Abstract
Peptidoglycan (PG) is a cell wall heteropolymer that is essential for cell integrity. PG hydrolases participate in correct assembly of the PG layer and have been shown to be required for cell division, cell daughter separation, and maintenance of bacterial morphology. In silico analysis of the Helicobacter pylori genome resulted in identification of three potential hydrolases, Slt, MltD, and AmiA. This study was aimed at determining the roles of the putative lytic transglycosylases, Slt and MltD, in H. pylori morphology, growth, and PG metabolism. Strain 26695 single mutants were constructed using a nonpolar kanamycin cassette. The slt and mltD mutants formed normal bacillary and coccoid bacteria in the exponential and stationary phases, respectively. The slt and mltD mutants had growth rates comparable to the growth rate of the parental strain. However, the mltD mutant exhibited enhanced survival in the stationary phase compared to the wild type or the slt mutant. PG was purified from exponentially growing bacteria and from bacteria in the stationary phase, and its muropeptide composition was analyzed by high-pressure liquid chromatography. This analysis revealed changes in the muropeptide composition indicating that MltD and Slt have lytic transglycosylase activities. Glycan strand analysis suggested that Slt and MltD have exo and endo types of lytic transglycosylase activity, indicating that Slt is involved mainly in PG turnover and MltD is involved mainly in rearrangement of the PG layer. In this study, we determined the distinct roles of the lytic transglycosylases Slt and MltD in PG metabolism.
Resistance to antibiotics has increased dramatically in the last few decades. Helicobacter pylori, the etiological agent of gastric diseases such as gastroduodenal ulcers and adenocarcinoma, is becoming increasingly resistant to the few antibiotics effective in vivo against this infection.
Hence, new therapeutic strategies are required to overcome resistance to known antibiotics. PG is an essential macromolecule that surrounds bacteria and is responsible for their shape and resistance to turgor pressure. Its central role in cell viability has made the biosynthesis of PG one of the most successful antibiotic targets in bacteria. However, little is known about PG metabolism in H. pylori. Detailed knowledge about the PG metabolism of H. pylori could lead to the development of new antibiotics. Based on genome sequences, H. pylori appears to have little redundancy of genes involved in PG metabolism (1, 4, 20). H. pylori has all of the genetic complement required for the synthesis of PG precursors. Assembly of these precursors in the periplasm requires synthetases and PG hydrolases. H. pylori has three synthetases, PBP1, PBP2, and PBP3, and three PG hydrolases, including two lytic transglycosylases, Slt (HP0645) and MltD (HP1572), and an N-acetylmuramoyl-l-alanyl amidase, AmiA (HP0772).
The aim of this work was to characterize the two lytic transglycosylases, Slt and MltD. We constructed single and double mutants and studied their growth and morphological phenotypes. Using reverse-phase HPLC, we analyzed the PG muropeptide composition and glycan strand distribution in the mutants. Our results indicate that Slt and MltD are nonredundant lytic transglycosylases with exo and endo types of activity, respectively.
MATERIALS AND METHODS
Abbreviations:
PG, peptidoglycan; PBP, penicillin-binding protein; HPLC, high-pressure liquid chromatography; LPS, lipopolysaccharide; PBS, phosphate-buffered saline; G, N-acetyl-d-glucosamine; M, N-acetylmuramic acid; anhM, N-acetyl-anhydromuramic acid; G-M-dipeptide, N-acetyl-d-glucosaminyl-β(1,4)-N-acetylmuramyl-l-Ala-d-Glu; G-M-tripeptide, N-acetyl-d-glucosaminyl-β(1,4)-N-acetylmuramyl-l-Ala-γ-d-Glu-meso-diaminopimelic acid; G-anhM-tetrapeptide, N-acetyl-d-glucosaminyl-N-acetyl-anhydromuramyl-l-Ala-γ-d-Glu-meso-diaminopimelic acid-d-Ala; G-anhM-tetrapeptide-glycine, N-acetyl-d-glucosaminyl-N-acetyl-anhydromuramyl-l-Ala-γ-d-Glu-meso-diaminopimelic acid-d-Ala-Gly; G-M-pentapeptide, N-acetyl-d-glucosaminyl-β(1,4)-N-acetylmuramyl-l-Ala-γ-d-Glu-meso-diaminopimelic acid-d-Ala-d-Ala.
Bacteria, cells, and growth conditions.
Escherichia coli MC1061 (8) and DH5α were used as hosts for construction and preparation of plasmids. These strains were cultivated in Luria-Bertani solid and liquid media supplemented as appropriate with spectinomycin (100 μg/ml), kanamycin (40 μg/ml), or both of these antibiotics. H. pylori strains 26695 (20) and N6 (11) were used to construct mutants. PG was extracted from strain 26695 and isogenic mutants of this strain. These bacteria were grown microaerobically at 37°C on blood agar plates or in liquid medium consisting of brain heart infusion (Oxoid) containing 0.2% β-cyclodextrin (Sigma) supplemented with an antibiotic-antifungal agent mixture (7). H. pylori mutants were selected with 20 μg/ml kanamycin or 10 μg/ml gentamicin.
Construction of mutants.
