo lesions in much greater numbers. It should be noted that cpa expression may be aberrant in the emm18 strain, which has a defect in its nra regulatory gene (56).
TABLE 6.
Distribution of FCT region loci encoding cell wall-anchored surface proteins among emm pattern-defined strains
| emm pattern | No. of isolates | No. (%) of isolates positive
|
||||
|---|---|---|---|---|---|---|
| prtF1 | cpa | fctAa | fctB | prtF2 | ||
| A-C | 20 | 15 (75) | 5 (25) | 10 (50) | 10 (50) | 10 (50) |
| D | 38 | 8 (21) | 35 (92) | 35 (92) | 35 (92) | 35 (92) |
| E | 55 | 51 (93) | 30 (55) | 38 (69) | 38 (69) | 38 (69) |
Includes isolates positive for the fctA bridge reaction.
In a somewhat parallel but inverse relationship, 75% of emm pattern A-C strains have prtF1, compared to only 21% of emm pattern D strains (Table 6). However, prtF1 is not exclusive to pattern A-C strains, and a substantial minority (25%) of pattern A-C strains lack prtF1 entirely. The positive correlation between prtF1 and emm patterns A-C is weaker than the relationship between cpa and the emm pattern D genotype. Thus, although prtF1 may facilitate infection of the throat, it appears that a prtF1-independent mechanism must also exist.
Three other surface protein genes—fctA, fctB, and prtF2—are also found in high numbers among emm pattern D strains, 92% of which have all three genes plus cpa. However, fctA, fctB, and prtF2 are also present among 50% of the emm pattern A-C strains (Table 6). Although it is possible that the fctA, fctB, and/or prtF2 gene is necessary for skin infection, they do not appear to be sufficient to cause disease at this site, because each is present in many of the throat specialists strains as well. As a group, emm pattern E strains are recovered in high numbers from both throat and skin sites of infection, so it might be expected that these organisms have virulence determinants directed toward both tissues. A high proportion (93%) of emm pattern E strains harbor prtF1 (Table 6). However, only 55% of the pattern E strains have cpa. Therefore, if cpa is a determinant for infection of the skin, it would appear that many pattern E strains may utilize an alternative molecular strategy.
The findings of this study provide supporting evidence that Cpa may, in large part, be necessary for S. pyogenes infection of the skin, whereas PrtF1 may be a key virulence determinant in the throat. However, there are several emm pattern D strains harboring prtF1 and several emm pattern A-C strains having cpa. Thus, virulence determinants in addition to PrtF1 may be required for throat infection. Likewise, Cpa may not be sufficient for skin infection. For those few pattern A-C strains lacking prtF1 and those few pattern D strains lacking cpa, there may exist alternative strategies for causing infection of the designated tissue.
Molecular epidemiologic findings often provide a sound basis for formulating testable hypotheses. In experimental studies, the role of cpa in the virulence of an emm pattern D strain was investigated using a highly sensitive, humanized mouse model of impetigo. The experiments show that cpa is essential for skin infection under certain growth conditions (42). Prior reports had demonstrated that three additional factors, whose genes lie outside the FCT region, were also required for maximal virulence at the skin (57, 59). Importantly, two of the factors—secreted cysteine protease and plasminogen-binding M protein—were largely lacking among the emm pattern A-C strains. Thus, the population findings of this report, supporting a role of Cpa in skin infection as being necessary but not sufficient, confirms the combined experimental findings. A critical role for PrtF1 in throat infection remains to be established via a refined model of streptococcal pharyngitis.
