Abstract
Enteropathogenic Escherichia coli (EPEC) infections are a leading cause of infantile diarrhea in developing nations. Typical EPEC isolates are differentiated from other types of pathogenic E. coli by two distinctive phenotypes, attaching effacement and localized adherence. The genes specifying these phenotypes are found on the locus of enterocyte effacement (LEE) and the EPEC adherence factor (EAF) plasmid. To describe how typical EPEC has evolved, we characterized a diverse collection of strains by multilocus sequence typing (MLST) and performed restriction fragment length polymorphism (RFLP) analysis of three virulence genes (eae, bfpA, and perA) to assess allelic variation. Among 129 strains representing 20 O-serogroups, 21 clonal genotypes were identified using MLST. RFLP analysis resolved nine eae, nine bfpA, and four perA alleles. Each bfpA allele was associated with only one perA allele class, suggesting that recombination has not played a large role in shuffling the bfpA and perA loci between separate EAF plasmids. The distribution of eae alleles among typical EPEC strains is more concordant with the clonal relationships than the distribution of the EAF plasmid types. These results provide further support for the hypothesis that the EPEC pathotype has evolved multiple times within E. coli through separate acquisitions of the LEE island and EAF plasmid.
Enteropathogenic E. coli (EPEC) infections are a leading cause of infantile diarrhea in developing nations (31, 37). A key characteristic of EPEC strains is the ability to intimately attach to intestinal epithelial cells and create attaching and effacing (AE) lesions (24). The AE phenotype is specified by genes of the locus of enterocyte effacement (LEE), a ∼35-kb pathogenicity island located in the bacterial chromosome (23, 41). The LEE island comprises approximately 40 genes and encodes the components of a type III secretion system, various effector molecules, and the intimin adhesin (23, 33, 68). Intimin plays a crucial role in AE lesion formation (15) and is encoded by the highly polymorphic eae gene (1, 72), which can be divided into periplasmic, transmembrane, and extracellular domains (39). To date, more than 25 major allelic variants of eae have been described (36).
Most typical EPEC strains fall into one of two phylogenetically distinct groups or clonal lineages, designated EPEC 1 and EPEC 2 (69), and differ from atypical EPEC and other types of pathogenic E. coli by their ability to form microcolonies on the surface of intestinal epithelial cells (4). This phenotype, termed localized adherence (LA), correlates with the presence of a large virulence plasmid called the EPEC adherence factor (EAF) plasmid (18). The EAF plasmids from different EPEC strains show considerable variation in size (∼85 to 110 kb) (48) and, presumably, gene content. Comparison of the complete EAF plasmid sequences from two prototypical EPEC strains (O127:H6 EPEC 1 strain E2348/69 and O111:NM EPEC 2 strain B171) indicates that the EPEC 2 plasmid of B171 carries fewer genes (80 versus 115 open reading frames) and a greater percentage of intact or partial insertion sequence elements (33% versus 19%) than the pMAR7 plasmid of EPEC 1 strain E2348/69 (11, 64). Nevertheless, certain parts of the plasmid show a high degree of sequence conservation among typical EPEC strains (47), particularly in the region encoding the bundle-forming pilus (BFP), a type IV fimbria whose production is associated with the LA phenotype. An operon of 14 genes is necessary for expression of the BFP (20, 63), with bfpA encoding the major structural subunit (bundlin). Sequence comparisons of nine bfpA alleles have provided compelling evidence for the action of diversifying selection at the molecular level (7, 8). A second locus on the EAF plasmid implicated in the full virulence of EPEC is the plasmid-encoded regulator (Per), consisting of three genes (perA, perB, and perC). Per has been shown to activate genes within the bfp operon (65) and the LEE pathogenicity island (22, 42).
Little is known about the allelic distributions of eae, bfpA, and perA among the EPEC 1, EPEC 2, and other clonal lineages of typical EPEC. In this study we characterized a diverse collection of 129 EPEC, including strains of the classical EPEC serotypes (52), through multilocus sequence typing (MLST) and restriction fragment polymorphism (RFLP) analysis to elucidate the extent to which horizontal transfer of the LEE island and EAF plasmid have contributed to the evolution and diversification of EPEC clones.
MATERIALS AND METHODS
Strains.
A collection of 95 EPEC strains was assembled based on serotype or their inclusion in one of two studies examining bfpA allelic variation (7, 8). These strains represent a variety of serotypes originally isolated between 1947 and 1998 from different regions around the world and were obtained from a number of sources, including the Centers for Disease Control and Prevention, Alejandro Cravioto, Helge Karch, Frits and Ida Ørskov, Phillip I. Tarr, and Luis Trabulsi (Table 1; see also Table S1 in the supplemental material). An additional 34 eae+ bfpA+ strains were selected from a cohort study of childhood diarrheal disease in Guinea-Bissau, West Africa (67) (Table 1; see also Table S1). Each strain was grown overnight at 37°C in 10 ml of Luria-Bertani broth with moderate shaking. Genomic DNA was isolated using the Puregene DNA isolation kit (Gentra Systems Inc., Minneapolis, MN). DNA concentrations were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc., Rockland, ME), which were diluted to 25 ng/μl for PCR.
TABLE 1.
Summary of EPEC strains investigated
| Serogroup | No. of isolates | Yr(s) of isolation | Locale(s) | Reference(s) |
|---|---|---|---|---|
| Classical serogroups | ||||
| O55 | 21 | 1947-1998 | Brazil, Congo, Dutch Guiana, France, Germany, Guinea-Bissau, Mexico, Scotland, United States | 8, 44, 47, 52, 56, 57, 67 |
| O86 | 10 | 1950-1997 | Brazil, Bulgaria, Denmark, Germany, Guinea-Bissau, United Kingdom, United States | 8, 9, 34, 52, 56, 67 |
| O111 | 16 | 1947-1996 | Austria, Brazil, Dutch Guiana, Germany, Mexico, Peru, Scotland, United Kingdom, United States | 8, 17, 38, 44, 45, 47, 52, 53, 56 |
| O114 | 4 | 1969-1997 | Guinea-Bissau, United Kingdom, United States | 16, 67 |
| O119 | 28 | 1960-1998 | Brazil, Chile, Guinea-Bissau, Mexico, Peru, United Kingdom, United States | 8, 16, 28, 38, 46, 47, 58, 61, 67 |
| O127 | 4 | 1969-1997 | Guinea-Bissau, United Kingdom | 16, 67 |
| O142 | 11 | <1960-1997 | Brazil, Canada, Guinea-Bissau, Indonesia, Peru, Portugal, Scotland, United States | 21, 32, 45, 56, 60, 67 |
| Other serogroups | ||||
| O2 | 1 | 1997 | Guinea-Bissau | 67 |
| O33 | 2 | 1997 | Guinea-Bissau | 67 |
| O34 | 1 | 1997 | Guinea-Bissau | 67 |
| O49 | 2 | 1997 | Guinea-Bissau | 67 |
| O51 | 1 | 1997 | Guinea-Bissau | 67 |
| O73 | 1 | 1997 | Guinea-Bissau | 67 |
| O76 | 1 | 1982 | Peru | 45 |
| O110 | 1 | 1993 | Germany | 59 |
| O126 | 3 | 1962-1964 | Egypt, Iran, Pakistan | 52 |
| O128 | 15 | 1953-1991 | Denmark, Germany, Pakistan, United Kingdom, United States | 9, 52, 55, 56 |
| O157 | 3 | 1983-1998 | Brazil, United States | 7, 70 |
| OX9 | 1 | 1997 | Guinea-Bissau | 67 |
| O- | 3 | 1996-1997 | Guinea-Bissau | 67 |
MLST.
