Abstract
Lipoprotein processing by the type II signal peptidase (SPase II) is known to be critical for intracellular growth and virulence for many bacteria, but its role in rickettsiae is unknown. Here, we describe the analysis of lspA, encoding a putative SPase II, an essential component of lipoprotein processing in gram-negative bacteria, from Rickettsia typhi. Alignment of deduced amino acid sequences shows the presence of highly conserved residues and domains that are essential for SPase II activity in lipoprotein processing. The transcription of lspA, lgt (encoding prolipoprotein transferase), and lepB (encoding type I signal peptidase), monitored by real-time quantitative reverse transcription-PCR, reveals a differential expression pattern during various stages of rickettsial intracellular growth. The higher transcriptional level of all three genes at the preinfection time point indicates that only live and metabolically active rickettsiae are capable of infection and inducing host cell phagocytosis. lspA and lgt, which are involved in lipoprotein processing, show similar levels of expression. However, lepB, which is involved in nonlipoprotein secretion, shows a higher level of expression, suggesting that LepB is the major signal peptidase for protein secretion and supporting our in silico prediction that out of 89 secretory proteins, only 14 are lipoproteins. Overexpression of R. typhi lspA in Escherichia coli confers increased globomycin resistance, indicating its function as SPase II. In genetic complementation, recombinant lspA from R. typhi significantly restores the growth of temperature-sensitive E. coli Y815 at the nonpermissive temperature, supporting its biological activity as SPase II in prolipoprotein processing.
Rickettsiae are gram-negative, obligate intracellular bacteria that are transmitted to their mammalian hosts by arthropod vectors such as ticks, fleas, and lice. Some members of the genus Rickettsia are responsible for the most-severe bacterial diseases of humans and have been classified as select agents with potential use as tools for bioterrorism. Currently, diagnosis of rickettsial infections is difficult. No reliable protective vaccine and only a few antibiotics to treat rickettsiae are available (4, 13, 33). Little is known about rickettsial gene function because of the intrinsic difficulty in working with these obligate intracellular bacteria and their genetic intractability (31, 40).
The genomes of several rickettsial species have been sequenced and have provided many insights into their biology (3, 19, 21-23); however, the molecular basis of pathogenesis remains an important area of research for rickettsiae. Several bacterial virulence factors point to the importance of secreted proteins in pathogenesis. Secreted and extracytoplasmic proteins are of particular importance, because this subset of proteins are involved in many essential cellular processes, including adherence, virulence, immunogenicity, environmental sensing, and host-pathogen interactions, and are potential targets of therapeutic interest (10, 17).
The major route of bacterial protein secretion from the cytoplasm is the Sec pathway (7). Bacterial proteins transported through the Sec translocon are synthesized as preproteins with an amino-terminal extension known as the signal or leader peptide. The signal peptide is required for the targeting of preproteins to the membrane for translocation by Sec machinery. During the translocation process, the signal peptides of nonlipoproteins are cleaved off by the type I signal peptidase (SPase I), while those of lipoproteins are cleaved off by the type II signal peptidase (SPase II) (9, 24, 25, 29). Thus, SPases play a key role in the transport of proteins across membranes in all organisms (24, 25). The major difference between signal peptides of lipoproteins and nonlipoproteins is the presence of a conserved consensus sequence called a “lipobox” of four amino acids in lipoprotein signal peptides. The carboxyl-terminal amino acid of the lipobox is a universally conserved cysteine at position +1. The cysteine residue of the lipobox is modified by diacylglyceryl transferase (Lgt). Diacylglyceryl modification is a prerequisite for the specific cleavage of the signal peptide from prolipoprotein by SPase II (25, 35, 36).
Like the SPase I family, SPase II members are membrane-bound proteases. Type II signal peptidases have been identified in many gram-negative and gram-positive bacteria, including Mycoplasma. However, SPase II has not been found in archaea or eukaryotes (25). Lipoprotein processing by SPase II has been reported to be essential for the viability of gram-negative bacteria (11, 41), but it has been shown to be dispensable for the growth of many gram-positive bacteria (34, 37, 38). Processing of lipoproteins by SPase II plays an important role in the virulence of Streptococcus pneumoniae and Mycobacterium tuberculosis (27, 34) and has been shown to be critical for the intracellular growth of many bacterial pathogens (32, 34). In addition, bacterial lipoproteins are reported to initiate both innate and adaptive immune responses in humans (1). Based on these findings, LspA, a membrane-bound protease, is considered an attractive target for chemotherapy (34).
The annotation of published rickettsial genome sequences reveals the presence of bacterial Sec translocon homologs (3, 19, 21-23). However, the genes specifically involved in rickettsial protein secretion remain uncharacterized. In order to elucidate the mechanism of protein secretion and their role in rickettsial virulence, we are interested in characterizing the genes involved in the rickettsial Sec pathway. In this effort, we reported (30, 31) our work on the putative lepB and secA genes, encoding the SPase I and ATPase, respectively, of rickettsiae. In this communication, we report a detailed transcriptional and functional characterization of lspA, which encodes a putative SPase II from Rickettsia typhi, the causative agent of murine typhus.