Several investigators have screened numerous isolates, defined according to emm or M protein type, for a variety of FCT region genes (5, 25, 38, 47, 62), primarily prtF1, prtF2, and cpa. However, direct comparisons to data presented in this report are limited, because technical approaches with various degrees of sensitivity and specificity were employed and many emm types are found in association with distant genetic backgrounds, as defined by the ST (reference 21 and unpublished data). The degree to which an emm type is associated with different FCT region forms is not known, and it is probably not correct to simply assume that a particular emm type has a strict correlation with the FCT region form assigned to the 113 isolates of this study. In addition, a given emm type is often found in association with numerous T serotypes (33). However, much remains to be uncovered about the molecular relationship between the T serotype and the FCT region genotype. The recent finding that at least three FCT region gene products are essential for measuring the T type of one strain (42) may signify that extensive nucleotide sequence determination is required to relate a T type to an FCT region genotype. Conceivably, the molecular determinants of the T serotype lie within the highly heterogeneous portions of just a few genes, such as cpa or fctA. The modes of genetic diversification that give rise to different T types are unknown and may involve mutation, intragenic recombination, and/or en bloc transfer of the entire FCT region. The many non-T-typeable strains that exist (33) may also pose difficulties in relating the T type to the FCT region genotype. The fact that FCT region genes are either positively or negatively regulated under the same growth conditions according to strain (35, 37, 48) may undermine the phenotypic approach used for T-serotype determination. For the molecular epidemiologic analysis of S. pyogenes, the sequenced-based typing methods of emm typing and MLST should be regarded as the gold standards.
Despite the potential for variance in the relationship between individual emm types and FCT region genotypes, the data analysis of this report groups emm types together in accordance with their emm patterns, and comparisons are made between the emm pattern group and either the FCT region form or specific FCT region genes. The emm pattern group sizes range from 20 (A-C) to 55 (E) isolates and consist of genetically distant strains, so in many ways, the sample set is quite robust. The degree to which more extensive sampling will alter the general findings of this report remains to be determined.
Pathways of evolution.
Structural analysis of the FCT regions of the S. pyogenes population supports a key role in evolution for recombination leading to gene replacements. The gain or loss of genes can be assessed by maximum parsimony, a character state-based method for inferring phylogenetic trees. The characters (i.e., genes) are assigned a state (i.e., presence or absence, or lineage), and the ordered series of character states yields a taxon. It is assumed that the relative chromosomal orders of the genes under study are the same in all strains, which seems justified, since there are no examples of gene order reversal (Fig. 1).
The nine FCT region genes targeted for PCR amplification yield 13 distinct taxa among the 113 strains (based on data in Table S1 in the supplemental material). A phylogenetic tree was constructed by maximum parsimony by including one representative of each taxon in the data matrix (Fig. 3). Although some of the FCT region forms are represented by multiple taxa (FCT-3, -4, and -7), they lie along adjacent branches. Each strain is denoted by a symbol at one of the nodes represented by the 13 taxa, in accordance with the emm pattern genotype. Most emm pattern D strains cluster at one end of the tree, whereas emm pattern A-C strains are scattered at many nodes throughout, without strong clustering in any one region. Interspersed among the branches leading to pattern A-C strains are the emm pattern E strains, although the latter group forms two dense clusters that are separated by four genetic changes. The data provide evidence that the majority of FCT regions present within emm pattern E strains evolved along two discrete pathways.
FIG. 3.
Phylogenetic tree based on FCT region genes. Nine FCT region genes provided the characters for the unrooted tree shown, which is based on maximum parsimony of 13 taxa. The character states correspond to gene presence/absence or lineage. The branch-and-bound search method was used; 24 trees were found, and the strict consensus is shown. The consistency index was 0.600, and the retention index was 0.806. The 113 S. pyogenes strains are indicated by symbols placed next to the nodes corresponding to their taxa as follows: emm patterns A-C (filled), D (open), and E (stippled). The progenitor FCT region form and direction of evolution along the branches of the tree are unknown.
