Abstract
Simple sequence repeats located within reading frames mediate phase-variable ON/OFF switches in gene expression by generating frameshifts. Multiple translation initiation codons in different reading frames are found upstream of most Haemophilus influenzae tetranucleotide repeat tracts, raising the possibility of multiple active reading frames and more than two levels of gene expression for these loci. Phase variation between three levels of gene expression (strong, weak, and none) was observed when lic2A was fused to a lacZ reporter gene. The lic2A 5′ CAAT repeat tract is preceded by four 5′ ATG codons (x, y, z1, and z2) in two reading frames. Each of these initiation codons was inactivated by site-directed mutagenesis. Strong expression from frame 1 was associated with x but not y. Weak expression from frame 2 was mainly dependent on the z2 codon, and there was no expression from frame 3. Using monoclonal antibodies specific for a digalactoside epitope of lipopolysaccharide whose synthesis requires Lic2A, two levels (strong and undetectable) of antibody reactivity were detected, suggesting that weak expression of lic2A is not discernible at the phenotypic level. Inactivation of the x initiation codon resulted in loss of strong expression of the digalactoside epitope and elevated killing by human serum. The failure to detect more than two phenotypes for lic2A, despite clear evidence of weak expression from the z1/z2 initiation codons, leaves open the question of whether or not multiple initiation codons are associated with more complex patterns of phenotypic variation rather than classical phase-variable switching between two phenotypes.
The capacity for generating extensive structural diversity of surface molecules is a feature of many microbial pathogens and facilitates their potential to colonize new niches, to adapt to variations in their microenvironments, or to survive attack from other organisms. An important process utilized by some species of bacteria for the generation of diversity is “phase variation” (PV), that is, stochastic, high-frequency switches in expression of structures present on the surface of an organism (1). PV increases the fitness of a population by generating diversity in the absence of selective pressure (3, 27). This allows a population to “preempt” a catastrophic collapse in numbers, such as might occur after an immune response to a surface antigen or during the establishment of a founding population after transfer between genetically distinct hosts. A major molecular mechanism of PV involves mutations in simple sequence repeat tracts (microsatellites). This mechanism occurs in multiple genes of a number of important bacterial pathogens including Haemophilus influenzae, Neisseria meningitidis, Helicobacter pylori, and Campylobacter jejuni.
H. influenzae is a gram-negative commensal bacterium of the human upper respiratory tract which is capable of causing severe diseases such as otitis media, septicemia, and meningitis. Several molecules on the surface of H. influenzae are subject to phase-variable expression including numerous epitopes of lipopolysaccharide (LPS), the major surface glycolipid of this bacterium (21). LPS in H. influenzae consists of lipid A linked via 2-keto-3-deoxyoctulosonic acid to a triheptosyl backbone (21). Oligosaccharide extensions, consisting of mainly hexose sugars, protrude from this backbone and are subject to substitution with additional groups, such as sialic acid (20) or phosphorylcholine (35). Many of these extensions and substitutions are subject to PV. One example is the α-Gal-(1-4)-β-Gal epitope, whose synthesis requires two phase-variable glycosyl transferases, Lic2A (16) and LgtC (19), and can be influenced by a third, Lex2A (12).
The repeat tracts of many phase-variable loci are located within the reading frames of genes (2). Changes in repeat number affect gene expression by placing the reading frame in or out of frame with the translation initiation codon, usually resulting in two levels of gene expression, ON and OFF (e.g., siaD in N. meningitidis [14]). The potential for more than two levels of differential expression is raised when there are multiple initiation codons located upstream of the repeat tract, and changes in the N-terminal region do not alter protein activity. In H. influenzae, tetranucleotide repeat tracts (microsatellites) located within the N-terminal region of the reading frame are the major mechanism of PV (19, 33). Previous studies using monoclonal antibodies (MAbs) specific for surface-exposed epitopes of LPS have suggested that phase-variable genes in H. influenzae are translated from initiation codons located in different frames (16, 33). The lic2A gene has a 5′ CAAT repeat tract that is preceded by putative initiation codons in two reading frames, x/y in frame 1 and z1/z2 in frame 2. Expression of the digalactoside has been assigned to both these reading frames (23, 34) even though convincing evidence for the association between genotype and phenotype is only available for the x/y reading frame (17). Also, three levels of reactivity or expression were detected and correlated with different repeat numbers and reading frames for the lic1 locus using MAb 6A2 (33) and by fusion of lacZ in frame with lic1D (26). Similarly, differential levels of expression were observed and correlated with use of initiation codons in different reading frames when lacZ was inserted downstream of the repeat tract in the lic3A locus (30). Finally, three levels of gene expression were reported for the mod gene, with weak expression being 40-fold lower than maximal expression (29). Most of these studies were preliminary in nature and did not eliminate the possibility that the lower level of expression in a population of bacteria was due to small numbers of highly expressing phase variants. Additionally, these studies did not clarify the mechanism of differential expression and the correlation between gene expression and phenotype.
It is shown herein that lic2A is associated with three levels of gene expression due to repeat-mediated switching between the three reading frames, of which two contain multiple potential initiation codons. Site-directed mutagenesis demonstrated that alternate functional initiation codons give rise to three levels of differential gene expression of lic2A but to only two detectable levels of phenotypic expression of the Lic2A-dependent digalactoside epitope in the LPS.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
H. influenzae strain RM118, herein termed Rd, is an acapsular serotype d-derived strain and is equivalent to strain KW20, for which a genome sequence has been derived (10). Strain RM7004 is a serotype b capsular strain (31). H. influenzae strains were grown in brain heart infusion (BHI) broth supplemented with either hemin (10 μg ml−1) and NAD (2 μg ml−1) for liquid medium or with Levinthal supplement (10%) for solid medium. When required, kanamycin was added to a concentration of 10 μg ml−1, and X-Gal (5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside) was added at a concentration of 40 μg ml−1.
Escherichia coli strain DH5α [λ− Φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK− mK−) supE44 thi-1 gyrA relA1] was used to propagate cloned plasmids and was grown at 37°C in Luria-Bertani broth supplemented, when required, with either ampicillin (100 μg ml−1) or kanamycin (50 μg ml−1).
Construction of a lic2A-lacZ fusion.
A lic2A-lacZ fusion was constructed on a plasmid in E. coli and then moved into the H. influenzae genome by transformation. Two separate regions encompassing the 5′ and 3′ ends of the lic2A gene and 1 kb of flanking sequence, to aid recombination into the H. influenzae genome, were amplified from genomic DNA of strain Rd by PCR. To amplify the upstream region, primer lic2Eco (5′-CTGAATTCAGCAGTGAGCGAGAGCATAG-3′) was used in conjunction with lic2Kpn (5′-AAGGTACCTCGCCAAGAATTACATCATCTTC-3′), and to amplify the downstream region, primer lic2Bam (5′-ACGGATCCAGAAATGCAAGGTAAAGAAAT T-3′) was used with lic2Sal (5′-CTGTCGACACTCAATTTGGCATAACCTAAC-3′). The resultant products were cloned using the TOPO TA cloning kit (Invitrogen). To form the plasmid plic2AlacZ, the lic2A regions were excised with the restriction enzymes whose sites were present in the PCR primers (EcoRI, KpnI, BamHI, and SalI) and cloned in a stepwise procedure into the plasmid pUC19 either side of a lacZ gene which was excised from pCH110 (Pharmacia) using the KpnI and BamHI restriction sites. A Tn903 kanamycin resistance cassette (Kanr) was excised from pUC4K (Pharmacia) using BamHI and cloned into the corresponding site downstream of the lacZ open reading frame. The resultant plasmid, plic2AlacZkan, was linearized using SphI prior to transformation into H. influenzae strain Rd as previously described (15).