MATERIALS AND METHODS
Bacterial strains and host cells.
R. typhi strain Wilmington (ATCC VR-144) was used in this study. E. coli strain Y815 was used for functional complementation. E. coli strain Y815 is temperature sensitive due to a point mutation in the signal peptidase II gene and also carries an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible lpp gene for the major outer membrane lipoprotein (also known as Braun's lipoprotein) on the low-copy-number plasmid pHY001, which contains a tetracycline resistance marker (41).
Mouse subcutaneous connective tissue NCTC clone 929 cells (ATCC CCL-1) were used as host cells for rickettsial culture. The NCTC clone 929 cells were grown in Dulbecco's modified minimal essential medium with 4.5 g of glucose/liter with glutamine (Biofluids Inc., Rockville, MD) supplemented with 5% fetal bovine serum (Gemini, Calabasas, CA) at 37°C and 5% CO2 in an air atmosphere.
Genomic DNA extraction.
Propagation and partial purification of R. typhi from host (NCTC clone 929) cells were performed as previously described (31). Genomic DNA of R. typhi was extracted with the Wizard genomic DNA purification kit (Promega, Madison, WI).
Cloning of lspA gene.
The lspA gene, encoding SPase II from R. typhi and E. coli, was cloned by PCR into expression vector pMW119 under a lac promoter (2). Primers used in PCR are shown in Table 1 and were designed based on the genome sequences available in the GenBank database (www.ncbi.nlm.nih.gov). Primers AZ3321 (BamHI) and AZ2793 (EcoRI) were used to amplify a 723-bp fragment of R. typhi lspA with Herculase DNA polymerase (Stratagene, La Jolla, CA) and cloned into pMW119 to generate pMWRTlspA5. A 533-bp fragment containing the entire open reading frame (ORF) of lspA from E. coli strain MC4100 (30) was amplified by using primers AZ2357 (the BamHI) and AZ2358 (EcoRI) and cloned into pMW119 to generate pMWEClspA19. The constructed plasmids pMWRTlspA5 and pMWEClspA19 were confirmed by sequencing.
TABLE 1.
Primers used in PCR
| Primer | Sequencea (5′ to 3′) | Nucleotide positionb |
|---|---|---|
| AZ2357 | TAAggatccCTGATGAGTCAATCG | 427-450* |
| AZ2358 | GTCAGCATCgaattcGGCAGGG | 938-957* |
| AZ2793 | gaattcTGGTCCTGCTGTTGATAGAC | 501542-501567† |
| AZ3321 | ggatccATGAATGATCTCATTATTTAAAAAAC | 500845-500876† |
| AZ3598 | ATTTAAGATGGAAGCCAGGC | 500961-500980† |
| AZ3599 | GTCAAAGACAGCCCCTCTAC | 501213-501232† |
| AZ3593 | CCACCAGGTTATTTTTGTTG | 21833-21852† |
| AZ3594 | GAAAGGACAGAGGTTGGGAC | 22113-22132† |
| AZ3617 | GTCCATTTTCAATCTCTTGG | 109638-109657† |
| AZ3618 | TGAAATGACATACCGCCTTC | 109871-109890† |
| AZ3641 | GTTCGGAATTACTGGGCGTA | 780639-780658† |
| AZ3642 | AATTAAACCGCATGCTCCAC | 780252-780271† |
| AZ3458 | ggatccTCTATGATCTCATTATTTAAAAAAC | 500846-500876† |
The sequence of the lspA gene and deduced amino acid sequence from R. typhi were analyzed with MacVector 7.1.1 software (Genetics Computer Group, Inc., Madison, WI). Sequence comparisons to those available in GenBank were performed with BLAST (www.ncbi.nlm.nih.gov).
Complementation assay and expression of rickettsial SPase II in E. coli.
The constructed plasmids pMWRTlspA5 and pMWEClspA19 and the vector pMW119 were transformed into E. coli Y815 (temperature-sensitive) cells. Transformants were selected on antibiotic medium 3 (AM3) agar containing ampicillin (100 μg ml−1) and tetracycline (10 μg ml−1) and incubated at 30°C for 3 days. For the complementation by CFU assay, the transformants actively growing at 30°C in AM3 broth containing ampicillin (100 μg ml−1) and tetracycline (10 μg ml−1) were plated onto two series of AM3 agar containing ampicillin (100 μg ml−1), tetracycline (10 μg ml−1), and IPTG (0.6 mM). The plates were incubated at either the permissive (30°C) or the nonpermissive (42°C) temperature for colony formation. The colonies were counted after 72 h of incubation to determine the percentage of growth at 42°C with respect to growth at 30°C.