Since the emm pattern genotypes appear to serve as reliable markers for tissue site preferences for infection by S. pyogenes, it was of interest to construct an evolutionary model that included the genes most likely to be direct determinants of niche specificity. Six genotypes derived from the FCT and emm regions were selected for construction of a mixed FCT- and emm region-based genotype tree (Fig. 4). Included were four genes encoding MSCRAMMs (prtF1, cpa, prtF2, and sof) and two loci encoding transcriptional regulators (rofA/nra and mga1/mga2). The mga locus lies immediately upstream of emm and is comprised of two discrete lineages, whereas sof lies ∼10 kb upstream of emm in some strains and encodes a multifunctional protein that can bind human fibronectin (36). Mga can regulate the transcription of emm, sof, and rofA/nra, whereas RofA/Nra can effect the transcription of mga, prtF1, the series of cpa to fctB genes, and prtF2 (2, 7, 24, 35, 37, 48, 61). Furthermore, both the mga and rofA/nra loci are subject to autoregulation. Some of the gene-to-gene interactions may be strain dependent. The emm and FCT regions are separated by ∼0.3 Mb and do not lie in close proximity on the chromosome.
FIG. 4.
Phylogenetic tree based on select emm and FCT region genes. Four FCT region genes (rofA/nra, prtF1, cpa, and prtF2) and two emm region genes (mga and sof) provided the six characters for the unrooted tree shown, which is based on maximum parsimony. The character states correspond to gene presence/absence or lineage. Taxa (n = 11) representing at least 1% of the 113 strains were used to generate the tree by the branch-and-bound search method; one tree was found. The consistency index was 0.600, and the retention index was 0.800. All nodes of the tree are represented by taxa corresponding to known extant strains. The 113 S. pyogenes strains are indicated by symbols placed next to the nodes corresponding to their taxa as follows: emm patterns A-C (filled), D (open), and E (stippled). The progenitor FCT region form and direction of evolution along the branches of the tree are unknown.
The FCT region forms and emm pattern-defined strains associated with each node are indicated in Fig. 4. In contrast to the FCT region-only tree (Fig. 3), the mixed FCT and emm region tree shows splitting of an FCT region form (FCT-7) between distant nodes. The emm pattern A-C and E strains that have FCT-7 are separated by four genetic changes. These two taxa may have arisen by convergent evolution of FCT-7 or by horizontal transfer of a very large block of genes, such as the entire FCT region or a block encompassing the sof-to-emm region.
A striking feature of the mixed FCT and emm region tree (Fig. 4) is the placement of most emm pattern E strains (generalists) between most throat specialist (pattern A-C) and skin specialist (pattern D) strains, so that the specialists cluster at opposite ends. This model is consistent with a fundamental concept of ecology and evolution, which states that specialists tend to emerge from generalists (20). Even if one of the specialist genotypes was the S. pyogenes progenitor, organisms would need to have evolved through the generalist stage in order to spawn new populations of specialists.
Concluding remarks.
The FCT region of S. pyogenes appears to be important in pathogenesis, because several of its products give rise to MSCRAMMs and pilus-like appendages that may facilitate interactions between the bacterium and host. The FCT region also displays a wide variety of structural forms and contains signatures of past recombination events, indicative of selective pressures that result in adaptation of the organism to a variety of environmental conditions.
Supplementary Material
Acknowledgments
We are grateful to Alicia Bennett for expert technical assistance, Ashley Robinson for assistance with maximum-parsimony tree construction, and Dee Jackson and Bernie Beall (CDC) for T-serotype determination.
This work was supported by funding from the National Institutes of Health (AI053826, AI061454, and GM060793) and the American Heart Association (to D.E.B.).
Footnotes
Published ahead of print on 6 October 2006.
Present address: Department of Medicine Unit I and Infectious Diseases, Christian Medical College, Vellore, India.