A kanamycin-resistant H. influenzae transformant which produced blue colonies on X-Gal-containing medium was designated Rdlic2AlacZ (data available upon request) and selected for use in PV experiments. The integrity of this transformant (and all the other transformants described below) was confirmed by automated DNA sequencing (ABI) of PCR products and by Southern blot analysis. DNA sequencing was performed with primer lic2A-ATG2 (5′TGACCGCTCTTTTATAAATAAATTTTG-3′), which binds 116 nucleotides upstream of the x initiation codon, a region that is likely to include the promoter elements for lic2A in addition to the four 5′ ATG initiation codons and the DNA repeat tract. No mutations were noted in this region for this transformant (or for any of the other transformants described below). This transformant had 22 5′ CAAT repeats, as in the wild-type strain, putting the lacZ open reading frame in frame with the x and y 5′ ATG initiation codons.
Construction of initiation codon mutants.
Translation initiation codons (Fig. 1) were mutated using a QuickChange site-directed mutagenesis kit (Stratagene). Complementary forward and reverse primers were designed to alter the DNA sequence from 5′ ATG to 5′ CTT. The sequences of these forward primers are as follows: ATGx Forward, 5′-GTTATTTCCATTTTTATTTAAATCTTAGTGCTATTGAAAATATTGTC-3′; ATGy Forward, 5′-GAAAATATTGTCATTAGTCT TGAAAATGCAACTGAACG-3′; ATGz1 Forward, 5′-GTCATTAGTATGGAAACTTC AACTGAACGTCGC-3′; and ATGz2 Forward, 5′-CTTTCATTTAGTTTCTTTACTTCGTACACTTATCAATCAATC-3′). Mutagenesis was carried out on the plic2AlacZ plasmid due to size constraints for efficient PCR amplification. Plasmids carrying the appropriate mutation were selected and Kanr was inserted as described above to form the constructs plic2AΔxlacZkan, plic2AΔylacZkan, plic2AΔz1lacZkan and plic2AΔz2lacZkan with altered x, y, z1, and z2 codons, respectively. In addition to these plasmids, a spontaneous mutation in codon y was also obtained that allowed construction of an x and y double mutant, which was designated plic2AΔxylacZkan. These mutated genes were transformed into H. influenzae strain Rd as described above (strain data available upon request).
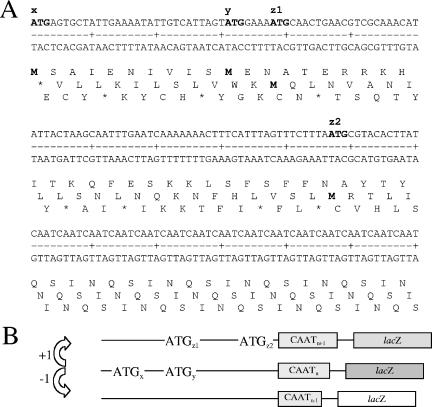
FIG. 1.
Putative initiation codons and potential for differential expression of the lic2A gene of H. influenzae strain Rd. Panel A shows the nucleotide sequence of the region of lic2A upstream of the repeat tract and part of the 5′ CAAT repeat tract. The four putative 5′ ATG initiation codons are highlighted in bold, and their designations are indicated above the sequence. Note that x and y are in one frame while z1 and z2 are in an alternative frame. The translated protein sequences for each frame are shown below the nucleotide sequence. The methionine codons in these sequences are also highlighted in bold. Panel B shows a schematic representation of the mechanism for differential expression from lic2A-lacZ reporter constructs. Loss or gain of a repeat unit places the reading frame of lacZ into alignment with different putative initiation codons, leading to changes in the level of expression. The nucleotide sequence is represented as a line with the relative positions of the four putative initiation codons being marked as ATGn. The 5′ CAAT repeat tract and lacZ gene are marked as rectangles with the intensity of shading of the lacZ rectangle representing differing levels of expression. In strains containing 22 repeats, the lacZ gene is in frame with the x/y reading frame.
Measurement of lic2A reporter construct phase variation rates.
PV rates (mutations/cell/generation) were calculated as previously described (4, 8). Briefly, a single colony was selected from an overnight BHI-X-Gal plate and serially diluted before plating. A further eight colonies were selected from this new plate and diluted again. Two 50-μl aliquots of the 10−3 dilution and one aliquot of the 10−4 dilution were plated onto BHI-X-Gal plates. The PV frequency was determined as the mean number of variant colonies from the two high-density plates divided by 10 times the total number of colonies on the low-density plate. To permit comparisons with previously published data, mutation rates were then calculated using the mutant accumulation method of Drake (9). The median mutation rate was calculated using the combined data from at least two independent experiments. Differences in the median mutation rates of strains were compared using a Mann-Whitney test for statistical significance. Statistical analyses were carried out using the Prism 3.0 biostatistics package (Graphpad, San Diego, CA).
Determination of variant colony frame switches.
To determine the frame into which variant colonies had switched, the region of the lic2A gene containing the repeat tract was amplified by PCR from the parental colony and two variants from each set of eight serial dilutions. The upstream primer lic2A-121F (5′ACTGAACGTCGCAAACAT-3′) was used in conjunction with three identical downstream primers (5′-TCCCAGTCACGACGTTGT-3′) lacZB1-F, lacZB1-T, and lacZB1-H, fluorescently labeled with the dyes 6-carboxyfluorescein, tetrachloro-6-carboxyfluorescein, and hexachloro-6-carboxyfluorescein, respectively. The resultant products were sized on a polyacrylamide gel using an ABI 377 DNA sequencer and the Genescan software package (Perkin Elmer). Frameshifts were inferred from the differences in sizes of PCR products derived from the variant and parental colonies. The DNA sequence of one set of parental and variant colonies from each experiment was determined by automated sequencing with a Big Dye kit (Perkin Elmer) to confirm the Genescan data.
β-Galactosidase assay of initiation codon mutants.
A total of 40 μl of overnight culture was used to inoculate 5 ml of fresh medium which was aerated at 37°C until it reached an optical density at 600 nm (OD600) of between 0.28 to 0.70. The cultures were cooled on an ice and water mixture for 20 min to stop growth before the culture was pelleted, and 1 ml (nonconcentrated) or 5 ml (concentrated) of culture was resuspended in 1 ml of phosphate-buffered saline (PBS) and the absorbance at OD600 was recorded. Aliquots (100 μl and 500 μl) of each sample were made up to 1 ml in a Bijou container using Z buffer (0.06 M Na2HPO4 · 7H2O, 0.04 M NaH2PO4 · H2O, 0.01 M KCl, 0.001 M MgSO4 · 7H2O, 0.05 M β-mercaptoethanol, pH 7.0 [28]). The cells were permeabilized by the addition of 20 μl of chloroform and 10 μl of 10% sodium dodecyl sulfate (SDS) before being vortexed briefly. The tubes were incubated in a water bath at 28°C for 5 min before the addition of 200 μl of o-nitrophenyl-β-d-galactopyranoside (4 mg ml−1) to begin the reaction. Reactions were stopped, using 0.5 ml 1 M Na2CO3, once sufficient yellow coloration was observed. Absorbances at OD420 and OD550 were recorded and used in conjunction with reaction time (minutes) and cell volume (milliliters) in the following equation to determine relative β-galactosidase activities: activity = 1,000 × [OD420 − 1.75(OD550)]/[time × volume × OD600]. A mean level of activity was calculated from three independent experiments.
Construction of initiation codon mutations in the lic2A gene.
The lic2A gene and flanking sequence were amplified by PCR with primers lic2Eco and lic2Sal and cloned using the TOPO TA (Invitrogen) system. The enzyme XbaI cleaves the lic2A gene at a site 3′ of the repeat tract. Used in conjunction with SalI, this digestion excises a fragment containing the section of lic2A gene replaced in the reporter constructs and 1 kb of downstream flanking sequence. The enzymes EcoRI and XbaI were used to digest the plasmids plic2AΔxlacZkan, plic2AΔylacZkan, plic2AΔz1lacZkan, and plic2AΔz2lacZkan, excising a fragment containing 1 kb of upstream sequence and the 5′ end of the lic2A gene, including the start codons and repeat tract. The upstream EcoRI/XbaI and downstream XbaI/SalI fragments were then ligated together into pUC19, previously digested with EcoRI and SalI, to form constructs containing the full-length lic2A gene and corresponding mutated initiation codons. The Kanr cassette was ligated into each of these constructs as described above to form the plasmids plic2AΔxkan, plic2AΔykan, plic2AΔz1kan, and plic2AΔz2kan. A control plasmid containing no mutations was also created. These plasmids were linearized, using SphI, and transformed into H. influenzae strains RM7004 and RM7004lgtCΔ5′GACA as described above (strain data available upon request).