Genes were disrupted as previously described (18). H. pylori mutants were constructed by allelic exchange after transformation with a suicide plasmid carrying the gene of interest interrupted by a nonpolar aphA-3 cassette (18) or the mini-Tn3-Km transposon (14). A double mutant was constructed by disrupting the slt gene with the nonpolar gentamicin aacC4 (6) cassette as described below for the nonpolar kanamycin cassette. PCR was used to confirm that correct allelic exchange occurred. Gene replacements were confirmed by sequencing to ensure sequence fidelity. All reagents, enzymes, and kits were used according to manufacturers' recommendations. Midiprep (HiSpeed plasmid midi kit) and DNA extraction kits (QIAamp DNA extraction kit) were purchased from QIAGEN.
Plasmids pILL2001 and pILL2002 were used to construct the slt and mltD mutants, respectively. pILL570ΔNot carrying the hp0645 (slt) and hp1572 (mltD) genes was used as the template for an Expand High Fidelity PCR (Amersham) with oligonucleotides 645-1 (5′-GAUGAUGAUGGTACCGTGTCTGTTGTTTCTAGCATC-3′ (the underlined sequence is a KpnI site) and 645-2 (5′-AUCAUCAUCGGATCCCTAAACGACATGTTTAACCCCAACATC-3′ (the underlined sequence is a BamHI site) for the slt gene and with oligonucleotides 1572-1 (5′-GAUGAUGAUGGTACCTTTTCCTGCTATAAGCCCTTGATG-3′ (the underlined sequence is a KpnI site) and 1572-2 (5′-AUCAUCAUCGGATCCCTTGGAAACCTTAAAATCCTACAACCAC-3′ (the underlined sequence is a BamHI site) for the mltD gene. PCR products were digested with BamHI (Amersham) and KpnI (Amersham) and ligated (T4 DNA ligase; Amersham) with the aphA-3 or aacC4 nonpolar cassette digested with the same endonucleases.
Extraction and analysis of lipopolysaccharide.
H. pylori LPS was extracted from plate cultures by the proteinase K method (10). LPS samples were separated by Tricine-sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis as described by Lesse and colleagues (16). The LPS was visualized by silver staining (21).
Peptidoglycan extraction and analysis.
The growth of liquid cultures of the H. pylori parental strain and isogenic mutant strains was stopped after various times, and the cultures were chilled in an ice-ethanol bath. The crude murein sacculus was immediately extracted in boiling sodium dodecyl sulfate (final concentration, 4%). Purification procedures and HPLC analyses were performed as previously described (12). Mutanolysin (Sigma)-digested samples were analyzed by HPLC using a Hypersil ODS18 reverse-phase column (250 by 4.6 mm; particle size, 3 μm) with a 0 to 15% methanol (HPLC grade; Fischer) gradient in sodium phosphate buffer (pH 4.3 to 5.0). Chromatograms were obtained by monitoring at 206 nm. Each peak was collected, desalted, and identified by matrix-assisted laser desorption ionization mass spectrometry as described previously (2).
A glycan chain analysis was performed as previously described (5, 13). Briefly, H. pylori PG was digested with purified human serum amidase kindly provided by Waldemar Vollmer. The digestion was done in 50 mM Tris-HCl (pH 7.9), 5 mM MgCl2, 0.02% NaN3. Soluble material was first purified on a MonoS (HR5/5) column (Amersham Pharmacia) using 10 mM sodium phosphate buffer (pH 2). Glycans eluted with the flowthrough and were collected. Free peptides were eluted in one step using 1 M NaCl-10 mM sodium phosphate buffer (pH 2). The experiments were performed at room temperature using a flow rate of 1 ml/min. An amino acid and amino sugar analysis was performed with the purified glycan fraction and the free peptide fraction to ensure the purity of each fraction, which confirmed that complete digestion had occurred. Purified glycans were analyzed by reverse-phase HPLC using a 5-μm Nucleosil 300 C18 column (250 by 4.6 mm) at 50°C. A convex gradient ranging from 0 to 10.5% acetonitrile (−4 curve of the Shimadzu CLASS-VP software) in 100 mM sodium phosphate buffer (pH 2) was used for 90 min at a flow rate of 0.5 ml/min. Unresolved glycan material was eluted after the convex gradient in a single step with 30% acetonitrile in 100 mM sodium phosphate buffer (pH 2). Glycan material was detected at 202 nm.
Slt70 digestion of H. pylori PG.
PG from strain 26695 slt mltD was incubated in 300 mM sodium acetate buffer (pH 4.5) with His6-tagged Slt70 (1 ng/ml) for different times (1 and 5 min and 48 h) at 37°C. The reaction was stopped by boiling a sample for 5 min. Muropeptides were separated as indicated above and were identified by matrix-assisted laser desorption ionization mass spectrometry as described previously (2).
Electron microscopy.
For transmission electron microscopy, samples were washed in PBS and prefixed in 2.5% glutaraldehyde in PBS for 30 min. After postfixation in 2% molybdate (in PBS), bacteria were examined with a JEOL Jem 1010 electron microscope.
RESULTS
Characterization of slt and mltD mutants.