MLST was performed on seven conserved housekeeping genes (aspC, clpX, fadD, icdA, lysP, mdh, and uidA). A detailed protocol of the MLST procedure, including allelic type and sequence type (ST) assignment methods, can be found at the EcMLST website (http://www.shigatox.net/mlst). Sequences were concatenated for phylogenetic analyses.
eae fRFLP.
Allelic variation in eae was resolved using fluorescent RFLP (fRFLP) as described previously (36).
bfpA PCR and DNA sequencing.
PCR primers were designed to target conserved flanking regions of bfpA based on nine available allelic sequences (7, 8). Each 25-μl reaction mixture contained 2.5 μl 10× buffer II (Applied Biosystems, Foster City, CA), 2.5 μl 2 mM deoxynucleoside triphosphate, 2.0 μl 25 mM MgCl2, 0.5 μl 10 μM bfpA_-52F primer (5′-AGATTATTCCGTGACCTATT-3′), 0.5 μl 10 μM bfpG_9R primer (5′-TGTCCTCACATATACCTCCC-3′), 1.5 U AmpliTaq Gold (Applied Biosystems), 1 μl of 25-ng/μl genomic DNA template, and 15.7 μl distilled H2O (dH2O). Amplification of the approximately 700-bp fragment utilized an initial denaturing step at 94°C for 10 min, followed by 35 cycles of 92°C for 1 min, 52°C for 1 min, and 72°C for 30 s. A final step of 72°C for 5 min was used to complete any partially extended product. PCR products (5 μl) were visualized on ethidium bromide-stained 1.5% agarose gels by illumination with UV light, purified using the QIAquick PCR purification kit (QIAGEN Inc., Valencia, CA), and quantified. Cycle sequencing reaction mixtures contained 4.0 μl CEQ DTCS Quick Start premix (Beckman Coulter Inc., Fullerton, CA), 1.0 μl 20 μM bfpA_-52F or bfpG_9R primer, approximately 70 ng of bfpA PCR product, and dH2O to a final volume of 10 μl. Amplification utilized an initial denaturing step at 94°C for 1 min, followed by 35 cycles of 96°C for 30 s, 52°C for 30 s, and 60°C for 2 min. Upon completion of cycle sequencing, samples were purified with Sephadex G-50 fine columns (Amersham Pharmacia Biotech Inc., Piscataway, NJ), dried under vacuum centrifugation (Savant Instruments Inc., Holbrook, NY), suspended in 40 μl of deionized formamide, and run on a CEQ2000XL instrument (Beckman Coulter Inc.). Samples were analyzed using the CEQ2000XL software and then exported for further analysis with the SeqMan module of the Lasergene software (DNASTAR Inc., Madison, WI).
bfpA PCR and RFLP.
PCR conditions were identical to those described above for bfpA except primers bfpA_114F (5′-GTCTGCGTCTGATTCCAATA-3′) and bfpA_521R (5′-TCAGCAGGAGTAATAGC-3′) were used to amplify a 408- to 414-bp internal fragment of the gene. Prior to digestion, each bfpA PCR product was purified using the QIAquick PCR purification kit. Three different restriction enzyme digests were used. Digestion with AluI and with BfaI was performed in separate 30-μl reaction mixtures with 10 U of enzyme, 3.0 μl 10× reaction buffer, and 26.0 μl purified PCR product, and samples were incubated overnight at 37°C, while digestion with 10 U of BstNI was performed in 30-μl reaction mixtures with 3.0 μl 10× reaction buffer, 0.3 μl 100× bovine serum albumin, and 25.7 μl purified PCR product followed by an overnight incubation at 60°C. All restriction enzymes were obtained from New England BioLabs Inc. (Ipswich, MA), and the reaction buffer provided with each enzyme was used. After incubation, 15 μl of the digests was visualized on ethidium bromide-stained 1.5% agarose gels by illumination with UV light.
perA PCR and DNA sequencing.
Primers were designed to target the conserved flanking and internal regions of perA based on 15 available sequences (27, 50, 65). PCR conditions are similar to those described above for bfpA except primers perA_-24F (5′-AACAAACGCGCATGAAGGTG-3′) and perB_222R (5′-TTCGCCGGTGATGTGGTCT-3′) were used with a 58°C annealing temperature and a 1-min extension time. The resulting PCR products (approximately 1.1 kb) were purified and quantified as described above. Cycle sequencing reaction mixtures were similar to those used for bfpA, except that 120 ng of perA PCR product and primers perA_-24F, perA_539F (5′-AAAACTGGAAACTAGGCGATGTCA-3′), perA_562R (5′-TGACATCGCCTAGTTTCCAGTTTT-3′), and perB_222R were used at a 58°C annealing temperature.
perA PCR and RFLP.
PCR conditions were similar to those described for bfpA except primers perA_-24F and perA_562R were used at an annealing temperature of 58°C. Digestion with DdeI and with Sau96I was performed in separate 30-μl reaction mixtures with 10 U of enzyme, 3.0 μl 10× reaction buffer, 20.0 μl unpurified PCR product, and dH2O to volume; samples were incubated overnight at 37°C and visualized on ethidium bromide-stained 1.5% agarose gels.
fliC typing.
Strains that were nonmotile or lacked flagellar serotype data were typed for the fliC locus. The entire fliC gene was amplified with primers fliC_1F (5′-ATGGCACAAGTCATTAATACCAA-3′) and fliC_1497R (5′-TTAACCCTGCAGCAGAGACA-3′) using the same PCR conditions described for bfpA except for an annealing temperature of 55°C and an extension time of 2 min. Amplicons (approximately 2 kb) were either digested with 5 U of DdeI under conditions similar to those described for perA or sequenced to determine the allele. H-types that were determined by fliC sequencing or RFLP are denoted with a lower case h and are enclosed in square brackets.
Phylogenetic analyses.
Sequences were aligned with the ClustalW algorithm using the MegAlign module of the Lasergene software. Neighbor-joining trees were constructed using the Kimura two-parameter model of nucleotide substitution with the MEGA3 software (35), and the inferred phylogenies were each tested with 500 bootstrap replications. Phylogenetic network analysis was conducted with the SplitsTree 4 program (30) using the neighbor-net algorithm (14) and untransformed distances (p distance). The φw recombination test (13) as implemented by SplitsTree 4 was used to distinguish recurrent mutation from recombination in generating genotypic diversity. The number of synonymous substitutions per synonymous site (dS) and the number of nonsynonymous substitutions per nonsynonymous site (dN) were estimated by the modified Nei-Gojobori method using MEGA3 (35). Allelic sequences were fit to a nucleotide substitution model using the Datamonkey website, and then either the single likelihood ancestor counting (SLAC) or random effects likelihood (REL) method was used to fit a codon model to detect selection on individual codons (54). The SLAC method was also used to calculate the ratio of dN to dS and estimate the 95% confidence interval.
Nucleotide sequence accession numbers.
The sequences for bfpA and perA were submitted to GenBank and given the accession numbers EF011027 to EF011059.
RESULTS
MLST analysis.
PCR amplification and sequencing of the seven MLST loci in 129 EPEC strains was successful in most (90%) cases. The notable exception was uidA, which failed to amplify in 13 strains, including a single O114:H2 strain (380/69) and 12 other strains, 11 of which are serotype O55:[h51]. These 12 uidA-negative strains were identical to each other at the six remaining MLST loci. PCR amplification with primers located in the genes flanking the uidA locus produced a truncated amplicon, suggesting that these strains belong to a clonal genotype that has lost most of the uidA gene (data not shown). For phylogenetic analysis, the sequenced internal fragments of the seven housekeeping genes were concatenated to yield 3,732 nucleotides. The uidA locus was treated as missing data and replaced with alignment gaps in the full concatenated sequence for the 13 uidA-negative strains.