Detection of the α-Gal-(1-4)-β-Gal digalactoside on surface-expressed LPS of H. influenzae.
Bacteria were grown overnight on agar plates and then transferred to nitrocellulose filters which were allowed to air dry before nonspecific binding sites were blocked with 0.05% (vol/vol) PBS-Tween 20 (PBST)-2% (wt/vol) bovine serum albumin (BSA) in a petri dish for 1 h. The filters were washed for three 2-min periods with PBST before being incubated with specific monoclonal antibody for 4 h, used at a dilution of 1/100 in PBST-BSA. Excess specific antibody was washed off as above using PBST, and the filter was incubated with a 1:2,000 dilution of an anti-mouse alkaline phosphatase conjugate antibody for 1 h. Filters were washed three times with PBST, and bound antibody was detected with 5-bromo-4-chloro-3-indolyl-phosphate-nitroblue tetrazolium (Perkin Elmer Life Sciences, Inc.). The color reaction was stopped by washing several times with water before allowing the blots to air dry.
Analysis of LPS by T-SDS-PAGE.
A 10-μl loopful of bacteria from an overnight plate-grown culture was resuspended in 1,000 μl of PBS. The relative concentration of this sample was determined by making a 1:50 dilution in 1% SDS (wt/vol)-0.1 M NaOH and measuring the absorbance at OD260. Suspensions were adjusted to equivalent concentrations in PBS prior to the addition of 12.5 μl of suspension to an equal volume of 2× dissociation buffer (125 mM Tris, pH 6.8, 20% [vol/vol] glycerol, 4% SDS, 10% mercaptoethanol, 0.004% bromophenol blue) and boiled for 5 min. Lysates were then resolved by tricine-SDS-polyacrylamide gel electrophoresis (T-SDS-PAGE) using a method described previously (25). Gels contained 16.5% acrylamide (wt/vol) and were run at 35 mA and 4°C overnight. Resolved bands were detected by staining with silver (Quicksilver; Amersham Pharmacia Biotechnology).
Survival of the bactericidal effects of human serum.
Bacteria cultured on BHI agar plates were assayed for survival in pooled human sera as described previously (20). Briefly, ∼1,000 cells were incubated for 45 min in serial dilutions of human sera in PBS containing 0.1% glucose and 0.1% fetal calf serum, and surviving bacteria were enumerated by plating samples on BHI agar plates.
RESULTS
Most H. influenzae phase-variable genes contain multiple initiation codons upstream of the repeat tract.
It was noted previously that several genes associated with tetranucleotide repeats have multiple potential translational initiation codons (33, 36). A systematic assessment of their prevalence was performed by searching for all the 5′ ATGs upstream of the repeat tract in genes within the published complete genome sequence of H. influenzae strain Rd that contained tetranucleotide repeats (19). Three cryptic genes could not be analyzed because they lacked the potential to be translated in any of the three frames owing to mutations in the sequence downstream of the repeat tract. Inspection of the DNA sequences upstream of the repeat tracts revealed that eight of these genes contained two or more putative 5′ ATG initiation codons in two or more of the three possible reading frames (Table 1). In all of these genes, except mod, these putative initiation codons were within 90 bp of the start of the repeat tract.
TABLE 1.
Number of initiation codon-containing frames and putative initiation codons of simple sequence contingency loci in H. influenzae strain Rd
| SSCL (HI no.)a | No. of initiation codon-containing framesb | No. of 5′ ATG codonsc |
|---|---|---|
| lic1A (1537) | 2 | 3 |
| lic2A | 2 | 4 |
| lic3A | 2 | 2 |
| lgtC | 1 | 1 |
| hgp (0635) | 2 | 2 |
| hgpB (0661) | 3 | 4 |
| hgpC (0712) | 2 | 3 |
| hgp (1565) | 2 | 3 |
| mod (1056/1058) | 2 | 2 |
SSCL, simple sequence contingency loci. HI numbers are from the H. influenzae strain Rd as annotated by The Institute for Genomic Research (www.tigr.org).
An initiation codon-containing frame is an open reading frame which contains no stop codons between a 5′ ATG and the repeat tract. One or more 5′ ATGs may be present in each of these frames.
Indicates the combined total of the 5′ ATG codons found in the initiation codon-containing open reading frames.
Multiple initiation codons of lic2A are conserved in different strains.
To investigate whether the presence of multiple initiation codons in strain Rd is a general feature of H. influenzae strains, the 180 bp of DNA upstream of the lic2A repeat tract were sequenced from several strains. These strains were selected as being representative of the genetic diversity of different capsular serotypes and capsule-deficient, nontypeable isolates of H. influenzae (7). Alignment of these sequences indicated that all of the 5′ ATG codons in lic2A were invariant and that the sequences around these codons were highly conserved (data not shown). Although the Shine-Dalgarno sequence is a major determinant of translation initiation in many bacteria and can be used to identify functional initiation codons, no such sequences were identified adjacent to any of the initiation codons in the lic2A 5′ region.
Demonstration of phase variation using a lic2A-lacZ reporter gene construct.
Phenotypic analysis of lic2A is complicated because synthesis of the α-Gal-(1-4)-β-Gal epitope is dependent on three phase-variable enzymes. Therefore, the effect of the different lic2A 5′ ATG initiation codons (Fig. 1A) on expression of this gene was investigated at the genotypic level by fusing a lacZ gene, lacking an initiation codon, in frame with the lic2A initiation codons. This reporter construct was introduced by transformation into the genome of H. influenzae strain Rd to generate strain Rdlic2AlacZ (the wild-type [WT] strain). Changes in the number of 5′ CAAT repeats will cause frameshifts which place the lacZ gene either into one of the two frames with initiation codons, where x and y are in one frame and z1 and z2 are in another frame, or into the frame lacking initiation codons (Fig. 1B).
PV rates were determined for variants with 22 5′ CAAT repeats which place lacZ in frame with the x/y codons. These colonies displayed an ON phenotype as evidenced by the production of blue colonies on plates supplemented with X-Gal. The rate for ON-to-OFF switching (blue to white) was 1.13 × 10−4 (Table 2, WT), which approximates to the rates previously determined for a 5′ AGTC repeat tract of similar length (8). The OFF-to-ON rate (white to blue) was determined using colony variants with 21 repeats (resulting in fusion of lacZ to the frame lacking initiation codons) and was 1.89 × 10−4 (Table 2, WT). This rate was significantly different from the ON-to-OFF rate (P > 0.0001 using a nonparametric Mann-Whitney Test). The ratio of ON-to-OFF:OFF-to-ON PV rates provides an indication of the number of frames associated with expression. A ratio of 2 indicates expression in one of the three frames, and a ratio of 0.5 indicates expression in two frames. The ratio of 0.6 observed for Rdlic2AlacZ indicated that expression of lic2A occurs in two of the three possible frames (Table 2).
TABLE 2.