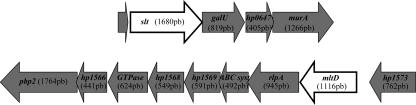
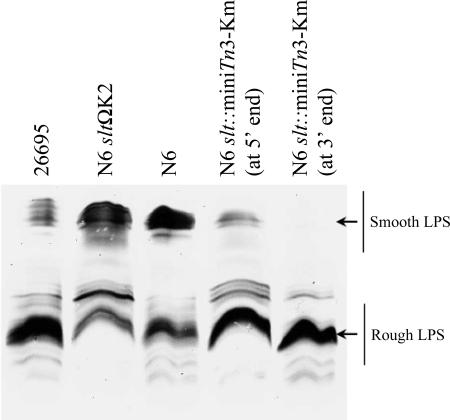
We constructed slt and mltD mutants having strain 26995 and N6 backgrounds using both the mini-Tn3-Km transposon (15) and the nonpolar kanamycin cassette (18). The slt and mltD genes are organized in putative operons (Fig. 1). Therefore, to study their role in H. pylori PG metabolism, we had to ensure that inactivation of these genes would not have polar effects on downstream genes, such as galU in the case of slt. GalU catalyzes the conversion of glucose-1-phosphate to UDP-glucose. UDP-glucose is a substrate of GalE, which generates UDP-galactose, an amino sugar precursor in the synthesis of LPS. Interference with GalU activity thus leads to a rough LPS. We analyzed the LPS phenotype of strain 26995 (Fig. 2) and found that this strain had a rough LPS. Therefore, to observe eventual polar effects of either the mini-Tn3 transposon or the nonpolar kanamycin cassette, we constructed slt mutants of strain N6, which has smooth LPS (Fig. 2). As shown in Fig. 2, while the two individual mini-Tn3 mutants with insertions either at the 5′ end or at the 3′ end of the slt gene affected the smooth LPS phenotype, the nonpolar kanamycin cassette had no effect on the LPS phenotype, showing the nonpolar nature of the mutant.
FIG. 1.
Schematic diagram of the genomic regions surrounding the slt and mltD genes in strain 26695. By PCR analysis, we confirmed the conservation of the two regions in different H. pylori strains.
FIG. 2.
Silver staining of LPS extracted from different H. pylori strains and slt mutants. While the mini-Tn3-Km transposon insertion into the slt gene had a polar effect on the galU gene resulting in a rough LPS phenotype, the nonpolar K2 cassette did not affect the smooth LPS genotype of the N6 strain. Inactivation of the mltD gene had no effect on the LPS phenotype (data not shown).
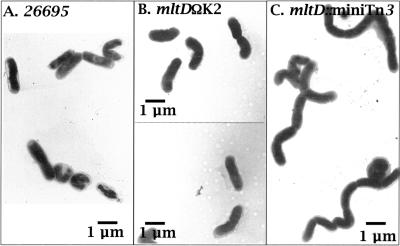
The same approach was used for analyzing the mltD mutants. The mltD gene appears to be the first gene of an eight-gene operon (Fig. 1). It includes homologues of the rare lipoprotein A (HP1571), the inner membrane component of an ABC transporter system (HP1570), a GTPase involved in cell division (HP1567), and penicillin-binding protein 2 (HP1565). An expected polar effect on downstream genes would affect cell division. The results of a morphological analysis of the mini-Tn3 and kanamycin mutants are shown in Fig. 3. While mini-Tn3 insertions at the 3′ end of mltD led to a chaining phenotype, mltD inactivation with the kanamycin cassette resulted in normal bacillary morphology. Hence, we confirmed the nonpolar nature of both slt and mltD mutants.
FIG. 3.
Impact of the polar effect on the mini-Tn3-Km transposon on the morphology of the H. pylori mltD mutant. Insertion of the transposon into the mltD gene resulted in a chaining phenotype, probably due to a polar effect on hp1567 encoding a putative GTPase. Inactivation of mltD with the nonpolar K2 cassette had no effect on the mutant morphology, reinforcing the nonpolar nature of the K2 cassette. An sltΩK2 mutant had normal morphology (data not shown).
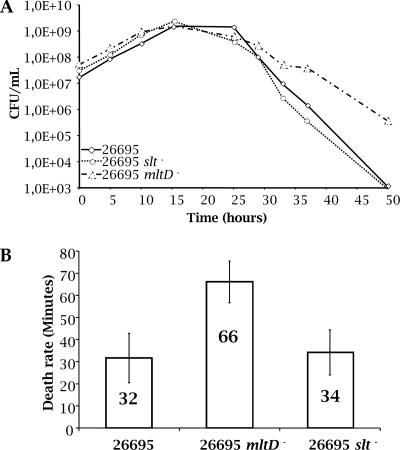
Next, we were interested in studying any growth defects that the nonpolar mutants might have. We determined the numbers of CFU during exponential growth and the stationary phase for wild-type strain 26695 and the isogenic slt and mltD mutants. As shown in Fig. 4, the three strains had the same growth rate. However, after entry into the stationary phase, the mltD mutant remained viable longer. This result was reflected by the lower rate of death (Fig. 4B) of the mltD mutant.
FIG. 4.
