Abstract
The switch complex at the base of the bacterial flagellum is essential for flagellar assembly, rotation, and switching. In Escherichia coli and Salmonella, the complex contains about 26 copies of FliG, 34 copies of FliM, and more then 100 copies of FliN, together forming the basal body C ring. FliG is involved most directly in motor rotation and is located in the upper (membrane-proximal) part of the C ring. A crystal structure of the middle and C-terminal parts of FliG shows two globular domains connected by an α-helix and a short extended segment. The middle domain of FliG has a conserved surface patch formed by the residues EHPQ125-128 and R160 (the EHPQR motif), and the C-terminal domain has a conserved surface hydrophobic patch. To examine the functional importance of these and other surface features of FliG, we made mutations in residues distributed over the protein surface and measured the effects on flagellar assembly and function. Mutations preventing flagellar assembly occurred mainly in the vicinity of the EHPQR motif and the hydrophobic patch. Mutations causing aberrant clockwise or counterclockwise motor bias occurred in these same regions and in the waist between the upper and lower parts of the C-terminal domain. Pull-down assays with glutathione S-transferase-FliM showed that FliG interacts with FliM through both the EHPQR motif and the hydrophobic patch. We propose a model for the organization of FliG and FliM subunits that accounts for the FliG-FliM interactions identified here and for the different copy numbers of FliG and FliM in the flagellum.
Bacterial flagella are built from about 25 proteins, most of which serve structural roles in forming the basal body, hook, and filament and only a few of which function in rotation (2, 23, 32). The stator is formed from the membrane proteins MotA and MotB, which form complexes with the composition MotA4MotB2 (9, 11, 25, 42), in the membrane surrounding the basal body (21). Each motor contains several MotA4MotB2 complexes, which can function independently to produce torque (4, 5, 41). The rotor proteins involved in rotation are FliG, FliM, and FliN. These form a large (ca. 4-MDa) assemblage termed the switch complex that is essential for flagellar assembly, rotation, and clockwise/counterclockwise (CW/CCW) switching (12, 57, 59). The switch complex is attached to the cytoplasmic face of the basal body MS ring, a large membrane-embedded structure formed from about 26 copies of the protein FliF (19, 53) (Fig. 1). FliF has two membrane-traversing segments and includes sizable domains in both the periplasm and cytoplasm. A conserved segment near the C terminus of FliF, located in the cytoplasm, forms the site of attachment for FliG (16).
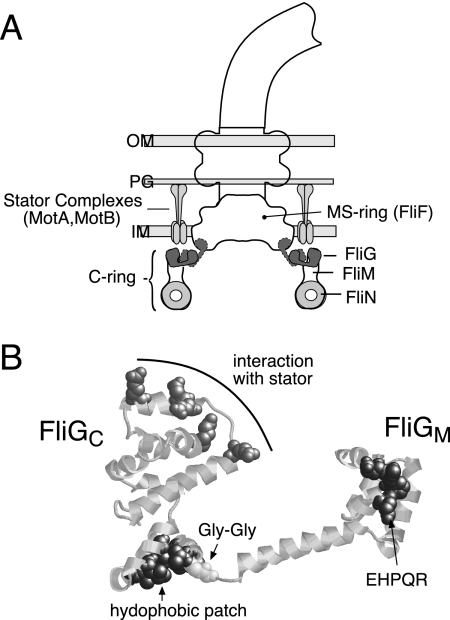
FIG. 1.
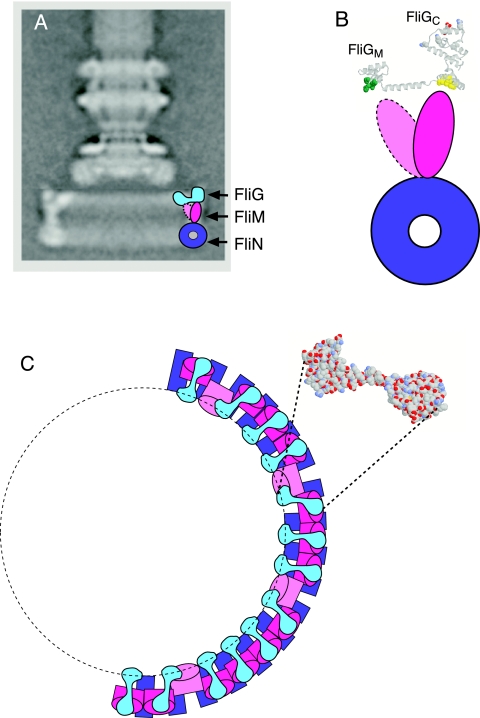
(A) Locations of proteins involved in flagellar rotation. The location of FliN is deduced from targeted cross-linking studies (39) and electron microscopic reconstructions (12, 48, 50, 58). Although FliG is known to be in the upper part of the C ring (16, 22, 26, 37, 61) and some features of its organization have been deduced from cross-linking (31), its exact location is not yet certain. Accordingly, two possible locations for FliG are indicated. OM, outer membrane; PG, peptidoglycan; IM, inner membrane. (B) Structure of residues 115 to 327 of T. maritima FliG (FliGMC) (7), highlighting conserved surface features.
Although each of the switch complex proteins performs multiple functions, mutational studies indicate that each is specialized to some extent. FliM is closely involved in direction switching and contains a segment near its N terminus that binds to the chemotaxis-signaling molecule phospho-CheY (6, 43). FliN is also involved in switching and makes a particularly important contribution to flagellar assembly, probably by binding to the flagellar export protein FliH (8, 15, 36a, 40, 55). FliG is involved most directly in rotation (20, 29). The C-terminal domain of FliG (FliGC) in particular functions specifically in rotation; deletion of most of this domain prevents rotation but allows flagellar assembly (29). Conserved charged residues in FliGC have been found to be important for flagellar rotation in a number of species (28, 56) and for control of speed modulation (chemokinesis) in the unidirectional rotary motor of Sinorhizobium meliloti (1). These charged residues of FliG were shown to interact with conserved charged residues in the cytoplasmic domain of the stator protein MotA (56, 60, 61). FliG also interacts with FliM (22, 34, 36, 47) and with the nucleoid-associated DNA-binding protein H-NS (35).
