The article by Brown et al. (6) in this issue of Journal of Bacteriology makes it possible to integrate a series of investigations that have given us a high-resolution picture of the rotational flagellar motors of Escherichia coli and Salmonella enterica. Although details will surely differ, it should also serve as a model for the flagellar architectures found within a wide spectrum of prokaryotes.
The beauty of the bacterial flagellum is captured in reconstructions of the basal body (21) and the external rod, hook, and filament, which comprise the propeller (18, 24). The output of this device is also impressive: E. coli flagella spin at hundreds of rotations per second, either clockwise (CW) or counterclockwise (CCW), and rotation is driven by a transmembrane proton current that brings up to a million H+ ions per motor into the cell per second. (For reviews of bacterial flagella and motility, see references 2, 11, and 13.) The reversibility of the motor generates alternating straight runs (CCW rotation) and reorienting tumbles (CW rotation) to create a three-dimensional random walk. During chemotaxis, the random walk is biased so that runs become longer when a cell happens to run up a gradient of an attractant chemical or down a gradient of a repellent chemical. (For reviews of bacterial chemotaxis, see references 1, 15, and 23.) The CheY protein, activated by phosphorylation at the chemoreceptor patch, binds to the motor to promote CW rotation and therefore tumbling.
Much of the bacterial flagellum can be extracted from the cell envelope as an intact basal body/hook/filament complex (8). This portion of the flagellum consists of three rings that encompass a central rod attached to a flexible hook that in turn is attached to a long, left-handed helical filament. The rings are, from the cell-proximal to cell-distal end of the basal body, MS, P, and L. Each is a polymer of one polypeptide. The rings associate with the cell membrane, the peptidoglycan wall, and the lipopolysaccharide-containing outer membrane, respectively. The P and L rings constitute a bushing that allows the rotating rod to penetrate the cell envelope. The diameter of the M portion of the MS ring is about 25 nm.
This description ignores two key components of a functional flagellum: the cytoplasmic C ring, which must firmly attach to the MS ring, and the membrane-spanning Mot protein complexes, which supply the transmembrane proton channel and anchor Mot complexes to the peptidoglycan cell wall to serve as stator elements for the motor. The symmetry of the MS ring is between 24- and 26-fold, and the symmetry of the C ring is 32- to 36-fold (21), at least for a CW-locked motor. These are not average values; they represent numbers for individual MS and C rings. What is more, there is no fixed relationship between the symmetries in the MS ring and the C ring of a given motor. This chaos is compounded by the fact that an individual rotating motor can contain from as few as 1 to as many as 12 (3, 17) MotA4MotB2 complexes (4, 19).
A clever approach using a chimeric Vibrio alginolyticus/E. coli Mot complex that employs Na+ rather than H+ ions has demonstrated that the 26-fold symmetry of the MS ring is reflected in 26 steps per single 360o rotation of the motor (20). The data leading to this conclusion were collected under conditions in which the sodium motive force was maintained at a low value and the number of Mot complexes per motor was manipulated to approach one. As a result, the motor turns very slowly, and individual steps can be counted. The 26 steps may reflect that the motors observed contained only one functional Mot complex.
The MS ring is made up of the FliF protein, which associates at its cytoplasmic face with the FliG protein. FliG connects the MS ring to the C ring, and it also interacts with the cytoplasmic loops of MotA to generate rotation in response to transmembrane proton flow. The partial crystal structure of FliG from Thermotoga maritima (5, 12) strongly suggests that two distinct and rather distant domains are responsible for interactions with FliF and MotA, respectively. A long helix and a flexible linker connect these two domains. The bulk of the C ring is made up of the FliM and FliN proteins, with 32 to 36 FliM monomers, most of which probably bind a FliN tetramer (16). (FliN exists in ≥100 copies per basal body.)
The structure of the large middle domain of FliM was recently solved (14). It is rather compact, with dimensions of 5 by 3.5 by 3 Å. At one end of the long axis there is a poorly resolved flexible GGXG-containing loop that joins the two pseudosymmetric domains of the folded polypeptide. At the other end is the C-terminal region that binds the FliN tetramer. The N-terminal sequence that binds to phospho-CheY is also not resolved. The C ring is ∼44 nm in diameter and can accommodate 32 to 36 FliM subunits if the long axis of FliM is perpendicular to the ring and the intermediate axis is parallel to the circumference of the ring. Cross-linking studies using introduced cysteine residues are consistent with that organization (14).
The work of Brown et al. (6) ties all this information together. Tryptophan-scanning mutagenesis of FliG implicates two regions, which flank the connecting helix, as being important for flagellar assembly, motility, and directional control. The region in the domain closer to the MS ring contains an EHPQ…R sequence that is conserved in FliG proteins from a wide range of bacteria. The second region includes a hydrophobic patch that is on the opposite side of the motility domain from the ridge of charged residues that interact with the cytoplasmic loop of MotA to drive flagellar rotation (25). So far, so good. But what about the numerical mismatch between the MS and C rings, and how can one region on a FliM subunit simultaneously interact with two distinct and rather distant sites on FliG?