For the globomycin resistance assay, E. coli Top10 cells (Invitrogen-Life Technologies, Carlsbad, CA) were transformed with the constructed plasmids pMWRTlspA5 and pMWEClspA19 or the vector pMW119. Transformants were grown to stationary phase in Luria-Bertani (LB) medium containing ampicillin (100 μg ml−1). Stationary-phase cultures were diluted 50-fold in LB medium containing ampicillin (100 μg ml−1) and incubated at 37°C for 8 h with shaking. The exponentially growing cells were diluted 10-fold in LB medium containing ampicillin (100 μg ml−1) and IPTG (1 mM) with various concentrations of globomycin and were incubated at 37°C for 18 h with shaking. The cell density was measured by spectrophotometer at 600 nm.
All experiments were performed at least three times, and statistical analyses were performed with Microsoft Excel software.
To analyze the synthesis of R. typhi SPase II in E. coli, the 722-bp fragment containing the entire ORF of lspA from R. typhi was amplified by using primers AZ3458 (BamHI) and AZ2793 (EcoRI) and cloned at the BamHI and EcoRI sites of pTrcHisA vector, which contains an N-terminal His6 tag under the trc (trp-lac) promoter (Invitrogen-Life Technologies, Carlsbad, CA), to generate the plasmid pTrcHisRTlspA127. As a positive control, a 533-bp fragment containing the entire ORF of lspA from E. coli was amplified by using primers AZ2357 (BamHI) and AZ2358 (EcoRI) and was cloned similarly in pTrcHisA vector to generate the plasmid pTrcHisEClspA7. The constructed plasmids pTrcHisRTlspA127 and pTrcHisEClspA7 were confirmed by sequencing. The synthesis of recombinant SPase II proteins in the transformed E. coli Top10 cells (Invitrogen-Life Technologies, Carlsbad, CA) was detected by Anti-HisG monoclonal antibody (Invitrogen-Life Technologies) with a WesternBreeze chemiluminescent immunodetection kit (Invitrogen-Life Technologies), as described previously (30, 31).
Isolation of RNA from rickettsiae grown at various time points.
To titer the partially purified R. typhi from infected host cells (monolayer of NCTC clone 929), freshly isolated (partially purified) R. typhi was mixed with Live/Dead BacLight bacterial viability stain (Molecular Probes, Eugene, OR), according to the manufacturer's instructions. Rickettsiae were counted with a hemacytometer under fluorescence (excitation, 480 nm; emission, 500 nm) on a Nikon H550L. A monolayer of NCTC clone 929 cells was infected with R. typhi at a multiplicity of infection of (approximately) 90 rickettsiae per cell. Rickettsia-infected NCTC clone 929 cells were incubated in Dulbecco's modified minimal essential medium supplemented with 5% fetal bovine serum at 37°C and 5% CO2 for various time points (0, 1, 4, 8, 24, 48, and 120 h). In our experimental design, 0 h indicates the preinfection time point of rickettsiae to host cells. Rickettsia-infected host cells were harvested by scraping the culture and mechanically rupturing them by passage through a 27-gauge needle to release the intracellular rickettsiae. To enrich for rickettsiae, large host cell fragments were removed by centrifugation (500 × g for 5 min at 4°C). The supernatant was centrifuged at 14,000 × g for 10 min at 4°C to pellet the partially purified rickettsiae. The rickettsial pellet was immediately resuspended in TRIzol reagent (Invitrogen, Carlsbad, CA) for RNA isolation, according to manufacturer's instructions. The rickettsial RNA was DNase treated to remove contaminating DNA by using Turbo DNA-free (Ambion, Austin, TX), according to the manufacturer's recommendations. The DNase-treated rickettsial RNA was purified by the RNeasy MinElute cleanup kit (QIAGEN, Valencia, CA), according to the manufacturer's protocol.
Real time qRT-PCR for gene expression analyses.
To analyze rickettsial gene expression during the various stages of the infection cycle, two-step real-time quantitative reverse transcription-PCR (qRT-PCR) was performed on RNA samples isolated from rickettsiae at different time points of infection to host cells. First-strand cDNA was synthesized with 1 μg of RNA isolated at different time points postinfection by using the SuperScript III First-Strand Synthesis Supermix for qRT-PCR kit (Invitrogen) according to the manufacturer's recommendations. For real-time quantitative PCR (qPCR), the cDNA synthesized was amplified with the SYBR greenER qPCR Supermix universal kit (Invitrogen) according to the instructions of the manufacturer. The specific primers used (Table 1) in qPCR for R. typhi genes were AZ3593 and AZ3594 for lepB, AZ3598 and AZ3599 for lspA, AZ3617 and AZ3618 for lgt, and AZ3641 and AZ3642 for 16S rRNA. Cycling conditions used were as follows: 1 cycle at 50°C for 2 min; 1 cycle at 95°C for 10 min; 40 cycles at 95°C for 15 s, 56°C for 30 s, and 60°C for 30 s; and 1 cycle for the dissociation curve. qPCR amplification and detection were performed on an MX3000P thermal cycler (Stratagene, La Jolla, CA).
Controls for the RT-PCRs included amplifications without reverse transcriptase to account for genomic DNA contamination. Controls to detect reagent contamination included amplifications without template.
