Abstract
The two-component system TCS08 is one of the regulatory systems that is important for virulence of Streptococcus pneumoniae. In order to investigate the TCS08 regulon, we have analyzed transcription profiles of mutants derived from S. pneumoniae R6 by microarray analysis. Since deletion mutants are often without a significant phenotype, we constructed a mutation in the histidine kinase HK08, T133P, in analogy to the phosphatase mutation T230P in the H box of the S. pneumoniae CiaH kinase described recently (D. Zähner, K. Kaminski, M. van der Linden, T. Mascher, M. Merai, and R. Hakenbeck, J. Mol. Microbiol. Biotechnol. 4:211-216, 2002). In addition, a deletion mutation was constructed in rr08, encoding the cognate response regulator. The most heavily suppressed genes in the hk08 mutant were spr0276 to spr0282, encoding a putative cellobiose phosphoenolpyruvate sugar phosphotransferase system (PTS). Whereas the R6 Smr parent strain and the Δrr08 mutant readily grew on cellobiose, the hk08 mutant and selected mutants with deletions in the PTS cluster did not, strongly suggesting that TCS08 is involved in the catabolism of cellobiose. Homologues of the TCS08 system were found in closely related streptococci and other gram-positive cocci. However, the genes spr0276 to spr0282, encoding the putative cellobiose PTS, represent a genomic island in S. pneumoniae and homologues were found in Streptococcus gordonii only, suggesting that this system might contribute to the pathogenicity potential of the pneumococcus.
Streptococcus pneumoniae is a major human pathogen that is capable of causing serious infections such as pneumonia, meningitis, and septicemia. Invasive pneumococcal disease results in a high mortality rate among the young, the elderly, and the immunosuppressed (24). In third-world countries the pneumococcus is the single most common cause of these pneumonia deaths in children under the age of 5 and accounts for up to 25% of all deaths in this age group (3).
Colonization and invasion of the host require the pneumococcus to regulate gene expression to adapt to the various environmental conditions experienced (11, 19, 29). Bacterial two-component signal-transducing systems are one mechanism employed to regulate adaptive responses to environmental systems. They typically consist of two proteins: (i) a histidine kinase, which is usually located in the cell membrane, that acts as a sensor for stimuli and (ii) a cognate cytoplasmic response regulator that changes the expression profiles of target genes (27, 36).
S. pneumoniae contains 13 two-component regulatory systems (TCS) and one orphan response regulator (18, 39). The first TCS in S. pneumoniae to be described and characterized was the CiaR/CiaH system (8), which belongs to the EnvZ/OmpR family like the majority of the pneumococcal two-component systems (18, 39). The most widely studied pneumococcal two-component signal-transducing system is the ComDE system, which is involved in genetic competence in response to competence-stimulating peptide (5, 28). Other systems that have been partially characterized include BlpR/BlpS (TCS13), which is induced by the autoinducible peptide BlpC and controls bacteriocin production (6, 32); TCS04, which is involved in the regulation of PsaA (23); and TCS06, which regulates expression of cbpA (35). The only essential response regulator, RR02, regulates expression of PspA (25), and the orphan response regulator, RitR, regulates iron transport (41). Eight of the regulatory systems have been shown to be critical for growth in a mouse respiratory tract infection model, suggesting that they are important for pathogenesis of S. pneumoniae (39). Among those was TCS08 (or 484 hk/rr), whose function is yet unknown.
Apparent activating point mutations in histidine kinases have previously been reported and presumably result in the continuous phosphorylation of the response regulator, such as an Asp299-to-Asn mutation in the transmembrane histidine kinase ComD that results in constitutive competence for transformation (17). Also, a mutation in the histidine kinase CiaH that apparently result in the activation of the cia system has been observed in laboratory mutants (8, 42). Activation has indeed been shown to be mediated by the T230P mutation in strain C306, which is located in the conserved H box of the transmembrane histidine kinase CiaH and results in a strong phenotype, including increased resistance to cell wall damage, reduced autolysis, and complete suppression of the development of genetic competence (8, 42). It has been suggested that this mutation results in a continuously elevated phosphorylation of CiaR due to a phosphatase deficiency in CiaH (8, 43), since this mutation is similar to a mutation in the Escherichia coli EnvZ histidine kinase that apparently inactivates the phosphatase activity of the enzyme (33, 40). Indeed, transcriptional analyses in comparison to an R6 Smr parent strain indicate opposing transcription profiles between ΔciaR and CiaH T230P (21, 43).
This study examines the pneumococcal TCS08 system first described by Lange et al. (18), which has also been referred to as TCS484 (39). We have analyzed a specific point mutation in the histidine kinase protein HK08 in R6, which is believed to continuously activate TCS08, and have examined its role in gene regulation. Microarray analysis was used to examine the expression profile and to identify transcriptional changes in the HK08 mutant. In vivo characterization of the HK08 mutant revealed a role in cellobiose metabolism for this regulatory system.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains used in this study are listed in Table 1. Pneumococcal strains were grown without aeration at 37°C either in C medium (16) supplemented with 0.2% yeast extract (Difco) or in chemically defined media (C-den) (40a). Chemically defined medium was supplemented with sucrose (Merck), glucose (Merck), cellobiose (AppliChem GmbH), or hyaluronic acid from Streptococcus equi (Sigma-Aldrich). Carbohydrates were purchased from Sigma-Aldrich. Pneumococci were grown on D agar supplemented with 3% sheep blood (Oxoid) and containing the appropriate antibiotic, where necessary. Growth was monitored by nephelometry.
TABLE 1.
S. pneumoniae strains used in this study
| Strain | Genotype | Phenotype | Source or reference |
|---|---|---|---|
| R6 | Wild type | Sms Kns | 26 |
| R6 Smr | Parent strain | Smr Kns | 38 |
| R6 484HK | 484HK::Ery | Eryr Sms | 39 |
| R6 Δhk08 | hk08::Kn | Knr Sms | This study |
| hk08 mutant | hk08 A398C | Smr Kns | This study |
| R6 Δrr08 | rr08::Ery | Eryr Smr | This study |
| R6 Δspr0282 | spr0282::Kn | Knr Sms | This study |
| R6 Δspr0281 | spr0281::Sm | Smr Kns | This study |
| R6 Δspr0279 | spr0279::Sm | Smr Kns | This study |
Transformation procedure.