REFERENCES
- 1.Alberti, S., C. Garcia-Rey, M. A. Dominguez, L. Aguilar, E. Cercenado, M. Gobernado, A. Garcia-Perea, and the Spanish Surveillance Group for Respiratory Pathogens. 2003. Survey of emm gene sequences from pharyngeal Streptococcus pyogenes isolates collected in Spain and their relationship with erythromycin susceptibility. J. Clin. Microbiol. 41:2385-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almengor, A. C., M. S. Walters, and K. S. McIver. 2006. Mga is sufficient to activate transcription in vitro of sof-sfbX and other Mga-regulated virulence genes in the group A streptococcus. J. Bacteriol. 188:2038-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Anthony, B. F., E. L. Kaplan, L. W. Wannamaker, and S. S. Chapman. 1976. The dynamics of streptococcal infections in a defined population of children: serotypes associated with skin and respiratory infections Am. J. Epidemiol. 104:652-666. [DOI] [PubMed] [Google Scholar]
- 5.Barnett, T. C., A. R. Patel, and J. R. Scott. 2004. A novel sortase, SrtC2, from Streptococcus pyogenes anchors a surface protein containing a QVPTGV motif to the cell wall. J. Bacteriol. 186:5865-5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnett, T. C., and J. R. Scott. 2002. Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs. J. Bacteriol. 184:2181-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beckert, S., B. Kreikemeyer, and A. Podbielski. 2001. Group A streptococcal rofA gene is involved in the control of several virulence genes and eukaryotic cell attachment and internalization. Infect. Immun. 69:534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beres, S. B., E. W. Richter, M. J. Nagiec, P. Sumby, S. F. Porcella, F. R. Deleo, and J. M. Musser. 2006. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc. Natl. Acad. Sci. USA 103:7059-7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bessen, D. E., J. R. Carapetis, B. Beall, R. Katz, M. Hibble, B. J. Currie, T. Collingridge, M. W. Izzo, D. A. Scaramuzzino, and K. S. Sriprakash. 2000. Contrasting molecular epidemiology of group A streptococci causing tropical and non-tropical infections of the skin and throat. J. Infect. Dis. 182:1109-1116. [DOI] [PubMed] [Google Scholar]
- 10.Bessen, D. E., and A. Kalia. 2002. Genomic localization of a T-serotype locus to a recombinatorial zone encoding extracellular matrix-binding proteins in Streptococcus pyogenes. Infect. Immun. 70:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bessen, D. E., A. Manoharan, F. Luo, J. E. Wertz, and D. A. Robinson. 2005. Evolution of transcription regulatory genes is linked to niche specialization in the bacterial pathogen Streptococcus pyogenes. J. Bacteriol. 187:4163-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bessen, D. E., C. M. Sotir, T. L. Readdy, and S. K. Hollingshead. 1996. Genetic correlates of throat and skin isolates of group A streptococci. J. Infect. Dis. 173:896-900. [DOI] [PubMed] [Google Scholar]
- 13.Bisno, A. L., and D. Stevens. 2000. Streptococcus pyogenes (including streptococcal toxic shock syndrome and necrotizing fasciitis), p. 2101-2117. In G. L. Mandell, R. G. Douglas, and R. Dolin (ed.), Principles and practice of infectious diseases, 5th ed., vol. 2. Churchill Livingstone, Philadelphia, PA. [Google Scholar]
- 14.Brandt, C. M., B. Spellerberg, M. Honscha, N. D. Truong, B. Hoevener, and R. Lutticken. 2001. Typing of Streptococcus pyogenes strains isolated from throat infections in the region of Aachen, Germany. Infection 29:163-165. [DOI] [PubMed] [Google Scholar]