While the mechanism of flagellar rotation is not yet understood at a detailed level, some key features of the mechanism have been established. Energy for rotation comes from the transmembrane gradient of protons (14, 27, 33) or sodium ions (18). Recent studies support a mechanism in which the energizing ions flow through the stator complexes and drive conformational changes as they bind to and dissociate from a critical aspartate residue in MotB (3, 24, 62). We hypothesize that these conformational changes provide the power stroke that drives rotation, applying force to the rotor via the MotA-FliG interface identified in mutational studies (28, 56, 60, 61).
Although the stator appears to be the instigator of movement, the rotor has the major role in controlling the direction of rotation. Mutational replacements that affect the CW/CCW bias of the motor are fairly common in the switch complex proteins but not in MotA or MotB (13, 20, 43). The molecular events that underlie switching have not been defined but must presumably involve some movement of the C-terminal domain of FliG, to alter the way in which the rotor engages the stator.
Recent studies have begun to provide detailed structural information on the components of the switch complex and their overall organization. The basal body has been imaged in several electron microscopic studies (12, 48, 58), most recently with resolution sufficient to see domain-sized features within the C ring (49). Crystal structures have been solved for most of FliN (8), most of FliM (38), and most of FliG (the middle and C-terminal domains, termed FliGMC) (7, 30). The FliGMC structure shows two compact globular domains joined by an α-helix and a short linker with extended structure (7). The linker includes two well-conserved Gly residues and might therefore be flexible. The functionally important charged residues of FliG cluster on a ridge in the C-terminal domain. In the flagellum, this ridge must be positioned to allow interaction with the charged residues in the cytoplasmic domain of MotA (7, 61).
Besides the charge-bearing ridge, other conserved features of FliGMC include a surface hydrophobic patch on the end of the C-terminal domain opposite the charge-bearing ridge and the residues EHPQ125-128 and R160 (the EHPQR motif) on the surface of the middle domain (Fig. 1). These conserved features might provide binding sites for FliM or other binding partners of FliG. The structure of FliGMC led to the suggestion that switching might occur by relative movement of the two globular domains, with the Gly-Gly linker serving as a hinge (7). Mutations in the linker and nearby residues were subsequently shown to alter both the switching rate and CW/CCW bias of the motor (13, 54), in support of this model. We recently examined the arrangement of FliG subunits in the flagellum by targeted cross-linking experiments using introduced Cys residues (31). These experiments showed that FliG subunits are arranged adjacent to each other in the flagellum, with the charge-bearing ridge of the C-terminal domain oriented in an approximately radial direction (Fig. 1). However, the precise location of FliG in the flagellum and its relationship to the other switch complex proteins remain uncertain.
Here, we have undertaken a systematic mutational study of surface residues of FliGMC, to identify functionally important regions of the protein and examine their contributions to flagellar assembly and function. Tryptophan replacements were made at positions sampling the surface of FliGMC, and the effects on flagellar assembly and rotation were measured. Trp replacements were used because the large Trp side chain is likely to alter normal protein-protein associations (and thus compromise function) when it is inserted into a functionally important contact surface. Flagellar assembly and switching were affected strongly by mutations in the EHPQR motif or hydrophobic patch, confirming the functional importance of these regions. Pull-down assays showed that the EHPQR motif and hydrophobic patch are both involved in interactions with FliM. These results, together with previous mutational and electron microscopic studies, lead to a model for the arrangement of FliG subunits and their relationship to FliM subunits in the flagellar switch complex.
MATERIALS AND METHODS
Strains and mutagenesis.
Escherichia coli RP437 (wild type for motility and chemotaxis) and RP3098 (ΔflhDC) were gifts from J. S. Parkinson (University of Utah). FlhD and FlhC are master regulators of flagellar gene expression, and the deletion of these in strain RP3098 prevents the expression of all chromosomal flagellar genes. The fliG-null strain DFB225 contains an in-frame deletion of most of fliG (29). Site-directed mutagenesis used the Altered Sites procedure (Promega) on the fliG gene cloned in plasmid pSL27 (29), a derivative of pAlter-1 (Promega). The pSL27 derivatives encoding mutant variants of FliG confer ampicillin resistance. Plasmid pDFB96 is a pACYC184 derivative that expresses FliM and FliN from the tac promoter and confers chloramphenicol resistance (29). pDFB66 is a pACYC184 derivative that expresses CheY from the ara promoter and confers chloramphenicol resistance (40). pHT97 encodes a glutathione S-transferase (GST)-FliG fusion protein and confers kanamycin resistance, and pHT100 is the corresponding GST-only control (47).
Function of the mutant proteins.
To assay the effects of Trp replacement mutations, cells of DFB225 were transformed with pSL27 derivatives encoding the Trp mutant proteins, and fresh transformants were cultured with shaking at 32°C in TB-Ap (10 g/liter tryptone, 5 g/liter NaCl, 100 μg/ml ampicillin) to mid-exponential phase. One microliter of each culture was spotted onto swarm plates (TB solidified with 0.28% Bacto-agar) and incubated at 32°C. Swarm diameters were measured at regular intervals, and plots of diameter versus time were used to determine swarming rates. Swarming rates of the mutants are relative to wild-type controls. Plasmid pSL27 carrying wild-type fliG restored full swarming ability to the fliG-null strain DFB225 (29), indicating that the level of FliG expression from this plasmid is sufficient for normal flagellar assembly and function.
For the analysis of swimming behavior, cells were picked from plates and grown to saturation at 32°C in TB-Ap. The cultures were diluted 100-fold into fresh medium and shaken at 32°C for approximately 4 h. Cells were diluted into motility medium (67 mM NaCl, 10 mM potassium phosphate [pH 7], 0.1 mM EDTA, 1 μM methionine, 5 mM sodium lactate) on a microscope slide and observed under a microscope with the stage temperature controlled at 32°C. Motility was scored visually and compared with that of wild-type cells prepared in the same way.