Transformation of S. pneumoniae strains was performed essentially according to published procedures, with 30 min of incubation in the presence of DNA at 30°C followed by a 2-h phenotypic expression period at 37°C (8). Antibiotic concentrations used for the selection of transformants were as follows: streptomycin (Sm), 200 μg/ml; kanamycin (Kn), 250 μg/ml; and erythromycin (Ery), 0.2 μg/ml.
Construction of HK08 point mutation.
The hk08 mutant was constructed via a two-step process using the Janus cassette (38), which is shown in Fig. 1A. Janus is a Kn rpsL+ cassette that confers a Knr Sms phenotype in an Smr background. During a second round of mutagenesis, the Janus cassette is replaced with the desired DNA which restores the original Smr phenotype. Two PCR products were amplified using the primer pairs GATGGGAAAGACAATTTTACTCG-GAGACGGATCCCAGCACCGACAAG and TTGACCTGGAGTTACGCGG-GTCCAAAAGCATAAGGAAAGGGGCCCTAGGCGCCAGACGGAGAGGCTC, to obtain two fragments flanking the portion of hk08 that encodes the H box. These products were digested with the appropriate enzyme, ligated to the Janus cassette, and transformed into Smr S. pneumoniae R6. Knr Sms transformants (Δhk08) were obtained and screened for the correct mutation. Two PCR products were generated separately using the primer pairs GATAGGAGGCTTAATATCATGG-GATGGGAAAGACAATTTTACTCG and CGCATGATATTAAGCCTCCTATC-TTGACCTGGAGTTACGCGG to obtain fragments containing the point mutation (the mutated positions are underlined). These products were then subjected to further PCR amplification to obtain an overlapped product containing the point mutation and the flanking regions of the genes. The resulting fragment was transformed into Δhk08 competent cells. DNA from transformants displaying a Smr Kns phenotype was extracted, PCR amplified, and sequenced to confirm the mutation.
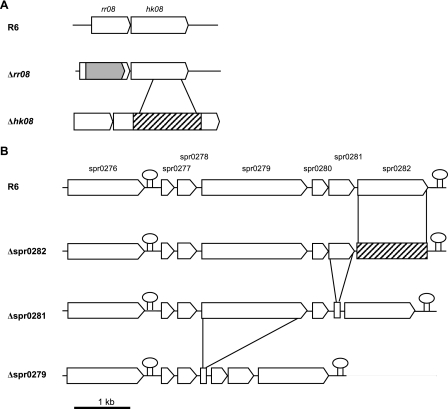
FIG. 1.
Diagrammatic representation of the strategy used for construction of the deletion mutants. (A) Deletion mutants constructed for the TCS08 locus; (B) deletion mutants constructed for the spr0276-0282 locus. The genes are represented by arrows, while the lollipops represent putative transcriptional terminators. Hatched areas, inserted Janus cassette; gray area, ermAB.
Pneumococcal DNA was extracted using the Wizard genomic DNA purification kit (Promega), and PCR products were purified using the JETquick spin column technique kit (Genomed). Restriction enzymes and T4 DNA ligase were purchased from Roche Applied Science and used according to the manufacturer's instructions. PCRs were performed using either Goldstar Red Taq polymerase (Eurogentec) or PfuTurbo Hotstart DNA polymerase (Stratagene) according to the manufacturer's instructions. Nucleotide sequencing was performed using the ABI Prism BigDye Terminator Ready Reaction cycle sequencing kit, version 3.1 (Perkin Elmer-ABI).
Construction of Δrr08 mutants.
DNA from the Δ484rr mutant constructed by Throup et al. (39) was extracted and PCR amplified using the primers CCACTTCCAGCTTGGTTCCTGG and GATGTTCCAGCAGCCTACTTAG to amplify the erythromycin cassette and its flanking regions; this was then transformed into the Smr R6 parent strain to ensure that all mutants investigated had the same genetic background. Eryr transformants were subsequently screened and sequenced to confirm the mutation.
Construction of the Δrr08 hk08 double mutant.
DNA from the Δ484rr mutant was amplified using a different combination of primers: S58, which hybridized before the region encoding the conserved histidine to ensure maintenance of the hk08 mutation described above, and S87, which primed upstream the erythromycin cassette in rr08. The PCR product was then transformed into the competent hk08 mutant cells. Proper insertion of the erythromycin cassette in Eryr Smr transformants was confirmed by PCR analysis.
Construction of Δspr0279, Δspr0281, and Δspr0282 mutants.
The S. pneumoniae Δspr0282 mutant was constructed by amplifying two PCR products by using the primers pairs S98-S106 and S100-S101, encoding the 5′ and 3′ flanking regions, respectively. These fragments were then purified, digested, and ligated to the Janus cassette (see above) (38). Transformants displaying a kanamycin-resistant, streptomycin-sensitive phenotype were verified by sequence analysis.
Construction of Δspr0279 and Δspr0281 mutants was achieved by back transformation of the Δspr0282 mutant. The Δspr0279 mutant was constructed by PCR amplifying the 5′ and 3′ portions of the gene and its flanking regions by using the primer pairs S104-S118 and S101-S119, respectively. Similarly, the Δspr0281 mutant was constructed by PCR amplifying the 5′ and 3′ portions of the gene and its flanking regions by using the primer pairs S101-S115 and S104-S114, respectively; these and all other primer sequences described here are listed at http://www.nbc.uni-kl.de/data/microarray/microarray_mckessar.zip. These products were digested with BamHI, ligated, and transformed into competent Δspr0282 cells. Transformants having an Smr Kns phenotype were screened and sequenced to confirm the mutation.
RNA extraction.