Phase variation rates of lic2A-lacZ reporter strains
| Relevant genotypeb | Direction of switching
|
ON-to-OFF/OFF-to-ON ratio | |||
|---|---|---|---|---|---|
| ON-to-OFF
|
OFF-to-ON
|
||||
| No. of 5′ CAAT repeats | Mutation rate (10−4 [95% CI])a | No. of 5′ CAAT repeats | Mutation rate (10−4 [95% CI])a | ||
| WT | 22 | 1.13 (0.99-1.24) | 21 | 1.89 (1.79-2.14) | 0.6 |
| ΔATG | 23 | 1.98 (1.49-2.78) | 22 | 1.05 (0.90-1.25) | 1.9 |
| ΔATG | 22 | 1.36 (0.96-1.63) | 21 | 2.30 (1.96-2.78) | 0.6 |
| ΔATG | 23 | 2.54 (2.07-2.92) | 22 | 1.26 (1.16-1.37) | 2.0 |
| ΔATG | 22 | 1.16 (0.94-1.46) | 21 | 2.20 (1.87-2.89) | 0.5 |
| ΔATG | 22 | 1.88 (1.56-2.60) | 21 | 0.95 (0.80-1.19) | 2.0 |
The mutation rate was calculated using the median frequency by the method of Drake (9). Confidence intervals (CIs) were calculated as described by Kokoska et al. (24).
WT, wild-type strain Rdlic2AlacZ. ΔATGx, Rdlic2AΔxlacZ; ΔATGy, Rdlic2AΔylacZ; ΔATGxy, Rdlic2AΔxylacZ; ΔATGz1, Rdlic2AΔz1lacZ; ΔATGz2, Rdlic2AΔzlacZ.
The repeat tract lengths of variant colonies were analyzed in order to determine the frames associated with expression of lic2A in Rdlic2AlacZ. For OFF-to-ON switching, ON variant colonies were associated in high proportions with both the x/y and z1/z2 frames (Table 3, WT). For ON-to-OFF switching, the majority of OFF variant colonies were, as expected, associated with the frame lacking initiation codons (Table 3, WT). However, a high proportion of OFF variants (29.3%) were also associated with the z1/z2 frame. Qualitative assessment of colony phenotypes indicated that, whereas ON variants in frame with the x/y codons were “strong blue,” those in frame with the z1/z2 codons were “light blue.” It was concluded that factors such as colony size and age resulted in variations in the intensity of the light blue such that detection of this phenotype was difficult, resulting in the association of the z1/z2 frame with colonies of both the ON (light blue) and OFF (white) phenotypes.
TABLE 3.
Detected mutation events and frames associated with expression in lic2A-lacZ reporter strains
| Relevant genotypeb | Parental reading frame | % of deletions (frame)a
|
% of insertions (frame)a
|
Total no.
|
||||
|---|---|---|---|---|---|---|---|---|
| >−2 | −2 | −1 | +1 | +2 | >+2 | |||
| ON-to-OFF | ||||||||
| WT | ATGx/ATGy | 55.2 (0) | 29.3 (z) | 15.5 (0) | 58 | |||
| ΔATGx | ATGz1/ATGz2 | 65.6 (x) | 28.1 (0) | 6.3 (x) | 32 | |||
| ΔATGy | ATGx/ATGy | 68.8 (0) | 15.6 (z) | 15.6 (0) | 32 | |||
| ΔATGz1 | ATGx/ATGy | 2.2 (z) | 75.6 (0) | 4.4 (z) | 17.8 (0) | 45 | ||
| ΔATGz2 | ATGx/ATGy | 3.3 (z) | 86.7 (0) | 10.0 (z) | 30 | |||
| OFF-to-ON | ||||||||
| WT | 0 | 1.1 (x) | 2.2 (x) | 52.2 (z) | 41.3 (x) | 2.2 (z) | 1.1 (x) | 92 |
| ΔATGx | ATGx/ATGy | 6.9 (z) | 93.1 (z) | 29 | ||||
| ΔATGy | 0 | 3.1 (x) | 43.8 (z) | 53.1 (x) | 32 | |||
| ΔATGz1 | 0 | 3.8 (z) | 7.7 (x) | 42.3 (z) | 42.3 (x) | 3.8 (z) | 26 | |
| ΔATGz2 | 0 | 3.3 (x) | 96.7 (x) | 30 | ||||
Mutation events were classified as either deletions or insertions of 5′ CAAT repeat units that consisted of 1, 2, or more than 2 repeat units. Each type of mutation event is expressed as a percentage of the total number of events analyzed. The initiation codon associated with each mutation event or frameshift is indicated in parentheses. The frame lacking any putative initiation codons is represented by zero.
WT, wild-type strain Rdlic2AlacZ. For other designations, see Table 2.
Phase variation rates of initiation codon mutants.
The above results indicated that both the x/y and z1/z2 frames of the lic2A-lacZ construct were associated with expression but did not identify which initiation codons were functional. To determine their precise roles, initiation codons x, z1, and z2 were altered, by site-directed mutagenesis of plic2AlacZ, from 5′ ATG to 5′ CTT. This codon was chosen because it maintained the percent G+C content of the DNA flanking the repeat tract, a possible factor affecting DNA stability and, hence, the stability of repeat tracts (6). A spontaneous mutation of the y codon from 5′ ATG to 5′ AGG was also obtained. This serendipity provided a y initiation codon mutation and also enabled construction of an x/y double mutant. A kanamycin resistance gene was cloned into these plasmids and used to select for transformants of H. influenzae strain Rd in which the plasmid had recombined into the chromosome.
Colonies of ON variants from the strains with mutations in the y, z1, z2, and x/y codons (Rd lic2AΔylacZ, Rdlic2AΔz1lacZ, Rdlic2AΔz2lacZ, and Rdlic2AΔxylacZ, respectively) were blue while those of the x codon mutant (Rdlic2AΔxlacZ) were light blue. The blue color observed for the x/y double mutant indicated that mutation of the y codon from 5′ ATG to 5′ AGG had increased expression from the z1/z2 frame (possibly due to generation of a more effective Shine-Dalgarno site).
For each of the above mutants, PV rates were determined for both directions of switching and these rates were in the same order of magnitude as those of the parental strain, Rdlic2AlacZ (Table 2). However, differences were apparent in the ON-to-OFF:OFF-to-ON ratios, which were 1.9, 2.0, and 2.0 for the x, x/y, and z2 mutants, respectively, and 0.5 and 0.6 for the z1 and y mutants, respectively. These results indicated that only one reading frame was active in the x, x/y, and z2 mutants.
Determination of frames associated with expression in variant colonies of initiation codon mutants.
Repeat tract lengths in variants of the initiation codon mutants were determined in order to identify the frames associated with expression (Table 3). While the pattern of mutations in Rdlic2AΔylacZ was unaltered relative to the strain containing the parental construct, mutation of the x codon led to a complete loss of expression associated with this frame; i.e., all of the ON variants of Rdlic2AΔxlacZ were in frame with the z1/z2 codons while many of the OFF variants were now in frame with the x/y frame.
For OFF-to-ON switching, mutation of z2 abrogated detection of ON variants associated with the z1/z2 frame while mutation of z1 produced no clear change relative to the parental construct. For ON-to-OFF switching, OFF variants in frame with the z1/z2 codons were detected for both the z1 and z2 mutants but at a lower level than for the parental strain. These results suggested that inactivation of either the z1 or z2 initiation codons reduced expression from this frame but that the effect of the z2 mutation was greater than the z1 mutation.
Quantification of expression levels associated with each initiation codon.