MLST analysis resolved an average of 25.4 variable nucleotide sites per locus, which defined a number of alleles, ranging from 7 to 12, at the seven MLST genes (Table 2). The synonymous rate of substitution (dS) ranged from a low of 3.44% for lysP to a high of 8.56% for fadD, with an average of 5.41 synonymous substitutions per 100 synonymous sites (Table 2). The nonsynonymous rate (dN) per 100 nonsynonymous sites was generally 2 orders of magnitude lower than dS, ranging from 0.00 for lysP to 0.63 for uidA. Tests for natural selection operating on the allelic variation at each MLST locus based on the SLAC method found no individual sites (codons) that were under significant positive selection and only six codons under negative selection at the 0.1 significance level (one in clpX and mdh and two in fadD and icdA). Thus, low values of dN/dS at the MLST loci reflect weak negative selection over many codons.
TABLE 2.
Sequence variation among alleles of seven MLST genes, eae, bfpA, and perA
| Locusa | No. of sites | No. of variable sites | No. of alleles | % GC | dS × 100 (mean ± SE) | dN × 100 (mean ± SE) | dN/dS (95% CI) |
|---|---|---|---|---|---|---|---|
| aspC | 513 | 21 | 9 | 51.4 | 5.14 ± 1.23 | 0.06 ± 0.06 | 0.035 (0.002, 0.154) |
| clpX | 567 | 34 | 11 | 53.4 | 5.51 ± 1.06 | 0.05 ± 0.05 | 0.014 (0.001, 0.066) |
| fadD | 483 | 32 | 11 | 50.7 | 8.56 ± 1.50 | 0.05 ± 0.05 | 0.008 (0.000, 0.036) |
| icdA | 567 | 30 | 8 | 51.3 | 6.44 ± 1.25 | 0.20 ± 0.15 | 0.037 (0.006, 0.120) |
| lysP | 477 | 10 | 8 | 53.9 | 3.44 ± 1.14 | 0.00 ± 0.00 | 0.000 (0.000, 0.061) |
| mdh | 549 | 25 | 12 | 52.6 | 3.92 ± 0.84 | 0.04 ± 0.04 | 0.015 (0.001, 0.065) |
| uidA | 576 | 26 | 7 | 51.9 | 4.84 ± 1.10 | 0.63 ± 0.25 | 0.171 (0.080, 0.326) |
| Avg | 533.1 | 25.4 | 9.4 | 52.2 | 5.41 ± 1.16 | 0.15 ± 0.09 | 0.040 (0.013, 0.118) |
| eae | 2,853 | 790 | 9 | 42.3 | 25.22 ± 1.25 | 10.89 ± 0.74 | 0.310 (0.298, 0.349) |
| eaePPD | 558 | 67 | 9 | 40.7 | 9.39 ± 1.47 | 2.74 ± 0.55 | 0.245 (0.171, 0.358) |
| eaeTMD | 1,092 | 84 | 8b | 44.4 | 8.48 ± 1.15 | 0.88 ± 0.21 | 0.132 (0.086, 0.204) |
| eaeECD | 1,203 | 639 | 9 | 41.1 | 59.69 ± 3.13 | 27.02 ± 1.84 | 0.284 (0.275, 0.343) |
| bfpA | 588 | 94 | 11 | 40.0 | 9.36 ± 1.46 | 6.46 ± 1.11 | 0.480 (0.384, 0.608) |
| perA | 825 | 99 | 20 | 28.0 | 4.91 ± 0.80 | 2.03 ± 0.33 | 0.400 (0.322, 0.522) |
Average values for seven MLST loci. PPD, periplasmic domain; TMD, transmembrane domain; ECD, extracellular domain.
The κ and μ eae alleles analyzed have identical transmembrane domain sequences.
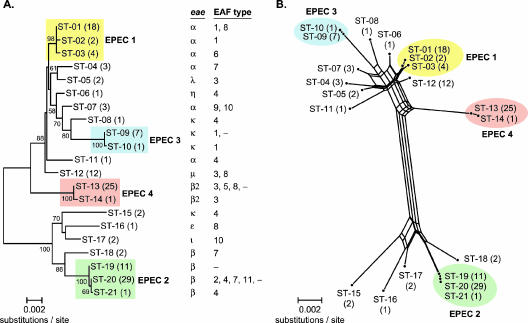
The distinct combinations of alleles across the seven MLST loci were used to define 21 multilocus genotypes or sequence types (STs) among the 129 EPEC strains. Classification of the strains based on the bootstrap analysis indicates that most (77%) of the strains belong to one of four main clonal groups, designated EPEC 1, EPEC 2, EPEC 3, and EPEC 4 (Fig. 1). With the exception of EPEC 3 strains, which were all O86:H34 (or nonmotile relatives), the EPEC groups based on the classification of STs included strains of various O-types (see complete list of serotypes in Table S1 in the supplemental material). There were strains representing three O-types in EPEC 1 (O55, O127, and O142), five O-types in EPEC 2 (O111, O114, O119, O126, and O128), and two O-types in EPEC 4 (O110 and O119). H-types (or the inferred H-type from the fliC allele) were conserved among strains of each group: EPEC 1 strains were H6, EPEC 2 were H2, EPEC 3 were H34, and EPEC 4 were H6. These four clonal groups were represented among both the worldwide and Guinea-Bissau strains. The 21 STs differed on average at 1.4% and 0.2% of the nucleotide and amino acid sites, respectively. ST-20 was the most common multilocus genotype (22.5%), followed by ST-13 (19.4%) and ST-1 (14.0%) (Fig. 1).
FIG. 1.
Phylogenetic relationships of 21 EPEC sequence types. (A) An unrooted phylogenetic tree was constructed by the neighbor-joining algorithm based on the Kimura two-parameter model of nucleotide substitution. (B) The phylogenetic (splits) network is based on the neighbor-net algorithm using a p distance matrix. The four main clonal groups are indicated by colored boxes or ellipses. The ST and number of isolates are given at the branch tips. Bootstrap values greater than 50% based on 500 replications are given at the internal nodes. The distributions of eae alleles and EAF plasmid types are shown on the right (see Table 5, below, for plasmid type definitions).
The splits network (Fig. 1B) revealed several parallel paths indicative of the presence of phylogenetic incompatibilities in the divergence of EPEC clones. Such incompatibilities could arise from recurrent mutation or recombination in the MLST loci. To detect recombination, we used the φw test, which has been shown to discriminate between recurrent mutation and recombination in a variety of circumstances (13). In application to the concatenated sequences of the 21 STs, there were 129 informative sites, and the φw test found statistically significant evidence of recombination (P < 0.001). The four main clonal groups, however, are separated and intact. Three of the four groups occur at the end of long branches without evidence of multiple paths, suggesting that recombination occurred early in the divergence of EPEC genotypes. With an EPEC phylogenetic framework in place, the allelic distributions of eae, bfpA, and perA were assessed.
Allelic variation in eae.
The eae locus was subtyped by fRFLP (36), and nine alleles (α, β, β2, ɛ, η, ι, κ, λ, and μ) were observed among the 129 EPEC strains (Fig. 1). Representative sequences of each fRFLP profile (36) were used for subsequent analyses. In comparison to the MLST loci, the synonymous rate for eae was 25.22%, more than 4 times greater than the mean for the seven MLST genes. The nonsynonymous rate of 10.89% for eae was more than 72 times greater the average dN across all seven MLST genes. When eae is divided into its three primary domains, the periplasmic and transmembrane domains have considerably lower synonymous and nonsynonymous rates than the extracellular region (Table 2). However, only a single codon within the extracellular domains of eae was found to be under significant positive selection using the SLAC method.
Allelic variation in bfpA.
