Abstract
We studied the roles of Streptococcus thermophilus phosphogalactosyltransferase (EpsE) (the priming enzyme), tyrosine kinase (EpsD), phosphatase (EpsB), and a membrane-associated protein with no known biochemical function (EpsC) in exopolysaccharide (EPS) synthesis. These proteins are well-conserved among bacteria and are usually encoded by clustered genes. Exopolysaccharide synthesis took place in the wild-type strain and a mutant lacking EpsB but not in mutants lacking EpsC, EpsD, or EpsE. The three mutants unable to synthesize EPS lacked the EpsE phosphogalactosyltransferase activity, while the two EPS-synthesizing strains possessed this activity, showing that EpsC and EpsD are required for EpsE function. An EpsD phosphorylated form was found in all strains except the epsC mutant, indicating that EpsC is necessary for EpsD phosphorylation. Moreover, the phosphorylated form of EpsD, a supposedly cytoplasmic protein, was found to be associated with the plasma membrane, possibly due to interaction with EpsC. Finally, the EpsD and EpsE elution profiles in a gel filtration chromatography assay were similar, suggesting that these two proteins colocalize in the membrane. Mutation of Tyr200, predicted to be a phosphorylation site and present in a conserved motif in bacterial phosphoglycosyltransferases, led to EpsE inactivation. In contrast, mutation of Tyr162 or Tyr199 had no effect. Taken together, these data show that EpsD controls EpsE activity. Possible mechanisms for this control are discussed.
Extracellular polysaccharides (EPS) are produced by a great variety of bacteria, including environmental bacteria, pathogens, and food bacteria (12, 27). These polymers may be assembled as capsular polysaccharides (CPS) tightly associated with the cell surface, or they may be liberated into the growth medium. In nature, bacterial EPS are thought to have diverse functions. Potential roles are often related to the fact that EPS form a capsule around the cell, leading to the formation of a microenvironment or a layer allowing biofilm formation. They are thought to protect bacteria against both environmental and host factors that may be detrimental to survival. In commensal or pathogenic bacteria, EPS are involved in host-bacterium interactions, and this is illustrated by the fact that acapsular Streptococcus pneumoniae is usually nonpathogenic (6).
EPS are complex polymers consisting of repeated units containing different simple sugars. Genes involved in EPS biosynthesis are usually clustered on the chromosome and encode proteins involved in sugar priming, formation of repeated units, export, and polymerization. The clusters may differ greatly among strains belonging to the same species, but they always contain a tyrosine phosphorylation regulatory system involved in modulation of capsule synthesis, such as Wzb-Wzc in Enterobacteriaceae, YwqCDE in Bacillus subtilis, or CpsABCD in streptococci (for a review, see reference 10). Streptococcus thermophilus, a food bacterium closely related to Streptococcus salivarius, an opportunistic pathogen colonizing the human oral cavity (5, 17), has an eps gene cluster (29). This cluster has the same organization as the cps gene clusters involved in capsule synthesis in pathogenic streptococci, such as S. pneumoniae or Streptococcus agalactiae. The first five genes are well conserved, while the subsequent genes are variable and encode different glycosyltransferases, transporters, and polymerases. The putative products of the 5′ conserved genes are phosphatase (EpsB), kinase (EpsD), a priming phosphogalactosyltransferase (EpsE), and two proteins with no known biochemical functions, EpsA and EpsC (29). Inactivation of CpsC or CpsD drastically decreases EPS production in S. agalactiae (9) and S. pneumoniae (25). However the role of CpsC is not known, and the role of CpsD is controversial, as its phosphorylated form was found to negatively regulate CPS production in S. pneumoniae Rx1 (25) but stimulate CPS production in S. pneumoniae D39 (1).
Here we report the role of EpsC and EpsD in S. thermophilus EPS synthesis. We show that EPS is not synthesized in epsC, epsD, and epsE mutants, that EpsC is required to visualize a phosphorylated form of EpsD, and that EpsC and EpsD modulate the activity of EpsE, the priming phosphogalactosyltransferase. We also discuss models for the regulation of EPS synthesis in S. thermophilus, which are presumably valid for other bacteria that have homologues of the epsB, epsC, epsD, and epsE genes.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and DNA manipulation.
The wild-type (WT) strain used in this work was the sequenced S. thermophilus strain CNRZ1066 (5). Escherichia coli TG1 was used to facilitate intermediate plasmid construction as described previously (14). S. thermophilus strains were grown on M17 containing 2% lactose at 42°C (31). When necessary, erythromycin was used at a final concentration of 5 μg/ml for S. thermophilus and at a final concentration of 100 μg/ml for E. coli. For measurement of EPS production, precultures in Elliker broth supplemented with 0.5% β-glycerophosphate were used to inoculate (2%) a chemically defined medium for S. thermophilus (20) supplemented with 7% lactose. Chromosomal and plasmid DNA were extracted as previously described (14). PCR amplification and manipulation of DNA were performed using standard methods (28) or according to the instructions of the manufacturer when a commercial kit was used.
Construction of eps nonpolar deletion mutants.