For mRNA analyses, cells were grown to the desired Nephelo units (N) in either C medium or C-den medium with the appropriate carbohydrate added (N = 40 or 80 corresponded to mid-exponential growth phase, and stationary phase was reached at around N = 130 in C-medium; in the case of cellobiose as the sole carbon source with C-den medium, cells were harvested at N = 30 since cells reached stationary phase at N = 70, or they were harvested at N = 40 with glucose or sucrose as the sole carbon source where stationary phase corresponded to N = 100). Approximately 108 cells were pelleted by centrifugation at 8,000 × g for 10 min at 4°C. The supernatant was removed and the cells resuspended in 4 ml of prewarmed (60°C) 50 mM sodium acetate-10 mM EDTA (pH 5.1)-saturated phenol and incubated at 60°C. After 5 min, 4 ml of prewarmed NAES (50 mM sodium acetate, 10 mM EDTA, 1% [wt/vol] sodium dodecyl sulfate [SDS] [pH 5.1] treated with diethylpyrocarbonate [DEPC]-treated water) was added to the lysate, mixed by inversion, and incubated at 60°C. After 5 min, the lysate was placed on ice for 5 min and centrifuged at 8,000 × g for 8 min at 4°C. The aqueous phase was removed and placed in a phase lock tube, and phenol-chloroform (5:1) extractions were performed twice. The layers were separated by centrifugation for 8 min at 8,000 × g. The aqueous layer was placed in a new tube, after which 400 μl of DEPC-treated sodium acetate and 4 ml of 100% isopropanol were added and mixed by inversion. All nucleic acids were then precipitated overnight at −70°C. Nucleic acids were pelleted by centrifuging at 8,000 × g for 40 min at 4°C. The pellet was washed with 4 ml 70% ethanol (0.7 volume 100% ethanol and 0.3 volume DEPC-treated water) and centrifuged for 10 min at 8,000 × g. The supernatant was removed, and any residual ethanol was removed by air drying for 15 min. The nucleic acids were redissolved in 440 μl DEPC-treated water by heating for 10 min at 60°C. RNase activity was inhibited by the addition 20 U RNasin RNase inhibitor (Promega). DNA was digested by addition of 90 U of RTQ1 RNase-free DNase in 50 μl of 10× DNase buffer (Promega) and incubation for 30 min at 37°C. The RNA was then purified further using the QIAGEN RNeasy midikit according to the manufacturer's instructions.
Microarray.
The microarray used included 50-mer oligonucleotide probes for all S. pneumoniae R6 annotated genes (2,045 genes) and was obtained from MWG Biotech AG. Each oligonucleotide was spotted in duplicate onto Schott Nexterion slide E. Fifty micrograms of purified mRNA was synthesized from random hexamers into labeled cDNA by using the LabelStar array kit (QIAGEN). The probes were then resuspended in 100 μl hybridization solution (Nexterion) and heated at 95°C for 5 min prior to hybridization.
Pretreatment, hybridization, and slide washing.
Hybridizations were performed using a Tecan HS400 hybridization station. Prehybridization consisted of one of each of the following steps: a 30-s wash with 0.5% SDS at 25°C, a 30-s water rinse at 25°C, and a 30-min incubation with prehybridization solution (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS, 0.5 mg/ml bovine serum albumin) at 42°C. The probe was then ready for injection. Hybridization was performed at 40°C with agitation for 16 h. Slide washing consisted of the following steps, all of which were performed at 30°C: a 1-min wash with 2× SSC and 0.1% SDS, a 1-min wash with 1× SSC, and a 1-min wash with 0.1× SSC. The slides were finally dried under nitrogen for 3 min.
Experimental data and replicates.
For each growth condition, two independently grown cultures were used. The mRNA preparations were divided into two parts, and each part was labeled with either Cy3 or Cy5 in order to eliminate any labeling bias during the following analyses. Since the oligonucleotides on the microarray were spotted in duplicate, a final number of eight spots per growth condition were analyzed. For each of the single mutants, two time points during exponential growth were taken and the results from two independent experiments pooled; the double mutant was harvested at N = 40 only. The chips were scanned using the ScanArray4000 microarray analysis system. The hybridization spots were analyzed using the EasyScan method in the ScanArrayExpress software, version 2.1, and the ArrayInformatics visualization tool software. A noise-to-background ratio of 3 was used as the cutoff. The microarray data were normalized using the LOWESS fit and the resulting ratios analyzed using Student's t test available at http://nbc3.biologie.uni-kl.de/ (P < 0.01). Only genes that had reproducible changes in the transcript amount of greater than a threefold threshold were considered further. Original raw data are listed at http://www.nbc.uni-kl.de/data/microarray/microarray_mckessar.zip.
Sugar fermentation.
Mid-exponential-phase cells growing in THY (Todd-Hewitt broth plus yeast extract) were harvested by centrifugation, washed once, and then resuspended in purple broth (Becton Dickinson Microbiology Systems) supplemented with 1% of the relevant sugar. The cultures were incubated at 37°C for up to 48 h and the color changes monitored regularly. For monitoring cellular growth with different carbon sources, mid-exponential-phase cells growing in C medium were harvested by centrifugation, washed once, and then subcultured into C-den medium containing the appropriate sugar.
Real-time PCR.
One microgram of total RNA, extracted from cells grown to N = 40 in C-den medium supplemented with the appropriate carbohydrate, was reverse transcribed into cDNA by using random hexamer primers and the Roche cDNA kit according to the manufacturer's instructions. After the cDNA was diluted 10-fold, 5 μl was used in a real-time PCR (RT-PCR) using the LightCycler Fast Start DNA Master Plus SYBR green kit (Roche). Five picomoles of each gene-specific primer was used in a total reaction volume of 20 μl according to the manufacturer's instructions. The reaction mixture was loaded into a 20-μl LightCycler capillary (Roche), and the capillaries were placed in an LC Carousel 2.0 (Roche) and centrifuged at 735 × g for 15 s. The capillaries were then loaded into a LightCycler 2.0 thermocycler (Roche). Amplification included an initial denaturation step at 95°C for 10 min, followed by 45 cycles, each consisting of 10 s of denaturation at 95°C (temperature transition, 20°C s−1), 10 s of annealing at 55°C (temperature transition, 20°C s−1), and 14 s of elongation at 72°C (temperature transition, 20°C s−1). Fluorescence acquisition was performed at the end of each cycle. The melting curve analysis was performed at 65 to 95°C (temperature transition, 20°C s−1) with continuous fluorescence acquisition. Each measurement was performed in duplicate, and the mean was taken for calculating the differences of expression levels, in Cp values; all experiments were performed with two independently grown cultures.
