Abstract
Kingella kingae is an emerging bacterial pathogen that is increasingly recognized as the causative agent of a variety of pediatric diseases, including septic arthritis and osteomyelitis. The pathogenesis of K. kingae disease is believed to begin with colonization of the upper respiratory tract. In the present study, we examined interactions between K. kingae and cultured respiratory epithelial cells and observed potent cytotoxicity, detected by both microscopy and lactic acid dehydrogenase (LDH) release assays. Experiments with synovial and macrophage cell lines revealed cytotoxicity for these cell types as well. Using mariner mutagenesis and a screen for loss of cytotoxicity, a genetic locus encoding an RTX toxin system was identified. Disruption of the K. kingae RTX locus resulted in a loss of cytotoxicity for respiratory epithelial, synovial, and macrophage cell lines. DNA sequence analysis demonstrated that the RTX locus is flanked by insertion elements and has a reduced G+C content compared to that of the whole genome. Two relatively less invasive Kingella species, K. oralis and K. denitrificans, were found to be noncytotoxic and to lack the RTX region, as determined by LDH release assays and Southern blotting. We concluded that K. kingae expresses an RTX toxin that has wide cellular specificity and was likely acquired horizontally. The possible roles for this toxin in the pathogenesis of K. kingae disease include breaching of the epithelial barrier and destruction of target tissues, such as synovium (joint lining).
Kingella kingae is a fastidious gram-negative bacterium and is a member of the Neisseriaceae family. Until recently, K. kingae was believed to be a rare pathogen and to exist primarily as a commensal organism in the upper respiratory tract. However, improvements in culture techniques and molecularly based detection methods have led to the recognition that K. kingae is an important cause of a variety of pediatric diseases, including osteomyelitis, septic arthritis, bacteremia, and endocarditis (8, 16, 19, 22, 25). In several recent studies, K. kingae has been identified as a leading cause of osteomyelitis and septic arthritis in children and the most common etiology of culture-negative cases of these infections in pediatric patients (8, 16, 19, 23).
Invasive disease due to K. kingae begins with asymptomatic colonization of the upper respiratory tract. One study demonstrated that more than 70% of young children are colonized with K. kingae at least once per year and that the same strain of K. kingae is able to persist in the respiratory tract of children for at least 2 months (22, 24, 25). In order to cause invasive disease, K. kingae must first breach the respiratory epithelium, which allows access to the underlying intravascular space and permits dissemination of the bacterium to distant sites, such as bones, joints, and endocardium. Infection at these sites is characterized by tissue destruction and inflammation.
To better understand the essential processes of respiratory tract colonization and breaching of the respiratory epithelium, we examined K. kingae in assays with respiratory epithelial cell lines. We observed potent cytotoxicity for cultured respiratory epithelial cells. Microscopy and lactic acid dehydrogenase (LDH) release experiments revealed that K. kingae was also cytotoxic to macrophage-like cells and synovial cells, two cell types that the organism encounters in the joint (4). Using the mariner element and transposon mutagenesis, we identified a locus encoding an RTX toxin system flanked by insertion elements. Disruption of this locus resulted in loss of toxicity for all cell types tested. Further analyses suggested that the RTX locus was acquired via horizontal gene transfer.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmid construction.
The strains used in this study are listed in Table 1. All strains were stored in either brain heart infusion (BHI) broth with 30% glycerol or Luria-Bertani (LB) broth with 15% glycerol at −80°C. Escherichia coli was routinely grown in LB broth or on LB agar plates supplemented with 30 μg/ml chloramphenicol or 100 μg/ml ampicillin as appropriate. K. kingae and K. oralis were grown on chocolate agar or TSA II blood agar plates (Becton-Dickson, Franklin Lakes NJ) at 37°C with 5% CO2. K. denitrificans was grown on TSA II blood agar plates. Chocolate agar was supplemented with 50 μg/ml kanamycin as appropriate. Plasmid pRTXAC was constructed by first amplifying the rtxAC region from K. kingae strain 269-492 using primers GATCGGATCCTTACACTGCGCTAGGTGCTAATACATTTTGCGG and GATCGAATTCATGGATAAATTTTCAGAACTAGGCAGCATCGCG containing BamHI and EcoRI restriction sites, respectively, and then ligating the fragment into pTrc99A.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| 269-492 | K. kingae clinical isolate | St. Louis Children's Hospital |
| ATCC 23330 | K. kingae nasal isolate | ATCC |
| ATCC 23332 | K. kingae blood isolate | ATCC |
| 97-982 | K. kingae clinical isolate | St. Louis Children's Hospital |
| 05-001-1818 | K. kingae clinical isolate | St. Louis Children's Hospital |
| KK01 | Nonspreading/noncorroding derivative of 269-492 | This study |
| KK03 | Spreading/corroding derivative of 269-492 | This study |
| 60H11T1 | Noncytotoxic transposon mutant of 269-492 | This study |
| 67A6T1 | Noncytotoxic transposon mutant of 269-492 | This study |
| 97G1T1 | Noncytotoxic transposon mutant of 269-492 | This study |
| ATCC 33394 | K. denitrificans type strain | ATCC |
| ATCC 51147 | K. oralis type strain | ATCC |
| DH5α | E. coli F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE441 thi-1 gyrA96 relA1 | 17 |
| RTXAC | DH5α containing rtxAC in pRTXAC | This study |
| WAM716 | DH5α containing rtxBD in pWAM716 | 5 |
| RTXAC/WAM716 | DH5α containing pRTXAC and pWAM716 | This study |
| Plasmids | ||
| pWAM716 | rtxB and rtxD from E. coli in pACYC184 | 5 |
| pRTXAC | rtxA and rtxC from K. kingae cloned into pTrc99A | This study |
Cell lines.