Nonpolar mutations in the eps cluster were obtained by allele exchange of the wild-type genes with genes containing in-frame internal deletions. Each deleted gene was constructed by ligating two ∼1-kb PCR fragments amplified by oligonucleotides designed to obtain the desired deletion and introduce convenient restriction sites. Each PCR fragment was first cloned into pGEM-T (Promega) and sequenced to check for the absence of secondary mutations. Regions flanking the deleted gene were then ligated by cloning the downstream DNA region of the truncated gene into the plasmid containing the cloned upstream sequence of the gene to be mutated using BamHI (newly created restriction site) and NotI (present in the vector). The resulting plasmid was then digested with PstI and ligated to pGhost9, a thermosensitive replicon (3) digested with the same enzyme. This construct was introduced into S. thermophilus CNRZ1066 by electroporation as described elsewhere. Allele exchange was performed by a two-step procedure described by Cieslewicz et al. (9), using 42°C as the nonpermissive temperature. The primers used for this work are shown in Table 1. The nonpolarity of the mutations was checked by complementation with a plasmid expressing the corresponding gene.
TABLE 1.
Oligonucleotides used for PCR amplificationa
| Oligonucleotide | Sequence (5′-3′) | Use |
|---|---|---|
| EpsB1F | GTTAAAGTTGATGATGTTGCC | epsB deletion mutant; left flanking region amplification |
| 1BR | CGGGATCCGTGAGTGAACGTCAATCAC | epsB deletion mutant; left flanking region amplification |
| 2BF | CGGGATCCCCAAAACATTACTAGAAAATC | epsB deletion mutant; right flanking region amplification |
| 2BR | CGTTTCATTTAGATCGGCATTTCCTGAA | epsB deletion mutant; right flanking region amplification |
| EpsC1F | ATAATCTCCAAGACTCTGTCC | epsC deletion mutant; left flanking region amplification |
| EpsC1R | CGGGATCCAGTGTTATCTTG | epsC deletion mutant; left flanking region amplification |
| EpsC2F | CGGGATCCCTTGGAATTGTC | epsC deletion mutant; right flanking region amplification |
| EpsC2R | TAGGTGCTGTTGGTTCAGAG | epsC deletion mutant; right flanking region amplification |
| 1DF | GCTCACATGTCCTCAAACCAAAGCTC | epsD deletion mutant; left flanking region amplification |
| 1DR | CGGGATCCCTCTTCCGTCTTTTTAGCA | epsD deletion mutant; left flanking region amplification |
| 2DF | CGGGATCCGTCGGTTGGAATTAACGCGT | epsD deletion mutant; right flanking region amplification |
| 2DR | CATATTTCGGTTTAATTTCTCTTACGAGAA | epsD deletion mutant; right flanking region amplification |
| 1EF | GCCAGAGTCACCATCTTCACC | epsE deletion mutant; left flanking region amplification |
| 1ER | GCGGATCCCTTTAGCTTGTGACATCTCATTC | epsE deletion mutant; left flanking region amplification |
| 2EF | CGGGATCCGAGTTGGAGCGCGTTAGTACTG | epsE deletion mutant; right flanking region amplification |
| 2ER | CCACTCTAAGTCAATCAAATAACC | epsE deletion mutant; right flanking region amplification |
| EHNF | GCGGATCCATGTCACAAGCTAAAGAGGAAATT | Insertion of epsE open reading frame into pQE-30 to produce His6-EpsE |
| EHNR | CCCAAGCTTCTAACGCGCTCCAACTCTCTTCAAAACAATC | Insertion of epsE open reading frame into pQE-30 to produce His6-EpsE |
| P-eps-F | GGAATTCGCAGACTAGTTTGTAAAAGGACG | eps promoter amplification to construct pJIM4843 |
| P-eps-R | CGCGGATCCCCATTACTCGTATGCTTTTGC | eps promoter amplification to construct pJIM4843 |
Oligonucleotide primers were derived from the eps gene sequence of CNRZ1066 (accession number CP000024). In some cases, additional nucleotides were added to create restriction sites (underlined).
Construction of a plasmid expressing histidine-tagged EpsE and tyrosine-substituted derivatives.
A fragment carrying the epsE coding sequence was amplified and cloned into E. coli plasmid pQE-30 (QIAGEN) digested with BamHI and HindIII downstream and in frame with a His6 tag sequence. The fused epsE gene with the His6 tag sequence was then released from the plasmid using XhoI and HindIII and cloned into pBSIISK (Stratagene) digested with the same enzymes. The resulting plasmid was digested with XhoI and PstI, and the recombinant gene was subcloned into pJIM4843 cleaved with SalI and PstI. pJIM4843 is an expression vector based on the pGKV210 plasmid backbone (32), carrying a ∼0.4-kb EcoRI-BamHI PCR fragment containing the promoter of the S. thermophilus eps operon (Table 1). This promoter allows constitutive expression according to procedures performed with luciferase genes as a reporter (not shown). His-tagged EpsE variants were constructed by targeted mutagenesis. Substitutions were introduced on oligonucleotides that were used to modify the His6-epsE sequence on pBSIISK to produce His6-epsE-Y162F, -Y199F and -Y200F. The resulting genes were then cloned into pJIM4843 as described previously. All constructions were verified by sequencing the modified gene and its promoter in the pJIM4843 derivatives.