- 15.Carapetis, J., B. Currie, and E. Kaplan. 1999. Epidemiology and prevention of group A streptococcal infections: acute respiratory tract infections, skin infections, and their sequelae at the close of the twentieth century. Clin. Infect. Dis. 28:205-210. [DOI] [PubMed] [Google Scholar]
- 16.Colman, G., A. Tanna, A. Efstratiou, and E. Gaworzewska. 1993. The serotypes of Streptococcus pyogenes present in Britain during 1980-1990 and their associations with disease. J. Med. Microbiol. 39:165-178. [DOI] [PubMed] [Google Scholar]
- 17.Dicuonzo, G., G. Gherardi, G. Lorino, S. Angeletti, M. DeCesaris, E. Fiscarelli, D. E. Bessen, and B. Beall. 2001. Group A streptococcal genotypes from pediatric throat isolates in Rome, Italy. J. Clin. Microbiol. 39:1687-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dillon, H., C. Derrick, and M. Dillon. 1974. M-antigens common to pyoderma and acute glomerulonephritis. J. Infect. Dis. 130:257-267. [DOI] [PubMed] [Google Scholar]
- 19.Dramsi, S., E. Caliot, I. Bonne, S. Guadagnini, M. C. Prevost, M. Kojadinovic, L. Lalioui, C. Poyart, and P. Trieu-Cuot. 2006. Assembly and role of pili in group B streptococci. Mol. Microbiol. 60:1401-1413. [DOI] [PubMed] [Google Scholar]
- 20.Elena, S. F., and R. Sanjuan. 2003. Climb every mountain? Science 302:2074-2075. [DOI] [PubMed] [Google Scholar]
- 21.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationship between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Espinosa, L. E., Z. Y. Li, D. G. Barreto, E. C. Jaimes, R. S. Rodriguez, V. Sakota, R. R. Facklam, and B. Beall. 2003. M protein gene type distribution among group A streptococcal clinical isolates recovered in Mexico City, Mexico, from 1991 to 2000, and Durango, Mexico, from 1998 to 1999: overlap with type distribution within the United States. J. Clin. Microbiol. 41:373-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feil, E. J., E. C. Holmes, D. E. Bessen, M.-S. Chan, N. P. J. Day, M. C. Enright, R. Goldstein, D. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fogg, G. C., C. M. Gibson, and M. G. Caparon. 1994. The identification of rofA, a positive-acting regulatory component of prtF expression: use of a mu-gamma-delta-based shuttle mutagenesis strategy in Streptococcus pyogenes. Mol. Microbiol. 11:671-684. [DOI] [PubMed] [Google Scholar]
- 25.Goodfellow, A. M., M. Hibble, S. R. Talay, B. Kreikemeyer, B. J. Currie, K. S. Sriprakash, and G. S. Chhatwal. 2000. Distribution and antigenicity of fibronectin binding proteins (SfbI and SfbII) of Streptococcus pyogenes clinical isolates from the Northern Territory, Australia. J. Clin. Microbiol. 38:389-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanage, W. P., C. Fraser, and B. G. Spratt. 2006. The impact of homologous recombination on the generation of diversity in bacteria. J. Theoret. Biol. 239:210-219. [DOI] [PubMed] [Google Scholar]
- 28.Hanski, E., and M. Caparon. 1992. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 89:6172-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hollingshead, S. K., T. Readdy, J. Arnold, and D. E. Bessen. 1994. Molecular evolution of a multi-gene family in group A streptococci. Mol. Biol. Evol. 11:208-219. [DOI] [PubMed] [Google Scholar]
- 30.Hollingshead, S. K., T. L. Readdy, D. L. Yung, and D. E. Bessen. 1993. Structural heterogeneity of the emm gene cluster in group A streptococci. Mol. Microbiol. 8:707-717. [DOI] [PubMed] [Google Scholar]
- 31.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, D., E. Kaplan, J. Sramek, R. Bicova, J. Havlicek, H. Havlickova, J. Motlova, and P. Kriz. 1996. Laboratory diagnosis of group A streptococcal infections. World Health Organization, Geneva, Switzerland.