RT-PCR.
One microgram of total RNA was extracted from cells grown to N = 40 in C medium and reverse transcribed into cDNA using random hexamers and the Roche cDNA kit (Roche Diagnostics). One hundred nanograms of cDNA was then PCR amplified using Goldstar Red Taq polymerase (Eurogentec) for 35 cycles, after which 5 μl of the amplified product was electrophoresed on 1.5% agarose.
RESULTS
Construction of hk08 and Δrr08 mutants.
The H box of HK08 with the conserved histidine (SH129DIKT*P134) shares four identical and three similar amino acid residues with the H box of the CiaH kinase (SH226ELRT*P231), where T* is the position of the mutation T230P in the CiaH mutant that has been described as an activated mutant (22). The corresponding mutation T133P in the HK08 was introduced into the laboratory strain R6 carrying a streptomycin resistance marker by using a two-step strategy with the Janus cassette as described recently (38) (see Materials and Methods for details) (Fig. 1A). The hk08 mutant thus obtained still contains an intact rr08 gene.
Second, a deletion in the response regulator rr08 was constructed by introducing into our laboratory R6 Smr strain the rr08 mutation described recently and named Δrr484 by Throup et al. (39). In this mutant, the Eryr cassette without the termination signals replaces most of the rr08 gene to minimize potential polarity effects on hk08 (39). In fact, the microarray analyses performed during these studies confirmed that the expression of hk08 in the Δrr08 mutant was approximately twofold higher than that in the R6 Smr strain. The Eryr cassette along with its flanking regions was amplified from Δrr484 and transformed into the R6 Smr strain (Fig. 1A), and correct integration was confirmed by PCR followed by DNA sequencing.
Cellular growth of both mutants was similar to that of the R6 Smr parent strain, with a doubling time of approximately 35 min when grown in C medium at 37°C. Examination of the mutants by phase-contrast microscopy did not reveal any noticeable changes in the cell morphology compared to the R6 Smr parent strain.
Comparative transcription analysis of TCS08 mutants.
Initial approaches to determine the genes regulated by the TCS08 involved comparison of the transcription profiles of TCS08 mutants and their respective parent strains (R6 and R6 Smr) by using microarray technology. For each strain, independently grown cultures were used for the preparation of RNA and were harvested at either N = 40 or N = 80 during exponential growth. The microarrays were probed with labeled cDNA obtained from the RNA samples. The data set from eight separate hybridizations was used for normalization and the ratios obtained for statistical analysis. The results are summarized in Table 2.
TABLE 2.
Summary of the microarray data for the HK08 T133P mutant compared to the R6 Smr wild-type straina
| Geneb | Avg relative fold change | Putative protein |
|---|---|---|
| spr0276 (bglA) | −3.5 | 6-Phospho-β-glucosidase |
| spr0277 | −20.0 | Conserved hypothetical protein |
| spr0278 | −47.2 | Phosphotransferase system sugar-specific EII |
| spr0279 (bglG) | −12.4 | Transcription antiterminator, BglG family |
| spr0280 | −7.3 | Phosphotransferase system sugar-specific EII |
| spr0281 | −8.1 | Hypothetical protein |
| spr0282 | −11.9 | Phosphotransferase system sugar-specific EII |
Only data for genes with a statistically significant average relative fold change of greater than 3 are shown.
Names in parentheses refer to the annotated genome (12).
The most significant changes in mRNA levels were seen in the hk08 mutant and concerned the spr0276-spr0282 locus, encoding the putative proteins involved in sugar metabolism (BglA and a phosphotransferase system [PTS]) (Table 2 and Fig. 2). In this case, expression was reduced between 3.5- and 47-fold for the individual genes of the locus. In contrast, this gene cluster was not affected in the Δrr08 mutant (Table 2). The expression of some genes differed between two- and threefold in individual experiments; however, they were not considered further since too much scattering was observed in independent experiments. Most of these genes belong to the competence regulon, the bacteriocin cluster blp/pnc, or phage-related integrase genes and were therefore not considered further.
FIG. 2.
Normalized log scatter plot showing the signal intensities of the hk08 mutant compared to the R6 Smr parent strain. Shown is the result obtained from one microarray. The two lines flanking the diagonal indicate differences of a factor of 3. The signals representing genes spr0276 to spr0282 are indicated by circles.
In order to eliminate the possibility that the difference in expression level between the hk08 mutant and the parental strain is mediated by the response regulator RR08 and not by an indirect effect of the mutated HK08 on another regulatory protein (e.g., by a cross talk mechanism), the Δrr08 hk08 double mutant was constructed, in which the mutant kinase was expressed in the absence of the rr08 gene. This double mutant gave results identical to those with the Δrr08 single mutant, with no effect on expression of spr0276 to spr0282 being apparent. This strongly suggests that RR08 directly acts upon this gene locus.
Sugar fermentation of the TCS08 mutants.
Blast searches revealed similarities of the putative PTS encoded by spr0276 to spr0282 with cellobiose and lactose PTSs of other organisms. In order to establish the specificity of this system and the link to the HK/RR08 system, S. pneumoniae R6 Smr and the Δrr08 and hk08 mutants were tested for their ability to grow on different carbon sources in a first set of experiments. The strains were grown in purple broth supplemented with the appropriate carbon source. The R6 Smr parent strain fermented most sugars tested but not arbutin, lichenan, or salicin (Table 3). This fermentation pattern remained unchanged in the Δrr08 mutant. In the hk08 mutant, however, one clear exception was noted, i.e., a significantly reduced ability to ferment the β-glucoside cellobiose.