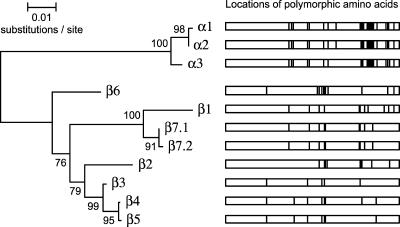
The entire bfpA gene was amplified and sequenced in 15 strains with STs for which bfpA allelic data were not previously available (STs 4, 5, 7, 8, 11, 15, 16, 17, and 18). Comparative sequence analysis revealed the existence of a 10th allele of bfpA, which we designated β7.1 (Fig. 2). A minor variant of this allele that differed by a single synonymous substitution was designated β7.2.
FIG. 2.
Eleven bfpA alleles cluster into two major groups. A phylogenetic tree constructed by the neighbor-joining algorithm based on the Kimura two-parameter model of nucleotide substitution is shown on the left. Bootstrap values based on 500 replications are given at the internal nodes. To the right is a graph of the locations of the 39 polymorphic amino acid sites (195 total), which are marked as vertical lines that indicate differences from the consensus of all 11 alleles.
In comparison to the MLST loci, the synonymous rate for bfpA was 9.36%, slightly greater than the range and mean for the seven MLST genes. The nonsynonymous rate of 6.46% for bfpA was more than 43 times greater the average dN across all seven MLST genes. There was no evidence for significant positive selection at individual codons within bfpA using the SLAC method; however, the less conservative REL method (54) detected 22 codon positions with positive selection, 12 of which were polymorphic for three or more amino acids.
Using the identified bfpA sequences, we devised an RFLP-based typing system to subtype bfpA alleles based on new PCR primers designed to target the conserved internal regions of the gene. We identified three restriction enzymes (AluI, BfaI, and BstNI) which, when used separately, produced digestion patterns that combined could resolve nine bfpA alleles (Table 3). In silico analysis with over 500 restriction endonucleases failed to identify an enzyme that could easily distinguish the β1 and β7 alleles. However, β1 and β7 bfpA strains can be easily differentiated based on their perA allele (see below). PCR amplification of bfpA was successful in all but 21 isolates. Of the bfpA-negative strains, O128:H2 was the most common serotype, with 13 isolates. RFLP analysis of the 108 bfpA-positive strains showed that the α1 (n = 23) and α3 (n = 24) alleles were the most common. The α2 (n = 15) and β5 (n = 17) alleles were also frequently identified, but the β2 (n = 1), β3 (n = 4), β4 (n = 7), and β6 (n = 2) alleles were rarely observed. Fourteen strains were classified as β1/β7 by RFLP, and DNA sequencing confirmed 11 as β1 and 3 as β7.
TABLE 3.
Expected restriction fragment length polymorphisms of bfpA PCR amplicons
| bfpA allele | Digestion pattern (bp)a
|
||
|---|---|---|---|
| AluI | BfaI | BstNI | |
| α1 | 408 | 408 | 155, 253 |
| α2 | 408 | 75, 333 | 155, 253 |
| α3 | 408 | 75, 333 | 408 |
| β1 | 16, 54, 113, 231 | 39, 375 | 414 |
| β2 | 16, 170, 228 | 414 | 414 |
| β3 | 16, 54, 344 | 39, 375 | 414 |
| β4 | 16, 177, 215 | 39, 369 | 408 |
| β5 | 16, 54, 123, 215 | 39, 369 | 408 |
| β6 | 16, 392 | 39, 369 | 408 |
| β7 | 16, 54, 113, 225 | 39, 369 | 408 |
Underlined fragments are not detectable under standard electrophoretic conditions.
Allelic variation in perA.
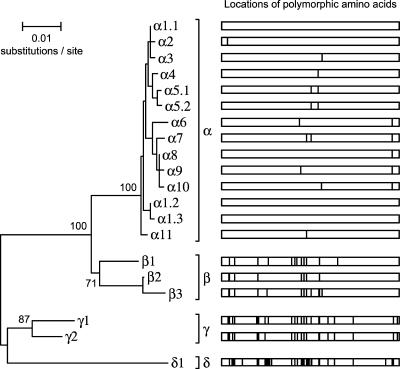
The entire perA gene was amplified and sequenced in 33 strains representing a diverse set of EPEC STs and bfpA alleles. One strain, 2309-77 (O111:H2), resulted in a PCR product approximately 1 kb larger than expected. DNA sequencing revealed the presence of a 1,055-bp IS element inserted into perA at position 414 with significant similarity to IS102 (86%) and IS903 (84%). Sequence analysis also identified eight strains that contained one or more frameshifts within mononucleotide repeats in perA that presumably inactivate the gene (see Table S2 in the supplemental material). The variability of these frameshifts among closely related alleles indicates their relatively recent occurrence, as there has not been sufficient time for the inactivated alleles to accumulate further mutations. These frameshifts were corrected, and the IS element sequence was excised in silico prior to allele assignment and phylogenetic analyses. The 33 sequences yielded 20 alleles, which clustered into four groups based on phylogenetic sequence analysis and, in keeping with the nomenclature for eae and bfpA, we designated these four allele classes α, β, γ, and δ (Fig. 3). As with bfpA, each distinct translated perA sequence was given an allele designation, resulting in 11 major α types, 3 β, 2 γ, and a single δ. Two of the α alleles had variants resulting from synonymous substitutions, and each variant was given its own subtype designation (α1.1, α1.2, α1.3, α5.1, and α5.2).
FIG. 3.
Twenty perA alleles cluster into four major groups. A phylogenetic tree constructed by the neighbor-joining algorithm based on the Kimura two-parameter model of nucleotide substitution is shown on the left. Bootstrap values for the major groups based on 500 replications are given at the internal nodes. To the right is a graph of the locations of the 46 polymorphic amino acid sites (274 total), which are marked as vertical lines that indicate differences from the consensus of all 20 alleles.
In comparison to the MLST loci, the synonymous rate for perA was 4.91%, within the range and slightly less than the mean for the seven MLST genes. The nonsynonymous rate of 2.03% for perA was more than 13 times greater the average dN across all seven MLST genes. However, only six codons within perA were found to be under significant negative selection, and there was no evidence for significant positive selection at individual codons using the SLAC method.
Based on the sequence data, an RFLP method using DdeI and Sau96I was designed to detect the four perA allele classes (Table 4). PCR amplification of perA was successful in all but 17 isolates. As with the bfpA-negative strains, O128:H2 was the most common serotype among the perA-negative isolates (n = 12). RFLP analysis of the 112 perA-positive strains showed that the α allele was the most common (n = 74), followed by β (n = 29), γ (n = 8), and δ (n = 1).
TABLE 4.
Expected restriction fragment length polymorphisms of perA PCR amplicons
| perA allele class | Digestion pattern (bp)a
|
|
|---|---|---|
| DdeI | Sau96I | |
| α | 13, 74, 82, 417 | 586 |
| β | 13, 74, 499 | 586 |
| γ | 82, 87, 417 | 174, 186, 226 |
| δ | 87, 499 | 226, 360 |
Underlined fragments are not detectable under standard electrophoretic conditions.
Association between EAF types, virulence factor alleles, and STs.
By combining the bfpA and perA allelic data, we found that each bfpA allele was associated with only one perA allele class, resulting in 11 distinct EAF plasmid types, which we have designated as EAF types 1 to 11 (Table 5). EAF plasmid types 4 and 8 appear to be the most promiscuous, being found in five and four clonal groups, respectively. Interestingly, the EAF type represented by the fully sequenced plasmid from O111:NM EPEC 2 strain B171 (64) is among the least promiscuous, being found in only one serotype (O111:H2) of a single sequence type (ST-20).
TABLE 5.