Purification, measurement, and molecular size analysis of EPS.
EPS were extracted from cultures grown at 42°C. The pH of each culture was neutralized using 36% (wt/vol) NaOH. Samples were heated at 80°C for 20 min. The cells were removed by centrifugation for 15 min at 16,000 × g, and the supernatants were poured onto Spectra/Por 6 membranes (Merck) with a 50,000-g/mol cutoff and dialyzed against distilled water until the residual lactose and galactose were removed. EPS concentrations were determined using the phenol-sulfuric acid method (13).
The molecular weights of the EPS samples were determined by using gel exclusion chromatography and light scattering detection (Summit System [Dionex, Sunnyvale, CA], PD2020 dual-angle laser light scattering detector [Precision Detectors Inc., Bellingham, MA], and RI-101 differential refractometer [Shodex; Showa Denko K.K., Tokyo, Japan]). The samples were separated by gel exclusion chromatography at a flow rate of 0.8 ml/min on a column system (Aquagel-OH 60, 50, and 40; 15 μm) equipped with a precolumn Aquagel-OH guard (Polymer Laboratories Inc., Amherst, MA). Elution was performed at 35°C with 100 mM sodium nitrate for 60 min. The average molecular weights (Mw) were obtained using the Rayleigh equation, which states that the intensity of the light scattered by molecules in solution is equal at an angle of zero to the concentration and Mw of the molecules times an optical constant: R(θ)|θ→0 ≈ KcMw, where θ is the scattering angle, R(θ) is the Rayleigh excess scattering ratio, K is the optical constant [4π2n2(δn/δc)2]/(λ04NA), n is the solvent refractive index, δn/δc is the specific refractive index increment (ml/g), NA is Avogadro's number, λ0 is the wavelength of the scattered light in vacuo (cm), and c the concentration of the eluting fraction determined by using the δn/δc value (0.150 ml/g in this case) and the differential refractive index signal.
The extrapolation model used to determine the average molecular weight was the Debye model, in which a plot of R(θ)/Kc versus sin2(θ/2) yields the Mw from the intercept.
Membrane protein extract from S. thermophilus.
Exponentially growing cells (optical density at 600 nm, 0.4) in M17 containing lactose were centrifuged at 6,000 × g for 10 min at 22°C and washed twice in TES buffer (25 mM Tris-HCl [pH 8.0], 10 mM EDTA, 25% saccharose). The pellets were resuspended in TES buffer containing 100 mg/ml lysozyme and 700 U/ml mutanolysin for 2 h. Cells were washed twice in buffer A (50 mM Tris-HCl [pH 8.0], 1 mM dithiothreitol, 1 mM EDTA, 10% [vol/vol] glycerol) and resuspended in 5 ml of buffer A containing 1 mM phenylmethylsulfonyl fluoride. Cells were broken on a mesh and blended for 5 min. Unbroken cells were removed by low-speed centrifugation at 10,000 × g for 10 min at 4°C. The supernatant was centrifuged at 100,000 × g for 60 min to separate the “soluble proteins” (fraction S) from the “membrane proteins” (fraction M). The pellet containing the membranes was resuspended in1 ml of ice-cold extraction buffer A and frozen at −40°C. The protein concentration was determined by the method of Lowry et al. (21), using bovine serum albumin dissolved in extraction buffer as the standard.
Galactosyltransferase assay.
The galactosyltransferase (GT) assay used in this study was a modified version of the assay described by Kolkman et al. (18) and Stingele et al. (30). The reaction mixture (total volume, 150 μl) contained 50 to 100 μl of membrane extract (0.3 to 1.0 mg of protein), 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 1 mM EDTA, and 1 mM UDP-[14C]galactose (25 nCi). The reaction was performed at 37°C for 1 h and stopped by addition of 2 ml of a chloroform-methanol mixture (2:1). The activity of phosphogalactosyltransferase was then determined under the standard conditions described previously by Stingele et al. (30).
The buffer system used to determine the pH dependence of the enzyme activities comprised 51 mM diethanolamine, 51 mM N-ethylmorpholine, and 100 mM morpholineethanesulfonic acid (MES). This three-buffer system covered the entire pH range used without a significant change in ionic strength.
SDS-PAGE and immunoblotting.
Proteins were separated by 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (19), electrotransferred to HybondP (Amersham) sheets (0.45 μm) for immunoblot analysis, and stained with Sypro Red (1/5,000 dilution) to verify that there was equal loading in all lanes. After destaining in 25 mM Tris-90 mM glycine-10% methanol-0.005% SDS, the blots were washed twice in Tris-buffered saline containing 0.05% (vol/vol) Tween 20 and incubated for 1 h in the same buffer containing 5% (wt/vol) defatted milk powder. Blots were then incubated with primary antibody (1/10,000 dilution for mouse antiphosphotyrosine monoclonal antibody purchased from Sigma-Aldrich and 1/5,000 dilution for mouse anti-His-RGS monoclonal antibody purchased from Sigma-Aldrich) overnight at room temperature in milk buffer and then detected according to the manufacturer's recommendations.