TABLE 3.
Fermentation results for the R6 Smr wild-type strain and the mutants tested with various carbohydrates
| Straina | Fermentation of:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glucose | Galactose | Fructose | Salicin | Cellobiose | Arbutin | Trehalose | Lichenan | Maltose | Lactose | Sucrose | GlcNAcb | |
| R6 | +++ | +++ | +++ | − | +++ | − | +++ | − | +++ | +++ | +++ | +++ |
| Δrr08 mutant | +++ | +++ | +++ | − | +++ | − | +++ | − | +++ | +++ | +++ | +++ |
| hk08 mutant | +++ | +++ | +++ | − | + | − | +++ | − | +++ | +++ | +++ | +++ |
| Δspr0279 mutant | +++ | +++ | +++ | − | + | − | +++ | − | +++ | +++ | +++ | +++ |
| Δspr0281 mutant | +++ | +++ | +++ | − | + | − | +++ | − | +++ | +++ | +++ | +++ |
| Δspr0282 mutant | +++ | +++ | +++ | − | + | − | +++ | − | +++ | +++ | +++ | +++ |
All mutants are derivatives of S. pneumoniae R6.
GlcNAc, N-acetylglucosamine.
In addition, hyaluronic acid was also included as a test compound. Hyaluronic acid is hydrolyzed by the S. pneumoniae hyaluronidases (hyaluronate lyases) (31), and the cleavage products might be utilized as structural analogues of cellobiose by the PTS. Hyaluronic acid was also utilized by R6, but the cells grew poorly, with a generation time of approximately 80 min. In this case, no difference between the parent and the mutant strains was observed (Fig. 3E and F). Nevertheless, this shows that S. pneumoniae R6 is capable of metabolizing hyaluronic acid into a readily available nutrient source.
FIG. 3.
Effects of TCS08, spr0279, spr0281, and spr0282 mutations. Growth in C-den medium was monitored by nephelometry. (A and B) Controls (C-den medium with glucose plus sucrose); (C and D) C-den medium with cellobiose; (E and F) C-den medium with hyaluronic acid. ▪, R6 Smr; (•), Δrr08 mutant; ▴, hk08 mutant; ○, Δspr0279 mutant; ▵, Δspr0281 mutant; ▿, Δspr0282 mutant.
A putative cellobiose PTS in S. pneumoniae.
In order to confirm the specificity of the putative PTS encoded by spr0276 to spr0282 for cellobiose, this locus was examined and mutants with mutations in some of its genes were analyzed in detail. In addition to the three putative PTS proteins (Spr0278, Spr0280, and Spr0282), the region includes the predicted 6-phospho-β-glucosidase BglA (Spr0276), the putative transcriptional antiterminator Spr0279, and the two hypothetical transmembrane proteins Spr0277 and Spr0281 (Fig. 1B). Deletion mutations in the genes encoding the transmembrane-spanning protein Spr0281 and the sugar-specific component Spr0282 were constructed with help of the Janus cassette as described above (see Materials and Methods and Fig. 1B). All constructs grew readily in the defined medium C-den (Fig. 3A to E). However, neither mutant grew in cellobiose-supplemented medium, in contrast to the R6 Smr strain, which grew with a generation time of 49 min. This demonstrates that cellobiose cannot be utilized by mutants with mutations in the putative PTS and therefore confirms that the PTS is involved in cellobiose uptake.
The mutants did grow on hyaluronic acid-containing medium, in agreement with the suggestion that the PTS is not (mainly) responsible for hyaluronic acid catabolism.
In a second set of experiments, the nature of the spr0279 product as a putative regulator of the PTS was investigated. BlastX and conserved amino acid motif analysis indicated that Spr0279 belongs to the BglG family of antiterminator regulatory proteins. The E. coli BglG antiterminator is part of the blg operon, which bears some similarities to the S. pneumoniae PST system. In E. coli, a constitutive bglGp promoter precedes the genes bglG, bglF (encoding IIBgl), and bglB (encoding a phospho-β-glucosidase). Transcription is attenuated by terminators which flank bglG, thereby allowing a residual transcription of bglG and bglF (13, 30). In the presence of substrate, the phosphate group from P-IIBgl is transferred to the β-glucoside. This causes the BglG to be dephosphorylated, which results in the dimerization of BglG to an active antiterminator (30).
The transcriptional antiterminator regulates transcription by binding to specific RNA sequences which are located upstream of transcriptional terminators, thus preventing termination and inducing the operon (13, 34). Similarly, a stem-loop acting as a potential terminator can be postulated to be present between spr0276 and spr0277 in the S. pneumoniae PTS operon (Fig. 1B) and might be targeted by the Spr0279 regulator. In order to test whether Spr0279 is required for efficient transcription of RNA over that stem-loop structure, transcription of the intergenic region between spr0276 and spr0277 in the deletion mutant spr0279 was determined by RT-PCR analysis and compared to that of the parental strain. Cells were grown in C-den medium, where the operon should be transcribed. The R6 Smr parent strain yielded a PCR product, while that of the Δspr0279 mutant was markedly decreased (Fig. 4), confirming the role of Spr0279 as a regulatory protein and putative antiterminator.
FIG. 4.
RT-PCR analysis. cDNAs were converted from RNAs isolated from the R6 Smr wild-type strain and the Δspr0279 mutant and subjected to RT-PCR amplification using primers specific to the flanking regions of the stem-loop structure located between spr0276 and spr0277. Lanes: M, 1-kb marker; a, DNA from R6 Smr; b, cDNA from R6 Smr; c, cDNA from the Δspr0279 mutant. The size of the predicted product is indicated.
Expression of spr0276-spr0282 locus.