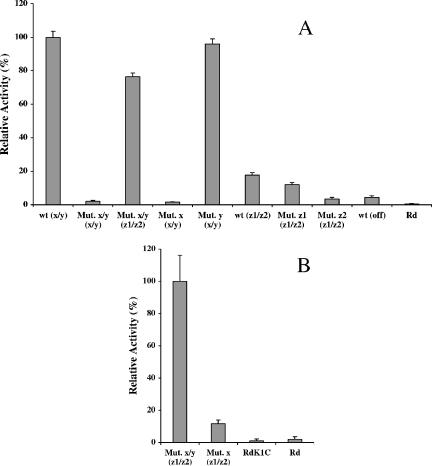
To quantify the differences in the translational activity associated with each initiation codon, β-galactosidase assays were performed on mid-log phase cultures of variants of the parental strain, Rdlic2AlacZ, and the mutant strains (Fig. 2). High levels of β-galactosidase activity were observed for the parental strain when the repeat number placed the lacZ gene in frame with the x/y reading frame [Fig. 2A, wt (x/y)]. Mutation of the x codon or the x and y codons, but not the y codon alone, resulted in undetectable levels of β-galactosidase activity [Fig. 2A, compare wt (x/y) with Mut. x (x/y), Mut. x/y (x/y), and Mut. y (x/y)], thus providing strong evidence that the x, but not the y, codon was the only functional codon in this frame. A sixfold lower level of activity was detected for the z1/z2 frame than for the x/y frame [Fig. 2A, compare wt (x/y) with wt (z1/z2)] in the parental strain. Intriguingly, a high level of activity associated with the z1/z2 frame was detected in the x/y double-mutant [Fig. 2A, Mut. x/y (z1/z2)], providing a further indication that the y-codon mutation had increased expression from this reading frame. The β-galactosidase activity associated with the z1/z2 frame was reduced to background levels when the z2 codon was mutated [Fig. 2A, Mut. z2 (z1/z2)] and was significantly reduced when the z1 codon was mutated [P < 0.01 for a pairwise comparison of β- galactosidase activities variants of strains Rdlic2AlacZ and Rdlic2AΔz1lacZ in frame with the z1/z2 codons wt (z1/z2) and Mut. (z1/z2) (Fig. 2A), respectively, using a Student t test). This result suggested that translation in the z1/z2 frame is primarily initiated from the z2 codon.
FIG. 2.
Relative β-galactosidase activities of lic2A-lacZ reporter strains with and without mutations in putative initiation codons. Assays were performed in triplicate using exponential phase cultures either nonconcentrated (A) or fivefold concentrated (B). Mean specific activities and standard deviations were calculated. Activities are expressed as a percentage of the strain with highest activity. The designations for each strain are as follows: wt, Rdlic2AlacZ; Mut. x/y, Rdlic2AΔxylacZ; Mut. x, Rdlic2AΔxlacZ; Mut. y, Rdlic2AΔylacZ; Mut. z1, Rdlic2AΔz1lacZ; Mut. z2, Rdlic2AΔz2lacZ; Rd, H. influenzae strain Rd (i.e., lacking a lacZ gene); and RdK1C, Rdlic2AK1ClacZ (a reporter strain containing a stop codon between the two genes preventing translation of the lacZ gene from any of the lic2A initiation codons). The frames, where relevant, are indicated in parentheses, with OFF indicating that the frame lacks initiation codons.
The β-galactosidase activities associated with the z1/z2 frame in the parental construct could be due to the presence of small numbers of phase variants in which the lacZ gene is in frame with the x/y initiation codons. Therefore, a further series of assays was performed with the x mutant and x/y double mutant (Fig. 2B) in which translational activity can only occur in the z1/z2 frame. As described above (Fig. 2A), a high level of activity was detected for the z1/z2 frame with the x/y double mutant. No activity was detected for the negative controls (strain Rd in which there is no lacZ gene and strain RdK1C in which there is a termination codon between the lic2A and lacZ genes abrogating expression in any reading frame). A low level of activity, ninefold lower than the positive control, was detected for variants of the x mutant in frame with the z1/z2 frame (Fig. 2B, Mut. x, z1/z2), providing an unequivocal demonstration that the z1/z2 reading frame is associated with an intermediate level of expression.
Influence of initiation codon mutations on expression of the native lic2A gene.
Given the results of the genotypic analysis with lacZ reporter constructs, lic2A phenotypes in wild-type and mutant strains were investigated by studying a digalactoside epitope of H. influenzae LPS whose expression is dependent on Lic2A (12, 16, 18). This digalactoside epitope is a component of the structures recognized by two monoclonal antibodies, 4C4 and MAHi5, and has been extensively studied in strain RM7004 (5, 13, 32). This strain, therefore, was chosen for studies of the native lic2A phenotypes, and transformants were obtained in which the native gene or the lic2A initiation codon mutations were transferred from the Rd background into strain RM7004. To enable selection of transformants, a kanamycin resistance cassette was inserted into an adjacent gene, ksgA, whose inactivation had previously been shown not to affect the LPS phenotypes of H. influenzae (17). A control construct was also generated in which ksgA was disrupted, but lic2A was unaltered. Expression of the digalactoside epitope is, in addition to lic2A, also dependent on the phase-variable genes lgtC and, to a lesser extent, lex2 (12, 16, 18). Thus, in addition to RM7004, the plasmid constructs were also transformed into an isogenic mutant (RM7004lgtCΔ5′GACA) in which expression of the lgtC gene is constitutive due to the absence of the tetranucleotide repeat tract (11).
The repeat numbers for lic2A, lgtC, and lex2 genes were determined for all transformants by PCR amplification and DNA sequencing. In each case, the lic2A gene had 22 repeats, which places the gene in frame with the x/y initiation codons. No changes were observed in the repeat numbers of the lgtC and lex2 genes relative to the parental strains. Phase variants showing reduced expression of the digalactoside epitope were obtained for the control strain and the strains with mutations in codons z1 and z2. These variants were identified by colony immunoblotting with MAbs 4C4 or MAHi5. Phase variants were selected for further analysis, which exhibited alterations in the lic2A repeat tract (i.e., resulting in a switch from the x/y frame to the z1/z2 frame or the frame lacking initiation codons) but no alterations in the lgtC or lex2 repeat tracts.
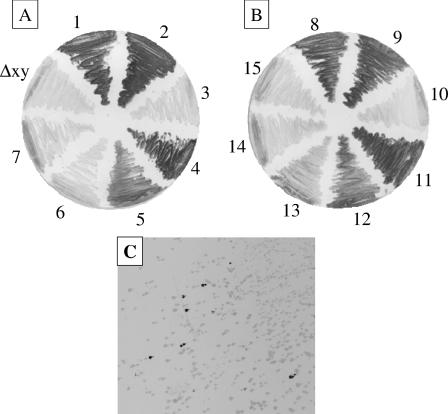
Expression of the digalactoside epitope was compared by performing immunoblotting and Western blotting with the MAbs 4C4 and MAHi5. Neither disruption of ksgA nor mutation of the y codon of lic2A affected expression of the digalactoside epitope, since the MAb MAHi5 showed strong reactivity against phase variants of RM7004lic2Akan and RM7004lic2AΔykan that were in frame with the x/y codons (Fig. 3A, sectors 2 and 4). Mutation of the x initiation codon abrogated the MAb reactivity associated with the x/y frame (Fig. 3A, sector 3). Similar results were found in the strain lacking tetranucleotide repeats in lgtC, RM7004lgtCΔ5′GACA (Fig. 3B, sector 10) and with MAb 4C4 (data not shown), although in this case reactivity was significantly less than that observed with MAb MAHi5.
FIG. 3.
Reactivity of H. influenzae lic2A initiation codon mutants with monoclonal antibody MAHi5. Phase variants or initiation codon mutants of strains RM7004lic2Akan (A) or RM7004lgtCΔGACAlic2Akan (B and C) were streaked on BHI agarose plates and probed with MAb MAHi5. Sectors 1 and 8 are both RM7004; 2 and 9 have 22 repeats (x/y frame) and a wild-type lic2A gene; 3 and 10 have 22 repeats (x/y frame) and a lic2AΔx gene; 4 and 11 have 22 repeats (x/y frame) and a lic2AΔy gene; 5 and 12 have 23 repeats (z1/z2 frame) and a wild-type lic2A gene; 6 and 13 have 23 repeats (z1/z2 frame) and a lic2AΔz1 gene; 7 and 14 have 23 repeats (z1/z2 frame) and a lic2AΔz2 gene; 15 has 21 repeats (frame lacking initiation codons) and a wild-type lic2A gene. The sector Δxy indicates an xy double mutant with 22 repeats (x/y frame). RM7004 and all RM7004lic2Akan strains have an identical number of repeats in lgtC. All strains have an identical number of repeats in lex2. Panel C is a colony blot of the culture used for sector 12 in panel A.