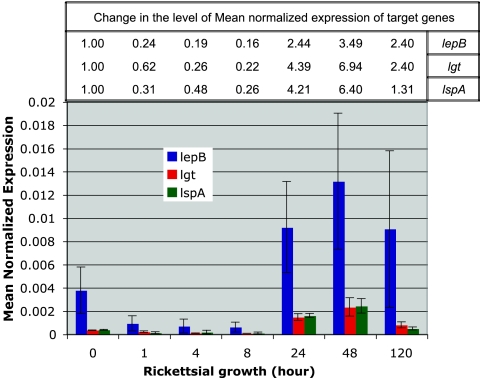
To analyze and quantify the gene expression data generated by real-time qRT-PCR, we used the Q-Gene software tool with amplification efficiency correction (20). For standard curve plotting, a gel-purified standard DNA template representing each target and reference gene was diluted in series from 1010 to 103 copies and used for real-time qPCR as described above. The amplification efficiency for each target (lepB, lspA, or lgt) and reference (16S rRNA) gene was derived from a standard curve plotted as the cycle threshold (CT) versus copies. To account for target gene transcript fluctuations between time points that might arise due to rickettsial burden (rickettsia/host cell and total number of rickettsia), which would confound the accuracy of our reported measurements, we normalized each target gene raw CT value by its corresponding 16S raw CT value. The means for the normalized CT values are reported as mean normalized expression (MNE) and plotted as MNE versus rickettsial growth (in hours) (see Fig. 2).
FIG. 2.
Kinetics of lepB, lgt, and lspA transcription during the R. typhi life cycle in in vitro host (NCTC clone 929) cells determined by two-step real-time qRT-PCR. The MNEs (± standard errors) of the target genes (lepB, lgt, and lspA) were calculated relative to the expression of the reference (16S rRNA) gene by Q-Gene software with amplification efficiency correction. At the top, the change in the level of MNE at different times of growth for each target gene is given with respect to that of the preinfection time point (0 h), set at 1.00. The expression levels of lgt and lspA at 24 and 48 h postinfection were found to be significantly different (P < 0.05 [single-factor ANOVA]) from those at 1 to 8 h postinfection. However, there is a lack of statistical significance to support the same for lepB, which may be due to variations in expression levels.
The time course experiment for rickettsial growth in host cells was performed three times, and the real time qRT-PCR on RNA isolated in each time course experiment was repeated twice. Statistical analyses were performed with Microsoft Excel software with the XLStat add-in. Briefly, a single-factor analysis of variance (ANOVA) with Tukey's (HSD) post hoc multiple comparisons procedure was used to test for significant differences at the 5% level for each gene over time.
RESULTS
Sequence analysis of rickettsial SPase II homolog.
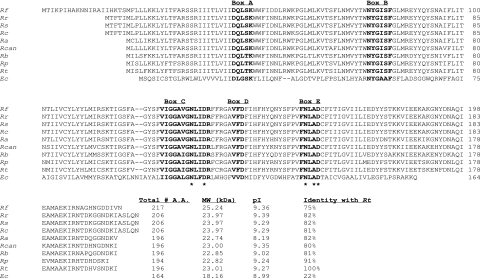
The deduced amino acid sequences of lspA among rickettsiae showed a very high degree of identity (ranging from 75% to 91%); however, the identity with the SPase II of E. coli was very low (around 22%) (Fig. 1). Alignment of the deduced amino acid sequences of LspA shown in Fig. 1 revealed the presence of five highly conserved domains (boxes A, B, C, D, and E) that are considered to be essential for the enzymatic activity of bacterial SPase II (6, 25, 38). The first conserved domain (box A) contains the aspartic acid (D) that is necessary for enzyme stability and function. Box B carries the invariable asparagine (N) and glycine (G) residues. The third domain (box C) contains asparagines and an aspartic acid that are critical for the SPase II activity. Additionally, aspartic acid is proposed to be one of the catalytic residues at the active sites. In box E, asparagine, alanine (A), and aspartic acid are considered to play important roles in prelipoprotein processing.
FIG. 1.
Alignment of deduced amino acid sequences of the lspA genes from R. typhi (Rt) (GenBank accession no. NC_006142), R. prowazekii (Rp) (GenBank accession no. AJ235271), R. bellii (Rb) (GenBank accession no. NZ_AARC01000001), R. canadensis (Rcan) (GenBank accession no. NZ_AAFF01000001), R. akari (Ra) (GenBank accession no. NZ_AAFE01000001), R. conorii (Rc) (GenBank accession no. NC_003103), R. sibirica (Rs) (GenBank accession no. AABW01000001), R. rickettsii (Rr) (GenBank accession no. NZ_AADJ01000001), R. felis (Rf) (GenBank accession no. NC_007109), and E. coli (Ec) (GenBank accession no. X00776). Molecular masses and isoelectric points were computed with MacVector 7.1.1 software. The conserved amino acid domains (boxes A through E) are shown in bold. The catalytic residues are marked by asterisks.
Kinetics of rickettsial lspA expression during different stages of growth in host cells.