- 33.Johnson, D. R., E. L. Kaplan, A. VanGheem, R. R. Facklam, and B. Beall. 2006. Characterization of group A streptococci (Streptococcus pyogenes): correlation of M-protein and emm-gene type with T-protein agglutination pattern and serum opacity factor. J. Med. Microbiol. 55:157-164. [DOI] [PubMed] [Google Scholar]
- 34.Johnson, D. R., D. L. Stevens, and E. L. Kaplan. 1992. Epidemiological analysis of group A streptococcal serotypes associated with severe systemic infections, rheumatic fever, or uncomplicated pharyngitis. J. Infect. Dis. 166:374-382. [DOI] [PubMed] [Google Scholar]
- 35.Kreikemeyer, B., S. Beckert, A. Braun-Kiewnick, and A. Podbielski. 2002. Group A streptococcal RofA-type global regulators exhibit a strain-specific genomic presence and regulation pattern. Microbiology 148:1501-1511. [DOI] [PubMed] [Google Scholar]
- 36.Kreikemeyer, B., D. R. Martin, and G. S. Chhatwal. 1999. SfbII protein, a fibronectin binding surface protein of group A streptococci, is a serum opacity factor with high serotype-specific apolipoproteinase activity. FEMS Microbiol. Lett. 178:305-311. [DOI] [PubMed] [Google Scholar]
- 37.Kreikemeyer, B., K. S. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224-232. [DOI] [PubMed] [Google Scholar]
- 38.Kreikemeyer, B., M. Nakata, S. Oehmcke, C. Gschwendtner, J. Normann, and A. Podbielski. 2005. Streptococcus pyogenes collagen type I-binding Cpa surface protein—expression profile, binding characteristics, biological functions, and potential clinical impact. J. Biol. Chem. 280:33228-33239. [DOI] [PubMed] [Google Scholar]
- 39.Kreikemeyer, B., S. Oehmcke, M. Nakata, R. Hoffrogge, and A. Podbielski. 2004. Streptococcus pyogenes fibronectin-binding protein F2—expression profile, binding characteristics, and impact on eukaryotic cell interactions. J. Biol. Chem. 279:15850-15859. [DOI] [PubMed] [Google Scholar]
- 40.Lauer, P., C. D. Rinaudo, M. Soriani, I. Margarit, D. Maione, R. Rosini, A. R. Taddei, M. Mora, R. Rappuoli, G. Grandi, and J. L. Telford. 2005. Genome analysis reveals pili in group B streptococcus. Science 309:105. [DOI] [PubMed] [Google Scholar]
- 41.Lawrence, J. G. 2003. Gene organization: selection, selfishness, and serendipity. Annu. Rev. Microbiol. 57:419-440. [DOI] [PubMed] [Google Scholar]
- 42.Lizano, S., F. Luo, and D. E. Bessen. 2007. Role of streptococcal T antigens in superficial skin infection. J. Bacteriol. 189:1426-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin, D. R., and K. S. Sriprakash. 1996. Epidemiology of group A streptococcal disease in Australia and New Zealand. Recent Adv. Microbiol. 4:1-40. [Google Scholar]
- 44.McGregor, K. F., B. G. Spratt, A. Kalia, A. Bennett, N. Bilek, B. Beall, and D. E. Bessen. 2004. Multi-locus sequence typing of Streptococcus pyogenes representing most known emm types and distinctions among subpopulation genetic structures. J. Bacteriol. 186:4285-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mora, M., G. Bensi, S. Capo, F. Falugi, C. Zingaretti, A. G. O. Manetti, T. Maggi, A. R. Taddei, G. Grandi, and J. L. Telford. 2005. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. USA 102:15641-15646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakata, M., A. Podbielski, and B. Kreikemeyer. 2005. MsmR, a specific positive regulator of the Streptococcus pyogenes FCT pathogenicity region and cytolysin-mediated translocation system genes. Mol. Microbiol. 57:786-803. [DOI] [PubMed] [Google Scholar]
- 47.Natanson, S., S. Sela, A. E. Moses, J. M. Musser, M. G. Caparon, and E. Hanski. 1995. Distribution of fibronectin-binding proteins among group A streptococci of different M types. J. Infect. Dis. 171:871-878. [DOI] [PubMed] [Google Scholar]
- 48.Podbielski, A., M. Woischnik, B. A. B. Leonard, and K.-H. Schmidt. 1999. Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31:1051-1064. [DOI] [PubMed] [Google Scholar]
- 49.Potter, E. V., M. Svartman, I. Mohammed, R. Cox, T. Poon-King, and D. P. Earle. 1978. Tropical acute rheumatic fever and associated streptococcal infections compared with concurrent acute glomerulonephritis. J. Pediatr. 92:325-333. [DOI] [PubMed] [Google Scholar]