Mutants that failed to swarm or swim were stained by using the wet-mount procedure of Heimbrook et al. (17) to determine whether flagella were present. Wild-type controls were included, and staining experiments were done at least twice.
Dominance and overexpression effects.
To assay the dominance of the fliG mutations, pSL27 variants expressing the mutant FliG proteins were transformed into the wild-type strain RP437 and swarming rates were measured as described above. The control was RP437 transformed with wild-type pSL27.
To measure effects of overexpressed FliM and FliN on the nonflagellate fliG mutants, cells of strain DFB225 were transformed with pSL27 plasmids carrying the fliG variants that gave nonflagellate phenotype, with pDFB96 present to express FliM and FliN. Controls were transformed with the pSL27 variants and pACYC184. Swarming was examined in plates containing ampicillin, chloramphenicol, and 100 μM IPTG (isopropyl-β-d-thiogalactopyranoside) to induce expression of FliM and FliN.
To examine effects of CheY overexpression on the smooth-swimming fliG mutants, DFB225 was transformed with the fliG plasmids that conferred aberrantly smooth swimming and with pDFB66 to express CheY. Controls were transformed with the pSL27 variants and pACYC184, the parent plasmid of pDFB66. Swarming was examined in plates containing ampicillin, chloramphenicol, and 1.0 mM arabinose to induce CheY expression.
Binding assays.
Binding of FliG to FliM was measured using a GST pull-down assay, essentially as described by Tang et al. (47) and Mathews et al. (36) with minor modification. Levels of some of the mutant FliG proteins were found to be decreased by coexpression of the GST-FliM fusion protein in the same cells, and so the experiments used two strains, one expressing GST-FliM from plasmid pHT86 (47) and another expressing FliG or its mutant variants from plasmid pSL27 (29). Control experiments used GST only, expressed from plasmid pHT100 (47). The strain was BL21(DE3) (44). Cells were cultured overnight at 32°C in 40 ml of TB containing the appropriate antibiotics and 400 μM IPTG. Cells were harvested and resuspended in lysozyme containing buffer as described previously (36). Following a 1-h incubation on ice, the cells were further disrupted by sonication (Branson model 450 Sonifier; power of 3, duty cycle of 50%, 180 s). Debris was pelleted (16,000 × g, 30 min, 4°C), and 50 μl of the supernatant was saved for use in estimating the amount of FliM present before addition of affinity beads. The rest (∼1 ml) was transferred to a clean tube, mixed with 150 μl of a 50% slurry of glutathione-Sepharose 4B (Pharmacia) prepared according to the manufacturer's directions, and incubated for 75 min at room temperature with gentle rotation to allow binding. The Sepharose beads were then pelleted by a 10-s microcentrifuge spin, washed with 1 ml of phosphate-buffered saline, and pelleted again by a brief spin. The beads were then incubated with 50 μl of elution buffer (50 mM reduced glutathione in 50 mM Tris-HCl [pH 8]) for 10 min at room temperature with gentle rotation to release the GST-FliM and associated proteins. Beads were then pelleted, and the supernatant was collected for analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting using anti-FliG antibody (29).
RESULTS
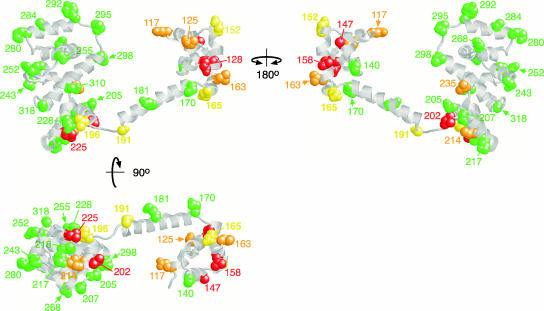
Trp replacements were made at 33 positions distributed over the surface of FliGMC. Plasmids expressing the mutant variants of FliG were transformed into the fliG-null strain DFB225, and function was assayed by measuring swarming in soft agar and motility in liquid. Immotile mutants were stained to see if flagella were assembled. The results are summarized in Table 1, and swarming phenotypes are mapped onto the FliG structure in Fig. 2. The structure is that of Thermotoga maritima FliG but the numbering used is for residues in the E. coli protein.
TABLE 1.
Effects of Trp replacements in FliGa
| Mutationb | Swarming ratec | Swimmingd | Flagellatione | Dominancef |
|---|---|---|---|---|
| None (wild type) | 1.0 | R/T | ||
| S117W | 0.02 | R | NM | 0.06 |
| E125W | 0.03 | R | NM | 0.05 |
| Q128W | 0.00 | Imm | − | 0.03 |
| A140W | 0.93 | R/T | NM | NM |
| A147W | 0.00 | Imm | −/+ | 0.04 |
| R152W | 0.28 | R | NM | 0.57 |
| M158W | 0.00 | Imm | − | 0.003 |
| T163W | 0.03 | R/t, w | +/− | 0.04 |
| G165W | 0.25 | r/T, w | +/− | 0.61 |
| A170W | 0.92 | R/T | NM | NM |
| G181W | 1.02 | R/T | NM | NM |
| S191W | 0.13 | R | NM | 0.41 |
| V196W | 0.30 | T | NM | 0.66 |
| I202W | 0.00 | Imm | − | 0.02 |
| I205W | 0.93 | R/T | NM | NM |
| K207W | 0.94 | R/T | NM | NM |
| V214W | 0.02 | R | NM | 0.09 |
| A217W | 0.97 | R/T | NM | NM |
| V218W | 0.93 | R/T | NM | NM |
| L225W | 0.0 (trails) | Imm | − | 0.39 |
| K228W | 0.75 | R/T | NM | NM |
| L235W | 0.06 | T | NM | 0.21 |
| D243W | 0.88 | r/T | NM | NM |
| Q252W | 0.83 | R/T | NM | NM |
| D255W | 0.73 | R/t | NM | NM |
| Q268W | 0.93 | R/T | NM | NM |
| Q280W | 0.75 | R/T | NM | NM |
| D284W | 1.04 | R/T | NM | NM |
| N292W | 0.78 | R/T | NM | NM |
| P295W | 1.00 | R/T | NM | NM |
| L298W | 1.02 | R/T | NM | NM |
| L310W | 1.06 | R/T | NM | NM |
| T318W | 1.09 | r/T | NM | NM |
Motility and flagellation were measured in cells of the fliG-null strain DFB225 transformed with mutant variants of pSL27.