To understand the role of SPase II in rickettsial host cell infection, transcription pattern of lspA at various stages of growth in vitro was investigated by two-step real-time qRT-PCR. In this study of the transcription pattern of R. typhi lspA, we also monitored the expression kinetics of R. typhi lgt (encoding prolipoprotein diacylglyceryl transferase) and lepB (encoding SPase I) for the comparative analyses of transcription of the key genes involved in lipoprotein and nonlipoprotein processing and transportation through the Sec pathway. It is observed from the transcription pattern shown in Fig. 2 that all three genes were differentially expressed at various stages of growth of R. typhi in host cells. The level of transcription of all three genes at the preinfection time point is higher and is followed by a decrease until 8 h postinfection. After the doubling time of rickettsiae (after 8 h postinternalization), the expression level of all three genes increased and peaked at 48 h postinfection. At 120 h postinfection, the infected host cells begin to detach from the surface of the culture flask, indicating lysis of host cells by R. typhi, and at that time all three genes show a decrease in expression. It is also observed from Fig. 2 that the transcription patterns of lspA and lgt, which are involved in lipoprotein secretion, are similar over the entire time course monitored in this study. However, the expression pattern of lepB is higher than those of the other two genes (lspA and lgt), suggesting that bacterial SPase I is the major signal peptidase for the processing of secretory proteins through the Sec pathway (24, 25).
Overexpression and functional activity of R. typhi lspA in E. coli.
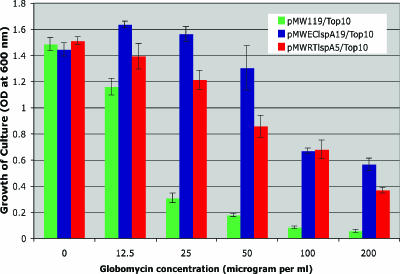
The functional activity of R. typhi lspA, encoding SPase II, was tested by a globomycin resistance assay in E. coli. Globomycin is a cyclic peptide antibiotic that inhibits the growth of gram-negative bacteria, such as E. coli. Gram-negative organisms are sensitive to globomycin due to inhibition of murein prolipoprotein processing to lipoprotein. The accumulation of unprocessed prolipoprotein in the inner membrane is considered to be the cause of the bacterial inability to grow in presence of this antibiotic (8, 14, 15). Globomycin inhibits prolipoprotein processing by acting as a substrate analog of the signal sequence, specifically binding and inhibiting SPase II activity in a noncompetitive manner (8, 15). The overexpression of lspA genes from both gram-negative and gram-positive bacteria has been reported to increase the level of globomycin resistance in E. coli and is generally used to demonstrate the functional activity of lspA homologs from many bacterial species (6, 26, 28). We used a globomycin resistance assay in E. coli to demonstrate the functional activity of the R. typhi lspA homolog. The SPase II activity of lspA from R. typhi was assayed by using the plasmids pMWEClspA19 (positive control, carrying the full-length ORF of E. coli lspA) and pMWRTlspA5 (carrying the full-length R. typhi lspA) and the vector plasmid pMW119 (negative control). It is observed from Fig. 3 that the growth of E. coli cells harboring pMW119 plasmid (negative control) rapidly decreased in the presence of a concentration of globomycin higher than 12.5 μg ml−1. The growth of E. coli cells carrying pMWEClspA19 or pMWRTlspA5 over that of E. coli cells harboring pMW119 for globomycin treatment from 25 μg ml−1 to 200 μg ml−1 was found to be statistically significant (P < 0.05) by Student's t test. The significant globomycin resistance growth demonstrates that R. typhi lspA, encoding SPase II, is functionally active in E. coli.
FIG. 3.
Globomycin resistance assay for R. typhi lspA. E. coli cells carrying the designated plasmids were incubated in the presence of various concentrations of globomycin, and growth of the cultures was measured as the optical density at 600 nm (OD600). The mean OD600 ± standard error is plotted against the globomycin concentration.
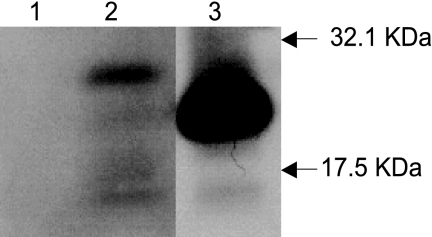
The expression of the recombinant R. typhi SPase II in E. coli was detected by Western blot analysis (Fig. 4). A band of approximately 27.13 kDa was detected for E. coli Top10 cells carrying plasmid pTrcHisRTlspA127 (Fig. 4, lane2); that band was not detected for negative control expression (Fig. 4, lane1). For the positive control, the higher expression of E. coli SPase II (Fig. 4, lane3, a band of 22.29 kDa) may have resulted from codon bias for protein translation in E. coli.
FIG. 4.
Western blot analysis of the expression of R. typhi SPase II in E. coli Top10 cells. Total proteins from E. coli cells carrying the appropriate plasmid were probed with anti-HisG monoclonal antibody. Lane 1, total proteins from E. coli Top10/pTrcHisA; lane2, total proteins from E. coli Top10/pTrcHisRTlspA127; lane3, total proteins from E. coli Top10/pTrcHisEClspA7. Arrows indicate the sizes of Bio-Rad Kaleidoscope prestained marker bands (32.1 and 17.5 kDa). The minor bands below the 17.5-kDa bands in lanes 2 and 3 may have resulted from the degradation of recombinant SPase II or nonspecific binding in the total proteins.