The cell lines used in this study were obtained from the American Tissue Culture Collection or the Duke Comprehensive Cancer Center cell culture facility and are listed in Table 2. Most cell lines were maintained at 37°C with 7.5% CO2; the only exception was SW982 cells, which were maintained at 37°C without CO2.
TABLE 2.
Cell lines used in this study
| Cell line | Mediuma | Description |
|---|---|---|
| Chang | MEM + 1× NEAA + 10% FCS | Human conjunctival cells |
| A549 | MEM + 10% FCS | Human type II pneumocytes |
| HEp-2 | DMEM + 10 % FCS | Human laryngeal cells |
| Hig-82 | Ham's F12 + 10% FCS | Rabbit synoviocytes |
| RAW 264.7 | DMEM + 10% FCS | Murine macrophage-like cells |
Abbreviations: MEM, Eagle minimal essential medium; NEAA, nonessential amino acids; FCS, fetal calf serum; DMEM, Dulbecco's modified Eagle's medium.
Cytotoxicity assays.
Bacteria were cultured on appropriate agar media overnight and were then resuspended in BHI broth to an optical density at 600 nm of 0.32. For microscopy studies, ∼3 × 106 bacteria were added to confluent cell monolayers in 24-well tissue culture plates. To quantitate cytotoxicity, we used LDH release as a surrogate marker for cell death. For LDH release assays, ∼7.5 × 105 bacteria were added to confluent cell monolayers in 96-well tissue culture plates. Each sample was assayed in triplicate. For microscopy and LDH release assays, plates were centrifuged for 5 min at 1,000 rpm and then incubated for 10 min at 37°C. Samples were then either fixed and stained with Giemsa stain for examination by microscopy or incubated for 20 min at room temperature and assayed for LDH release. LDH release was assayed using a Cytotox One kit (Promega, Madison, WI) according to the manufacturer's instructions, using excitation and emission wavelengths of 540 nm and 590 nm, respectively. The maximal LDH release was defined as 100% and was determined by adding the lysis solution (Cytotox One kit) to uninfected monolayers, determining the absorbance, and then subtracting the background value. In complementation assays LDH release was assayed as described above, except that bacteria were grown overnight in LB medium at 37°C with the appropriate antibiotics, back diluted 1:10, and then grown for two additional hours at 37°C. In these assays ∼2 ×106 bacteria were added to each well.
Transposon library construction and screening.
Chromosomal DNA was prepared from K. kingae strain 269-492 using a Wizard genomic DNA kit (Promega, Madison, WI) according to the manufacturer's instructions. To create a transposon library, chromosomal DNA from K. kingae 269-492 was mutagenized using the Himar1 transposase and pFalcon2, a plasmid that contains a mariner transposon derivative called Solo, which carries the aphA3 kanamycin resistance gene (11). Mutagenesis was performed as described by Hendrixson et al. (11), except that the buffer exchanges described in the protocol were accomplished with a Qiaex II gel extraction kit (QIAGEN, Valencia CA) used according to the manufacturer's instructions instead of using DNA precipitation. To transform K. kingae, bacteria were grown for 12 h on chocolate agar and were resuspended to an optical density at 600 nm of 0.8 in BHI medium supplemented with 2% bovine serum albumin and 0.5 mM CaCl2. Aliquots of bacteria and mutagenized DNA were mixed in 24-well plates. The plates were incubated for 30 min at room temperature and were then supplemented with an equal volume of BHI medium plus 2% yeast extract and 4% horse plasma and incubated for 1 h at 37°C. Subsequently, the transformation reaction mixtures were plated onto chocolate agar containing kanamycin. In order to assess the randomness of transposon insertion, chromosomal DNA was extracted from individual transformants, digested with ClaI, and examined by Southern hybridization, using the aphA3 cassette from pFalcon2 as a probe.