EAF plasmid types and distribution among EPEC clones
| EAF type | bfpA allele | perA allele | No. of isolates | No. of STs | No. of clonal groups |
|---|---|---|---|---|---|
| 1 | α1 | α | 23 | 4 | 2 |
| 2 | α2 | α | 15 | 1 | 1 |
| 3 | α3 | β | 24 | 4 | 3 |
| 4 | β1 | α | 11 | 6 | 5 |
| 5 | β2 | δ | 1 | 1 | 1 |
| 6 | β3 | γ | 4 | 1 | 1 |
| 7 | β4 | α | 7 | 3 | 3 |
| 8 | β5 | α | 18 | 4 | 4 |
| 9 | β6 | β | 2 | 1 | 1 |
| 10 | β7 | β | 3 | 2 | 2 |
| 11 | Nega | γ | 4 | 1 | 1 |
| Neg | Neg | Neg | 17 | 4 | 3 |
Neg, negative by PCR.
The α1 and α2 alleles of bfpA, as well as β4 and β5, differ by only one nonsynonymous nucleotide substitution. These two sets of closely related bfpA alleles were found in divergent EPEC lineages: EPEC 1 contains α1 and β5, whereas EPEC 2 has α2 and β4 (Table 6). In addition, multiple bfpA alleles were found within the same sequence type: α3, β2, and β5 within ST-13, and α2, β1, and β4 within ST-20 (Table 6).
TABLE 6.
Characteristics of four common EPEC clonal groups
| Clonal group and ST | eae allele | EAF typea | Serotypeb | No. of isolates |
|---|---|---|---|---|
| EPEC 1 STs | ||||
| 1 | α | 1 | O55:H6 | 8 |
| 1 | α | 1 | O127:[h6] | 4 |
| 1 | α | 1 | O142:[h6] | 1 |
| 1 | α | 1 | O-:[h6] | 1 |
| 1 | α | 8 | O142:H6 | 4 |
| 2 | α | 1 | O55:H6 | 2 |
| 3 | α | 6 | O142:H6 | 4 |
| EPEC 2 STs | ||||
| 19 | β | O128:H2 | 11 | |
| 20 | β | 2 | O111:[h2] | 15 |
| 20 | β | 4 | O114:[h2] | 4 |
| 20 | β | 4 | O126:H2 | 1 |
| 20 | β | 7 | O128:H2 | 2 |
| 20 | β | Neg | O111:H2 | 1 |
| 20 | β | Neg | O126:H2 | 1 |
| 20 | β | Neg | O128:H2 | 1 |
| 20 | β | 11 | O119:H2 | 3 |
| 20 | β | 11 | O128:H2 | 1 |
| 21 | β | 4 | O126:H2 | 1 |
| EPEC 3 STs | ||||
| 9 | κ | 1 | O86:[h34] | 6 |
| 9 | κ | Neg | O86:[h34] | 1 |
| 10 | κ | 1 | O86:H34 | 1 |
| EPEC 4 STs | ||||
| 13 | β2 | 3 | O119:H6 | 20 |
| 13 | β2 | 5 | O110:H6 | 1 |
| 13 | β2 | 8 | O119:H6 | 2 |
| 13 | β2 | Neg | O119:H6 | 2 |
| 14 | β2 | 3 | O119:H6 | 1 |
Neg, negative for bfpA and perA by PCR.
Lowercase H-types in square brackets were inferred from the fliC allele.
As shown previously (1), the EPEC 1 and EPEC 2 clonal groups possess the α and β alleles of eae, respectively. In contrast to the distribution of the EAF types, strains of the same ST had identical eae alleles as resolved by fRFLP. The α-eae allele had the widest distribution among the EPEC clones, being found in strains with O51:[h49], O73:[h34], O142:[h34], and O157:[h45] serotypes in addition to the EPEC 1 group (O55:H6, O127:H6, and O142:H6). The rarest eae alleles were ɛ, η, ι, and λ and, combined, these alleles account for less than 5% of the strains examined.
DISCUSSION
Common EPEC clones.
This is the first study to take a comprehensive look at the evolution of typical EPEC by combining clonal relatedness based on MLST with the allelic distributions of three important virulence factors. Previous clonal studies of EPEC have focused primarily on two main groups, EPEC 1 and EPEC 2, which were first described and defined based on the genetic relatedness of strains as determined by multilocus enzyme electrophoresis (69). EPEC 1 was described to comprise strains with O55:H6, O86:H34, O127:H6, and O142:H6 serotypes, while EPEC 2 included O111:H2, O114:H2, O126:H2, and O128:H2. O119:H6 strains have also been regarded as members of EPEC 1, because they share a number of genetic traits with the group (H6 flagellar antigen, eae+, and EAF+) (66), even though multilocus enzyme electrophoresis places them just outside of EPEC 1 (69). However, we feel that there are sufficient genetic differences to warrant the removal of O86:H34 and O119:H6 from EPEC 1, and we have reclassified them as EPEC 3 and EPEC 4, respectively (Fig. 1; Table 6). Our data indicate that EPEC 1 strains (O55:H6, O127:H6, and O142:H6) all possess α-eae, whereas the κ and β2 eae alleles are associated with EPEC 3 and EPEC 4, respectively. O86:H34 strains have also been shown to possess cytolethal distending toxin, while EAF+ O55:H6, O119:H6, O127:H6, and O142:H6 strains are negative (29). O119:H6 strains, on the other hand, are negative for espC, which encodes an enterotoxin, whereas EPEC 1 strains are positive (43).
Other EPEC clones.
Thirty of the 129 strains (23%) that were analyzed did not belong to any of the above-mentioned major lineages, and most of these had unusual serotypes for EPEC. Of these serotypes, only O2:[h2], O49:[h10], and O51:[h49] have previously been reported to possess eae and/or express the AE phenotype (2, 5, 12), and only O33:[h34], O142:[h34], and O157:[h45] have been previously described as eae+ bfpA+ and therefore classified as typical EPEC (5, 25, 26, 40, 51, 62). Literature searches on the remaining serotypes, including O34:[h45], O73:[h34], O76:H51, O86:[h8], O142:[h21], OX9:[h7], O-:[h7], and O-:[h34], failed to find any association with typical EPEC.
An interesting finding of this study was the prevalence of the O55:[h51] clone among the strains isolated in Guinea-Bissau. Strains with this serotype have previously been described as relatively minor members of the O55 serogroup and have been reportedly isolated only in South America (6, 57). It is possible that O55:[h51] strains are simply common among children in Guinea-Bissau, or their higher prevalence may be due to sampling bias. Alternatively, the abundance of these strains among the West African isolates could indicate that O55:[h51] is an emerging clone which is increasing in frequency and spreading geographically, possibly because of a distinct combination of μ intimin and EAF type 8.
Virulence gene distribution.
Recombination appears to have played a role in the initial generation of the EAF plasmid types. The highly divergent α1/α2 and β4/β5 bfpA alleles are all associated with α-perA, whereas the closely related β1 and β7 bfpA alleles are each found with different perA alleles. However, since bfpA and perA are in complete linkage disequilibrium (each bfpA allele is associated with only one perA allele class), it does not appear that recombination has played a large role in assorting allele combinations. Our results also indicate varying degrees of promiscuity among the different EAF types (Table 5). Recently, the complete sequence of a derivative of the wild-type EAF plasmid (pMAR7) from prototypical EPEC 1 strain E2348/69 was determined (11). In comparison to the EAF of O111:NM strain B171, the primary difference is the presence of the tra locus in pMAR7 (EPEC 1, EAF type 1) and its absence from pB171 (EPEC 2, EAF type 2). The tra genes, which are responsible for conjugal transfer in plasmids F and R100, were found to have varying degrees of conservation among the EAF plasmids from different EPEC strains (11). This finding has intriguing implications for our results. In addition to EAF type 2, plasmid types 5, 6, 9, and 11 were all found in only a single ST. Like pB171, these plasmids may lack the entire tra locus or possess defective conjugation machinery, thereby preventing their transfer to other EPEC clones. In contrast to the distribution of the EAF plasmid types, the distribution of eae alleles among EPEC strains is more consistent with their clonal relationships. This suggests that the EAF plasmid is more mobile than the LEE island.