The in vivo functional activity of R. typhi lspA in E. coli was further investigated by genetically complementing the E. coli strain Y815. E. coli strain Y815, which has a point mutation in the SPase II gene, is temperature sensitive in growth in the presence of IPTG due to the accumulation of prolipoprotein. Strain Y815 has been used to demonstrate the complementation ability of many bacterial SPase II (26, 28, 38). E. coli strain Y815 was transformed with pMWEClspA19 (positive control), pMWRTlspA5, and pMW119 (negative control). The temperature-sensitive growth of the resulting transformants was tested by CFU assay. In this assay, E. coli strain Y815 showed no growth at the nonpermissive temperature of 42°C, and the vector plasmid pMW119 (negative control) was unable to restore growth. However, E. coli strain Y815 carrying pMWEClspA19 (positive control, carrying the full-length ORF of E. coli lspA) or pMWRTlspA5 (carrying full-length R. typhi lspA) showed a significant (P < 0.05 [Student's t test]) restoration of growth at 42°C by 21.5 ± 5.3% or 4.7 ± 1.3% (compared to that at 30°C), respectively, indicating the functional activity of R. typhi SPase II in E. coli.
DISCUSSION
In continuation of our efforts to elucidate the mechanism of rickettsial protein secretion through the Sec translocon and the genes involved in this multicomponent biological process of bacteria, we report in this communication the sequence, transcriptional analysis, and functional characterization of lspA from R. typhi, encoding a putative SPase II. Type II SPases have been identified in all rickettsial species in which the genomes have been sequenced and are found to possess highly conserved domains required for functional activity. SPase II members are considered to be an unusual class of aspartic acid proteases (25). Sequence alignment of rickettsial SPase II with that of E. coli showed the presence of Asn, Asp, and Ala residues in boxes C and D (marked by asterisks in Fig. 1), which are critical for the catalytic activity of bacterial LspA. R. typhi SPase II shows maximum identity (91%) with the sequence of R. prowazekii SPase II, which is a member of the typhus group rickettsiae. However, the SPase II from R. typhi and little known R. canadensis, which is considered to be another member of the typhus group (18), showed 80% identity, similar to that observed between R. typhi and non-typhus-group rickettsiae.
Rickettsiae enter host cells by induced phagocytosis in nonphagocytic cells in an unknown mechanism, followed by rapid escape from the phagosome (within 30 min postinfection) into the host cytoplasm. Rickettsiae undergo massive replication (8 to 48 h postinfection) in the host cytoplasm, and further growth results in host cell death and release of rickettsiae (12, 13, 39). The higher MNE level at the preinfection time point for all three target genes, lepB, lgt, and lspA, involved in protein secretion supports the biological finding that only live and metabolically active rickettsiae are capable of inducing host cell phagocytosis (13) and escape from the phagosome into host cytosol within 30 min postinfection (32, 34). During intracellular living (1 to 8 h postinfection) of rickettsiae, the expression of the target genes remains relatively lower. The increase in the transcription levels of the target genes after 8 h and the peak at 48 h postinfection suggests that the lepB, lgt, and lspA genes are involved in protein secretion during rickettsial replication in host cells.
Using the SignalP, version 3.0, neural network and hidden Markov model tools (5) and the LipoP, version 1.0 (16), software package, we identified a total of 89 secretory proteins with a putative signal peptide sequence out of the 838 annotated ORFs from the R. typhi genome (19). Of the 89 predicted secretory proteins, 14 are recognized as putative lipoproteins (data not shown). This in silico prediction of secretory nonlipoproteins and lipoproteins and the higher transcriptional level of lepB compared to lgt and lspA at all of the growth points examined in this study suggest that the SPase I encoded by lepB is involved in the processing of more secretory proteins than is SPase II.
The overexpression of lspA from E. coli or R. typhi in E. coli cells results in increased globomycin resistance, indicating that both SPases II were properly synthesized and are functional in E. coli. In genetic complementation experiments, the E. coli lspA gene restores the growth of temperature-sensitive E. coli strain Y815 at the nonpermissive temperature (42°C) at a rate approximately fivefold higher than that by R. typhi lspA. The complementation assay indicates that the biological activity of the recombinant SPase II from R. typhi is relatively low but significantly functional in E. coli for prolipoprotein processing. The globomycin assay showed that the SPase II from either R. typhi or E. coli confers a similar level of globomycin resistance to E. coli. These data suggest that globomycin binding to SPase II in the globomycin resistance assay and the processing of prolipoprotein by SPase II in the genetic complementation assay are two independent cellular activities (8, 15, 26). Although the functional domains are conserved, the overall low identity (22%) of R. typhi SPase II with that of E. coli may explain the low genetic complementation in the complex multicomponent lipoprotein secretion pathway of E. coli.