To identify mutants lacking cytotoxic activity, individual transformants were screened using LDH release assays. To confirm that the loss of cytotoxicity was due to the transposon insertion, chromosomal DNA was isolated from noncytotoxic mutants and retransformed into the parent K. kingae strain 269-492. The resulting transformants were then examined using LDH release assays. Following confirmation of the noncytotoxic phenotype, the sequence surrounding the transposon insertion site was determined using arbitrary PCR. The initial PCR was performed using arbitrary primer ARB1 (5′ GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT 3′) or ARB6 (5′ GGCCACGCGTCGACTAGTACNNNNNNNNNNACGCC 3′) and specific primer Solo5′Arb#1 (5′ GCCCGGGAATCATTTGAAGGTTG 3′) or Solo3′Arb#1 (5′ CGCGTCGCGACGCGTCAATTCGAGG 3′). Solo5′Arb#1 anneals at the 5′ end of the Solo transposon, and Solo3′Arb#1 anneals at the 3′ end of the Solo transposon. In the second round of amplification we utilized ARB2 (5′ GGCCACGCGTCGACTAGTAC 3′), which anneals to the 5′ end of ARB 1 and ARB 6, and Solo5′outN (5′ AATATGCATTTAATACTAGCGACGCC 3′) or Solo3′outN (5′ CGCTCTTGAAGGGAACTATGTTG 3′), which are external to Solo5′Arb#1 and Solo3′Arb#1, respectively. The PCR products from the second round of amplification were gel purified and sequenced (Seqwright, Houston, TX) using either Solo5′outN or Solo3′outN, as appropriate.
Electron microscopy.
Bacteria were allowed to absorb onto Formvar/carbon-coated grids for 1 min and were then washed in distilled H2O and stained with 1% aqueous uranyl acetate (Ted Pella Inc., Redding, CA) for 1 min. Excess liquid was gently wicked off, and the grids were allowed to air dry. Samples were viewed with a JEOL 1200EX transmission electron microscope (JEOL USA, Peabody, MA) at an accelerating voltage of 80 kV.
Southern analysis for rtx locus.
Approximately 1 μg of chromosomal DNA from each strain was digested with BspHI, separated by agarose gel electrophoresis, and then transferred to a nitrocellulose membrane. The DNA probe was generated by PCR amplifying a 0.9-kb region containing rtxC and 0.3 kb of the 5′ end of rtxA from K. kingae strain 269-469 using primers GCAGAAACGGCTACACCAGTTTGAG and CAGAACTAGGCAGCATCGCGTGG. The probe was labeled using the ECL direct nucleic acid labeling system (GE Healthcare, Piscataway, NJ) and was then incubated with the membrane at 42°C in blocking solution (ECL direct nucleic acid labeling system). Subsequently, the membrane was washed two times with 0.5× SSC and 0.4% (wt/vol) sodium dodecyl sulfate at 55°C for 10 min and then rinsed two times with 2× SSC at room temperature for 5 min (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Hybridization was detected using Supersignal West Pico (Pierce, Rockford, IL).
Nucleotide sequence accession number.
The sequence corresponding to the K. kingae RTX locus has been deposited in the GenBank under accession number EF067866.
RESULTS
K. kingae is cytotoxic to a range of cell types.
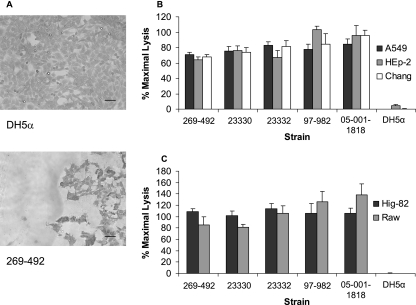
To examine the interaction between K. kingae and respiratory epithelial cells, we inoculated wild-type strain 269-492 onto monolayers of Chang (human conjunctiva), HEp-2 (human larynx), and A549 (human type II pneumocyte) cells, incubated the preparations for various times, and then viewed them by light microscopy. E. coli DH5α was included as a negative control. As shown in Fig. 1A, we observed that K. kingae induced rounding and detachment of cells from the monolayer. In an effort to confirm and quantitate this apparent cytotoxicity, we used an LDH release assay. As shown in Fig. 1B, incubation of K. kingae strain 269-492 and four additional K. kingae strains with Chang, A549, and HEp-2 cells for 10 min at 37°C resulted in 70% of the maximal LDH release.
FIG. 1.
K. kingae cytotoxicity. (A) Light microscopy evidence of K. kingae cytotoxicity for Chang cells. The top panel shows an intact monolayer after incubation for 10 min with E. coli DH5α, and the bottom panel shows a destroyed monolayer after incubation for 10 min with K. kingae strain 269-492. Bars = 100 μM. (B and C) LDH release assays with K. kingae strains 269-492, 23330, 23332, 97-982, and 05-001-1818 and E. coli DH5α with respiratory epithelial cells (B) and synovial and macrophage-like cells (C).
To extend these observations and to study the cellular specificity of K. kingae cytotoxicity, we examined whether K. kingae was cytotoxic to Hig-82 rabbit synoviocytes and RAW 264.7 murine macrophage-like cells. All five K. kingae strains tested were highly cytotoxic to these cells, and the LDH release was more than 80% of the maximal LDH release (Fig. 1C). These results demonstrate that K. kingae expresses a potent cytotoxin with broad cell specificity, including human and nonhuman cells.
Cytotoxicity is independent of colony morphology.