Gel filtration chromatography.
Protein extracts were freshly prepared before the gel filtration analysis. Frozen cells (∼1 g) were suspended in 2 ml of ice-cold extraction buffer A in the presence of 0.4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) and 30 μg/ml phenylmethylsulfonyl fluoride. The homogenate was then centrifuged at 8,000 × g for 15 min. The resulting 0.5-ml supernatant (2.8 mg protein) was injected into a column (1 by 80 cm) of Sephacryl S-200 precalibrated with the following molecular weight markers: thyroglobulin (molecular weight, 669,000), ferritin (440,000), aldolase (158,000), albumin (67,000), and cytochrome c (12,400). Equilibration and elution were performed with 30 mM Tris-HCl (pH 8.0)-0.150 M NaCl-0.4% CHAPS. Fractions (500 μl) were collected, and 30 μl of each fraction was used for SDS-PAGE.
RESULTS
Effects of epsB, epsC, epsD, and epsE deletion mutations on EPS production.
To study the role of the conserved genes present at the 5′ end of the eps gene cluster in S. thermophilus, we constructed mutant strains with internal deletions in the epsB, epsC, epsD, and epsE genes. The concentrations and lengths of EPS produced by the WT and mutant strains were examined. The concentrations of EPS detected in the WT and epsB mutant strains were 31 ± 2 and 28 ± 3 mg/liter, respectively, and the molecular masses were 4.0 ± 0.2 and 4.4 ± 0.1 MDa, respectively. In contrast, the concentrations of EPS were not above the threshold of detection in the epsC, epsD, and epsE mutants. These data show that the products of the latter three genes are required for EPS synthesis in S. thermophilus CNRZ1066.
Galactosyltransferase activities in epsB, epsC, epsD, and epsE mutants.
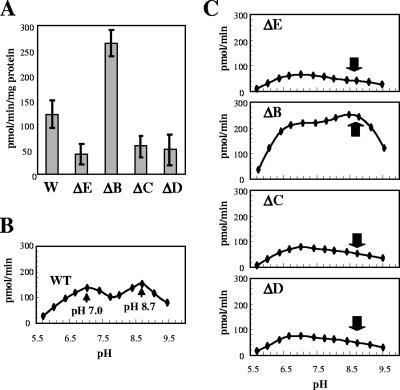
The lack of EPS in the epsC, epsD, and epsE mutants indicated that the cognate gene products are required for EPS synthesis. We first studied epsE, encoding the priming GT, which catalyzes the first step in EPS biosynthesis by transfer of galactose to the membrane lipid carrier (30). We tested the GT activity in membrane protein fractions of the WT and mutant strains. Significant GT activity was observed in the epsE mutant (Fig. 1A), suggesting that there is at least one other GT enzyme. In order to measure EpsE activity more specifically, we determined the GT activities in our strains over a range of pH values (pH 5.5 to 9.5) (“pH activity profile”). The GT pH activity profile revealed that there were two pH optima for the WT strain, pH 7.0 and pH 8.7 (Fig. 1B). Remarkably, the pH activity profile of the epsE mutant strain differed significantly, as there was only one pH optimum, pH 7.0 (Fig. 1C). This indicated that the second pH optimum in the WT pH activity profile was due to EpsE activity.
FIG. 1.
GT activities in S. thermophilus WT and mutant strains. (A) Global GT activities. (B and C) Effect of pH on GT activities in the WT strain (B) and in strains lacking EpsE, EpsB, EpsC, and EpsD (C). Samples (0.3 to 1.0 mg) of membrane protein extracts of S. thermophilus CNRZ1066 and the epsB, epsC epsD, and epsE mutant strains were incubated with 1 mM UDP-galactose at pH 8.0 (A) or various pHs (B and C) for 1 h at 37°C. See Materials and Methods for the experimental details. The results shown for the GT assay are the results for three independent experiments. The arrows indicate the peak corresponding to EpsE activity. W, wild type; ΔE, EpsE mutant; ΔB, EpsB mutant; ΔC, EpsC mutant; ΔD, EpsD mutant.
The GT pH activity profiles of the epsC and epsD mutant strains were similar to that of the epsE mutant, indicating that EpsE activity is dependent on both EpsC and EpsD. In contrast, the GT pH activity profile of the epsB mutant strain was qualitatively similar to that of the WT strain, but the overall GT activity was twofold higher (Fig. 1A, B, and C). Taken together, these results indicate that EpsE activity requires EpsC and EpsD but is negatively affected by EpsB. It should be noted that the alternative GT activity, whose optimum pH is pH 7.0, also appears to be negatively affected by EpsB, even though it does not require EpsC and EpsD, and is not sufficient for EPS synthesis.
Phosphorylation of EpsD and association of EpsD with the membrane fraction.