Residue numbering is for E. coli FliG; numbers for T. maritima FliG are greater by one (for positions 117 to 181) or by two (for positions 191 and higher).
Rates are relative to that of a wild-type control strain included on the plates.
R/T, normal pattern of runs and tumbles; R, smooth swimming; T, tumbly; R/t, run biased but some tumbles noted; r/T, tumble biased but short runs noted; w, motility weak; Imm, immotile.
−, nonflagellate; +/−, one or two flagella per cell; −/+, the majority of cells are nonflagellate but a rare cell has one flagellum; NM, not measured.
Swarming rate of wild-type (RP437) cells transformed with the mutant FliG plasmids, relative to controls expressing wild-type FliG from the plasmid.
FIG. 2.
Locations of the Trp replacement mutations on FliGMC and their swarming phenotypes. Green, swarming rate (relative to wild type) of 0.7 or better; yellow, relative rate of between 0.1 and 0.3; orange, rate nonzero but less than 0.1; red, nonswarming.
Regions important for flagellar assembly.
The wild type had an average of about five flagella per cell. Flagellar assembly appeared to be completely disrupted by Trp replacements at positions 128 and 158 in the middle domain and at positions 202 and 225 in the C-terminal domain (Table 1). Cells of these mutants were immotile in liquid media and were nonflagellate. On soft-agar plates, the 128W, 158W, and 202W mutants failed to swarm, while the 225W mutant gave rise to small numbers of satellite microcolonies after prolonged incubation (not shown), indicating the rare occurrence of motile cells. Residue 128 is Q of the EHPQR motif, and residues 202 and 225 are in the hydrophobic patch on the C-terminal domain. Met158 is in a shallow hydrophobic cleft in the middle domain, on a face distant from the EHPQR motif (Fig. 2).
Flagellation was decreased but not eliminated by Trp replacements in residues 147, 163, and 165. Most cells of these mutants lacked flagella, but a few had one or two. Consistent with the poor flagellation, these mutants appeared to be either immotile (position 147) or weakly motile (positions 163 and 165) in liquid medium. Residue 163 is adjacent to the Arg residue of the EHPQR motif. Residues 147 and 165 are more distant from the EHPQR motif, on the same face as residue 158 (Fig. 2).
Defects in switching.
Several of the Trp mutants were flagellate and motile but swarmed poorly owing to an aberrantly CCW or CW motor bias (Table 1). Smooth swimming, which is characteristic of exclusively CCW motor rotation, was seen for the Trp replacements at positions 117, 125, and 214. Residue 125 is E of the EHPRQ motif, and residue 214 is a V in the hydrophobic patch. Residue 117 is in the middle domain but somewhat (about 10 Å) apart from the EHPQR motif (Fig. 2). Milder swarming defects associated with a less severe CCW bias occurred for the mutations at position 152, which is near the margin of the EHPQR motif, and position 191, on the interdomain helix near the Gly-Gly linker. Tumbly motility indicative of CW motor bias was seen for mutations at position 196 in the hydrophobic patch and position 235 in the relatively narrow “waist” between the upper and lower parts of the C-terminal domain (Fig. 2).
Regions that tolerate Trp replacements.
Most (19 out of 33) Trp replacement mutants swarmed at 70% of the wild-type rate or better, indicating nearly normal function of the mutant FliG proteins in assembly, rotation, and switching. Positions where Trp was tolerated occur in the interdomain helix at the end attaching to the middle domain, on and around the charge-bearing ridge, and in several positions near the bottom of FliGC but outside the hydrophobic patch. The functionally important region at the bottom of FliGC is delineated by five positions that tolerated Trp replacement (205, 207, 217, 218, and 228) surrounding four positions that did not (196, 202, 214, and 225) (Fig. 2). In the middle domain, Trp was fully tolerated at only one position, residue 140, on the face of the domain opposite the EHPQR motif.
Dominance and overexpression effects.
To examine dominance of the Trp replacement mutants, plasmids encoding the FliG mutant variants were introduced into wild-type cells and swarming rates were measured. All of the strong switch bias mutations (at positions 117, 125, 214, and 235) exhibited strong dominance in this assay, indicating that the mutant proteins can be incorporated into flagellar motors and impose an aberrant switch bias (Table 1). Most of the Fla− mutations also were dominant, reducing swarming rates of the wild type to less than 5% of normal. One exception was the mutation at position 225, which as noted above also allowed infrequent flagellar assembly (as evidenced by satellite colonies in swarm plates).
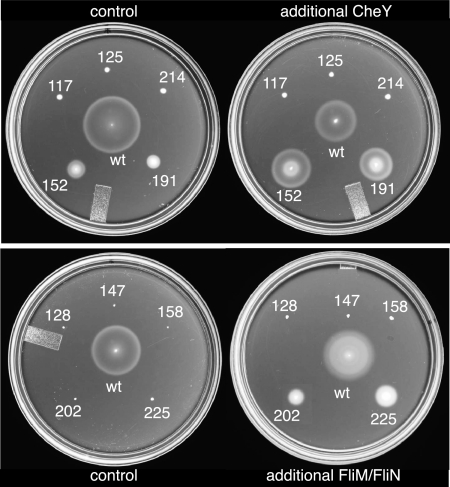
FliG mutations might affect flagellar assembly or function by impeding the installation of FliM. To determine whether flagellar assembly in the mutants could be improved by increasing the amount of the other switch complex proteins, cells of the poorly flagellated and nonflagellated mutants were transformed with a plasmid that allowed IPTG-induced overexpression of FliM and FliN. (The two proteins were coexpressed because overexpression of FliM alone causes severe motility impairment owing to an imbalance between FliM and FliN levels [10, 46].) Additional FliM and FliN markedly improved the swarming of the 202W and 225W mutants (Fig. 3). The other assembly-defective mutants (128W, 147W, and 158W) were not helped by additional FliM and FliN. Several CCW-biased mutants were also tested, but none showed any improvement in swarming upon overexpression of FliM and FliN (data not shown; the mutants tested were 117W, 125W, 191W, and 214W).