(A) Growth curves for H. pylori 26695 and slt and mltD mutants of this strain. Growth in liquid cultures was monitored by determining the optical density at 600 nm, and bacterial viability was monitored by counting the number of CFU. Growth experiments were done six times (the results of a representative experiment are shown). The three strains had identical growth rates. (B) Survival of the mltD strain in the stationary phase was enhanced, as illustrated by a lower death rate. On average, it took 66 min for one-half of the mltD population to die, while on average one-half of the populations of both the 26695 strain and the slt mutant died in 32 to 34 min.
Muropeptide composition.
Since Slt and MltD are predicted to be involved in PG metabolism, in particular in PG degradation, we purified the PG of the parental strain and of the two mutants in order to analyze their muropeptide compositions by reverse-phase HPLC. The results are shown in Tables S1, S2, and S3 in the supplemental material, which show the muropeptide compositions of the three strains at different times during growth (8 h, 24 h, and 48 h, respectively). A difference that was growth dependent but strain independent involved the increase in the level of the G-M-dipeptide motif when H. pylori entered the stationary phase. This modification is discussed elsewhere (9).
Globally, the mltD mutant had a muropeptide composition similar to that of parental strain 26695. Some differences were apparent, such as a modest decrease in the level of anhydromuropeptides (Table 1). Another difference involved the proportions of monomeric and dimeric muropeptides. The mltD mutant had a modest increase in the proportion of the monomeric muropeptides compared to the parent strain, indicating that there was a decrease in the degree of cross-linking in the mltD mutant. The slt mutant exhibited the same global trend in terms of changes in muropeptide composition as the mltD mutant, but to a much greater extent. The decrease in the level of anhydromuropeptides and degree of cross-linking (decrease in the proportion of dimers) were more pronounced in the slt mutant. Furthermore, in the slt mutant there was a marked accumulation of muropeptides having a tripeptide as a stem peptide. The increase was inversely proportional to the decrease in the level of muropeptides having tetrapeptides and tetraglycine peptides (Table 2). The differences in the degrees of cross-linking, in the levels of anhydromuropeptides, and in the levels of G-M-tripeptide were even greater in the slt mltD double mutant. Most importantly, despite the fact that slt and mltD are the only two homologues of genes encoding known lytic transglycosylases in the H. pylori genome, the double mutant still had anhydromuropeptide structures in its muropeptide. These structures were composed mainly of the G-anhM-penta, G-anhM-tri-tetra-G-M, and G-anhM-penta-tetra-G-M muropeptides (see Tables S1, S2, and S3 in the supplemental material). Only trace amounts of the other anhydromuropeptides were found.
TABLE 1.
Proportions of monomers, dimers, and anhydromuropeptides and average glycan chain lengths after 8, 24, and 48 h of incubation
| Time (h) | Strain | % (mean ± SD) of:
|
Avg glycan chain length (±SD) | ||
|---|---|---|---|---|---|
| Monomers | Dimers | Anhydromuropeptides | |||
| 8 | 26695 | 70.7 ± 1.8 | 29.3 ± 1.8 | 14.2 ± 1.1 | 9.1 ± 0.3 |
| mltD | 73.1 ± 1.6 | 26.9 ± 1.6 | 12.0 ± 0.9 | 10.7 ± 0.7 | |
| slt | 76.3 ± 2.0 | 23.7 ± 2.0 | 9.9 ± 1.4 | 13.1 ± 0.1 | |
| slt mltD | 78.8 | 21.2 | 5.7 | 15.7 | |
| 24 | 26695 | 68.0 ± 0.8 | 32.0 ± 0.8 | 14.9 ± 0.8 | 8.9 ± 0.4 |
| mltD | 71.2 ± 0.4 | 28.8 ± 0.4 | 14.5 ± 0.7 | 8.9 ± 0.4 | |
| slt | 73.3 ± 0.4 | 26.7 ± 0.4 | 10.6 ± 1.5 | 11.6 ± 1.4 | |
| slt mltD | 77.9 | 22.1 | 5.4 | 16.6 | |
| 48 | 26695 | 68.9 ± 2.2 | 31.1 ± 2.2 | 13.3 ± 4.9 | 7.9 |
| mltD | 70.5 ± 2.9 | 29.0 ± 2.3 | 13.1 ± 0.1 | 9.8 | |
| slt | 74.5 | 25.5 | 9.8 | 12.8 | |
| slt mltD | 79.7 | 20.3 | 4.2 | 20.0 | |
TABLE 2.