In conclusion, this work describes, for the first time, a rickettsial LspA homolog with highly conserved domains for bacterial SPase II activity. Our data show the differential expression of R. typhi lspA and its functional activity. Further work is necessary to elucidate the role of LspA in the secretion of rickettsial virulence factors.
Acknowledgments
The research presented in this article was supported by funds from the National Institutes of Health (R37 17828 and R01 A1059118).
We gratefully acknowledge Shunichi Miyakoshi, Sankyo Co., Ltd., Tokyo, Japan, for the generous gift of globomycin and J. D. H. Jongbloed, Department of Molecular Genetics, University of Groningen, Groningen, The Netherlands, for generously providing E. coli strain Y815.
Footnotes
Published ahead of print on 10 November 2006.
REFERENCES
- 1.Aliprantis, A. O., R. B. Yang, M. R. Mark, S. Suggett, B. Devaux, J. D. Radolf, G. R. Klimpel, P. Godowski, and A. Zychlinsky. 1999. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science 285:736-739. [DOI] [PubMed] [Google Scholar]
- 2.Al Mamun, A. A., R. S. Yadava, L. Ren, and M. Z. Humayun. 2000. The Escherichia coli UVM response is accompanied by an SOS-independent error-prone DNA replication activity demonstrable in vitro. Mol. Microbiol. 38:368-380. [DOI] [PubMed] [Google Scholar]
- 3.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 4.Azad, A. F., and C. B. Beard. 1998. Rickettsial pathogens and their arthropod vectors. Emerg. Infect. Dis. 4:179-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 6.De Greeff, A., A. Hamilton, I. C. Sutcliffe, H. Buys, L. Van Alphen, and H. E. Smith. 2003. Lipoprotein signal peptidase of Streptococcus suis serotype 2. Microbiology 149:1399-1407. [DOI] [PubMed] [Google Scholar]
- 7.Desvaux, M., N. J. Parham, A. Scott-Tucker, and I. R. Henderson. 2004. The general secretory pathway: a general misnomer? Trends Microbiol. 12:306-309. [DOI] [PubMed] [Google Scholar]
- 8.Dev, I. K., R. J. Harvey, and P. H. Ray. 1985. Inhibition of prolipoprotein signal peptidase by globomycin. J. Biol. Chem. 260:5891-5894. [PubMed] [Google Scholar]
- 9.Economou, A. 1999. Following the leader: bacterial protein export through the Sec pathway. Trends Microbiol. 7:315-320. [DOI] [PubMed] [Google Scholar]
- 10.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gan, K., S. D. Gupta, K. Sankaran, M. B. Schmid, and H. C. Wu. 1993. Isolation and characterization of a temperature-sensitive mutant of Salmonella typhimurium defective in prolipoprotein modification. J. Biol. Chem. 268:16544-16550. [PubMed] [Google Scholar]
- 12.Gaywee, J., S. Radulovic, J. A. Higgins, and A. F. Azad. 2002. Transcriptional analysis of Rickettsia prowazekii invasion gene homolog (invA) during host cell infection. Infect. Immun. 70:6346-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hackstadt, T. 1996. The biology of rickettsiae. Infect. Agents Dis. 5:127-143. [PubMed] [Google Scholar]
- 14.Inukai, M., M. Nakajima, M. Osawa, T. Haneishi, and M. Arai. 1978. Globomycin, a new peptide antibiotic with spheroplast-forming activity. II. Isolation and physico-chemical and biological characterization. J. Antibiot. (Tokyo) 31:421-425. [DOI] [PubMed] [Google Scholar]
- 15.Inukai, M., M. Takeuchi, K. Shimizu, and M. Arai. 1978. Mechanism of action of globomycin. J. Antibiot. (Tokyo) 31:1203-1205. [DOI] [PubMed] [Google Scholar]
- 16.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewenza, S., J. L. Gardy, F. S. Brinkman, and R. E. Hancock. 2005. Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. Genome Res. 15:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKiel, J. A., E. J. Bell, and D. B. Lackman. 1967. Rickettsia canada: a new member of the typhus group of rickettsiae isolated from Haemaphysalis leporispalustris ticks in Canada. Can. J. Microbiol. 13:503-510. [DOI] [PubMed] [Google Scholar]
- 19.McLeod, M. P., X. Qin, S. E. Karpathy, J. Gioia, S. K. Highlander, G. E. Fox, T. Z. McNeill, H. Jiang, D. Muzny, L. S. Jacob, A. C. Hawes, E. Sodergren, R. Gill, J. Hume, M. Morgan, G. Fan, A. G. Amin, R. A. Gibbs, C. Hong, X. J. Yu, D. H. Walker, and G. M. Weinstock. 2004. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J. Bacteriol. 186:5842-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller, P. Y., H. Janovjak, A. R. Miserez, and Z. Dobbie. 2002. Processing of gene expression data generated by quantitative real-time RT-PCR. BioTechniques 32:1372-1379. [PubMed] [Google Scholar]