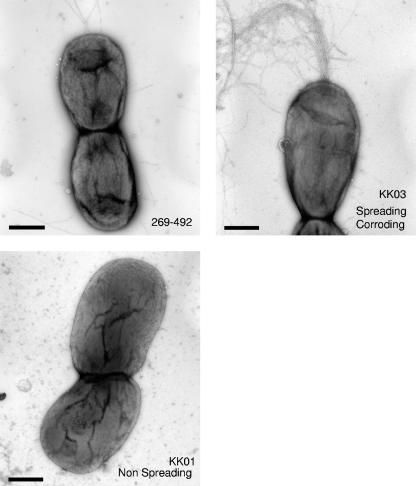
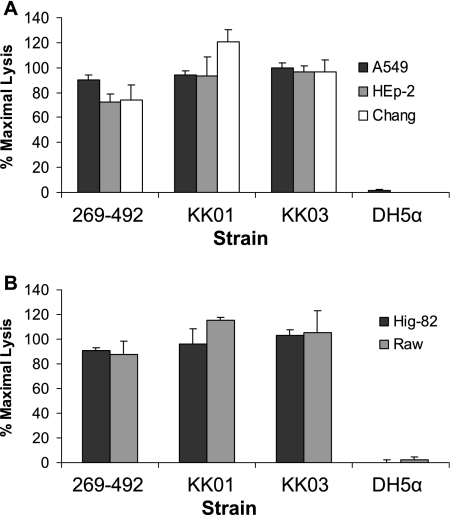
In previous work on K. kingae workers identified two interchanging colony morphologies, referred to as spreading/corroding and nonspreading/noncorroding, which are believed to reflect phase-variable expression of pili (7, 12, 13). The spreading/corroding morphology (SC type) correlates with expression of pili, and the nonspreading/noncorroding (N type) morphology correlates with reduced expression of pili (7, 12, 20). To determine if colony morphology and the density of pili influence K. kingae cytotoxicity, we recovered two morphological variants of strain 269-492, designated KK01 and KK03. As predicted, examination by negative staining transmission electron microscopy confirmed that KK01 has reduced numbers of pili and that KK03 expresses abundant pili (Fig. 2). Based on colony morphology, the phenotypes of the two variants appear to be stable, with more than 99% of colonies maintaining the parental morphology. As shown in Fig. 3, strains 269-492, KK01, and KK03 all displayed similar levels of cytotoxicity in assays with respiratory, synovial, and macrophage-like cells, suggesting that cytotoxicity is independent of colony morphology and the level of piliation.
FIG. 2.
Transmission electron micrographs of K. kingae strain 269-492 (top left), an N-type variant (bottom left) designated KK01, and an SC-type variant (top right) designated KK03 after negative staining with uranyl acetate. Bars = 100 nM.
FIG. 3.
Cytotoxicity of spreading/corroding and nonspreading/noncorroding variants of K. kingae. K. kingae strains KK01, KK03, and 269-492 were assayed for cytotoxicity for respiratory epithelial cells (A) and synovial and macrophage-like cells (B) by the LDH release assay. E. coli DH5α was used as a negative control.
K. kingae mutants 60H11T1, 69A6T1, and 97G1T1 lack cytotoxicity.
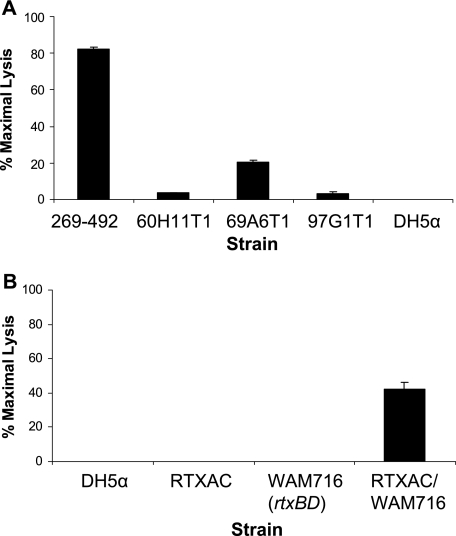
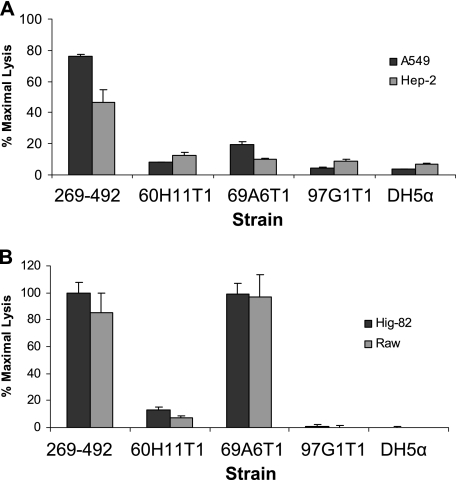
To identify the factors responsible for K. kingae cytotoxicity, we used the Himar1 transposase and pFalcon2 to generate a transposon library in strain 269-492. Assessment of 10 transformants by Southern hybridization confirmed that there was random insertion of the Solo transposon (data not shown). Using the LDH release assay, the library was screened for mutants that lacked cytotoxicity for Chang cells. Utilizing this approach, we recovered three independent noncytotoxic mutants, designated 60H11T1, 69A6T1, and 97G1T1 (Fig. 4A).