An intermediate level of MAb MAHi5 reactivity was exhibited by phase variants of both RM7004lic2Akan (Fig. 3A, sector 5) and RM7004lgtCΔ5′GACA (Fig. 3B, sector 12) that were in frame with the z1/z2 initiation codons. Antibody reactivity was reduced to background levels in the z1 and z2 mutants of both strains (Fig. 3, sectors 6, 7, 13, and 14), suggesting that the z1/z2 frame was associated with an intermediate level of expression. However, populations with intermediate levels of reactivity contained a small number (Fig. 3B, ∼1/60 for sector 12) of highly reactive phase variants (Fig. 3C and data not shown), indicating that this reactivity may be due to the presence of variants in frame with the x/y frame.
Functional characterization of the lic2A initiation codon mutants.
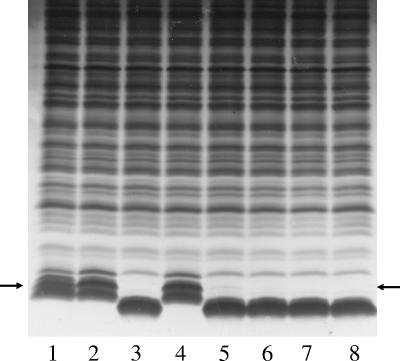
Further investigation of the LPS glycoforms was undertaken using SDS gel electrophoresis (Fig. 4) and Western blotting (data not shown). Phase variants of strains RM7004, RM7004lgtCΔ5′GACAlic2Akan, and RM7004lgtCΔ5′GACAlic2AΔykan (Fig. 4, lanes 1, 2 and 4), in which lic2A was in frame with the x/y initiation codons, had equivalent LPS profiles, consistent with the presence of digalactoside extensions from the first and/or second heptoses. In other phase variants and mutant strains, most notably the x initiation codon mutant (Fig. 4, lane 3), truncated glycoforms were the major LPS component. Small amounts of the larger glycoforms were observed when lic2A was in frame with the z1/z2 codons but not in the frame lacking initiation codons (Fig. 4, lanes 5 and 8). The presence of the digalactoside epitope in the larger LPS glycoforms of RM7004, RM7004lic2Akan, and RM7004lic2AΔykan was confirmed by Western blotting with MAb 4C4 (data not shown). No MAb 4C4-reactive bands were detected in the other mutants. Attempts to demonstrate differences in bactericidal sensitivity between these strains were unsuccessful, possibly due to the dominant effect of the capsule as a determinant of bacterial survival in this assay (data not shown).
FIG. 4.
LPS profiles of lysates of lic2A initiation codon mutants of H. influenzae strain RM7004lgtCΔGACA. Cell lysates were subjected to T-SDS-PAGE and stained with silver. The positions of the LPS glycoforms with an α-Gal-(1-4)-β-Gal extension from heptoses 1 and 2 are indicated by arrows. Lanes are as follows: lane 1, RM7004; lanes 2, 5, and 8, RM7004lgtCΔGACAlic2Akan with 22 (x/y frame), 23 (z1/z2 frame), and 21 (no initiation codons) repeats in lic2A; lane 3, RM7004lgtCΔGACAlic2AΔxkan with 22 (x/y frame) repeats in lic2A; lane 4, RM7004lgtCΔGACAlic2AΔykan with 22 (x/y frame) repeats in lic2A; lane 6, RM7004lgtCΔGACAlic2AΔz1kan with 23 (z1/z2 frame) repeats in lic2A; lane 7, RM7004lgtCΔGACAlic2AΔz2kan with 23 (z1/z2 frame) repeats in lic2A. All strains have an identical number of repeats in lex2.
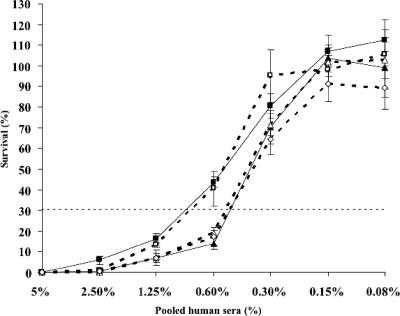
Some of the lic2A initiation codon mutations were then transferred into an RdlgtCΔ5′GACA strain background. This acapsulate strain is more sensitive to bactericidal killing than RM7004 and may permit small effects on bactericidal sensitivity to be detected. The mutations were transferred into this strain using chromosomal DNA from the RM7004 mutants, resulting in the concomitant transfer of genes lic2B and lic2C in all cases. LPS profiles of the Rdlic2Akan and Rdlic2AΔykan strains exhibited multiple glycoforms and a slightly more complex pattern than strain Rd (data not shown), consistent with previous studies (22) indicating that transfer of lic2B/lic2C leads to low levels of glucose extension from the second heptose as well as the third heptose (the normal position of oligosaccharide extensions in strain Rd). The similar profiles of Rdlic2Akan and Rdlic2AΔykan and the truncated profile observed with strain Rdlic2AΔxkan (data not shown) again indicated that lic2A expression was abrogated by mutation of the x but not the y codon. Variants of Rdlic2Akan in frame with either the z1/z2 codons or no initiation codons also exhibited truncated LPS profiles (data not shown). Bactericidal assays with these mutants and variants demonstrated that strains exhibiting a truncated LPS profile were more sensitive to killing by normal human serum than strains expressing a more extended profile (Fig. 5).
FIG. 5.
Survival in pooled human serum of lic2A initiation codon mutants of H. influenzae strain Rd. Strains were incubated with twofold serial dilutions of pooled human sera. Each experiment was repeated in triplicate. The mean and standard deviations from the mean are depicted. Strains are indicated by symbols: filled squares, Rd; open squares, Rdlic2Akan (22 repeats; x/y frame); open triangles, Rdlic2Akan (23 repeats; z1/z2 frame); open diamonds, Rdlic2Akan (22 repeats; out-of-frame); filled triangles, Rdlic2AΔxkan (22 repeats; x/y frame). Thirty percent survival indicates a level at which the serum sensitivity of strains can reproducibly be differentiated (20).
DISCUSSION
Bacteria can adapt to new environments through selection of genetic variants. These variants may contain multiple mutations and exhibit alterations in the expression of several genes. The presence of multiple contingency loci within a genome enables a bacterial species to rapidly explore numerous combinations of mutations and phenotypes. Thus, if the nine known functional contingency loci of H. influenzae strain Rd (Table 1) (18) exist in two states, ON and OFF, then this organism has access to 512 genotypes. Table 1 indicates that eight of these loci have initiation codons in two or more readings frames. If these reading frames were associated with three levels of expression (or indeed different functional variants of the protein), then there are 13,122 potential genotypes. Multiple levels of differential expression from phase-variable loci could, therefore, be a powerful driver of genetic diversity. We have examined this phenomenon by investigating the expression associated with multiple initiation codons in lic2A, a phase-variable gene of H. influenzae.
Three lines of evidence were obtained with a reporter strain, Rdlic2AlacZ, indicating expression from two reading frames of lic2A and differential expression levels for each of the three reading frames: (i) a low ON-to-OFF:OFF-to-ON ratio (0.6) indicating expression in two frames (Table 2), (ii) ON variants containing repeat numbers placing either the x/y or z1/z2 initiation codons in frame with lacZ (Table 3), and (iii) qualitative assessment of colonies indicating three levels of lacZ expression. These results with the lic2A-lacZ reporter construct were then confirmed and extended to an identification of the active initiation codons by site-specific mutagenesis of the lic2A 5′ ATG initiation codons. Expression from two reading frames in the wild-type gene was confirmed by the loss of this phenotype in the x, x/y, and z2 mutants which exhibited an increase in the ON-to-OFF:OFF-to-ON ratios to ∼2.0 (Table 3; i.e., a ratio of 2.0 indicates that ON-to-OFF switching is twice as likely as OFF-to-ON switching because there are two OFF reading frames for every one ON reading frame) and an association of ON variants in these mutants with only one reading frame (Table 3, OFF-to-ON switching). Differential expression of the x/y and z1/z2 reading frames was indicated by the light blue phenotype of x-mutant ON variants (i.e., in frame with the z1/z2 codons) and the dark blue phenotype of ON variants of z2 mutants (i.e., in frame with x/y codons) (data not shown). The x codon was identified as the functional codon in the x/y frame by the opposing phenotypes of the x and y mutants (compare the ON-to-OFF:OFF-to-ON ratios, the association of ON variants with x/y frame, and the qualitative assessment of the ON variants expression levels) and by the lack of activity associated with the x/y frame in the x/y mutant (Fig. 2). Finally, the z2 codon was identified as the major functional initiation codon in the z1/z2 frame by the higher ON-to-OFF:OFF-to-ON ratio of the z2 mutant relative to the z1 mutant (Table 2) and the inability to detect ON variants associated with the z1/z2 frame during OFF-to-ON switching of the z2 mutant in contrast to the frequent detection of such variants with the z1 mutant (Table 3).