FIG. 3.
Enhancement of swarming by overexpression of CheY (top) or FliM and FliN (bottom) in selected FliG mutants. CheY or FliM-FliN were expressed from plasmids, as described in Materials and Methods. Control strains contained a second plasmid that encoded the relevant antibiotic resistance but no flagellar genes. Swarms were allowed to develop for approximately 9 h for all of the mutants and for 5 h for the wild type (wt).
CheY is the signaling protein that, when phosphorylated, promotes CW rotation of the motor and tumbling of cells. To determine whether additional CheY could improve the function of the CCW-biased, smooth-swimming mutants, the mutants were transformed with a plasmid allowing arabinose-regulated overexpression of CheY. The 117W, 125W, and 214W mutants, which exhibited the strongest swarming defects, were not helped by extra CheY. The 152W and 191W mutants, which exhibited milder swarming defects, were much improved by the extra CheY, swarming at about one-third of the wild-type rate (Fig. 3).
Binding to FliM.
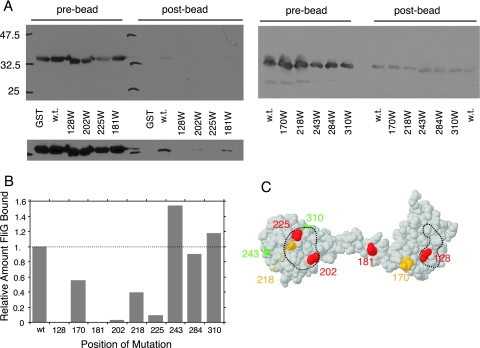
A GST pull-down assay was used to determine whether the EHPQR motif and hydrophobic patch function to bind FliM. The experiments employed a fusion of glutathione S-transferase to the amino terminus of FliM and procedures used previously in a study of FliG-FliM interaction (47). Wild-type FliG was reproducibly coisolated with GST-FliM in this assay. Three mutations in regions of the C-terminal domain apart from the hydrophobic patch (residues 243, 284, and 310) did not measurably weaken the FliG-FliM interaction, as evidenced by a similar yield of coisolated FliG. Binding was greatly reduced (to 10% or less of the wild-type level) by the Trp replacements at positions 128, 181, 202, and 225 and was partially reduced (to about half of the wild-type level) by the replacements at positions 170 and 202 (Fig. 4).
FIG. 4.
(A) Coisolation of FliG with GST-FliM and effects of FliG mutations on the binding. Coisolated FliG was detected using anti-FliG immunoblots. Prebead controls show the relative levels of FliG present in each sample prior to treatment with the beads, and postbead samples show levels of FliG coisolated with GST-FliM. Positions of the Trp mutations are shown at the bottom. Some of the mutations weakened binding so much that coisolated FliG could be seen only in a longer exposure, shown at the bottom for the left-hand gel. w.t., wild type. (B) Relative levels of coisolated wild-type and mutant FliG proteins. Results of a typical binding experiment are shown. (C) Mutations mapped onto the FliG structure. Red, binding reduced to 10% or less of wild-type level; orange, binding reduced to about half of wild-type level; green, binding similar to wild-type level. Dashed lines indicate the hydrophobic patch (left) and EHPQR motif (right). Position 284 (which did not affect the binding to FliM and so would be green) is not visible in this view but is on the charge-bearing ridge of the C-terminal domain (Fig. 2).
DISCUSSION
Regions that tolerate mutation.
Trp replacements were tolerated in many positions in the C-terminal domain of FliG, including the charge-bearing ridge that has been implicated in electrostatic interactions with the stator (56, 61). Previous mutational studies showed that while the charged residues of the ridge are collectively important, no single residue is indispensable for rotation (28, 60, 61). The ability of the ridge to tolerate Trp replacements indicates that the detailed topography of the ridge also is not critical. Several Trp replacements on the sides of the domain also had no measurable effect on flagellar assembly or function, as evidenced by normal rates of swarming. We conclude that these side surfaces of the C-terminal domain do not participate in functionally important binding interactions. This finding is consistent with recent cross-linking results, which support a model in which adjacent FliGC domains are spaced somewhat apart from each other but with no other protein in between (31).
Importance of the EHPQR motif and hydrophobic patch.
As expected from their high degree of conservation, both the EHPQR motif and the hydrophobic patch were found to be important for flagellar assembly and function. Tryptophan replacements in these regions either disrupted flagellar assembly or altered the CW/CCW bias of the motor. These findings are in agreement with previous studies of spontaneous fliG mutants, initially isolated in Salmonella (20) and subsequently characterized further in E. coli (29, 62). Most spontaneous fla mutations in E. coli fliG encoded replacements in or near the EHPQR motif (at positions 125, 128, 129, and 132) or in the hydrophobic patch (at positions 201, 202, and 219) (29, 62). Defects in flagellar assembly were also seen for some Trp replacements in the middle domain on the face opposite the EHPQR motif (positions 147, 158, and 165) (Fig. 2). These fall along a shallow cleft lined by several residues with conserved hydrophobic character (Leu 146, Met 158, Ile 161, and Phe 164 in E. coli, corresponding to Leu 147, Leu 159, Ile 162, and Leu 165 in T. maritima). This surface of the middle domain might also participate in functionally important contacts. However, because a substantial portion of this domain (>100 residues) is missing from the crystal structure, this surface of the domain could be buried in the intact protein and the replacements in this region might affect function by altering the overall domain structure rather than by disrupting an important interaction. By contrast, the EHPQR motif is polar and is almost certain to be on the protein surface, where it could engage in functionally important interactions.