FIG. 4.
Loss of cytotoxicity of K. kingae mutants 60HllT1, 69A6T1, and 97G1T1. (A) LDH release assays with K. kingae strain 269-492 and mutants 60H11T1, 69A6T1, and 97G1T1 using Chang epithelial cells. E. coli DH5α was used as a negative control. (B) LDH release assays with E. coli strains DH5α, RTXAC, WAM716, and RTXAC/WAM716 using Chang epithelial cells.
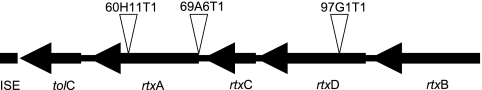
To identify the sites of transposon insertion in 60H11T1, 69A6T1, and 97G1T1, the transposon junctions were amplified using arbitrary PCR and were then sequenced. The resulting sequences were examined for homology to the results from the K. kingae sequencing project (T. E. Kehl-Fie et al., unpublished data). All three transposon insertions were located on an approximately 10-kb contig. In mutant 60H11T1, the transposon inserted into a gene encoding a protein with a high level of homology to MbxA, an RTX toxin from Moraxella bovis. In mutant 69A6T1, the transposon inserted immediately upstream of the predicted start site of the mbxA homolog. In mutant 97G1T1, the transposon inserted into a gene encoding a protein with a high level of homology to the M. bovis MbxD protein, a component of the type I secretion apparatus for export of MbxA. Further analysis of the contig revealed the presence of genes with homology to the genes encoding MbxC, MbxB, and TolC (Table 3 and Fig. 5), indicating that K. kingae possesses all of the genes necessary for production and secretion of an active RTX toxin (designated rtxA, rtxB, rtxC, rtxD, and tolC). To confirm that the RTX locus is responsible for the observed cytotoxicity, the K. kingae rtxA and rtxC genes (the structural and lipidating components of the locus) were cloned into pTrc99A, creating pRTXAC. This construct was introduced into WAM716 (DH5α carrying a plasmid that contains rtxB and rtxD, encoding the E. coli type I secretion system), generating strain RTXAC/WAM716. As shown in Fig. 4B, RTXAC/WAM716 was found to be cytotoxic to Chang cells, providing strong evidence that the K. kingae locus is responsible for the observed K. kingae cytotoxicity.
TABLE 3.
K. kingae RTX locus homology
| K. kingae gene product | Best homolog | % Identitya | % Similaritya |
|---|---|---|---|
| TolC | N. meningitidis TolC | 64 | 82 |
| RtxA | M. bovis MbxA | 72 | 85 |
| RtxC | M. bovis MbxC | 73 | 87 |
| RtxD | N. meningitidis RtxD | 81 | 91 |
| RtxB | M. bovis MbxB | 83 | 91 |
Levels of identity and similarity were determined by comparing the entire K. kingae gene product and the entire sequence of the closest homolog.
FIG. 5.
Diagram of the K. kingae RTX locus of strain 269-492, showing the locations of transposon insertions in mutants 60H11T1, 69A6T1, and 97G1T1.
To extend our characterization of mutants 60H11T1, 69A6T1, and 97G1T1, we examined these strains using cytotoxicity assays with respiratory epithelial, synovial, and macrophage-like cells and hemolysis assays on sheep blood agar plates. In assays with HEp-2 and A549 cells, mutants 60H11T1 and 97G1T1 were virtually devoid of cytotoxic activity and mutant 69A6T1 had markedly reduced cytotoxic activity (Fig. 6A). In assays with Hig-82 and RAW 264.7 cells, mutants 60H11T1 and 97G1T1 were found to be noncytotoxic, while mutant 69A6T1 remained highly cytotoxic, comparable to strain 269-492 (Fig. 6B). The observation that mutant 69A6T1 remained cytotoxic to Hig-82 and RAW 264.7 cells suggests that this strain produces reduced levels of functional toxin, a conclusion consistent with the finding that the insertion in this strain is immediately upstream of rtxA. When grown on sheep blood agar plates, 60H11T1 and 97G1T1 were found to be nonhemolytic and 69A6T1 was minimally hemolytic (data not shown).
FIG. 6.
Cytotoxicity of K. kingae mutants 60HllT1, 69A6T1, and 97G1T1 for different cell types. LDH release assays were performed by using K. kingae strain 269-492 and mutants 60H11T1, 69A6T1, and 97G1T1 with respiratory epithelial cells (A) and synovial and macrophage-like cells (B). E. coli DH5α was used as a negative control.
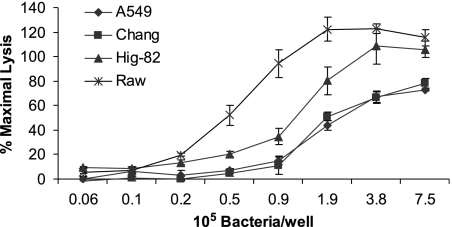
To further explore the apparent increased K. kingae cytotoxicity for synovial and macrophage-like cells compared to the cytotoxicity for respiratory epithelial cells, we prepared serial dilutions of wild-type strain 269-492 and then measured LDH release. As shown in Fig. 7, Hig-82 and RAW 264.7 cells were found to be more sensitive than Chang and A549 cells, exhibiting similar levels of cytotoxicity with two- to fourfold fewer bacteria.