Proportions of dipeptides, tripeptides, tetrapeptides, tetraglycinepeptides, and pentapeptides after 8, 24, and 48 h of incubation
| Time (h) | Strain | % (mean ± SD) of:
|
||||
|---|---|---|---|---|---|---|
| Dipeptides | Tripeptides | Tetrapeptides | Tetraglycinepeptides | Pentapeptides | ||
| 8 | 26695 | 3.2 ± 1.0 | 19.1 ± 0.8 | 45.4 ± 1.7 | 6.6 ± 1.9 | 54.9 ± 1.7 |
| mltD | 3.7 ± 1.5 | 23.0 ± 0.3 | 42.6 ± 2.1 | 5.5 ± 0.8 | 52.2 ± 2.3 | |
| slt | 4.7 ± 1.0 | 28.3 ± 0.9 | 32.0 ± 1.8 | 5.2 ± 0.7 | 53.4 ± 1.5 | |
| slt mltD | 6.4 | 36.6 | 30.1 | 6.2 | 41.8 | |
| 24 | 26695 | 9.5 ± 0.6 | 18.9 ± 0.1 | 47.8 ± 1.2 | 5.9 ± 0.3 | 49.8 ± 0.6 |
| mltD | 15.4 ± 0.6 | 18.3 ± 0.5 | 44.8 ± 0.5 | 5.4 ± 0.5 | 45.0 ± 0.3 | |
| slt | 8.2 ± 1.1 | 28.5 ± 0.7 | 34.7 ± 0.2 | 5.3 ± 0.5 | 50.1 ± 0.0 | |
| slt mltD | 10.5 | 36.2 | 34.5 | 7.6 | 33.2 | |
| 48 | 26695 | 16.6 ± 7.6 | 9.9 ± 0.6 | 45.5 ± 4.2 | 6.5 ± 0.1 | 52.6 ± 0.5 |
| mltD | 13.8 ± 0.7 | 16.0 ± 0.3 | 50.6 ± 4.9 | 5.8 ± 1.4 | 42.3 ± 0.8 | |
| slt | 18.3 | 17.0 | 35.7 | 6.1 | 48.5 | |
| slt mltD | 18.7 | 24.8 | 39.2 | 8.6 | 28.9 | |
Since anhydromuropeptides represent the ends of the glycan chains, we estimated the overall average lengths of the glycan chains of parental strain 26695, the two single mutants, and the double mutant. While the parental strain and the mltD mutant had average glycan chain lengths of around 8 to 10 disaccharide repeating units, with a moderate decrease in the stationary phase, the slt mutant had markedly greater average chain lengths, which ranged from 11.6 to 13.1 disaccharide repeating units. The increase in average length was clearly greater in the double mutant (range, 15.7 to 20 disaccharide repeating units).
Glycan chain length analysis.
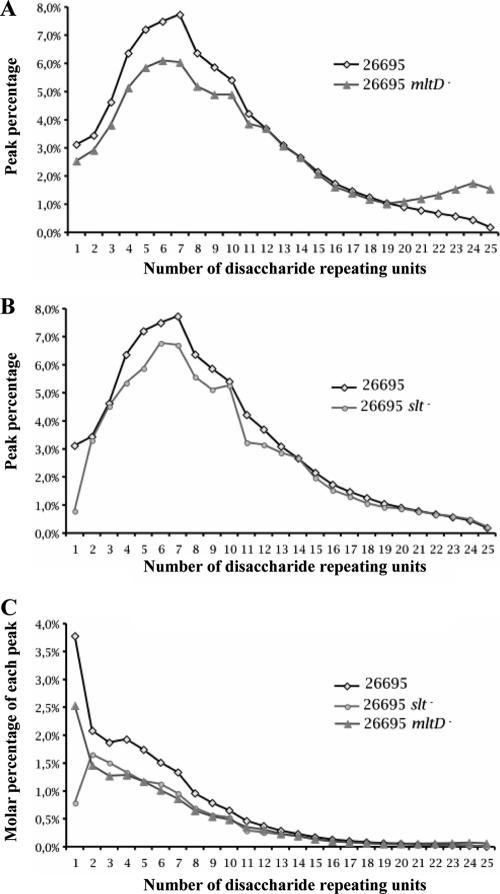
Next, we were interested in characterizing by a more detailed method the glycan strands of the parental strain and the single mutants. We digested the purified peptidoglycan of each strain with human serum amidase and separated the glycan strands from the free peptides by chromatography using a MonoS column. The glycan fraction eluted exclusively with the flowthrough, while the free peptides were retained on the column. The purified glycan fraction was analyzed by reverse-phase HPLC (see Fig. S1 in the supplemental material). The profile is reminiscent of the results of a glycan strand analysis of E. coli (13). The analysis of the glycan strand distribution was restricted to an analysis after 8 h of growth since the human serum amidase was not able to cleave the G-M-dipeptide that accumulated in the stationary phase (24 h and 48 h), as previously described (23). The proportion of each peak was calculated based on the total amount of UV-absorbing material (Fig. 5; see Fig. S1 in the supplemental material). Several differences were observed between the parental strain and the two single mutants. Both mutants showed marked increases in the proportion of very long glycan chains (≥26 disaccharide repeating units) (see Fig. S1 in the supplemental material). The proportion of these glycan species increased from 17.4% in the parental strain to 23.6% and 28.3% in the mltD and slt mutants, respectively. From the analysis of the area percentage of each glycan species and the corresponding molar percentages, we observed that in the mltD mutant there was a marked decrease in the short glycan species (1 to 11 disaccharide repeating units) (Fig. 5A and C) and an inverse increase of the glycan species with more than 19 disaccharide repeating units (Fig. 5A). These results suggest that mltD might have an endotransglycosylase activity.
FIG. 5.