The T133P point mutation in HK08, which possibly results in activation of the kinase and hence of the TCS08 system, leads to strong repression of the spr0276-spr0282 locus in growth medium supplemented with glucose and sucrose, and the hk08 mutant does not grow on cellobiose. This strongly suggests that the PTS is induced by glucose and/or sucrose and that induction is required with cellobiose as the only carbon source. In order to test this hypothesis, we compared the relative transcription levels of this locus, in medium supplemented with either glucose, sucrose, or cellobiose as the sole carbon source, in the R6 Smr parent strain (where the activation state of the TCS08 system could influence the transcription level) and the Δrr08 mutant derivative (where TCS08 could not manifest itself). Primers S107 and S108, specific for spr0282, were used for examining induction in conjunction with control primers specific for ldh (spr1100). The results obtained by real-time PCR show that spr0282 is expressed equally well under all of these conditions, and no difference was apparent (not shown). If this locus is controlled only by TCS08, then this also indicates that the TCS08 system is not activated under any of these conditions.
TCS08 and the cellobiose PTS in other gram-positive organisms.
The HK/RR08 regulatory system is present in all S. pneumoniae strains, according to genomic comparison on oligonucleotide microarrays, although some minor variation was noted (9). In contrast, the putative cellobiose PTS gave very low hybridization signals with DNAs from strains of major penicillin-resistant clones, such as the Spanish 23F clone Spain23F-1 and an Hungarian 19A clone, documenting that this locus is not part of the core genome of S. pneumoniae. Blast searches with TCS08 against microbial databases showed that this system is conserved in the closely related species Streptococcus mitis (with over 90% identical amino acids), Streptococcus agalactiae (55% identity), and Streptococcus gordonii (>30%). In Staphylococcus aureus, the highest degree of similarity was found with the SaeRS locus (33% identity and 55% similarity).
On the other hand, no homologues of the PTS with amino acid identity of above 90% were detected in the two S. mitis genomes available, those for S. mitis NCTC 12261 (www.tigr.org) and S. mitis B6 (our unpublished results). Closely related proteins, however, were represented by the deduced products of the bfp locus in the S. gordonii strain Challis NCTC 7688 (www.tigr.org) (Lin Tao, personal communication), which is involved in biofilm formation (data not shown) (15).
DISCUSSION
The characterization of two-component regulatory systems is often hampered by the lack of appropriate mutants. Loss-of-function mutants are often without a phenotype (10), and we have therefore investigated whether a mutation in the kinase CiaH, shown previously to activate the CiaRH TCS (21), also results in detectable phenotypes and changes in the transcription profile of the mutant when introduced into the homologous site in the histidine kinase HK08. The active-site region where this mutation, T230P (close to the active site H226 in CiaH), is located is well conserved in HK08 (T133, close to H129). Indeed, whereas a mutant with a deletion in the response regulator did not show significant changes in the transcription pattern, the hk08 mutant did, with the genes spr0276 to spr0282 being downregulated between 3.5- and over 47-fold compared to the wild-type situation. This region encodes a putative cellobiose PTS, and its function was confirmed by demonstrating that the hk08 mutant as well as mutants with mutations generated in genes within this region (encoding the regulatory protein Spr0279, the transmembrane-spanning protein Spr0281 of unknown function, and the PTS membrane protein Spr0282) could barely ferment the β-glucoside cellobiose. The fact that expression of this PTS was restored in the Δrr08 hk08 double mutant strongly suggests that in the HK08 mutant the TCS08 system is activated (i.e., the response regulator is phosphorylated) and therefore links the activated (i.e., phosphorylated) state of the response regulator RR08 to the repression of the cellobiose PTS.
The expression of the cellobiose PTS operon in S. pneumoniae appears to involve a variety of regulatory proteins, implying a new complexity for the regulation of PTSs in low-GC gram-positive bacteria (for a review, see reference 7). The operon includes the gene spr0279, encoding a putative transcriptional antiterminator homologous to the E. coli BglG antiterminator. In E. coli, these proteins are phosphorylated by Hpr and the respective enzyme IIBgl (7, 37), resulting in their inactivation in the presence of β-glucosides (1). As expected, a mutant with a deletion in spr0279, which should not be able to eliminate the impact of the potential stem-loop or terminator signal located between genes spr0276 and spr0277, cannot grow on cellobiose as the sole carbon source. However, unlike the situation reported for E. coli, where BlgG is inactive in the absence of β-glucosides (7), the cellobiose PTS in S. pneumoniae was expressed equally well in the presence of cellobiose as with glucose or sucrose as the sole carbon source. Also, there is no homologue of the IIBgl component in S. pneumoniae. Perhaps the products of spr0281 and/or spr0282 which encode proteins containing 4 and 12 transmembrane domains, respectively, both of which are required for growth on cellobiose, could be involved in Hpr-dependent phosphorylation of the antiterminator Spr0279.
There are other studies that have suggested that some two-component regulatory systems are involved in the regulation of carbohydrate metabolism. The putative sugar metabolism locus manLMN system in S. pneumoniae R6 is upregulated in a ΔciaR mutant, indicating that the response regulator represses transcription when phosphorylated (22). Furthermore, the gom regulon in S. gordonii for the metabolism of α-1,6-d-mannobiose appears to be regulated by the putative GomH-GomI two-component regulatory system (15). A previous study has shown that catabolite control protein A (CcpA) functions in carbon catabolite repression of lactose-inducible β-galactosidase activity in S. pneumoniae. However, that study showed that CcpA was not involved in the regulation of the cellobiose-inducible β-glucosidase (14), which suggested that the β-glucosidase is regulated by another, as-yet-uncharacterized mechanism. An interesting mechanism in the gram-negative bacterium Acidovorax was discussed recently, where the authors suggested the possibility of phosphotransfer between HprK/P and the response regulator BphQ to relate an apparent activated state of BphQ to the regulation of the catabolite-controlled operon bph (37).
It is curious that the PTS, the genes for which are the only genes apparently controlled by TCS08, is not part of the common genome in S. pneumoniae, whereas TCS08 is even conserved in closely related streptococci such as S. mitis (4, 9). It is possible that the regulatory system controls other (PTS?) loci in the strains that do not contain this cellobiose PTS, which would not be detectable on microarrays based on the R6 or the TIGR4 genome.