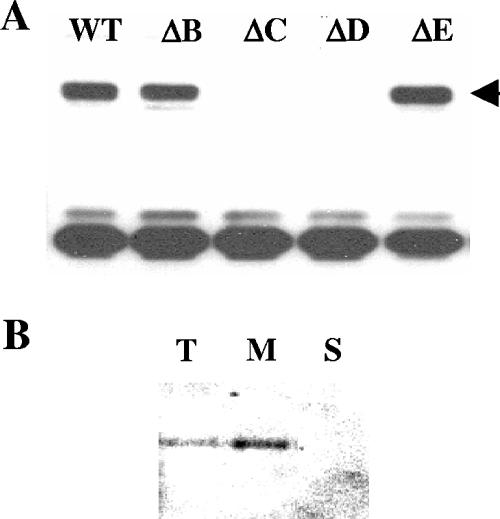
As EpsD is a putative kinase, we searched for phosphorylation of this molecule at tyrosine residues, using anti-P-Tyr antibodies (Fig. 2A). The phosphorylated form of EpsD was identified as a band present in the WT strain but absent in the epsD strain. A similar band was found in the epsB and epsE mutant strains but not in the epsC mutant strain. This result shows that phosphorylation of EspD requires EpsC.
FIG. 2.
Detection of Tyr-phosphorylated proteins by immunoblotting in S. thermophilus WT and mutant strains. (A) Presence of the 25-kDa Tyr-phosphorylated proteins (EpsD) as determined by Western immunoblotting using mouse antiphosphotyrosine monoclonal antibody. Thirty micrograms of proteins prepared from the WT, epsB, epsC, epsD, or epsE mutant strain was loaded in each lane. The arrow indicates the expected location of EpsD. (B) Cellular localization of Tyr-phosphorylated proteins as determined by immunoblotting. Total (lane T), membrane (lane M), and soluble (lane S) protein extracts were used for analysis. Twenty micrograms of protein was loaded in each lane. ΔE, EpsE mutant; ΔB, EpsB mutant; ΔC, EpsC mutant; ΔD, EpsD mutant.
A bioinformatic analysis of EpsC using Tmpred (http://www.ch.embnet.org) and SOSUI (www.sosui.proteome.bio.tuat.ac.jp) indicated that this protein contains two transmembrane domains, suggesting that it is located in the cytoplasmic membrane, while EpsD is predicted to be a cytoplasmic soluble protein (data not shown). If these predictions are correct, the interaction between EpsC, EspD, and the phosphorylated form of EspD takes place in the membrane. To test this possibility, we analyzed tyrosine-phosphorylated proteins using total cell extracts, fractionated membranes, and soluble extracts of the WT strain. In agreement with our hypothesis, a band corresponding to the tyrosine-phosphorylated form of EpsD was detected in the total free extract and in the membrane protein fractions but not in the soluble fraction (Fig. 2B). This result indicates that the phosphorylated form of EspD is bound to the membrane.
Protein complex containing EpsE and EpsD.
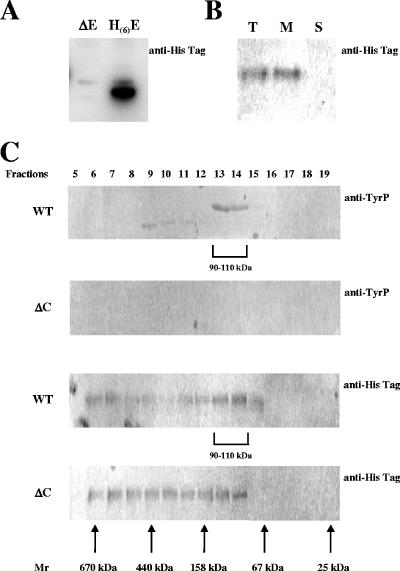
As (i) EpsE and the phosphorylated form of EspD are bound to the membrane, (ii) EpsC is predicted to be a membrane protein, and (iii) functional EpsC and EpsD are required for EpsE activity, it is possible that these proteins form a complex. To test this possibility, we first analyzed the localization of EpsE, using an N-terminally His-tagged protein form (His6-EpsE) able to complement the epsE mutant for EPS synthesis (not shown). His6-EpsE was visualized by Western blot analysis using antibody against the His tag (Fig. 3A). As expected, it was localized at the plasma membrane (Fig. 3B).
FIG. 3.
EpsE and EpsD protein analysis by immunoblotting and gel filtration. (A) Detection of the 25-kDa His6-EpsE recombinant proteins on a Western immunoblot probed with mouse anti-His6 monoclonal antibody in the epsE mutant strain and the same strain carrying the expression vector pJIM4843-epsHE. (B) Cellular localization of His6-EpsE by immunoblotting. Total (lane T), membrane (lane M), and soluble (lane S) protein extracts were used for analysis. Twenty micrograms of protein was loaded in each lane. (C) Localization of EpsE and EpsD in gel filtration analysis fractions. His6-EpsE and the tyrosine-phosphorylated form of EpsD were revealed on Western immunoblots probed with mouse anti-His6 monoclonal antibody and with mouse antiphosphotyrosine monoclonal antibody, respectively, after SDS-PAGE gel separation. The membrane extracts were prepared from a WT strain and an epsC mutant strain. Thirty-microliter aliquots of each fraction were assayed by blotting. ΔE, EpsE mutant; H(6)-E, His6-EpsE; ΔC, EpsC mutant.