(A and B) Glycan strand length distribution for parental strain 26695 compared to the glycan strand length distributions for the slt mutant (A) and the mltD mutant (B). The level of each glycan strand species is expressed as a percentage of the total UV absorbing material (see Fig. S1 in the supplemental material). (C) Level of each glycan strand species expressed as a molar percentage, taking into account the abundance of each species. The data show that the majority of the H. pylori glycan strands are short and that the two mutants have decreased amounts of short glycan strands. Furthermore, the slt mutant is characterized by an almost complete absence of G-anhM disaccharide (peak 1 in Fig. S1 in the supplemental material).
In the slt mutant there was also a marked decrease in the very short glycan strands (Fig. 5B and C). However, the major decrease was due to an almost complete absence of the disaccharide species (peak 1) from the slt mutant, which was present in the mltD mutant. This suggests that in contrast to MltD, Slt appears to cut preferentially at the ends of the glycan strands to generate free disaccharide units, suggesting that Slt has an exo type of activity.
Slt70 digestion and G-M-tripeptide localization.
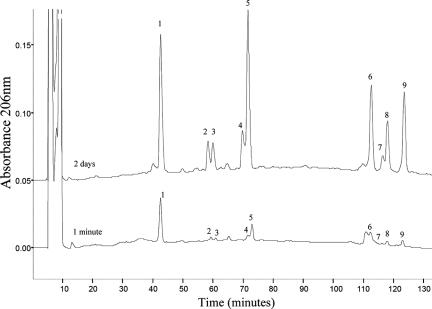
In the muropeptide and glycan strand analysis, we observed that the slt mutant accumulated the G-M-tripeptide motif and generated significantly less of the disaccharide G-anhM glycan species. Since we did not observe the monomeric G-anhM-tripeptide in the PG under any conditions tested, this suggests that the G-M-tripeptides are either at the nonreducing ends of the glycan strands or in the middle of the glycan strands. However, since the slt mutant generates less G-anhM, this suggests that Slt functions as an exoenzyme. Hence, the accumulation of the G-M-tripeptides was probably due to a preferential location at the nonreducing ends of the glycan strands. To test this hypothesis, we used the E. coli Stl70 lytic transglycosylase, which has been shown to be an exoenzyme from the nonreducing end (3, 17). We digested the same amount of H. pylori PG with Slt70 for very brief periods (1 and 5 min) and after 2 days analyzed the nature of the Slt70-generated muropeptides by HPLC. As shown in Fig. 6, Slt70 generated the G-anhM-tripeptide preferentially after 1 min of incubation, clearly indicating that the G-M-tripeptides are located preferentially at the nonreducing ends of the glycan strands. Similar results were obtained after 5 min of digestion (data not shown).
FIG. 6.
Digestion of H. pylori PG with exo-type lytic transglycosylase Slt70 from E. coli. H. pylori PG was digested either completely with Slt70 (2 days) or for brief periods (1 and 5 min). Each peak was collected, desalted, and analyzed by matrix-assisted laser desorption ionization—time of flight mass spectrometry to confirm the muropeptide nature of each peak. Peaks 1 to 9 correspond to G-anhM-tripeptide, G-anhM-tetrapeptide, G-anhM-tetrapeptide-glycine, G-anhM-dipeptide, G-anhM-pentapeptide, G-anhM-tri-tetra-G-anhM, G-anhM-tetra-tetra-glycine-G-anhM, G-anhM-tetra-tetra-G-anhM, and G-anhM-penta-tetra-G-anhM, respectively.
DISCUSSION
Based on the genome analysis, two genes, slt and mltD, are the only genes that encode proteins having a lytic transglycosylase domain (4). The slt gene is predicted to encode a 560-amino-acid protein with a classical signal peptide. Slt has an SLT domain at the C-terminal end with 34% identity to E. coli Slt70. The rest of the protein has no homologue in the databases. In contrast, the mltD gene encodes a shorter protein (372 amino acids) with a classical signal peptide, an SLT domain at its N-terminus, and a single LysM domain at its C terminus. The SLT domain exhibits 31% identity to Slt70. Hence, both proteins were predicted to function as lytic transglycosylases.
Analyses of the muropeptide compositions and the glycan strand distributions of the single and double mutants suggested that both proteins are lytic transglycosylases with nonredundant functions. Inactivation of each gene resulted in a substantial decrease in the level of anhydromuropeptides. Since these muropeptides species represent the products of lytic transglycosylase activities, the results suggested that both proteins function as transglycosylases. The muropeptide composition analysis of the single mutants and the double mutant (Table 1; see Tables S1 to S3 in the supplemental material) showed that the total percentage of anhydromuropeptides results from the additive effects of Slt and MltD. Hence, the decrease in the level of anhydromuropeptides in the double mutant compared to the level in the parental strain corresponds to the difference in the level of the anhydromuropeptides in the slt mutant plus the difference in level in the mltD mutant. This suggests that Slt and MltD each generate anhydromuropeptides independent of the other lytic transglycosylase and that the two proteins have different physiological roles.