Inferred evolution of EAF plasmids and EPEC clones.
Based on the MLST and EAF plasmid type data, ancestral or primitive clonal types within the four main EPEC groups can be inferred under the parsimony principle, that is, positing a simple evolution model based on minimizing the number of evolutionary genetic events. The principal events in the evolutionary change of an ancestral EPEC clone are EAF plasmid recombination, plasmid replacement, and plasmid loss. Here, we define recombination of the EAF plasmid as a change in bfpA allele, since each bfpA allele is associated with a single perA allele class. Plasmid replacement, presumably resulting from the horizontal transfer of an EAF plasmid, is believed to have occurred when both bfpA and perA differ from the primitive condition. Plasmid loss is inferred when an isolate is PCR negative for both bfpA and perA.
Under the parsimony principle, we have deduced the types of genetic events underlying the evolution of each of the EPEC groups (Table 7). The EPEC 3 group is the most homogenous, with only plasmid loss being inferred. Two of the three possible plasmid changes were found in the EPEC 1 and EPEC 4 clonal groups. For EPEC 1, the inferred ancestral type possessed EAF type 1 (α1-bfpA, α-perA). Plasmid loss was not observed, but plasmid recombination (EAF type 8; β5-bfpA, α-perA) and replacement (EAF type 6; β3-bfpA, γ-perA) have occurred. For EPEC 4, plasmid recombination was not observed, but plasmid loss and replacement were detected. EAF type 3 (α3-bfpA, β-perA) was replaced with EAF type 5 (β2-bfpA, δ-perA) and with EAF type 8 (β5-bfpA, α-perA). EPEC 2 is the most variable clonal group. With an inferred ancestral state of EAF type 4 (β1-bfpA, α-perA), all three types of plasmid changes were observed. Two different recombination events involving EAF type 2 (α2-bfpA, α-perA) and EAF type 7 (β4-bfpA, α-perA), plasmid replacement with EAF type 11 (bfpA negative, γ-perA), and plasmid loss have shaped the diversity of this clonal group.
TABLE 7.
EAF plasmid changes within four common EPEC clonal groups
| Clonal group | Recombination (change in bfpA) | Plasmid replacement (change in bfpA and perA) | Plasmid loss |
|---|---|---|---|
| EPEC 1 | EAF 1 to EAF 8 | EAF 1 to EAF 6 | Not observed |
| EPEC 2 | EAF 4 to EAF 2 or EAF 7 | EAF 4 to EAF 11 | Observed |
| EPEC 3 | Not observed | Not observed | Observed |
| EPEC 4 | Not observed | EAF 3 to EAF 5 or EAF 8 | Observed |
EAF type 11 is unusual in that it was the only plasmid type that did not contain bfpA according to our PCR screening. Bortolini and colleagues (10) reported a similar finding when they described EAF+ O119:H2 and O128:H2 strains in which most (∼13 kb) of the bfp operon had been deleted and replaced with an IS66-like element. We suspected that since the deletion encompasses the 3′ end of bfpA, this could explain why plasmid type 11 is bfpA negative. To confirm this, we designed new primers to target the 5′ end of bfpA and the IS66-like element. All EAF type 11 strains yielded the expected amplicon, indicating that this plasmid type possesses a similar bfp operon structure as that described by Bortolini and colleagues (10). Aside from the EAF type 11 strains, 17 additional isolates were bfpA negative. These strains, however, were also perA negative, suggesting that they did not possess the EAF plasmid. Of the EAF-negative isolates, most (82%) were part of the EPEC 2 clonal group, with the O128:H2 serotype being the most common. This finding was not unexpected, as O128:H2 strains are often reported to be EAF negative and, therefore, classified as atypical EPEC (66).
Relationship between typical and atypical EPEC.
Our work shows that at least some atypical EPEC strains, such as those that lost both bfpA and perA (and presumably the whole EAF plasmid), evolved from typical EPEC, rather than typical EPEC evolving from atypical EPEC by acquisition of the plasmid. It has been shown that typical EPEC can lose the EAF plasmid at a surprisingly high rate during passage through adult volunteers (19, 38), and so there appears to be selective pressure to lose the plasmid and convert from typical to atypical EPEC. This is interesting, given the recent reports of atypical EPEC in human clinical isolates, some of which belong to typical EPEC O-serogroups (3, 5, 49).
Concluding remarks.
The evolution of EPEC appears to be a dynamic process involving repeated acquisition of the LEE island and transfer of the EAF plasmid. The work presented here is the first to classify EAF plasmids into types based on bfpA and perA allelic data, and we identified 11 distinct plasmid types among the EPEC strains investigated. Nevertheless, it remains unclear what level of conservation exists among plasmids of the same EAF type. Given the amount of IS elements present within the two fully sequenced EAF plasmids (11, 64), there could be considerable variation within each plasmid type, and further characterization of the EAF types is warranted. Another area of future research could focus on the insertion site of the LEE island. Although it is known that the EPEC 1 LEE is within the selC site, whereas EPEC 2 has LEE inserted into pheU (61, 71), the insertion sites of the LEE island in the other EPEC clonal types remain to be elucidated. The further characterization of pathogenic strains will improve our understanding of the processes that underlie microbial evolution. The identification of unique genetic determinants in these strains may then be used to facilitate the detection of specific epidemic clones during outbreaks of disease.
Supplementary Material
Acknowledgments
We thank Lindsey Ouellette and Weihong Qi for technical assistance and Shannon Manning for comments on a previous version of the manuscript.
This project has been funded in part with federal funds from the NIAID, NIH, DHHS, under NIH research contract N01-AI-30058 (T.S.W.) and NIH research grant AI-37606 (M.S.D.).