Since the cellobiose PTS was the only region highlighted by our transcriptome analysis, other genes controlled by this system are not known. We did not find any obvious binding sites such as direct repeats for the regulator RR08 in the putative regulatory regions of the cellobiose PTS operon, and therefore a genome-wide search for such motifs was not possible. The TCS08 operon has been shown to be among the 126 genes responding to acidification of the growth medium (20), but the significance of this is unclear.
The region encompassing spr0276 through spr0282 is absent from major multiple-antibiotic-resistant clones, such as Spain23F-1 and a serotype 19A clone from Hungary. It has been suggested that the regulatory protein Spr0279 is it is essential for lung infection in certain S. pneumoniae strains (11, 19). Curiously, the related bdp locus described for S. gordonii strain Challis NCTC 7688 is involved in biofilm production, a property that relates to the capacity for colonization and hence for virulence. All these data suggest that the cellobiose PTS could contribute to different virulence potentials of different pneumococcal strains and affect the outcome of an infection at some sites. This may even be the reason why conflicting results have been obtained for the role of RR08/HK08 in pneumococcal virulence (11, 18, 19, 39). It is possible that cellobiose is not the only sugar that is transported by the cellobiose system. We have investigated whether the utilization of degradation products of hyaluronic acid that are readily produced by the pneumococcal hyaluronidases (2) depends on a functional cellobiose PTS. The results indicate that while S. pneumoniae strains are capable of either utilizing hylauronic acid or its degradation disaccharide as a carbon source, this is not related to TCS08 or to the protein products of spr0279, spr0281, and spr0282.
In conclusion, we have shown that the T230P mutation in the H box of CiaH, which results in the continuous activation of the histidine kinase, can be transferred to other related histidine kinases to elucidate the phosphorylation of the response regulator. Microarray analyses of the constructed hk08 mutant indicated that the cellobiose metabolism is downregulated by this two-component regulatory system. Furthermore, the previously uncharacterized conserved hypothetical transmembrane protein Spr0281 has been shown to be essential for cellobiose metabolism. Finally, the presence of the cellobiose PTS may play a part in the modulation of the pneumococcal virulence potentials of different strains.
Acknowledgments
We thank Martin K. R. Burnham for providing TCS484 mutants, Lin Tao for providing the sequence data for the bfp locus in S. gordonii prior to publication, Sonja Schröck for technical assistance in DNA sequencing, and the Nano+Bio center at the University of Kaiserslautern for bioinformatic support.
Sequencing of the S. gordonii and S. mitis NCTC 12261 genomes was accomplished with support from the National Institutes of Health, National Institute of Dental and Craniofacial Research, Bethesda, MD (www.tigr.org). This work was supported by the BMBF (grant PTJ-BIO/0313134), the DFG (grant Ha 1011 9-1/2), the EU (grant 83 15-38 51 04), the Stiftung Rheinland Pfalz für Innovation (grant 15202-38 62 61/580), and the Helmholtz Gesellschaft (grant 1800002_1).
Footnotes
Published ahead of print on 6 October 2006.
REFERENCES
- 1.Amster-Choder, O., F. Houman, and A. Wright. 1989. Protein phosphorylation regulates transcription of the beta-glucoside utilization operon in E. coli. Cell 58:847-855. [DOI] [PubMed] [Google Scholar]
- 2.Boulnois, G. J. 1992. Pneumococcal proteins and the pathogenesis of disease caused by Streptococcus pneumoniae. J. Gen. Microbiol. 138:249-259. [DOI] [PubMed] [Google Scholar]
- 3.Broome, C. 1996. Meningococcal and pneumococcal disease vaccines, p. 28-32. In Progress of vaccine research and development—1996. WHO document VRD/GEN/96.02. World Health Organization, Geneva, Switzerland.
- 4.Brückner, R., M. Nuhn, P. Reichmann, B. Weber, and R. Hakenbeck. 2004. Mosaic genes and mosaic chromosomes—genomic variation in Streptococcus pneumoniae. Int. J. Med. Microbiol. 294:157-168. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, Q., E. A. Campbell, A. M. Naughton, S. Johnson, and H. R. Masure. 1997. The com locus controls genetic transformation in Streptococcus pneumoniae. Mol. Microbiol. 23:683-692. [DOI] [PubMed] [Google Scholar]
- 6.de Saizieu, A., C. Gardes, N. Flint, M. T. J., K. E. Amrein, and R. Lange. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 182:4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Görke, B., and B. Rak. 1999. Catabolite control of Escherichia coli regulatory protein BglG activity by antagonistically acting phosphorylations. EMBO J. 18:3370-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guenzi, E., A. M. Gasc, M. A. Sicard, and R. Hakenbeck. 1994. A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol. Microbiol. 12:505-515. [DOI] [PubMed] [Google Scholar]
- 9.Hakenbeck, R., N. Balmelle, B. Weber, C. Gardes, W. Keck, and A. de Saizieu. 2001. Mosaic genes and mosaic chromosomes: intra- and interspecies variation of Streptococcus pneumoniae. Infect. Immun. 69:2477-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hakenbeck, R., and J. B. Stock. 1996. Approaches to the analysis of novel two component signal transduction systems involved in transcriptional regulation. Methods Enzymol. 273:281-300. [DOI] [PubMed] [Google Scholar]
- 11.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 12.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D.-J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. J. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. Q. Mundy, T. I. Nicas, F. H. Norris, J. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P.-M. Sun, J. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. J. Rosteck, P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houman, F., M. R. Diaz-Torres, and A. Wright. 1990. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell 62:1153-1163. [DOI] [PubMed] [Google Scholar]