A previous study using the two-hybrid system in yeast indicated that both the middle and C-terminal domains of FliG participate in binding to FliM (34). The present mutational results identify the EHPQR motif and hydrophobic patch as likely candidates for interaction with FliM. Trp replacements in these regions gave strong phenotypes and caused a substantial reduction in binding to FliM as assayed by GST pull-down experiments (Fig. 4). The overexpression experiment also points to an interaction through the hydrophobic patch; two hydrophobic-patch mutants that showed strong motility impairment when FliM levels were wild type swarmed well when FliM and FliN were overexpressed (Fig. 3). The FliM-binding site may also include parts of the interdomain helix in FliG, because a mutation in residue 181 also weakened the interaction. Interactions in this region of FliG appear less critical for flagellar assembly, however, because the 181W mutation did not impair swarming (Table 1).
Model for subunit organization in the switch complex.
A variety of evidence indicates that FliG is located in the upper part of the C ring (16, 45, 48, 61). As discussed previously (31), previous data were consistent with either of two locations for FliG, one with the C-terminal domain at the outer edge of the C ring and the other with FliG in a more “inboard” location (Fig. 1). In the more inboard location, the middle domain of FliG would be located at the lower edge of the MS ring, quite distant from the parts of the C ring that might contain FliM. The binding of FliM to the EHPQR motif therefore argues against the inboard location and instead supports the “outboard” location shown in Fig. 5A. Figure 5 also presents a model for the overall organization of FliG, FliM, and FliN. This model accounts for the present results and appears to be consistent with all available mutational, cross-linking, and electron microscopic data.
FIG. 5.
Model for subunit organization in the switch complex. (A) Proposed locations of FliG, FliM, and FliN in the C ring. FliG is placed in the more outboard of the two locations shown in Fig. 1, to allow interaction between FliM and the middle domain of FliG (the EHPQR motif). (B) Enlarged view of the hypothesized FliG-FliM interactions. FliM is pictured in two orientations, one interacting with the EHPQR motif in the FliG middle domain (green) and the other interacting with the hydrophobic patch in the FliG C-terminal domain (yellow). (C) View from the “top” (the membrane-proximal side) of the C ring. Subunit organization is illustrated for half of the ring, using the same coloring as in panel A. The shape of the FliG subunits is based on the FliG structure, with the relative orientations of the domains adjusted slightly to account for targeted cross-linking results reported previously (31). The structure of a FliG subunit, viewed from membrane-proximal side, is shown for comparison. In the model, 26 copies of FliM attach to the C-terminal domain of FliG through the hydrophobic patch, while the approximately eight “extra” FliM subunits tilt inward to interact with the middle domain of FliG.
A major feature of the model is that FliM is positioned between FliG and FliN, to account for the finding that FliM interacts with both FliG and FliN (34, 35, 47, 51, 52, 57). The space occupied by FliM in the model has a height of about 5 nm. This is similar to the ca. 5.3-nm long dimension of the major domain of FliM, determined in a recent crystal structure of the T. maritima protein (38). The crystal structure shows that the short dimension of the domain is about 2.5 nm, which is comparable to the thickness of the C ring wall in this region. The intermediate dimension of the FliM domain, ca. 4 nm, is very close to the spacing between adjacent units seen in bottom views of the C ring, and targeted cross-linking experiments showed that adjacent FliM subunits are in contact along this intermediate dimension (38).
Given its shape and size, a single FliM subunit appears to be unable to contact both the EHPQR motif and the hydrophobic patch simultaneously. Although the two domains of FliG might be arranged somewhat differently in the flagellum than in the crystal, there does not appear to be an accessible conformation that brings the EHPQR and hydrophobic patch close together, and the structures seen by electron microscopy show the domains separated by about 5 nm (48, 49). Accordingly, we suggest that some of the FliM subunits in the C ring interact with the EHPQR motif while others interact with the hydrophobic patch. The C ring contains about 34 copies of FliM but only about 26 copies of FliG. This mismatch implies that the FliM subunits in the flagellum cannot all occur in strictly equivalent, symmetry-related environments. The proposed subunit arrangement is consistent with (and might be the reason for) this difference in FliG and FliM copy numbers. We propose that 26 copies of FliM bind to the hydrophobic patch on the C-terminal domain, while the remaining 8 are tilted inward to interact with the EHPQR motif on the middle domain, as detailed in Fig. 5B and C. This would account for the binding data and is also consistent with the electron microscopy images, which show most electron density in the C ring occurring under the C-terminal domain of FliG but some density also reaching inward toward the middle domain. We note that two molecules of FliM might act cooperatively to bind FliG, because adjacent FliM subunits are also likely to interact with each other.
Switching.
Previously, we proposed that CW/CCW switching might involve FliM-regulated movements of the FliG C-terminal domain relative to the middle domain (7). The subunit arrangement proposed here is consistent with such a switching mechanism. The phenotypes of Trp replacements are also in accord with the model. Motor bias was affected by certain Trp replacements in and near the EHPQR motif (positions 117, 152, and 125), in the hydrophobic patch (positions 196 and 214), and also in the waist between the upper and lower parts of the C-terminal domain (position 235). Mutations in the EHPQR or hydrophobic patch might alter the relative positions or orientations of the domains by directly altering the FliG-FliM interface. The mutation at position 235 might alter the orientation of the upper part of the C-terminal domain relative to the lower part, moving or reorienting the charge-bearing ridge. This region of the domain is fairly narrow and does not appear to impose any strong structural constraints between the upper and lower parts. As noted above, regions on the interdomain helix might also participate in contacts with FliM. One bias-altering Trp replacement occurred on the bottom of the interdomain helix, at the end nearest the hydrophobic patch (position 191), and several spontaneous mutations affecting switch bias also encoded replacements on the interdomain helix (43). While they might be important for switching, these contacts involving the interdomain helix are evidently less important for flagellar assembly.
In summary, the mutational results, in conjunction with data from previous mutational, binding, and structural studies, lead to a specific model for subunit arrangement in the switch complex. Although the molecular movements responsible for direction switching are not yet precisely defined, the available data are consistent with a model in which the C-terminal domain of FliG moves relative to the middle domain, altering the rotor-stator interface by altering the position or orientation of the charge-bearing ridge.