Footnotes
Published ahead of print on 10 November 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adu-Bobie, J., G. Frankel, C. Bain, A. G. Goncalves, L. R. Trabulsi, G. Douce, S. Knutton, and G. Dougan. 1998. Detection of intimins α, β, γ, and δ four intimin derivatives expressed by attaching and effacing microbial pathogens. J. Clin. Microbiol. 36:662-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert, M. J., K. Alam, M. Ansaruzzaman, J. Montanaro, M. Islam, S. M. Faruque, K. Haider, K. Bettelheim, and S. Tzipori. 1991. Localized adherence and attaching-effacing properties of nonenteropathogenic serotypes of Escherichia coli. Infect. Immun. 59:1864-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alikhani, M. Y., A. Mirsalehian, and M. M. Aslani. 2006. Detection of typical and atypical enteropathogenic Escherichia coli (EPEC) in Iranian children with and without diarrhoea. J. Med. Microbiol. 55:1159-1163. [DOI] [PubMed] [Google Scholar]
- 4.Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. [DOI] [PubMed] [Google Scholar]
- 5.Blanco, M., J. E. Blanco, G. Dahbi, M. P. Alonso, A. Mora, M. A. Coira, C. Madrid, A. Juarez, M. I. Bernardez, E. A. Gonzalez, and J. Blanco. 2006. Identification of two new intimin types in atypical enteropathogenic Escherichia coli. Int. Microbiol. 9:103-110. [PubMed] [Google Scholar]
- 6.Blanco, M., J. E. Blanco, G. Dahbi, A. Mora, M. P. Alonso, G. Varela, M. P. Gadea, F. Schelotto, E. A. Gonzalez, and J. Blanco. 2006. Typing of intimin (eae) genes from enteropathogenic Escherichia coli (EPEC) isolated from children with diarrhoea in Montevideo, Uruguay: identification of two novel intimin variants (μB and ξR/β2B). J. Med. Microbiol. 55:1165-1174. [DOI] [PubMed] [Google Scholar]
- 7.Blank, T. E., D. W. Lacher, I. C. Scaletsky, H. Zhong, T. S. Whittam, and M. S. Donnenberg. 2003. Enteropathogenic Escherichia coli O157 strains from Brazil. Emerg. Infect. Dis. 9:113-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blank, T. E., H. Zhong, A. L. Bell, T. S. Whittam, and M. S. Donnenberg. 2000. Molecular variation among type IV pilin (bfpA) genes from diverse enteropathogenic Escherichia coli strains. Infect. Immun. 68:7028-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bokete, T. N., T. S. Whittam, R. A. Wilson, C. R. Clausen, C. M. O'Callahan, S. L. Moseley, T. R. Fritsche, and P. I. Tarr. 1997. Genetic and phenotypic analysis of Escherichia coli with enteropathogenic characteristics isolated from Seattle children. J. Infect. Dis. 175:1382-1389. [DOI] [PubMed] [Google Scholar]
- 10.Bortolini, M. R., L. R. Trabulsi, R. Keller, G. Frankel, and V. Sperandio. 1999. Lack of expression of bundle-forming pili in some clinical isolates of enteropathogenic Escherichia coli (EPEC) is due to a conserved large deletion in the bfp operon. FEMS Microbiol. Lett. 179:169-174. [DOI] [PubMed] [Google Scholar]
- 11.Brinkley, C., V. Burland, R. Keller, D. J. Rose, A. T. Boutin, S. A. Klink, F. R. Blattner, and J. B. Kaper. 2006. Nucleotide sequence analysis of the enteropathogenic Escherichia coli adherence factor plasmid pMAR7. Infect. Immun. 74:5408-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broes, A., R. Drolet, M. Jacques, J. M. Fairbrother, and W. M. Johnson. 1988. Natural infection with an attaching and effacing Escherichia coli in a diarrheic puppy. Can. J. Vet. Res. 52:280-282. [PMC free article] [PubMed] [Google Scholar]
- 13.Bruen, T. C., H. Philippe, and D. Bryant. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryant, D., and V. Moulton. 2004. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21:255-265. [DOI] [PubMed] [Google Scholar]
- 15.Cleary, J., L. C. Lai, R. K. Shaw, A. Straatman-Iwanowska, M. S. Donnenberg, G. Frankel, and S. Knutton. 2004. Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: role of bundle-forming pili (BFP), EspA filaments and intimin. Microbiol. 150:527-538. [DOI] [PubMed] [Google Scholar]
- 16.Cravioto, A., R. J. Gross, S. M. Scotland, and B. Rowe. 1979. An adhesive factor found in strains of Escherichia coli belonging to the traditional infantile enteropathogenic serotypes. Curr. Microbiol. 3:95-99. [Google Scholar]
- 17.Cravioto, A., A. Tello, A. Navarro, J. Ruiz, H. Villafan, F. Uribe, and C. Eslava. 1991. Association of Escherichia coli HEp-2 adherence patterns with type and duration of diarrhoea. Lancet 337:262-264. [DOI] [PubMed] [Google Scholar]
- 18.Donnenberg, M. S., J. A. Giron, J. P. Nataro, and J. B. Kaper. 1992. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol. Microbiol. 6:3427-3437. [DOI] [PubMed] [Google Scholar]
- 19.Donnenberg, M. S., C. O. Tacket, S. P. James, G. Losonsky, J. P. Nataro, S. S. Wasserman, J. B. Kaper, and M. M. Levine. 1993. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J. Clin. Investig. 92:1412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donnenberg, M. S., H. Z. Zhang, and K. D. Stone. 1997. Biogenesis of the bundle-forming pilus of enteropathogenic Escherichia coli: reconstitution of fimbriae in recombinant E. coli and role of DsbA in pilin stability. Gene 192:33-38. [DOI] [PubMed] [Google Scholar]
- 21.Drolet, R., J. M. Fairbrother, J. Harel, and P. Helie. 1994. Attaching and effacing and enterotoxigenic Escherichia coli associated with enteric colibacillosis in the dog. Can. J. Vet. Res. 58:87-92. [PMC free article] [PubMed] [Google Scholar]
- 22.Elliott, S. J., V. Sperandio, J. A. Giron, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 24.Finlay, B. B., I. Rosenshine, M. S. Donnenberg, and J. B. Kaper. 1992. Cytoskeletal composition of attaching and effacing lesions associated with enteropathogenic Escherichia coli adherence to HeLa cells. Infect. Immun. 60:2541-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franzolin, M. R., R. C. Alves, R. Keller, T. A. Gomes, L. Beutin, M. L. Barreto, C. Milroy, A. Strina, H. Ribeiro, and L. R. Trabulsi. 2005. Prevalence of diarrheagenic Escherichia coli in children with diarrhea in Salvador, Bahia, Brazil. Mem. Inst. Oswaldo Cruz 100:359-363. [DOI] [PubMed] [Google Scholar]
- 26.Ghilardi, A. C., T. A. Gomes, W. P. Elias, and L. R. Trabulsi. 2003. Virulence factors of Escherichia coli strains belonging to serogroups O127 and O142. Epidemiol. Infect. 131:815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomez-Duarte, O. G., and J. B. Kaper. 1995. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 63:1767-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goncalves, A. G., L. C. Campos, T. A. Gomes, J. Rodrigues, V. Sperandio, T. S. Whittam, and L. R. Trabulsi. 1997. Virulence properties and clonal structures of strains of Escherichia coli O119 serotypes. Infect. Immun. 65:2034-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guth, B. E., R. Giraldi, T. A. Gomes, and L. R. Marques. 1994. Survey of cytotoxin production among Escherichia coli strains characterized as enteropathogenic (EPEC) by serotyping and presence of EPEC adherence factor (EAF) sequences. Can. J. Microbiol. 40:341-344. [DOI] [PubMed] [Google Scholar]
- 30.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 31.Kaper, J. B. 1994. Molecular pathogenesis of enteropathogenic Escherichia coli, p. 173-195. In V. L. Miller, J. B. Kaper, D. A. Portnoy, and R. R. Isberg (ed.), Molecular genetics of bacterial pathogenesis. American Society for Microbiology, Washington, D.C.