FIG. 7.
LDH release assays with K. kingae strain 269-492 comparing respiratory epithelial, synovial, and macrophage-like cells when different bacterial inocula were used.
K. kingae RTX region appears to have been horizontally acquired.
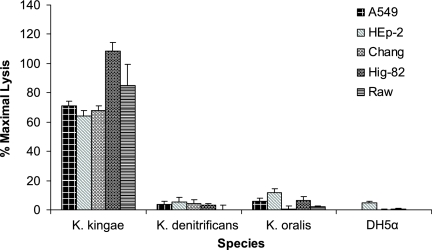
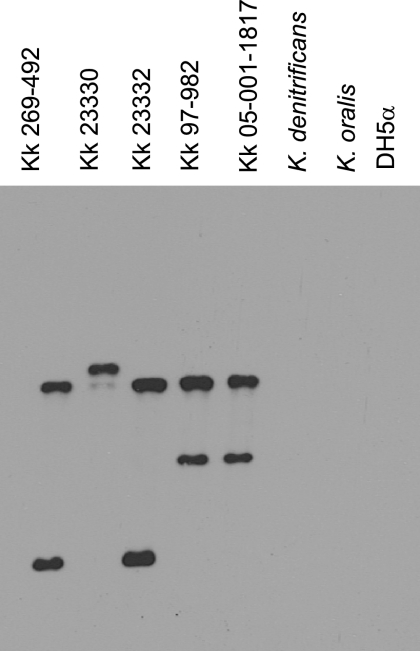
The presence of all five genes necessary for production and secretion of an RTX toxin in a single gene cluster on the chromosome raised the possibility that the K. kingae RTX locus was acquired via horizontal gene transfer, similar to speculation concerning other RTX toxins (6). To examine this hypothesis, we examined the flanking sequence and found that downstream of the RTX locus is a region with homology to insertion elements from M. bovis, Neisseria meningitidis, and Haemophilus influenzae. Further analysis revealed that the K. kingae RTX locus has a G+C content of 40%, which is significantly lower than the G+C content of the K. kingae genome (47%). To determine if the RTX locus is present in other Kingella species, K. oralis and K. denitrificans were examined to determine their cytotoxicity for respiratory, synovial, and macrophage-like cell lines using LDH release assays. Both K. oralis and K. denitrificans were found to be noncytotoxic to all cell types tested (Fig. 8). To assess whether this lack of cytotoxicity was due to inactivation or absence of the cytotoxin region, chromosomal DNA from K. oralis and K. denitrificans was examined by Southern hybridization using a 5′ fragment of rtxA and rtxC as a probe. As expected, all five K. kingae strains examined possessed the cytotoxin region, while both K. oralis and K. denitrificans did not react with the probe (Fig. 9).
FIG. 8.
LDH release assays examining K. kingae strain 269-492, K. denitrificans type strain, and K. oralis type strain cytotoxicity for respiratory epithelial cells, synovial cells, and macrophage-like cells. E. coli DH5α was used as a negative control.
FIG. 9.
Southern analysis of chromosomal DNA from K. kingae (Kk) strains 269-492, 23330, 23332, 97-982, and 05-001-1817, the K. denitrificans type strain, the K. oralis type strain, and E. coli DH5α, using a 0.9-k fragment containing rtxC and the 5′ end of rtxA from K. kingae strain 269-492 as a probe.
DISCUSSION
In this study we examined interactions between the emerging pediatric pathogen K. kingae and cultured respiratory epithelial cells. We observed that K. kingae expresses a potent cytotoxin capable of rapidly killing a variety of human respiratory epithelial cell lines. Additional investigation established that K. kingae is also cytotoxic to synovial cells and macrophage-like cells, two cell types that the bacterium is likely to encounter in the course of septic arthritis (4). By screening a K. kingae mariner transposon library for loss of cytotoxicity we identified a locus encoding an RTX toxin system. This locus is essential for cytotoxicity for respiratory epithelial, synovial, and macrophage-like cells.
RTX toxins have been divided into the following three categories based on cellular specificity: the hemolysins, which exhibit toxicity for a wide range of cell types (including erythrocytes); the cytotoxins, which exhibit toxicity for a wide but defined range of cell types; and the leukotoxins, which exhibit very narrow cell type and species specificity (21). The wide cellular specificity of the K. kingae RTX toxin suggests that this toxin belongs with the hemolysins. While the RTX hemolysins, cytotoxins, and leukotoxins are distinguished by different levels of specificity, all three classes possess several conserved motifs, including an amino-terminal hydrophobic domain implicated in pore formation, a species-specific lipid modification, a conserved calcium binding repeat domain, and a type I secretion signal (21). The K. kingae RTX toxin contains an amino-terminal hydrophobic domain, potential lipidation sites, and a calcium binding motif (data not shown). It is noteworthy that exposure of cells to sublytic doses of HlyA from E. coli or LktA from a variety of species results in a range of cellular reactions, including calcium fluxes, secretion of proinflammatory cytokines, and induction of apoptosis (2, 3, 10, 15, 18). Given the inflammatory nature of the disease caused by K. kingae, the possibility that the K. kingae cytotoxin may contribute to the inflammatory process is intriguing.