Further investigation of the potential interaction of the three proteins was carried out by determining the masses of the protein complexes containing the phosphorylated EpsD and/or His6-EpsE. For this purpose the whole-cell extracts were fractionated by gel filtration chromatography, and the fractions were analyzed by SDS-PAGE and immunoblotting, using antibodies recognizing tyrosine-phosphorylated proteins or His-tagged EpsE (Fig. 3C). In the WT strain, the majority of the tyrosine-phosphorylated proteins were in the size range from 90 to 110 kDa (Fig. 3C, top panel). Their mobility on the SDS-PAGE gel was the mobility expected for a protein that was the size of EpsD (about 27 kDa). Small amounts of tyrosine-phosphorylated proteins, which migrated fast on the SDS-PAGE gel, were found in the size range from 158 to 440 kDa. The nature of these proteins was not investigated further. No signal was detected in the epsC mutant strain, indicating that the protein visualized in the WT strain was EpsD, which is phosphorylated in the presence of EpsC but not in the absence of EpsC. Interestingly, in the WT strain, the majority of the His6-EpsE protein was also in the size range from 90 to 110 kDa (Fig. 3C), suggesting that EpsE and the phosphorylated form of EpsD are present in the same complex. A sizable fraction of EpsE was larger than 440 kDa, suggesting that the protein may form high-molecular-weight complexes. The His6-EpsE profile was markedly different in the epsC mutant (Fig. 3C, bottom panel), where the protein appeared to be almost uniformly distributed over the size range from 90 to 670 kDa. The preferential formation of the 90- to 110-kDa complex thus appears to require EpsC and/or the phosphorylated EpsD protein. It should be noted that a complex of this size could be composed of one molecule of EpsD, one molecule of EpsE, and one molecule of EpsC, as these three proteins have a total mass of 80 kDa, a value close to 90 to 110 kDa.
Effect of tyrosine substitution on EpsE activity in S. thermophilus.
The fact that EpsE could interact with the EpsD tyrosine kinase suggests that EpsE may be regulated by phosphorylation. We searched for potential phosphorylation sites in the EpsE sequence by using the NetPhos 2.0 program. Of the eight tyrosine residues present in EpsE, only two were found to be potential sites for phosphorylation; these residues were the residues at positions 162 (score, 0.63) and 200 (score, 0.82). Finally, multiple alignment of EpsE homologues in gram-positive and gram-negative bacteria showed that phosphogalactosyltransferases have common motifs around the region corresponding to the tyrosine residues at positions 162 and 200 in S. thermophilus (Fig. 4). Therefore, we constructed derivatives of His6-EpsE in which the tyrosine residues at positions 162 and 200 were replaced by phenylalanine residues in order to measure the effect of modification of these residues on EpsE activity. An additional variant in which the tyrosine residue at position 199 was replaced by serine was constructed to determine the role of this residue. Plasmids expressing the modified His6-EpsE enzymes were introduced into the S. thermophilus epsE mutant, and their abilities to produce EPS were compared to those of the wild-type and epsE mutant strains. No significant differences were found between strains producing the Y162F and Y199S variants and the wild-type strains, whereas the Y200F variant and epsE mutant strains did not produce detectable amounts of EPS. These results suggest that the tyrosine residue at position 200 is necessary for EpsE activity.
FIG. 4.
Conservation of tyrosine residues in EPS priming enzymes of different groups of bacteria: multiple alignment of EpsE homologues in bacteria around two conserved tyrosine residues located at positions 162 and 200 in EpsE from S. thermophilus CNRZ1066 (indicated by bold type). Other conserved residues are highlighted with a black or gray background. L. cremoris, Lactococcus cremoris; C. acetobutylicum, Clostridium acetobutylicum; H. actinomycetemcomitans, Actinobacillus actinomycetemcomitans; H. influenzae, Haemophilus influenzae; S. typhimurium, Salmonella enterica serovar Typhimurium; A. tumefaciens, Agrobacterium tumefaciens; R. sp NGR234, Rhizobium sp. strain NGR234; E. amylovora, Erwinia amylovora.
DISCUSSION
In this study we established that S. thermophilus EpsE, the phosphogalactosyltransferase involved in the first step of EPS biosynthesis, is not active in epsC and epsD mutant strains and that, as a consequence, these mutants do not produce EPS. These results show that EpsE activity requires functional EpsC and D proteins. In a different way, Wzc and YwqCD, which are EpsCD homologues in E. coli and B. subtilis, respectively, were reported to control UDP-glucose dehydrogenase activity (26, 23). This enzyme allows the synthesis of UDP-glucuronide, a precursor of EPS biosynthesis in these strains. However, glucuronide is not a constituent of S. thermophilus CNRZ1066 EPS, which, like the EPS of S. thermophilus Sfi6, contains only glucose and galactose (7, 30), nor is it present in many EPS-producing bacteria, as deduced from an in silico search for UDP-glucose dehydrogenase genes in complete genome sequences. Therefore, it is likely that EPS synthesis is controlled by other enzymatic steps in most bacteria. Interestingly, phosphoglycosyltransferases have common motifs in gram-positive and gram-negative bacteria, suggesting that EpsD homologues may play a general role in the regulation of EPS biosynthesis in bacteria by controlling the priming reaction (Fig. 4).