The solution offered by Brown et al. (6) is ingenious and compelling. If, for example, the MS ring of a particular basal body has 26-fold symmetry, implying that 26 FliF subunits attach to 26 FliG subunits, then 26 FliM subunits can contact the hydrophobic patches on the motility domains of FliG. The remaining 6 to 10 FliM subunits could face inward to interact with the EHPQ…R motif, which is close to the N-terminal region of FliG that attaches to FliF. Indeed, Thomas et al. (21) found that the most proximal part of the C ring has the same symmetry as the MS ring. Thomas et al. (21) also found that, at the level of FliM, there was a lower electron density inside the high-density outer wall of the C ring, a feature consistent with a 26 + 8 arrangement of FliM subunits.
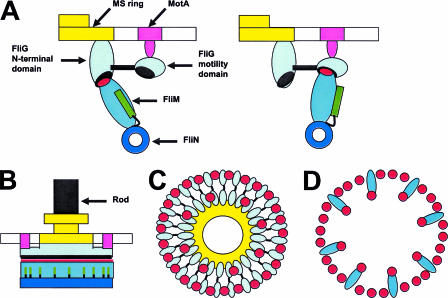
Figure 1 presents several views of a model for how the C ring may be configured in a CW-rotating motor, based on the most recent data. Phospho-CheY is not shown. It may bind only to the outward-facing FliM subunits that contact the FliG motility domain. CheY binding presumably generates a change in the conformation of FliM (and FliN) that is transmitted to FliG to reposition the motility domain with respect to the Mot protein complexes. The inward-facing FliM subunits might serve to stabilize the association of the MS and C rings.
FIG. 1.
Schematic model for C-ring architecture in a CW-locked flagellar motor. (A) Two alternative associations of a FliM monomer with FliG. The middle domain of FliM is in cyan, the flexible GGXG-containing loop that contacts FliG is in red, the N-terminal sequence that binds phospho-CheY is in green, and the flexible connector between the CheY-binding sequence and the middle domain is indicated by the black line. FliG is shown as two blue-gray ovals (the inner N-terminal and outer C-terminal domains) joined by the helical connector (black line), with the EHPQ…R region in the N-terminal domain and the hydrophobic patch in the C-terminal domain both shown as black ovals. The MS ring is in yellow, the Mot protein complex is in magenta, the FliN tetramer is a blue ring, and the membrane is indicated by the elongated white rectangle. (B) Side view of a complete flagellar basal body. The color scheme is as in panel A but with the flagellar rod shown in black. (C) The FliG layer of the C ring looking toward the membrane from the cytoplasm. The motor is shown with 26 FliG subunits and 34 FliM subunits. The color scheme is as in panel A, with the hollow center of the MS ring shown in white. (D) The FliM layer of the C ring. Eight FliM subunits tilt inward to contact the EHPQ…R motif in the N-terminal domain of FliG. The remaining 26 FliM subunits orient with their long axes perpendicular to the C ring and contact the C-terminal motility domains of the FliG subunits. The orientation of the C ring in panels C and D is the same, so that the two images can be superimposed to reconstruct a C ring lacking only FliN. Note that the outward-facing FliM subunits flanking the inward-facing FliM subunits would have to splay out in order to contact the FliG motility domains at equal spacing, as shown in panel C. Alternatively, there could be gaps in the C ring at the level of the FliG motility domains, as suggested by Brown et al. (6).
This model raises a number of questions. How can FliM within the C ring accommodate two orientations of FliM monomers, which must lead to nonequivalent subunit contacts? Does phospho-CheY bind only to the outward-facing or inward-facing FliM subunit or to both, and does CheY binding change the distribution of FliM between the two conformations? How does phospho-CheY binding modify the way that FliM interacts with the FliG motility domain? What coordinates the movements of FliM subunits within the ring (7, 9) to give essentially instantaneous switching from CCW to CW rotation and back? How do interactions between MotA and FliG change to produce the two different directions of rotation, and how are Mot complexes recruited to, and distributed around, the MS and C rings (10, 22)? The answers to these questions must be found before we can say that we have attained an in-depth understanding of the relationship between flagellar structure and flagellar function.
Acknowledgments
I thank David Blair and David DeRosier for extensive and very helpful discussion during the preparation of this commentary.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
Footnotes
Published ahead of print on 3 November 2006.