- 14.Iyer, R., N. S. Baliga, and A. Camilli. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 187:8340-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kilic, A. O., L. Tao, Y. Zhang, Y. Lei, A. Khammanivong, and M. C. Herzberg. 2004. Involvement of Streptococcus gordonii beta-glucoside metabolism systems in adhesion, biofilm formation, and in vivo gene expression. J. Bacteriol. 186:4246-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacks, S., and R. D. Hotchkiss. 1960. A study of the genetic material determining an enzyme activity in pneumococcus. Biochim. Biophys. Acta 39:508-517. [DOI] [PubMed] [Google Scholar]
- 17.Lacks, S. A., and B. Greenberg. 2001. Constitutive competence for genetic transformation in Streptococcus pneumoniae caused by mutation of a transmembrane histidine kinase. Mol. Microbiol. 42:1035-1045. [DOI] [PubMed] [Google Scholar]
- 18.Lange, R., C. Wagner, A. de Saizieu, N. Flint, J. Molnos, M. Stieger, P. Caspers, M. Kamber, W. Keck, and K. E. Amrein. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237:223-234. [DOI] [PubMed] [Google Scholar]
- 19.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 20.Martin-Galìano, A. J., K. Overweg, M. J. Ferrándiz, M. Reuter, J. M. Wells, and A. G. de la Campa. 2005. Transcriptional analysis of the acid tolerance response in Streptococcus pneumoniae. Microbiology 151:3935-3946. [DOI] [PubMed] [Google Scholar]
- 21.Mascher, T., M. Heintz, D. Zähner, M. Merai, and R. Hakenbeck. 2006. The CiaRH system of Streptococcus pneumoniae prevents lysis during stress induced by treatment with cell wall inhibitors and mutations in pbp2x involved in beta-lactam resistance. J. Bacteriol. 188:1959-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mascher, T., M. Merai, N. Balmelle, A. de Saizieu, and R. Hakenbeck. 2003. The Streptococcus pneumoniae cia regulon: CiaR target sites and transcription profile analysis. J. Bacteriol. 185:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCluskey, J., J. Hinds, S. Husain, A. Witney, and T. J. Mitchell. 2004. A two-component system that controls the expression of pneumococcal surface antigen A (PsaA) and regulates virulence and resistance to oxidative stress in Streptococcus pneumoniae. Mol. Microbiol. 51:1661-1675. [DOI] [PubMed] [Google Scholar]
- 24.Musher, D. M. 1992. Infections caused by Streptococcus pneumoniae: clinical spectrum, pathogenesis, immunity, and treatment. Clin. Infect. Dis. 14:801-809. [DOI] [PubMed] [Google Scholar]
- 25.Ng, W. L., H. C. Tsui, and M. E. Winkler. 2005. Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae. J. Bacteriol. 187:7444-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ottolenghi, E., and R. D. Hotchkiss. 1962. Release of genetic transforming agent from pneumococcal cultures during growth and disintegration. J. Exp. Med. 116:491-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkinson, J. S. 1993. Signal transduction schemes of bacteria. Cell 73:857-871. [DOI] [PubMed] [Google Scholar]
- 28.Pestova, E. V., L. S. Håvarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853-862. [DOI] [PubMed] [Google Scholar]
- 29.Polissi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pritchard, D. G., B. Lin, T. R. Willingham, and J. R. Baker. 1994. Characterization of the group B streptococcal hyaluronate lyase. Arch. Biochem. Biophys. 315:431-437. [DOI] [PubMed] [Google Scholar]
- 32.Reichmann, P., and R. Hakenbeck. 2000. Allelic variation in a peptide-inducible two component system of Streptococcus pneumoniae. FEMS Microbiol. Lett. 190:231-236. [DOI] [PubMed] [Google Scholar]
- 33.Russo, F. D., and T. J. Silhavy. 1991. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J. Mol. Biol. 222:567-580. [DOI] [PubMed] [Google Scholar]
- 34.Schnetz, K., C. Toloczyki, and B. Rak. 1987. Beta-glucoside (bgl) operon of Escherichia coli K-12: nucleotide sequence, genetic organization, and possible evolutionary relationship to regulatory components of two Bacillus subtilis genes. J. Bacteriol. 169:2579-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Standish, A. J., U. H. Strocher, and J. C. Paton. 2005. The two-component signal transduction system RR06/HK06 regulates expression of cbpA in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 102:7701-7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stock, J. B., M. G. Surette, M. Levit, and P. Park. 1995. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis, p. 25-51. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, DC.
- 37.Stülke, J., G. Arnaud, G. Rapoport, and I. Martin-Verstraete. 1998. PRD—a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol. Microbiol. 28:865-874. [DOI] [PubMed] [Google Scholar]
- 38.Sung, C. K., H. Li, J. P. Claverys, and D. A. Morrison. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67:5190-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. G. Wallis, J. R. Brown, D. J. Holmes, M. Rosenberg, and K. R. Burnham. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35:566-576. [DOI] [PubMed] [Google Scholar]
- 40.Tokishita, S., A. Kojima, H. Aiba, and T. Mizuno. 1991. Transmembrane signal transduction and osmoregulation in Escherichia coli. Functional importance of the periplasmic domain of the membrane-located protein kinase, EnvZ. J. Biol. Chem. 266:6780-6785. [PubMed] [Google Scholar]
- 40a.Tomasz, A. 1964. Bacteriol. Proc., p. 28.
- 41.Ulijasz, A. T., D. R. Andes, D. R. Glasner, and B. Weisblum. 2004. Regulation of iron transport in Streptococcus pneumoniae by RitR, an orphan response regulator. J. Bacteriol. 186:8123-8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zähner, D., T. Grebe, E. Guenzi, J. Krauβ, M. van der Linden, K. Terhune, J. B. Stock, and R. Hakenbeck. 1996. Resistance determinants for β-lactam antibiotics in laboratory mutants of Streptococcus pneumoniae that are involved in genetic competence. Microb. Drug Resist. 2:187-191. [DOI] [PubMed] [Google Scholar]
- 43.Zähner, D., K. Kaminski, M. van der Linden, T. Mascher, M. Merai, and R. Hakenbeck. 2002. The ciaR/ciaH regulatory network of Streptococcus pneumoniae. J. Mol. Microbiol. Biotechnol. 4:211-216. [PubMed] [Google Scholar]