Examination of the K. kingae RTX locus revealed five genes, designated rtxA, rtxC, rtxD, rtxB, and tolC. Further analysis demonstrated that downstream of the locus is a region with homology to insertion elements from M. bovis, Neisseria species, and Haemophilus species. The identification of tolC within the locus is notable, since in most instances this gene is not physically associated with the toxin locus (the M. bovis RTX locus and the Bordetella pertussis adenylate cyclase locus are exceptions) (1, 9). Although arranged in a noncanonical order, the K. kingae RTX locus exhibits marked homology with the M. bovis RTX locus. The K. kingae rtxA, rtxC, and rtxB genes encode proteins that have more then 70% identity with their M. bovis homologs (Table 3). The two remaining genes in the locus, tolC and rtxD, encode proteins with substantial homology to their M. bovis homologs but even greater homology to their N. meningitidis counterparts. These observations could be explained by acquisition of the RTX locus and then reacquisition of tolC and rtxD from either K. kingae itself or N. meningitidis. The results of the homology analysis combined with the observation that K. denitrificans and K. oralis lack an RTX locus suggest that the K. kingae RTX locus was acquired via horizontal gene transfer from either M. bovis or a common donor organism. The horizontal acquisition hypothesis is further supported by the identification of insertion elements flanking the M. bovis RTX locus (14).
In assays with synovial and macrophage-like cells, we observed two- to fourfold increases in sensitivity to K. kingae cytotoxic activity compared to the sensitivity observed with respiratory cells. This increased sensitivity is consistent with clinical observations that suggest that K. kingae colonizes the respiratory tract asymptomatically but causes significant tissue destruction in the joint. It is possible that reduced sensitivity and other factors, such as mucus and other barriers, could combine to produce asymptomatic carriage of K. kingae.
Given the broad cellular specificity of the K. kingae RTX toxin, there are a number of possible roles for the cytotoxin during colonization of the respiratory tract and development of invasive disease. First, expression of the cytotoxin in the respiratory tract may result in damage to epithelial cells and tight junctions, disrupting innate physical defenses, allowing binding to respiratory mucosa and invasion of the bloodstream. Second, expression of the cytotoxin in the joint may promote inflammation directly by damaging the synovium or indirectly by promoting the release of proinflammatory cytokines, resulting in the recruitment of immune cells and inflammation. Third, the cytotoxin may aid K. kingae in immune evasion by killing macrophages and neutrophils.
In this paper we report the identification of a K. kingae RTX toxin that may play multiple roles in the pathogenesis of K. kingae disease, including colonization of the respiratory tract, invasion of the bloodstream, and damage to the joints. Given that septic arthritis is probably a terminal path for K. kingae, it is intriguing to speculate that the toxin may have been acquired and maintained to increase the area of the respiratory tract that K. kingae is capable of colonizing, coincidentally increasing the pathogenic potential. Further study of the K. kingae RTX toxin may increase our understanding of the balance between commensal and pathogen that is common among microbial pathogens, including other members of the Neisseriaceae family.
Acknowledgments
We thank Shane Cotter for thoughtful discussions. Additionally, we thank Wandy Beatty for assistance with electron microscopy, Rodney Welch for providing strain WAM716, and Pablo Yagupsky for offering general advice about handling K. kingae.
This work was supported by NIH training grant T32-GM07067 to T.K.F.
Footnotes
Published ahead of print on 10 November 2006.