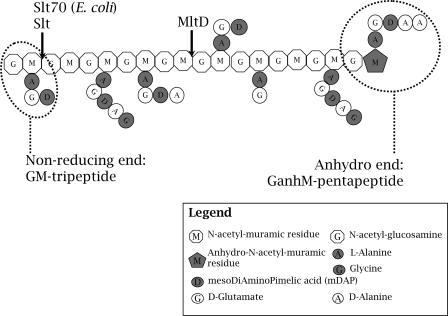
Glycan strand analysis of the slt and mltD mutants confirmed this hypothesis. While both mutants accumulate very long glycan strands, as would be expected for lytic transglycosylase-deficient mutants, the mutants seem to act by different mechanisms. While the slt mutant increases the length of its glycan strands by generating less of the very short glycan strands, in particular the disaccharide G-anhM, the mltD mutant increases the length of its glycan strands by reducing the amount of glycan strands that are up to 10 to 11 disaccharide units long and gradually increasing the amount of glycan strands with more than 19 disaccharide repeating units (Fig. 5). The distinct patterns of glycan strand distribution of the slt and mltD mutants suggest that Slt and MltD function as an exo type and an endo type of lytic transglycosylase, respectively (Fig. 7). The inferred types of activities of Slt and MltD are consistent with the increased fitness of the mltD mutant seen during the stationary phase growth. As an endo type of lytic transglycosylase, MltD would have a greater impact on PG layer stability than Slt would have. In contrast, Slt would function primarily in releasing anhydromuropeptides during PG turnover. However, the inferred exo and endo types of lytic transglycosylase activity of Slt and MltD, respectively, are based on indirect evidence. We are currently working on the biochemistry of the proteins to confirm their activities in vitro.
FIG. 7.
Schematic diagram of the preferential spatial organization of different stem peptides of H. pylori PG and substrate specificity of the lytic transglycosylases Slt and MltD.
The slt mutant has a marked effect on the proportion of G-M-tripeptide, massive amounts of which accumulate in the mutant PG layer. We can interpret this result either as a result of Slt substrate specificity or as a result of a particular localization of tripeptides along the glycan strands or both. Both hypotheses are possible, and the two processes might occur simultaneously. Based on the glycan strand analysis, Slt appears to be an exo type of enzyme. If the accumulation of G-M-tripeptide resulted exclusively from substrate specificity, we would expect to observe the presence of G-anhM-tripeptides in wild-type strains. However, these structures are completely absent from H. pylori PG. Therefore, we infer that these compounds are always generated as turnover products and immediately released from the PG layer. The only way to generate readily soluble G-anhM-tripeptides is to generate them when the G-M-tripeptide structures are exclusively at the nonreducing ends of glycan strands (Fig. 7). In the absence of Slt, these structures accumulate in the PG layer of the mutant exclusively as G-M-tripeptides. We confirmed the localization of the G-M-tripeptides at the ends of glycan strands by partially digesting H. pylori PG with the exo type of lytic transglycosylase (Slt70) from E. coli. Slt70 preferentially released G-anhM-tripeptide after very short incubation times (1 min) (Fig. 6).
Interestingly, in the double mutant, we still observed the presence of anhydromuropeptides. We can explain this result either (i) by the hypothesis that there is a novel class of lytic transglycosylases, which still have to be identified, or (ii) by the hypothesis that the glycosyltransferase domain of the bifunctional class A high-molecular-weight PBP1 is capable of generating the intramolecular anhydrous bond as a nascent glycan strand is released from the undecaprenylphosphate anchor. Further work is required to distinguish between these two hypotheses. Nevertheless, the double mutant contains almost exclusively the G-anhM-pentapeptide and the G-anhM-penta-tetra-G-M dimeric muropeptide. This indicates that the anhydro ends of glycan strands are enriched in intact stem pentapeptides (Fig. 7), consistent with de novo synthesis favoring a role for PBP1 in the generation of the anhydromuropeptides.
Finally, our results indicate that in H. pylori synthesis of new glycan strands is initiated by a G-M-tripeptide and terminates with a G-M-pentapeptide. The G-M-tripeptide might originate from a classical lipid II precursor immediately processed from a pentapeptide to a tripeptide. Processing might occur either with an ld-endopeptidase or by consecutive digestion with dd- and ld-carboxypeptidases. H. pylori lacks classical dd-carboxypeptidases, although we cannot exclude the possibility that the three high-molecular-weight PBPs function as dd-carboxypeptidases. However, no homologue of known ld-peptidase has been found in the H. pylori genome, which would require a novel class of ld-peptidases. Alternatively, the G-M-tripeptide might originate directly from the PG precursor pool as a lipid precursor (as shown for E. coli and Staphylococcus aureus [19, 22]) and be used to initiate glycan strand elongation. In fact, a precursor pool origin for the G-M-tripeptide could be an elegant mechanism to naturally regulate the glycan strand length. The glycan strand length distribution would be regulated by the precursor pool of UDP-M-tripeptide rather by the synthetases or by the PG hydrolases.
Supplementary Material
Acknowledgments
We thank Dominique Mengin-Lecreulx and Waldemar Vollmer for providing purified Slt70 and human serum amidase, respectively. We are grateful to Marie-Christine Prévost for access to the electron microscope.
C. Chaput was supported by a fellowship from the French Ministry of Science and from la Fondation pour la Recherche Médicale. I. G. Boneca was supported by a fellowship from the Fundação para a Ciência e Tecnologia (Portugal) and by a Bourse Roux from the Institut Pasteur and is an Institut National de la Santé et de la Recherche Médicale research scientist.