Acknowledgments
We thank Brian Crane and Donna Marykwas for valuable discussions and for sharing data in advance of publication.
This work was supported by grants 8R01-EB2041 from the National Institute of Biomedical Imaging and Bioengineering and grant 5R01-GH64664 from the National Institute of General Medical Sciences. P.N.B. received partial support from training grant 5T32-GM08537 from the National Institutes of Health. M.T. was supported by a minigrant from the Biosciences Undergraduate Research Program of the University of Utah. The protein-DNA core facility at the University of Utah receives support from the National Cancer Institute (grant 5P30 CA42014).
Footnotes
Published ahead of print on 3 November 2006.
REFERENCES
- 1.Attmannspacher, U., B. Scharf, and R. Schmitt. 2005. Control of speed modulation (chemokinesis) in the unidirectional rotary motor of Sinorhizobium meliloti. Mol. Microbiol. 56:708-718. [DOI] [PubMed] [Google Scholar]
- 2.Berg, H. C. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72:19-54. [DOI] [PubMed] [Google Scholar]
- 3.Berry, R. M., and H. C. Berg. 1999. Torque generated by the flagellar motor of Escherichia coli while driven backward. Biophys. J. 76:580-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blair, D. F., and H. C. Berg. 1988. Restoration of torque in defective flagellar motors. Science 242:1678-1681. [DOI] [PubMed] [Google Scholar]
- 5.Block, S. M., and H. C. Berg. 1984. Successive incorporation of force-generating units in the bacterial rotary motor. Nature 309:470-472. [DOI] [PubMed] [Google Scholar]
- 6.Bren, A., and M. Eisenbach. 1998. The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J. Mol. Biol. 278:507-514. [DOI] [PubMed] [Google Scholar]
- 7.Brown, P. N., C. P. Hill, and D. F. Blair. 2002. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 21:3225-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, P. N., M. A. A. Mathews, L. A. Joss, C. P. Hill, and D. F. Blair. 2005. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J. Bacteriol. 187:2890-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun, S. Y., and J. S. Parkinson. 1988. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science 239:276-278. [DOI] [PubMed] [Google Scholar]
- 10.Clegg, D. O., and D. E. J. Koshland. 1985. Identification of a bacterial sensing protein and effects of its elevated expression. J. Bacteriol. 162:398-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeMot, R., and J. Vanderleyden. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both Gram-positive and Gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333-334. [DOI] [PubMed] [Google Scholar]
- 12.Francis, N. R., G. E. Sosinsky, D. Thomas, and D. J. DeRosier. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261-1270. [DOI] [PubMed] [Google Scholar]
- 13.Garza, A. G., R. Biran, J. Wohlschlegel, and M. D. Manson. 1996. Mutations in motB suppressible by changes in stator or rotor components of the bacterial flagellar motor. J. Mol. Biol. 258:270-285. [DOI] [PubMed] [Google Scholar]
- 14.Glagolev, A. N., and V. P. Skulachev. 1978. The proton pump is a molecular engine of motile bacteria. Nature 272:280-282. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez-Pedrajo, B., T. Minamino, M. Kihara, and K. Namba. 2006. Interactions between C-ring proteins and export apparatus components: a possible mechanism for facilitating type III protein export. Mol. Microbiol. 60:984-998. [DOI] [PubMed] [Google Scholar]
- 16.Grunenfelder, B., S. Gehrig, and U. Jenal. 2003. Role of the cytoplasmic C terminus of the FliF motor protein in flagellar assembly and rotation. J. Bacteriol. 185:1624-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heimbrook, M. E., W. L. L. Wang, and G. Campbell. 1986. Easily made flagella stains, abstr. R-22, p. 240. Abstr. 86th Annu. Meet. Am. Soc. Microbiol. 1986. American Society for Microbiology, Washington, DC.
- 18.Hirota, N., M. Kitada, and Y. Imae. 1981. Flagellar motors of alkalophilic Bacillus are powered by an electrochemical potential gradient of Na+. FEBS Lett. 132:278-280. [Google Scholar]
- 19.Homma, M., S. I. Aizawa, G. E. Dean, and R. M. Macnab. 1987. Identification of the M-ring protein of the flagellar motor of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 84:7483-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irikura, V. M., M. Kihara, S. Yamaguchi, H. Sockett, and R. M. Macnab. 1993. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J. Bacteriol. 175:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan, S., M. Dapice, and T. S. Reese. 1988. Effects of mot gene expression on the structure of the flagellar motor. J. Mol. Biol. 202:575-584. [DOI] [PubMed] [Google Scholar]
- 22.Kihara, M., G. U. Miller, and R. M. Macnab. 2000. Deletion analysis of the flagellar switch protein FliG of Salmonella. J. Bacteriol. 182:3022-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kojima, S., and D. F. Blair. 2004. The bacterial flagellar motor: structure and function of a complex molecular machine. Int. Rev. Cytol. 233:93-134. [DOI] [PubMed] [Google Scholar]
- 24.Kojima, S., and D. F. Blair. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041-13050. [DOI] [PubMed] [Google Scholar]
- 25.Kojima, S., and D. F. Blair. 2004. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43:26-34. [DOI] [PubMed] [Google Scholar]
- 26.Kubori, T., S. Yamaguchi, and S.-I. Aizawa. 1997. Assembly of the switch complex onto the MS-ring complex of Salmonella typhimurium does not require any other flagellar proteins. J. Bacteriol. 179:813-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsen, S. H., J. Adler, J. J. Gargus, and R. W. Hogg. 1974. Chemomechanical coupling without ATP: the source of energy for motility and chemotaxis in bacteria. Proc. Natl. Acad. Sci. USA 71:1239-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd, S. A., and D. F. Blair. 1997. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J. Mol. Biol. 266:733-744. [DOI] [PubMed] [Google Scholar]
- 29.Lloyd, S. A., H. Tang, X. Wang, S. Billings, and D. F. Blair. 1996. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J. Bacteriol. 178:223-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lloyd, S. A., F. G. Whitby, D. F. Blair, and C. P. Hill. 1999. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature 400:472-475. [DOI] [PubMed] [Google Scholar]