Quantitative analysis of the expression levels associated with the different reading frames in a phase-variable locus is subject to a significant caveat, which is that the cultures/populations used for analysis may contain small numbers of phase variants of the highly expressing genotype (Fig. 2, OFF variants of Rdlic2AlacZ). In the β-galactosidase assays presented herein, different levels of expression were detected for each of the reading frames of lic2A: high expression from the x/y reading frame, ninefold lower expression from the z1/z2 reading frame, and no expression in the third reading frame. Expression from the z1/z2 frame was also quantified using Rdlic2AΔxlacZ. The β-galactosidase activity detected for this mutant cannot be due to small numbers of phase variants in frame with the x/y initiation codons as expression from this frame was abrogated by the mutation of the x initiation codon. While it is formally possible that the x codon mutation has activated expression from z1/z2 frame or that the absence of translation of the x/y frame increases translation of the z1/z2 frame, these results provided further evidence that the z1/z2 frame is associated with a low level of expression.
Expression of the digalactoside epitope has previously been assigned to both the x/y and z1/z2 frames of lic2A (23, 34), but this association has not been rigorously investigated. Expression of this epitope, therefore, was examined using monoclonal antibodies specific for this epitope and both wild-type and mutant strains. PV of lgtC and lex2 was controlled for in our experiments by examining strains lacking repeats (11) or by checking that repeat numbers of these genes had not altered (data not shown). Analysis in RM7004 demonstrated that strong expression of the digalactoside epitope was associated with only the x/y frame of lic2A (Fig. 3) and that this expression was lost in an x but not a y mutant as evidenced by loss of reactivity with MAbs and a truncated LPS profile for the former but not latter mutant (Fig. 4). The functional importance of this change was indicated by the increased serum sensitivity of the x mutant (Fig. 5). Some weak expression of the digalactoside epitope was detected for variants of the native lic2A gene in frame with the z1/z2 codons, but this reactivity was most likely due to small numbers of highly reactive variants in frame with the x codon (Fig. 3). Indeed, no phase variants exhibiting any reactivity with MAbs 4C4 and MAHi5 could be identified, despite repeated attempts, for x mutants of strain RM7004 (data not shown). Similarly, we were unable to detect changes in serum resistance associated with the z1/z2 frame for either the native or mutant genes of lic2A in strain Rd (Fig. 5). These results demonstrated that there was no detectable expression of the digalactoside epitope associated with the z1/z2 frame of lic2A and that, in contrast to the gene expression data, there were only two levels of phenotypic expression for this gene.
The inability to detect expression of the digalactoside epitope associated with the z1/z2 frame suggests that the findings relating to expression of lic2A (as assessed using the lacZ reporter constructs) do not correlate with expression of the tertiary gene product. Our analysis of phenotypic expression may indicate that there is no expression of the Lic2A protein from the z1/z2 frame or perhaps that a threshold level of Lic2A is required to allow effective synthesis and incorporation of this epitope into H. influenzae LPS. Alternatively, the N-terminal region of Lic2A encoded by the x/y frame may be required for Lic2A activity such that expression from the z1/z2 codons would result in a nonfunctional protein. It is notable that, in alignments between lic2A and related genes, conserved amino acids are present in the N-terminal sequences encoded from the x initiation codon up to the 5′ CAAT repeats (17). Finally, it is also possible that alterations in Lic2A expression results in changes to LPS epitopes not detectable with the monoclonal antibodies utilized in this study.
The observation of multiple initiation codons upstream of the repeat tracts of many of the H. influenzae phase-variable loci raises the possibility that these loci may exhibit gene expression profiles extending beyond the simple ON/OFF paradigm. The proposition is that more than one of these initiation codons is functional, that the functional codons are associated with different levels of gene expression, and that alterations in the repeat tract permit selection of variants with different levels of gene expression. This work has demonstrated that the lic2A gene phase varies between three levels of gene expression and has identified the codons responsible for these different levels of gene expression. This conclusion was reached using data from robust analyses of phase variation rates and repeat tracts with wild-type strains and strains carrying initiation codon mutations. One interesting extension of this work will be identification of the factors (e.g., ribosome binding sites) responsible for preferential usage of the x initiation codon. Previous reports of multiple levels of gene expression from three other phase-variable loci of H. influenzae, mod, lic3A, and lic1 (29, 30), were not supported by similar analyses such that the weak activity reported in these studies may have been due to small numbers of highly expressing variants or limited data sets. This work also showed, however, that only two levels of phenotypic variation can be detected for the lic2A phase variants. As the promoter (likely to be located 50 to 100 nucleotides upstream of the x initiation codon) and 5′ end of lic2A were conserved between strains (data not shown), it is likely that there will be a similar lack of weak expression of Lic2A-dependent glycoforms, particularly the digalactoside epitope, in other H. influenzae strains. It cannot be excluded, however, that minor amounts of other biologically important glycoforms will be produced in some strains, and, without further work, it cannot be concluded that the results obtained in the case of lic2A from one strain can be generalized to other phase-variable loci of H. influenzae, for example, the three levels of phenotypic variation proposed for lic1 (33). Thus, the demonstration of three levels of lic2A expression is provocative and suggests that there is the potential for phase-variable loci to produce more than two levels of phenotypic expression which, if applicable to other loci, could result in a significant repeat-mediated increase in the repertoire of variants in H. influenzae.
Acknowledgments
The authors thank Man-Suen Chan for assistance with bioinformatics and other members of the laboratory for their help and support.
K.D. was supported by an MRC Ph.D. studentship. C.D.B. was supported by Wellcome Trust program grant 070123/Z/02/Z entitled “Mutation rates of simple sequences and their contribution to the pathogenesis of Haemophilus influenzae and Neisseria meningitidis.” D.W.H. and K.M. were supported by an MRC program grant G0400426 entitled “The role of sialic acid in the communal and disease states of Haemophilus influenzae.”
Footnotes
Published ahead of print on 10 November 2006.
REFERENCES
- 1.Andrewes, F. W. 1922. Studies in group agglutination I. J. Path. Bacteriol. 25:505. [Google Scholar]
- 2.Bayliss, C. D., D. Field, and E. R. Moxon. 2001. The simple sequence contingency loci of Haemophilus influenzae and Neisseria meningitidis. J. Clin. Investig. 107:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayliss, C. D., and E. R. Moxon. 2005. Repeats and variation in pathogen selection, p. 54-76. In L. H. Caporale (ed.), The implicit genome. Oxford University Press, Oxford, United Kingdom.