EpsE enzymatic activity was higher in a strain lacking EpsB, a protein homologous to known phosphatases from various bacteria, particularly streptococcal CpsB, which is its closest homologue (2, 24). These phosphatases are involved in dephosphorylation of tyrosine kinases regulating bacterial EPS production (15, 22). This result is in agreement with a model in which tyrosine phosphorylation of CpsD triggers a limiting step in EPS production, whereas CpsB phosphatase acts as a modulator of its activity, as described previously for S. pneumoniae (2).
The regulation of EpsE activity by EpsC and EpsD could involve interaction of these three proteins, possibly in a complex. Our analyses showed that the sizes of EpsE and the phosphorylated form of EpsD are in the range from 90 to 110 kDa, a size compatible with the size of a complex that also includes EpsC. For B. subtilis, Mijakovic et al. (23) proposed a model in which YwqC might facilitate the interaction between YwqD and YwqF. Recently, Wzc has been shown to interact with Wza, the polymerase involved in the last step of EPS synthesis in E. coli (26). Thus, interaction of proteins involved in EPS synthesis seems to be a general process and could well take place within heteroprotein complexes.
The exact mechanism of regulation of EpsE is not known yet. Nevertheless, a model of regulation based on our observations and established data for streptococci can be proposed. EpsE could form high-molecular-weight complexes lacking phosphogalactosyltransferase activity. As shown using its homologues, EpsC, which is present in the membrane, recruits EpsD, which is then phosphorylated. The phosphorylated EpsD, possibly in conjunction with EpsC, could attract EpsE into a complex and activate the phosphogalactosyltransferase. A likely mechanism of activation is phosphorylation of EpsE by EpsD on a well-conserved Tyr200 residue, as UDP-glucose dehydrogenase is activated in this way by Wzc and YwqCD (16, 23). This hypothesis is supported by the fact that Tyr200 was shown to be essential for EpsE activity. The role of EpsB could be to dephosphorylate either EpsD, as shown for its homologue CpsD (2), or EpsE, as shown for UDP-glucose dehydrogenase in E. coli and B. subtilis (33, 22). Alternatively, the possibility that EpsD phosphorylation plays a role in the change in the EpsE conformation cannot be excluded; this could involve a mechanism of activation in which the enzyme is inactive in its oligomeric form and active in its monomeric form (4, 8, 11).
The discovery that the EpsD tyrosine kinase controls the EPS biosynthesis priming step strongly suggests that kinases belonging to this family have different targets in the cell. Interestingly, the kinase target found in this work is universally present in all bacteria synthesizing complex EPS. Moreover, the enzymes carry well-conserved motifs, including a potential tyrosine phosphorylation site. Further work is necessary to show that the products of genes homologous to epsBCDE interact in a similar manner and that the model for regulation of EPS synthesis, based on control of phosphoglycosyltransferase activity, may well be general. Since EPS play essential roles in bacterial pathogenicity, further insights in the biochemistry of EpsD-EpsE interaction could result in methods to develop a broad spectrum of new drugs to combat bacterial infections.
Acknowledgments
We thank Sandrine Petry for construction of the epsA mutant and J. Deutsher and I. Mijakovic for useful discussions.
This work was a joint effort of INRA and Danone-Vitapole and was supported by a grant to Z. Minic and C. Marie.
Footnotes
Published ahead of print on 15 September 2006.