REFERENCES
- 1.Angelos, J. A., J. F. Hess, and L. W. George. 2003. An RTX operon in hemolytic Moraxella bovis is absent from nonhemolytic strains. Vet. Microbiol. 92:363-377. [DOI] [PubMed] [Google Scholar]
- 2.Atapattu, D. N., and C. J. Czuprynski. 2005. Mannheimia haemolytica leukotoxin induces apoptosis of bovine lymphoblastoid cells (BL-3) via a caspase-9-dependent mitochondrial pathway. Infect. Immun. 73:5504-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhakdi, S., and E. Martin. 1991. Superoxide generation by human neutrophils induced by low doses of Escherichia coli hemolysin. Infect. Immun. 59:2955-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bremell, T., A. Abdelnour, and A. Tarkowski. 1992. Histopathological and serological progression of experimental Staphylococcus aureus arthritis. Infect. Immun. 60:2976-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felmlee, T., and R. A. Welch. 1988. Alterations of amino acid repeats in the Escherichia coli hemolysin affect cytolytic activity and secretion. Proc. Natl. Acad. Sci. USA 85:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frey, J., and P. Kuhnert. 2002. RTX toxins in Pasteurellaceae. Int. J. Med. Microbiol. 292:149-158. [DOI] [PubMed] [Google Scholar]
- 7.Froholm, L. O., and K. Bovre. 1972. Fimbriation associated with the spreading-corroding colony type in Moraxella kingii. Acta Pathol. Microbiol. Scand. B 80:641-648. [PubMed] [Google Scholar]
- 8.Gene, A., J. J. Garcia-Garcia, P. Sala, M. Sierra, and R. Huguet. 2004. Enhanced culture detection of Kingella kingae, a pathogen of increasing clinical importance in pediatrics. Pediatr. Infect. Dis. J. 23:886-888. [DOI] [PubMed] [Google Scholar]
- 9.Glaser, P., H. Sakamoto, J. Bellalou, A. Ullmann, and A. Danchin. 1988. Secretion of cyclolysin, the calmodulin-sensitive adenylate cyclase-haemolysin bifunctional protein of Bordetella pertussis. EMBO J. 7:3997-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gleason, T. G., C. W. Houlgrave, A. K. May, T. D. Crabtree, R. G. Sawyer, W. Denham, J. G. Norman, and T. L. Pruett. 1998. Hemolytically active (acylated) alpha-hemolysin elicits interleukin-1beta (IL-1beta) but augments the lethality of Escherichia coli by an IL-1- and tumor necrosis factor-independent mechanism. Infect. Immun. 66:4215-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendrixson, D. R., B. J. Akerley, and V. J. DiRita. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40:214-224. [DOI] [PubMed] [Google Scholar]
- 12.Henrichsen, J., L. O. Froholm, and K. Bovre. 1972. Studies on bacterial surface translocation. 2. Correlation of twitching motility and fimbriation in colony variants of Moraxella nonliquefaciens, M. bovis, and M. kingii. Acta Pathol Microbiol. Scand. B 80:445-452. [PubMed] [Google Scholar]
- 13.Henriksen, S. D. 1969. Corroding bacteria from the respiratory tract. 1. Moraxella kingii. Acta Pathol. Microbiol. Scand. 75:85-90. [PubMed] [Google Scholar]
- 14.Hess, J. F., and J. A. Angelos. 2006. The Moraxella bovis RTX toxin locus mbx defines a pathogenicity island. J. Med. Microbiol. 55:443-449. [DOI] [PubMed] [Google Scholar]
- 15.Ludwig, A., and W. Goebel. 2000. Dangerous signals from E. coli toxin. Nat. Med. 6:741-742. [DOI] [PubMed] [Google Scholar]
- 16.Moumile, K., J. Merckx, C. Glorion, P. Berche, and A. Ferroni. 2003. Osteoarticular infections caused by Kingella kingae in children: contribution of polymerase chain reaction to the microbiologic diagnosis. Pediatr. Infect. Dis. J. 22:837-839. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Uhlen, P., A. Laestadius, T. Jahnukainen, T. Soderblom, F. Backhed, G. Celsi, H. Brismar, S. Normark, A. Aperia, and A. Richter-Dahlfors. 2000. Alpha-haemolysin of uropathogenic E. coli induces Ca2+ oscillations in renal epithelial cells. Nature 405:694-697. [DOI] [PubMed] [Google Scholar]
- 19.Verdier, I., A. Gayet-Ageron, C. Ploton, P. Taylor, Y. Benito, A. M. Freydiere, F. Chotel, J. Berard, P. Vanhems, and F. Vandenesch. 2005. Contribution of a broad range polymerase chain reaction to the diagnosis of osteoarticular infections caused by Kingella kingae: description of twenty-four recent pediatric diagnoses. Pediatr. Infect. Dis. J. 24:692-696. [DOI] [PubMed] [Google Scholar]
- 20.Weir, S., and C. F. Marrs. 1992. Identification of type 4 pili in Kingella denitrificans. Infect. Immun. 60:3437-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welch, R. A. 2001. RTX toxin structure and function: a story of numerous anomalies and few analogies in toxin biology. Curr. Top. Microbiol. Immunol. 257:85-111. [DOI] [PubMed] [Google Scholar]
- 22.Yagupsky, P. 2004. Kingella kingae: from medical rarity to an emerging paediatric pathogen. Lancet Infect. Dis. 4:358-367. [DOI] [PubMed] [Google Scholar]
- 23.Yagupsky, P., R. Dagan, C. W. Howard, M. Einhorn, I. Kassis, and A. Simu. 1992. High prevalence of Kingella kingae in joint fluid from children with septic arthritis revealed by the BACTEC blood culture system. J. Clin. Microbiol. 30:1278-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yagupsky, P., R. Dagan, F. Prajgrod, and M. Merires. 1995. Respiratory carriage of Kingella kingae among healthy children. Pediatr. Infect. Dis. J. 14:673-678. [DOI] [PubMed] [Google Scholar]
- 25.Yagupsky, P., N. Peled, and O. Katz. 2002. Epidemiological features of invasive Kingella kingae infections and respiratory carriage of the organism. J. Clin. Microbiol. 40:4180-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]