- 31.Lowder, B. J., M. D. Duyvesteyn, and D. F. Blair. 2005. FliG subunit arrangement in the flagellar rotor probed by targeted crosslinking. J. Bacteriol. 187:5640-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 33.Manson, M. D., Tedesco, H. C. Berg, F. M. Harold, and C. van der Drift. 1977. A protonmotive force drives bacterial flagella. Proc. Natl. Acad. Sci. USA 74:3060-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marykwas, D. L., and H. C. Berg. 1996. A mutational analysis of the interaction between FliG and FliM, two components of the flagellar motor of Escherichia coli. J. Bacteriol. 178:1289-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marykwas, D. L., S. A. Schmidt, and H. C. Berg. 1996. Interacting components of the flagellar motor of Escherichia coli revealed by the two-hybrid system in yeast. J. Mol. Biol. 256:564-576. [DOI] [PubMed] [Google Scholar]
- 36.Mathews, M. A. A., H. L. Tang, and D. F. Blair. 1998. Domain analysis of the FliM protein of Escherichia coli. J. Bacteriol. 180:5580-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.McMurry, J. L., J. W. Murphy, and B. Gonzalez-Pedrajo. 2006. The FliN-Flitt interaction mediates localization of flagellar export ATPase FliI to the C ring complex. Biochemistry 45:11790-11798. [DOI] [PubMed] [Google Scholar]
- 37.Oosawa, K., T. Ueno, and S. I. Aizawa. 1994. Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J. Bacteriol. 176:3683-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, S. Y., B. J. Lowder, A. M. Bilwes, D. F. Blair, and B. R. Crane. 2006. Structure of FliM provides insight into assembly of the switch complex in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 103:11886-11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paul, K., and D. F. Blair. 2006. Organization of FliN subunits in the flagellar motor of Escherichia coli. J. Bacteriol. 188:2502-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paul, K., J. Harmon, and D. F. Blair. 2006. Mutational analysis of the flagellar rotor protein FliN: identification of surfaces important for flagellar assembly and switching. J. Bacteriol. 188:5240-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reid, S. W., M. C. Leake, J. H. Chandler, C. J. Lo, J. P. Armitage, and R. M. Berry. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl. Acad. Sci. USA 103:8066-8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato, K., and M. Homma. 2000. Functional reconstitution of the Na(+)-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 275:5718-5722. [DOI] [PubMed] [Google Scholar]
- 43.Sockett, H., S. Yamaguchi, M. Kihara, V. M. Irikura, and R. M. Macnab. 1992. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J. Bacteriol. 174:793-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki, H., K. Yonekura, and K. Namba. 2004. Structure of the rotor of the bacterial flagellar motor revealed by electron cryo-microscoy and single-particle image analysis. J. Mol. Biol. 337:105-113. [DOI] [PubMed] [Google Scholar]
- 46.Tang, H., S. Billings, X. Wang, L. Sharp, and D. F. Blair. 1995. Regulated underexpression and overexpression of the FliN protein of Escherichia coli and evidence for an interaction between FliN and FliM in the flagellar motor. J. Bacteriol. 177:3496-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang, H., T. F. Braun, and D. F. Blair. 1996. Motility protein complexes in the bacterial flagellar motor. J. Mol. Biol. 261:209-221. [DOI] [PubMed] [Google Scholar]
- 48.Thomas, D., D. G. Morgan, and D. J. DeRosier. 2001. Structures of bacterial flagellar motors from two FliF-FliG gene fusion mutants. J. Bacteriol. 183:6404-6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas, D. R., N. R. Francis, C. Xu, and D. J. DeRosier. 2006. Three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium J. Bacteriol. 188:7033-7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas, D. R., D. G. Morgan, and D. J. DeRosier. 1999. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc. Natl. Acad. Sci. USA 96:10134-10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toker, A. S., M. Kihara, and R. M. Macnab. 1996. Deletion analysis of the FliM flagellar switch protein of Salmonella typhimurium. J. Bacteriol. 178:7069-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toker, A. S., and R. M. Macnab. 1997. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J. Mol. Biol. 273:623-634. [DOI] [PubMed] [Google Scholar]
- 53.Ueno, T., K. Oosawa, and S. I. Aizawa. 1992. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J. Mol. Biol. 227:672-677. [DOI] [PubMed] [Google Scholar]
- 54.Van Way, S. M., S. G. Millas, A. H. Lee, and M. D. Manson. 2004. Rusty, jammed, and well-oiled hinges: mutations affectig the interdomain region of FliG, a rotor element of the Escherichia coli flagellar motor. J. Bacteriol. 186:3173-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vogler, A. P., M. Homma, V. M. Irikura, and R. M. Macnab. 1991. Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar, and archaebacterial ATPase subunits. J. Bacteriol. 173:3564-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yakushi, T., J. H. Yang, H. Fukuoka, M. Homma, and D. F. Blair. 2006. Roles of charged residues of rotor and stator in flagellar rotation: comparative study using H+-driven and Na+-driven motors in Escherichia coli. J. Bacteriol. 188:1466-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamaguchi, S., S.-I. Aizawa, M. Kihara, M. Isomura, C. J. Jones, and R. M. Macnab. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 168:1172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Young, H. S., H. Dang, Y. Lai, D. J. DeRosier, and S. Khan. 2003. Variable symmetry in Salmonella typhimurium flagellar motors. Biophys. J. 84:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao, R., N. Pathak, H. Jaffe, T. S. Reese, and S. Khan. 1996. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J. Mol. Biol. 261:195-208. [DOI] [PubMed] [Google Scholar]
- 60.Zhou, J., and D. F. Blair. 1997. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J. Mol. Biol. 273:428-439. [DOI] [PubMed] [Google Scholar]
- 61.Zhou, J., S. A. Lloyd, and D. F. Blair. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 95:6436-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou, J., L. L. Sharp, H. L. Tang, S. A. Lloyd, S. Billings, T. F. Braun, and D. F. Blair. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J. Bacteriol. 180:2729-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]