- 50.Ramachandran, V., J. D. McArthur, C. E. Behm, C. Gutzeit, M. Dowton, P. K. Fagan, R. Towers, B. Currie, K. S. Sriprakash, and M. J. Walker. 2004. Two distinct genotypes of prtF2, encoding a fibronectin binding protein, and evolution of the gene family in Streptococcus pyogenes. J. Bacteriol. 186:7601-7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson, D., J. Sutcliffe, W. Tewodros, A. Manoharan, and D. Bessen. 2006. Evolution and global dissemination of macrolide-resistant group A streptococci. Antimicrob. Agents Chemother. 50:2903-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosini, R., C. D. Rinaudo, M. Soriani, P. Lauer, M. Mora, D. Maione, A. Taddei, I. Santi, C. Ghezzo, C. Brettoni, S. Buccato, I. Margarit, G. Grandi, and J. L. Telford. 2006. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol. Microbiol. 61:126-141. [DOI] [PubMed] [Google Scholar]
- 53.Sakota, V., A. M. Fry, T. M. Lietman, R. R. Facklam, Z. Y. Li, and B. Beall. 2006. Genetically diverse group A streptococci from children in Far-Western Nepal share high genetic relatedness with isolates from other countries. J. Clin. Microbiol. 44:2160-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneewind, O., K. F. Jones, and V. A. Fischetti. 1990. Sequence and structural characterization of the trypsin-resistant T6 surface protein of group A streptococci. J. Bacteriol. 172:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shulman, S. 2004. Group A streptococcal pharyngitis serotype surveillance in North America, 2000-2002. Clin. Infect. Dis. 39:325-332. [DOI] [PubMed] [Google Scholar]
- 56.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Svensson, M. D., D. A. Scaramuzzino, U. Sjobring, A. Olsen, C. Frank, and D. E. Bessen. 2000. Role for a secreted cysteine proteinase in the establishment of host tissue tropism by group A streptococci. Mol. Microbiol. 38:242-253. [DOI] [PubMed] [Google Scholar]
- 58.Svensson, M. D., U. Sjöbring, and D. E. Bessen. 1999. Selective distribution of a high-affinity plasminogen binding site among group A streptococci associated with impetigo. Infect. Immun. 67:3915-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Svensson, M. D., U. Sjobring, F. Luo, and D. E. Bessen. 2002. Roles of the plasminogen activator streptokinase and plasminogen-associated M protein in an experimental model for streptococcal impetigo. Microbiology 148:3933-3945. [DOI] [PubMed] [Google Scholar]
- 60.Telford, J., M. Barocchi, I. Margarit, R. Rappuoli, and G. Grandi. 2006. Pili in gram-positive pathogens. Nat. Rev. Microbiol. 4:509-519. [DOI] [PubMed] [Google Scholar]
- 61.Terao, Y., S. Kawabata, E. Kunitomo, J. Murakami, I. Nakagawa, and S. Hamada. 2001. Fba, a novel fibronectin-binding protein from Streptococcus pyogenes, promotes bacterial entry into epithelial cells, and the fba gene is positively transcribed under the Mga regulator. Mol. Microbiol. 42:75-86. [DOI] [PubMed] [Google Scholar]
- 62.Terao, Y., S. Kawabata, M. Nakata, I. Nakagawa, and S. Hamada. 2002. Molecular characterization of a novel fibronectin-binding protein of Streptococcus pyogenes strains isolated from toxic shock-like syndrome patients. J. Biol. Chem. 277:47428-47435. [DOI] [PubMed] [Google Scholar]
- 63.Tewodros, W., and G. Kronvall. 2005. M protein gene (emm type) analysis of group a beta-hemolytic streptococci from Ethiopia reveals unique patterns. J. Clin. Microbiol. 43:4369-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Towers, R. J., P. K. Fagan, S. R. Talay, B. J. Currie, K. S. Sriprakash, M. J. Walker, and G. S. Chhatwal. 2003. Evolution of sfbI encoding streptococcal fibronectin-binding protein I: horizontal genetic transfer and gene mosaic structure. J. Clin. Microbiol. 41:5398-5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wannamaker, L. W. 1970. Differences between streptococcal infections of the throat and of the skin. N. Engl. J. Med. 282:23-31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.