REFERENCES
- 1.Baker, M. D., P. M. Wolanin, and J. B. Stock. 2006. Signal transduction in bacterial chemotaxis. Bioessays 28:9-22. [DOI] [PubMed] [Google Scholar]
- 2.Berry, R. M., and J. P. Armitage. 1999. The bacterial flagella motor. Adv. Microb. Physiol. 41:291-337. [DOI] [PubMed] [Google Scholar]
- 3.Blair, D. F., and H. C. Berg. 1988. Restoration of torque in defective flagellar motors. Science 242:1678-1681. [DOI] [PubMed] [Google Scholar]
- 4.Braun, T. F., L. Q. Al-Mawsawi, S. Kojima, and D. F. Blair. 2004. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry 43:35-45. [DOI] [PubMed] [Google Scholar]
- 5.Brown, P. N., C. P. Hill, and D. F. Blair. 2002. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 21:3225-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, P. N., M. Terrazas, K. Paul, and D. F. Blair. 2007. Mutational analysis of the flagellar protein FliG: sites of interaction with FliM and implications for organization of the switch complex. J. Bacteriol. 189:305-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cluzel, P., M. Surette, and S. Leibler. 2000. An ultrasensitive motor revealed by monitoring signaling proteins in single cells. Science 287:1652-1655. [DOI] [PubMed] [Google Scholar]
- 8.DePamphilis, M. L., and J. Adler. 1971. Fine structure and isolation of the hook-basal body complex of flagella from Escherichia coli and Bacillus subtillis. J. Bacteriol. 105:384-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duke, T. A., N. L. Novere, and D. Bray. 2001. Conformational spread in a ring of proteins: a stochastic approach to allostery. J. Mol. Biol. 308:541-553. [DOI] [PubMed] [Google Scholar]
- 10.Hosking, E. R., C. Vogt, E. P. Bakker, and M. D. Manson. 2006. The Escherichia coli MotAB proton channel unplugged. J. Mol. Biol. 364:921-927. [DOI] [PubMed]
- 11.Kojima, K., and D. F. Blair. 2004. The bacterial flagellar motor: structure and function of a complex molecular machine. Int. Rev. Cytol. 233:93-134. [DOI] [PubMed] [Google Scholar]
- 12.Lloyd, S. A., F. B. Whitby, D. F. Blair, and C. P. Hill. 1999. Structure of the C-terminal domain of FliG, a component in the rotor of the bacterial flagellar motor. Nature 400:472.475. [DOI] [PubMed] [Google Scholar]
- 13.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 14.Park, S.-Y., B. Lowder, A. M. Bilwes, D. F. Blair, and B. R. Crane. 2006. Structure of FliM provides insight into the assembly of the switch complex in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 103:11886-11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkinson, J. S., P. Ames, and C. A. Studdart. 2005. Collaborative signaling by bacterial chemoreceptors. Curr. Opin. Microbiol. 8:116-121. [DOI] [PubMed] [Google Scholar]
- 16.Paul, K., and D. F. Blair. 2006. Organization of FliN subunits in the flagellar motor of Escherichia coli. J. Bacteriol. 188:2502-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reid, S. W., M. C. Leake, J. H. Chandler, C.-J. Lo, J. P. Armitage, and R. M. Berry. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl. Acad. Sci. USA 103:8066-8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samatev, F. A., H. Matsunami, K. Imada, T. R. Shaikh, D. R. Thomas, J. Z. Chen, D. J. DeRosier, A. Kitao, and K. Namba. 2004. Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism. Nature 431:1062-1068. [DOI] [PubMed] [Google Scholar]
- 19.Sato, K., and M. Homma. 2000. Multimeric structure of PomA, a component of the Na+-driven polar flagellar motor of Vibrio alginolyticus. J. Biol. Chem. 275:20223-20228. [DOI] [PubMed] [Google Scholar]
- 20.Sowa, Y., A. D. Rowe, M. C. Leake, T. Yakushi, M. Homma, A. Ishijima, and R. M. Berry. 2006. Direct observation of steps in the bacterial flagellar motor. Nature 437:916-919. [DOI] [PubMed] [Google Scholar]
- 21.Thomas, D. R., N. R. Francis, C. Xu, and D. J. DeRosier. 2006. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:7033-7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Way, S. M., E. R. Hosking, T. F. Braun, and M. D. Manson. 2000. Mot protein assembly into the bacterial flagellum: a model based on mutational analysis of the motB gene. J. Mol. Biol. 297:7-24. [DOI] [PubMed] [Google Scholar]
- 23.Wadhams, G. H., and J. P. Armitage. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell. Biol. 5:1024-1037. [DOI] [PubMed] [Google Scholar]
- 24.Yonekura, K., S. Maki-Yonekura, and K. Namba. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424:643-650. [DOI] [PubMed] [Google Scholar]
- 25.Zhou, J., S. A. Lloyd, and D. F. Blair. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 95:6436-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]