- 21.Ogata, H., S. Audic, P. Renesto-Audiffren, P. E. Fournier, V. Barbe, D. Samson, V. Roux, P. Cossart, J. Weissenbach, J. M. Claverie, and D. Raoult. 2001. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 293:2093-2098. [DOI] [PubMed] [Google Scholar]
- 22.Ogata, H., B. La Scola, S. Audic, P. Renesto, G. Blanc, C. Robert, P. E. Fournier, J. M. Claverie, and D. Raoult. 2006. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogata, H., P. Renesto, S. Audic, C. Robert, G. Blanc, P. E. Fournier, H. Parinello, J. M. Claverie, and D. Raoult. 2005. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 3:e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paetzel, M., R. E. Dalbey, and N. C. Strynadka. 2000. The structure and mechanism of bacterial type I signal peptidases. A novel antibiotic target. Pharmacol. Ther. 87:27-49. [DOI] [PubMed] [Google Scholar]
- 25.Paetzel, M., A. Karla, N. C. Strynadka, and R. E. Dalbey. 2002. Signal peptidases. Chem. Rev. 102:4549-4580. [DOI] [PubMed] [Google Scholar]
- 26.Paitan, Y., E. Orr, E. Z. Ron, and E. Rosenberg. 1999. A nonessential signal peptidase II (Lsp) of Myxococcus xanthus might be involved in biosynthesis of the polyketide antibiotic TA. J. Bacteriol. 181:5644-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petit, C. M., J. R. Brown, K. Ingraham, A. P. Bryant, and D. J. Holmes. 2001. Lipid modification of prelipoproteins is dispensable for growth in vitro but essential for virulence in Streptococcus pneumoniae. FEMS Microbiol. Lett. 200:229-233. [DOI] [PubMed] [Google Scholar]
- 28.Pragai, Z., H. Tjalsma, A. Bolhuis, J. M. van Dijl, G. Venema, and S. Bron. 1997. The signal peptidase II (isp) gene of Bacillus subtilis. Microbiology 143:1327-1333. [DOI] [PubMed] [Google Scholar]
- 29.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahman, M. S., J. A. Simser, K. R. Macaluso, and A. F. Azad. 2005. Functional analysis of secA homologues from rickettsiae. Microbiology 151:589-596. [DOI] [PubMed] [Google Scholar]
- 31.Rahman, M. S., J. A. Simser, K. R. Macaluso, and A. F. Azad. 2003. Molecular and functional analysis of the lepB gene, encoding a type I signal peptidase from Rickettsia rickettsii and Rickettsia typhi. J. Bacteriol. 185:4578-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reglier-Poupet, H., C. Frehel, I. Dubail, J. L. Beretti, P. Berche, A. Charbit, and C. Raynaud. 2003. Maturation of lipoproteins by type II signal peptidase is required for phagosomal escape of Listeria monocytogenes. J. Biol. Chem. 278:49469-49477. [DOI] [PubMed] [Google Scholar]
- 33.Richards, A. L. 2004. Rickettsial vaccines: the old and the new. Expert Rev. Vaccines 3:541-555. [DOI] [PubMed] [Google Scholar]
- 34.Sander, P., M. Rezwan, B. Walker, S. K. Rampini, R. M. Kroppenstedt, S. Ehlers, C. Keller, J. R. Keeble, M. Hagemeier, M. J. Colston, B. Springer, and E. C. Bottger. 2004. Lipoprotein processing is required for virulence of Mycobacterium tuberculosis. Mol. Microbiol. 52:1543-1552. [DOI] [PubMed] [Google Scholar]
- 35.Sankaran, K., S. D. Gupta, and H. C. Wu. 1995. Modification of bacterial lipoproteins. Methods Enzymol. 250:683-697. [DOI] [PubMed] [Google Scholar]
- 36.Sankaran, K., and H. C. Wu. 1995. Bacterial prolipoprotein signal peptidase. Methods Enzymol. 248:169-180. [DOI] [PubMed] [Google Scholar]
- 37.Stoll, H., J. Dengjel, C. Nerz, and F. Gotz. 2005. Staphylococcus aureus deficient in lipidation of prelipoproteins is attenuated in growth and immune activation. Infect. Immun. 73:2411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venema, R., H. Tjalsma, J. M. van Dijl, A. de Jong, K. Leenhouts, G. Buist, and G. Venema. 2003. Active lipoprotein precursors in the Gram-positive eubacterium Lactococcus lactis. J. Biol. Chem. 278:14739-14746. [DOI] [PubMed] [Google Scholar]
- 39.Whitworth, T., V. L. Popov, X. J. Yu, D. H. Walker, and D. H. Bouyer. 2005. Expression of the Rickettsia prowazekii pld or tlyC gene in Salmonella enterica serovar Typhimurium mediates phagosomal escape. Infect. Immun. 73:6668-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wood, D. O., and A. F. Azad. 2000. Genetic manipulation of rickettsiae: a preview. Infect. Immun. 68:6091-6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamagata, H., C. Ippolito, M. Inukai, and M. Inouye. 1982. Temperature-sensitive processing of outer membrane lipoprotein in an Escherichia coli mutant. J. Bacteriol. 152:1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]