- 32.Kennedy, D. H., G. H. Walker, R. J. Fallon, J. F. Boyd, R. J. Gross, and B. Rowe. 1973. An outbreak of infantile gastroenteritis due to E. coli O142. J. Clin. Pathol. 26:731-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenny, B. 2002. Mechanism of action of EPEC type III effector molecules. Int. J. Med. Microbiol. 291:469-477. [DOI] [PubMed] [Google Scholar]
- 34.Knutton, S., T. Baldwin, P. H. Williams, and A. S. McNeish. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 36.Lacher, D. W., H. Steinsland, and T. S. Whittam. 2006. Allelic subtyping of the intimin locus (eae) of pathogenic Escherichia coli by fluorescent RFLP. FEMS Microbiol. Lett. 261:80-87. [DOI] [PubMed] [Google Scholar]
- 37.Levine, M. M., and R. Edelman. 1984. Enteropathogenic Escherichia coli of classic serotypes associated with infant diarrhea: epidemiology and pathogenesis. Epidemiol. Rev. 6:31-51. [DOI] [PubMed] [Google Scholar]
- 38.Levine, M. M., J. P. Nataro, H. Karch, M. M. Baldini, J. B. Kaper, R. E. Black, M. L. Clements, and A. D. O'Brien. 1985. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J. Infect. Dis. 152:550-559. [DOI] [PubMed] [Google Scholar]
- 39.Luo, Y., E. A. Frey, R. A. Pfuetzner, A. L. Creagh, D. G. Knoechel, C. A. Haynes, B. B. Finlay, and N. C. Strynadka. 2000. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature 405:1073-1077. [DOI] [PubMed] [Google Scholar]
- 40.Makino, S., H. Asakura, T. Shirahata, T. Ikeda, K. Takeshi, K. Arai, M. Nagasawa, T. Abe, and T. Sadamoto. 1999. Molecular epidemiological study of a mass outbreak caused by enteropathogenic Escherichia coli O157:H45. Microbiol. Immunol. 43:381-384. [DOI] [PubMed] [Google Scholar]
- 41.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 43.Mellies, J. L., F. Navarro-Garcia, I. Okeke, J. Frederickson, J. P. Nataro, and J. B. Kaper. 2001. espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect. Immun. 69:315-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moyenuddin, M., I. K. Wachsmuth, S. L. Moseley, C. A. Bopp, and P. A. Blake. 1989. Serotype, antimicrobial resistance, and adherence properties of Escherichia coli strains associated with outbreaks of diarrheal illness in children in the United States. J. Clin. Microbiol. 27:2234-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nataro, J. P., M. M. Baldini, J. B. Kaper, R. E. Black, N. Bravo, and M. M. Levine. 1985. Detection of an adherence factor of enteropathogenic Escherichia coli with a DNA probe. J. Infect. Dis. 152:560-565. [DOI] [PubMed] [Google Scholar]
- 46.Nataro, J. P., J. B. Kaper, R. Robins-Browne, V. Prado, P. Vial, and M. M. Levine. 1987. Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 cells. Pediatr. Infect. Dis. J. 6:829-831. [DOI] [PubMed] [Google Scholar]
- 47.Nataro, J. P., K. O. Maher, P. Mackie, and J. B. Kaper. 1987. Characterization of plasmids encoding the adherence factor of enteropathogenic Escherichia coli. Infect. Immun. 55:2370-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nataro, J. P., I. C. Scaletsky, J. B. Kaper, M. M. Levine, and L. R. Trabulsi. 1985. Plasmid-mediated factors conferring diffuse and localized adherence of enteropathogenic Escherichia coli. Infect. Immun. 48:378-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen, R. N., L. S. Taylor, M. Tauschek, and R. M. Robins-Browne. 2006. Atypical enteropathogenic Escherichia coli infection and prolonged diarrhea in children. Emerg. Infect. Dis. 12:597-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okeke, I. N., J. A. Borneman, S. Shin, J. L. Mellies, L. E. Quinn, and J. B. Kaper. 2001. Comparative sequence analysis of the plasmid-encoded regulator of enteropathogenic Escherichia coli strains. Infect. Immun. 69:5553-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okeke, I. N., A. Lamikanra, H. Steinruck, and J. B. Kaper. 2000. Characterization of Escherichia coli strains from cases of childhood diarrhea in provincial southwestern Nigeria. J. Clin. Microbiol. 38:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ørskov, F., T. S. Whittam, A. Cravioto, and I. Ørskov. 1990. Clonal relationships among classic enteropathogenic Escherichia coli (EPEC) belong to different O groups. J. Infect. Dis. 162:76-81. [DOI] [PubMed] [Google Scholar]
- 53.Paulozzi, L. J., K. E. Johnson, L. M. Kamahele, C. R. Clausen, L. W. Riley, and S. D. Helgerson. 1986. Diarrhea associated with adherent enteropathogenic Escherichia coli in an infant and toddler center, Seattle, Washington. Pediatrics 77:296-300. [PubMed] [Google Scholar]
- 54.Pond, S. L., and S. D. Frost. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531-2533. [DOI] [PubMed] [Google Scholar]
- 55.Reid, S. D., D. J. Betting, and T. S. Whittam. 1999. Molecular detection and identification of intimin alleles in pathogenic Escherichia coli by multiplex PCR. J. Clin. Microbiol. 37:2719-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robins-Browne, R. M., W. C. Yam, L. E. O'Gorman, and K. A. Bettelheim. 1993. Examination of archetypal strains of enteropathogenic Escherichia coli for properties associated with bacterial virulence. J. Med. Microbiol. 38:222-226. [DOI] [PubMed] [Google Scholar]
- 57.Rodrigues, J., I. C. Scaletsky, L. C. Campos, T. A. Gomes, T. S. Whittam, and L. R. Trabulsi. 1996. Clonal structure and virulence factors in strains of Escherichia coli of the classic serogroup O55. Infect. Immun. 64:2680-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rothbaum, R., A. J. McAdams, R. Giannella, and J. C. Partin. 1982. A clinicopathologic study of enterocyte-adherent Escherichia coli: a cause of protracted diarrhea in infants. Gastroenterology 83:441-454. [PubMed] [Google Scholar]
- 59.Schremmer, C., J. E. Lohr, U. Wastlhuber, J. Kosters, K. Ravelshofer, H. Steinruck, and L. H. Wieler. 1999. Enteropathogenic Escherichia coli in Psittaciformes. Avian Pathol. 28:349-354. [DOI] [PubMed] [Google Scholar]
- 60.Sohel, I., J. L. Puente, W. J. Murray, J. Vuopio-Varkila, and G. K. Schoolnik. 1993. Cloning and characterization of the bundle-forming pilin gene of enteropathogenic Escherichia coli and its distribution in Salmonella serotypes. Mol. Microbiol. 7:563-575. [DOI] [PubMed] [Google Scholar]
- 61.Sperandio, V., J. B. Kaper, M. R. Bortolini, B. C. Neves, R. Keller, and L. R. Trabulsi. 1998. Characterization of the locus of enterocyte effacement (LEE) in different enteropathogenic Escherichia coli (EPEC) and Shiga-toxin producing Escherichia coli (STEC) serotypes. FEMS Microbiol. Lett. 164:133-139. [DOI] [PubMed] [Google Scholar]
- 62.Stephan, R., N. Borel, C. Zweifel, M. Blanco, and J. E. Blanco. 2004. First isolation and further characterization of enteropathogenic Escherichia coli (EPEC) O157:H45 strains from cattle. BMC Microbiol. 4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stone, K. D., H. Z. Zhang, L. K. Carlson, and M. S. Donnenberg. 1996. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for the biogenesis of a type IV pilus. Mol. Microbiol. 20:325-337. [DOI] [PubMed] [Google Scholar]
- 64.Tobe, T., T. Hayashi, C. G. Han, G. K. Schoolnik, E. Ohtsubo, and C. Sasakawa. 1999. Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. Infect. Immun. 67:5455-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tobe, T., G. K. Schoolnik, I. Sohel, V. H. Bustamante, and J. L. Puente. 1996. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 21:963-975. [DOI] [PubMed] [Google Scholar]
- 66.Trabulsi, L. R., R. Keller, and T. A. Tardelli Gomes. 2002. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8:508-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valentiner-Branth, P., H. Steinsland, T. K. Fischer, M. Perch, F. Scheutz, F. Dias, P. Aaby, K. Molbak, and H. Sommerfelt. 2003. Cohort study of Guinean children: incidence, pathogenicity, conferred protection, and attributable risk for enteropathogens during the first 2 years of life. J. Clin. Microbiol. 41:4238-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vallance, B. A., and B. B. Finlay. 2000. Exploitation of host cells by enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 97:8799-8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Whittam, T. S., and E. A. McGraw. 1996. Clonal analysis of EPEC serogroups. Rev. Microbiol. 27(Suppl. 1):7-16. [Google Scholar]
- 70.Whittam, T. S., and R. A. Wilson. 1988. Genetic relationships among pathogenic Escherichia coli of serogroup O157. Infect. Immun. 56:2467-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wieler, L. H., T. K. McDaniel, T. S. Whittam, and J. B. Kaper. 1997. Insertion site of the locus of enterocyte effacement in enteropathogenic and enterohemorrhagic Escherichia coli differs in relation to the clonal phylogeny of the strains. FEMS Microbiol. Lett. 156:49-53. [DOI] [PubMed] [Google Scholar]
- 72.Zhang, W. L., B. Kohler, E. Oswald, L. Beutin, H. Karch, S. Morabito, A. Caprioli, S. Suerbaum, and H. Schmidt. 2002. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J. Clin. Microbiol. 40:4486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.