- 4.Bayliss, C. D., T. van de Ven, and E. R. Moxon. 2002. Mutations in polI but not mutSLH destabilize Haemophilus influenzae tetranucleotide repeats. EMBO J. 21:1465-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borrelli, S., K. Altmann, P. E. Jansson, and A. A. Lindberg. 1995. Binding specificity for four monoclonal antibodies recognizing terminal Galα1→4Gal residues in Haemophilus influenzae lipopolysaccharide. Microb. Pathog. 19:139-157. [DOI] [PubMed] [Google Scholar]
- 6.Brock, G. J., N. H. Anderson, and D. G. Monckton. 1999. Cis-acting modifiers of expanded CAG/CTG triplet repeat expandability: associations with flanking GC content and proximity to CpG islands. Hum. Mol. Genet. 8:1061-1067. [DOI] [PubMed] [Google Scholar]
- 7.Cody, A. J., D. Field, E. J. Feil, S. Stringer, M. E. Deadman, A. G. Tsolaki, B. Gratz, V. Bouchet, R. Goldstein, D. W. Hood, and E. R. Moxon. 2003. High rates of recombination in otitis media isolates of non-typeable Haemophilus influenzae. Infect. Gen. Evol. 3:57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Bolle, X., C. D. Bayliss, D. Field, T. van de Ven, N. J. Saunders, D. W. Hood, and E. R. Moxon. 2000. The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol. Microbiol. 35:211-222. [DOI] [PubMed] [Google Scholar]
- 9.Drake, J. W. 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. USA 88:7160-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. Kirkness, A. Kerlavage, C. Bult, J. Tomb, B. Dougherty, and J. Merrick. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 11.Griffin, R., C. D. Bayliss, M. A. Herbert, A. D. Cox, K. Makepeace, J. C. Richards, D. W. Hood, and E. R. Moxon. 2005. Digalactoside expression in the lipopolysaccharide of Haemophilus influenzae: role in intravascular survival. Infect. Immun. 73:7022-7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin, R., A. D. Cox, K. Makepeace, J. C. Richards, E. R. Moxon, and D. W. Hood. 2003. The role of lex2 in lipopolysaccharide biosynthesis in Haemophilus influenzae strains RM7004 and RM153. Microbiology 149:3165-3175. [DOI] [PubMed] [Google Scholar]
- 13.Gulig, P. A., C. F. Frisch, and E. J. Hansen. 1983. A set of two monoclonal antibodies specific for the cell surface-exposed 39K major outer membrane protein of Haemophilus influenzae type b defines all strains of this pathogen. Infect. Immun. 42:516-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammerschmidt, S., A. Muller, H. Sillmann, M. Muhlenhoff, R. Borrow, A. Fox, J. van Putten, W. D. Zollinger, R. Gerardy-Schahn, and M. Frosch. 1996. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialytransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol. Microbiol. 20:1211-1220. [DOI] [PubMed] [Google Scholar]
- 15.Herriott, R. M., E. M. Meyer, and M. J. Vogt. 1970. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J. Bacteriol. 101:517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.High, N. J., M. E. Deadman, and E. R. Moxon. 1993. The role of a repetitive DNA motif (5′-CAAT-3′) in the variable expression of the Haemophilus influenzae lipopolysaccharide epitope alpha Gal(1-4)β Gal. Mol. Microbiol. 9:1275-1282. [DOI] [PubMed] [Google Scholar]
- 17.High, N. J., M. P. Jennings, and E. R. Moxon. 1996. Tandem repeats of the tetramer 5′-CAAT-3′ present in lic2A are required for phase variation but not lipopolysaccharide biosynthesis in Haemophilus influenzae. Mol. Microbiol. 20:165-174. [DOI] [PubMed] [Google Scholar]
- 18.Hood, D. W., M. E. Deadman, T. Allen, H. Masoud, A. Martin, J. R. Brisson, R. Fleischmann, J. C. Venter, J. C. Richards, and E. R. Moxon. 1996. Use of the complete genome sequence information of Haemophilus influenzae strain Rd to investigate lipopolysaccharide biosynthesis. Mol. Microbiol. 22:951-965. [DOI] [PubMed] [Google Scholar]
- 19.Hood, D. W., M. E. Deadman, M. P. Jennings, M. Bisercic, R. D. Fleischmann, J. C. Venter, and E. R. Moxon. 1996. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 93:11121-11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hood, D. W., K. Makepeace, M. E. Deadman, R. F. Rest, P. Thibault, A. Martin, J. C. Richards, and E. R. Moxon. 1999. Sialic acid in the lipopolysaccharide of Haemophilus influenzae: strain distribution, influence on serum resistance and structural characterization. Mol. Microbiol. 33:679-692. [DOI] [PubMed] [Google Scholar]
- 21.Hood, D. W., and E. R. Moxon. 1999. Lipopolysaccharide Phase Variation in Haemophilus and Neisseria, p. 39-54. In H. Brude, S. M. Opal, S. N. Vogel, and D. C. Morrison (ed.), Endotoxin in health and disease. Marcel Dekker Inc., New York, NY.
- 22.Hood, D. W., G. Randle, A. D. Cox, K. Makepeace, J. Li, E. K. Schweda, J. C. Richards, and E. R. Moxon. 2004. Biosynthesis of cryptic lipopolysaccharide glycoforms in Haemophilus influenzae involves a mechanism similar to that required for O-antigen synthesis. J. Bacteriol. 186:7429-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosking, S. L., J. E. Craig, and N. J. High. 1999. Phase variation of lic1A, lic2A and lic3A in colonization of the nasopharynx, bloodstream and cerebrospinal fluid by Haemophilus influenzae type b. Microbiology 145:3005-3011. [DOI] [PubMed] [Google Scholar]
- 24.Kokoska, R. J., L. Stefanovic, H. T. Tran, M. A. Resnick, D. A. Gordenin, and T. D. Petes. 1998. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase δ (pol3-t). Mol. Cell. Biol. 18:2779-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesse, A. J., A. A. Campagnari, W. E. Bittner, and M. A. Apicella. 1990. Increased resolution of lipopolysaccharides and lipooligosaccharides utilising tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J. Immunol. Methods. 126:109-117. [DOI] [PubMed] [Google Scholar]
- 26.Moxon, E. R., and D. J. Maskell. 1992. Haemophilus influenzae lipopolysaccharide: the biochemistry and biology of a virulence factor, p. 75-96. In C. E. Hormaeche, C. W. Penn, and C. J. Smythe (ed.), Molecular biology of bacterial infection: current status and future perspectives. Society for General Microbiology Symposium 49. Cambridge University Press, Cambridge, United Kingdom.
- 27.Moxon, E. R., P. B. Rainey, M. A. Nowak, and R. Lenski. 1994. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 4:24-33. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Srikhanta, Y. N., T. L. Maguire, K. J. Stacey, S. M. Grimmond, and M. P. Jennings. 2005. The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc. Natl. Acad. Sci. USA 102:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szabo, M., D. Maskell, P. Butler, J. Love, and E. R. Moxon. 1992. Use of chromosomal gene fusions to investigate the role of repetitive DNA in regulation of genes involved in lipopolysaccharide biosynthesis in Haemophilus influenzae. J. Bacteriol. 174:7245-7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Alphen, L., T. Riemens, J. Poolman, C. Hopman, and H. Zanen. 1983. Homogeneity of cell envelope protein subtypes and biotypes among Haemophilus influenzae type b from patients with meningitis in The Netherlands. J. Infect. Dis. 148:75-81. [DOI] [PubMed] [Google Scholar]
- 32.Virji, M., J. N. Weiser, A. A. Lindberg, and E. R. Moxon. 1990. Antigenic similarities in lipopolysaccharides of Haemophilus and Neisseria and expression of a digalactoside structure also present on human cells. Microb. Pathog. 9:441-450. [DOI] [PubMed] [Google Scholar]
- 33.Weiser, J. N., J. M. Love, and E. R. Moxon. 1989. The molecular mechanism of phase variation of H. influenzae lipopolysaccharide. Cell 59:657-665. [DOI] [PubMed] [Google Scholar]
- 34.Weiser, J. N., and N. Pan. 1998. Adaption of Haemophilus influenzae to acquired and innate humoral immunity based on phase variation of lipopolysaccharide. Mol. Microbiol. 30:767-775. [DOI] [PubMed] [Google Scholar]
- 35.Weiser, J. N., N. Pan, K. L. McGowan, D. Musher, A. Martin, and J. C. Richards. 1998. Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J. Exp. Med. 187:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiser, J. N., A. Williams, and E. R. Moxon. 1990. Phase-variable lipopolysaccharide structures enhance the invasive capacity of Haemophilus influenzae. Infect. Immun. 58:3455-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]