Footnotes
Published ahead of print on 3 November 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Antignac, A., J. C. Rousselle, A. Namane, A. Labigne, M. K. Taha, and I. G. Boneca. 2003. Detailed structural analysis of the peptidoglycan of the human pathogen Neisseria meningitidis. J. Biol. Chem. 278:31521-31528. [DOI] [PubMed] [Google Scholar]
- 3.Beachey, E. H., W. Keck, M. A. de Pedro, and U. Schwarz. 1981. Exoenzymatic activity of transglycosylase isolated from Escherichia coli. Eur. J. Biochem. 116:355-358. [DOI] [PubMed] [Google Scholar]
- 4.Boneca, I. G., H. de Reuse, J. C. Epinat, M. Pupin, A. Labigne, and I. Moszer. 2003. A revised annotation and comparative analysis of Helicobacter pylori genomes. Nucleic Acids Res. 31:1704-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boneca, I. G., Z. H. Huang, D. A. Gage, and A. Tomasz. 2000. Characterization of Staphylococcus aureus cell wall glycan strands, evidence for a new beta-N-acetylglucosaminidase activity. J. Biol. Chem. 275:9910-9918. [DOI] [PubMed] [Google Scholar]
- 6.Bury-Moné, S., S. Skouloubris, C. Dauga, J. M. Thiberge, D. Dailidiene, D. E. Berg, A. Labigne, and H. De Reuse. 2003. Presence of active aliphatic amidases in Helicobacter species able to colonize the stomach. Infect. Immun. 71:5613-5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bury-Mone, S., J. M. Thiberge, M. Contreras, A. Maitournam, A. Labigne, and H. De Reuse. 2004. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53:623-638. [DOI] [PubMed] [Google Scholar]
- 8.Casadaban, M. J., J. Chou, and S. N. Cohen. 1980. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J. Bacteriol. 143:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaput, C., C. Ecobichon, N. Cayet, S. E. Girardin, C. Werts, S. Guadagnini, M.-C. Prevost, D. Mengin-Lecreulx, A. Labigne, and I. G. Boneca. 2006. Role of AmiA in the morphological transition of Helicobacter pylori and in immune escape. PLoS Pathogens 2:844-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards, N. J., M. A. Monteiro, G. Faller, E. J. Walsh, A. P. Moran, I. S. Roberts, and N. J. High. 2000. Lewis X structures in the O antigen side-chain promote adhesion of Helicobacter pylori to the gastric epithelium. Mol. Microbiol. 35:1530-1539. [DOI] [PubMed] [Google Scholar]
- 11.Ferrero, R. L., V. Cussac, P. Courcoux, and A. Labigne. 1992. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J. Bacteriol. 174:4212-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glauner, B. 1988. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem. 172:451-464. [DOI] [PubMed] [Google Scholar]
- 13.Harz, H., K. Burgdorf, and J. V. Holtje. 1990. Isolation and separation of the glycan strands from murein of Escherichia coli by reversed-phase high-performance liquid chromatography. Anal. Biochem. 190:120-128. [DOI] [PubMed] [Google Scholar]
- 14.Jenks, P. J., C. Chevalier, C. Ecobichon, and A. Labigne. 2001. Identification of nonessential Helicobacter pylori genes using random mutagenesis and loop amplification. Res. Microbiol. 152:725-734. [DOI] [PubMed] [Google Scholar]
- 15.Labigne, A., P. Courcoux, and L. Tompkins. 1992. Cloning of Campylobacter jejuni genes required for leucine biosynthesis, and construction of leu-negative mutant of C. jejuni by shuttle transposon mutagenesis. Res. Microbiol. 143:15-26. [DOI] [PubMed] [Google Scholar]
- 16.Lesse, A. J., A. A. Campagnari, W. E. Bittner, and M. A. Apicella. 1990. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J. Immunol. Methods 126:109-117. [DOI] [PubMed] [Google Scholar]
- 17.Romeis, T., W. Vollmer, and J. V. Holtje. 1993. Characterization of three different lytic transglycosylases in Escherichia coli. FEMS Microbiol. Lett. 111:141-146. [DOI] [PubMed] [Google Scholar]
- 18.Skouloubris, S., J. M. Thiberge, A. Labigne, and H. De Reuse. 1998. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect. Immun. 66:4517-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sobral, R. G., A. M. Ludovice, H. de Lencastre, and A. Tomasz. 2006. Role of murF in cell wall biosynthesis: isolation and characterization of a murF conditional mutant of Staphylococcus aureus. J. Bacteriol. 188:2543-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 21.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 22.van Heijenoort, Y., M. Gomez, M. Derrien, J. Ayala, and J. van Heijenoort. 1992. Membrane intermediates in the peptidoglycan metabolism of Escherichia coli: possible roles of PBP 1b and PBP 3. J. Bacteriol. 174:3549-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang, Z. M., X. Li, R. R. Cocklin, M. Wang, K. Fukase, S. Inamura, S. Kusumoto, D. Gupta, and R. Dziarski. 2003. Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-l-alanine amidase. J. Biol. Chem. 278:49044-49052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.