REFERENCES
- 1.Bender, M. H., R. T. Cartee, and J. Yother. 2003. Positive correlation between tyrosine phosphorylation of CpsD and capsular polysaccharide production in Streptococcus pneumoniae. J. Bacteriol. 185:6057-6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender, M. H., and J. Yother. 2001. CpsB is a modulator of capsule-associated tyrosine kinase activity in Streptococcus pneumoniae. J. Biol. Chem. 276:47966-47974. [DOI] [PubMed] [Google Scholar]
- 3.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blond-Elguindi, S., A. M. Fourie, J. F. Sambrook, and M. J. Gething. 1993. Peptide-dependent stimulation of the ATPase activity of the molecular chaperone BiP is the result of conversion of oligomers to active monomers. J. Biol. Chem. 268:12730-12735. [PubMed] [Google Scholar]
- 5.Bolotin, A., B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyprides, B. Purnelle, D. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau, and P. Hols. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brueggemann, A. B., D. T. Griffiths, E. Meats, T. Peto, D. W. Crook, and B. G. Spratt. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187:1424-1432. [DOI] [PubMed] [Google Scholar]
- 7.Cerning, J., C. Bouillanne, and M. J. Desmazeaud. 1988. Exocellular polysaccharide production by Streptococcus thermophilus. Biotechnol. Lett. 10:255-260. [Google Scholar]
- 8.Chakraborty, A., I. Das, R. Datta, B. Sen, D. Bhattacharyya, C. Mandal, and A. K. Datta. 2002. A single-domain cyclophilin from Leishmania donovani reactivates soluble aggregates of adenosine kinase by isomerase-independent chaperone function. J. Biol. Chem. 277:47451-47460. [DOI] [PubMed] [Google Scholar]
- 9.Cieslewicz, M. J., D. L. Kasper, Y. Wang, and M. R. Wessels. 2001. Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J. Biol. Chem. 276:139-146. [DOI] [PubMed] [Google Scholar]
- 10.Cozzone, A. J., C. Grangeasse, P. Doublet, and B. Duclos. 2004. Protein phosphorylation on tyrosine in bacteria. Arch. Microbiol. 181:171-181. [DOI] [PubMed] [Google Scholar]
- 11.de Vienne, D., and F. Rodolphe. 1985. Biochemical and genetic properties of oligomeric structures: a general approach. J. Theor. Biol. 116:527-568. [DOI] [PubMed] [Google Scholar]
- 12.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28:350-356. [Google Scholar]
- 14.Goupil-Feuillerat, N., G. Corthier, J. J. Godon, S. D. Ehrlich, and P. Renault. 2000. Transcriptional and translational regulation of alpha-acetolactate decarboxylase of Lactococcus lactis subsp. lactis. J. Bacteriol. 182:5399-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grangeasse, C., P. Doublet, C. Vincent, E. Vaganay, M. Riberty, B. Duclos, and A. J. Cozzone. 1998. Functional characterization of the low-molecular-mass phosphotyrosine-protein phosphatase of Acinetobacter johnsonii. J. Mol. Biol. 278:339-347. [DOI] [PubMed] [Google Scholar]
- 16.Grangeasse, C., B. Obadia, I. Mijakovic, J. Deutscher, A. J. Cozzone, and P. Doublet. 2003. Autophosphorylation of the Escherichia coli protein kinase Wzc regulates tyrosine phosphorylation of Ugd, a UDP-glucose dehydrogenase. J. Biol. Chem. 278:39323-39329. [DOI] [PubMed] [Google Scholar]
- 17.Hols, P., F. Hancy, L. Fontaine, B. Grossiord, D. Prozzi, N. Leblond-Bourget, B. Decaris, A. Bolotin, C. Delorme, S. Dusko Ehrlich, E. Guedon, V. Monnet, P. Renault, and M. Kleerebezem. 2005. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 29:435-463. [DOI] [PubMed] [Google Scholar]
- 18.Kolkman, M. A. B., B. A. M. vanderZeijst, and P. J. M. Nuijten. 1997. Functional analysis of glycosyltransferases encoded by the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniae serotype 14. J. Biol. Chem. 272:19502-19508. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Letort, C., and V. Juillard. 2001. Development of a minimal chemically-defined medium for the exponential growth of Streptococcus thermophilus. J. Appl. Microbiol. 91:1023-1029. [DOI] [PubMed] [Google Scholar]
- 21.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 22.Mijakovic, I., L. Musumeci, L. Tautz, D. Petranovic, R. A. Edwards, P. R. Jensen, T. Mustelin, J. Deutscher, and N. Bottini. 2005. In vitro characterization of the Bacillus subtilis protein tyrosine phosphatase YwqE. J. Bacteriol. 187:3384-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mijakovic, I., S. Poncet, G. Boel, A. Maze, S. Gillet, E. Jamet, P. Decottignies, C. Grangeasse, P. Doublet, P. Le Marechal, and J. Deutscher. 2003. Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J. 22:4709-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morona, J. K., R. Morona, D. C. Miller, and J. C. Paton. 2002. Streptococcus pneumoniae capsule biosynthesis protein CpsB is a novel manganese-dependent phosphotyrosine-protein phosphatase. J. Bacteriol. 184:577-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morona, J. K., J. C. Paton, D. C. Miller, and R. Morona. 2000. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 35:1431-1442. [DOI] [PubMed] [Google Scholar]
- 26.Reid, A. N., and C. Whitfield. 2005. Functional analysis of conserved gene products involved in assembly of Escherichia coli capsules and exopolysaccharides: evidence for molecular recognition between Wza and Wzc for colanic acid biosynthesis. J. Bacteriol. 187:5470-5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts, I. S. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50:285-315. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 29.Stingele, F., J. R. Neeser, and B. Mollet. 1996. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J. Bacteriol. 178:1680-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stingele, F., J. W. Newell, and J. R. Neeser. 1999. Unraveling the function of glycosyltransferases in Streptococcus thermophilus Sfi6. J. Bacteriol. 181:6354-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terzaghi, B., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van der Vossen, J. M., D. van der Lelie, and G. Venema. 1987. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl. Environ. Microbiol. 53:2452-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vincent, C., B. Duclos, C. Grangeasse, E. Vaganay, M. Riberty, A. J. Cozzone, and P. Doublet. 2000. Relationship between exopolysaccharide production and protein-tyrosine phosphorylation in gram-negative bacteria. J. Mol. Biol. 304:311-321. [DOI] [PubMed] [Google Scholar]