Abstract
Transcription of the biotin (bio) biosynthetic operon of Escherichia coli is negatively regulated by the BirA protein, an atypical repressor protein in that it is also an enzyme. The BirA-catalyzed reaction involves the covalent attachment of biotin to AccB, a subunit of acetyl coenzyme (acetyl-CoA) carboxylase. The two functions of BirA allow regulation of the bio operon to respond to the intracellular concentrations of both biotin and unbiotinylated AccB. We report here that bio operon expression is down-regulated by overproduction of AccC, another acetyl-CoA carboxylase subunit known to form a complex with AccB. This down-regulation is eliminated when AccB and AccC are coordinately overexpressed, but only when the AccB partner is competent to bind AccC. Under AccC overexpression conditions AccB is underbiotinylated. These findings can be explained by a model in which excess AccC sequesters AccB in a complex that is a poor substrate for biotinylation. The observed disruption of biotin synthesis and attachment provides an excellent rationale for the observation that in the vast majority of sequenced bacterial genomes AccB and AccC are encoded in a two-gene operon.
Expression of the Escherichia coli biotin synthetic (bio) operon is controlled by a remarkably sophisticated regulatory system in which the rate of transcription of the operon responds not only to the supply of biotin but also to the supply of proteins (called biotin acceptor proteins) that become modified by covalent attachment of biotin (Fig. 1) (2-6, 10, 16, 17). The response to the supply of biotin acceptor proteins is readily rationalized by the fact that biotin has a biological function only when the vitamin is covalently attached to such proteins; the free vitamin is inactive (18). The enzymes of E. coli biotin synthesis are encoded (with one exception) by a cluster of genes located adjacent to the attachment site of phage λ called the biotin (bio) biosynthetic operon. Transcription of these bio genes occurs from two partially overlapping face-to-face promoters controlled by a common operator site that binds a dimer of the BirA protein (2-6, 29). It is the properties of BirA that allow the system to respond to the intracellular concentrations of both biotin and biotin acceptor proteins (5, 16). This is because BirA is the enzyme (biotin protein ligase) that catalyzes attachment of biotin to acceptor proteins, as well as the repressor of the bio operon (2, 16).
FIG. 1.
Biotin regulatory system of E. coli. BirA is represented by green ovals, biotin is represented by black circles, the AMP moiety is represented by red pentagons, AccB is represented by dark blue ovals, and AccC is represented by light blue crescents. The arrows indicate transcription from the leftward and rightward bio promoters. (A to C) BirA switches from the biotin ligation function to the repressor function in response to the intracellular biotin requirement, which is indicated by the level of unbiotinylated AccB. If unbiotinylated AccB levels are high, the protein functions as a biotin ligase. Once the unbiotinylated AccB has been converted to the biotinylated form, the biotinoyl-5′-AMP is no longer consumed and remains bound to BirA. The liganded form of BirA accumulates to levels sufficiently high that the bio operator is fully occupied, resulting in transcriptional repression of the biotin biosynthetic genes. (D) Overproduction of AccC ties up unbiotinylated AccB in a complex that is a poor biotinylation substrate. Therefore, high levels of the liganded form of BirA accumulate, resulting in repression of bio operon transcription.
The biotin attachment activity of BirA proceeds through a biotinoyl-5′-AMP intermediate. Biotinoyl-5′-AMP is then attacked by the ɛ-amino group of a specific lysine of the acceptor protein to give the biotinylated acceptor protein (9) (Fig. 2A). In the absence of an appropriate acceptor protein the biotinoyl-5′-AMP intermediate remains firmly bound within the BirA active site, where it is quite stable (33). It is this liganded form of BirA that binds the bio operator. Therefore, increased bio operon transcription is triggered either by inhibition of biotinoyl-5′-AMP synthesis by intracellular biotin limitation (Fig. 1B) or by increased consumption of biotinoyl-5′-AMP due to high levels of unmodified acceptor proteins (Fig. 1C). The two conditions act by a common mechanism in that both of them decrease the levels of liganded BirA available to bind the bio operator (Fig. 1B and C). Hence, the degree of repression of bio operon transcription can be viewed most simply as due to antagonism between retention of biotinoyl-5′-AMP in the BirA active site and consumption of the BirA-bound biotinoyl-5′-AMP by transfer of the biotinyl moiety to unmodified acceptor proteins (16). A structural context for this antagonism is provided by the model of Beckett and coworkers (30, 32), in which the unmodified acceptor protein binds monomeric BirA and thereby inhibits formation of BirA dimers, the species required for effective repression.
FIG. 2.
BirA and acetyl-CoA carboxylase reactions. (A) BirA reaction, which is the general reaction of biotin protein ligases (9). (B) Acetyl-CoA carboxylase reaction. The functional enzyme is thought to have the composition 2AccA:4AccB:2AccC:2AccD, with the AccB-AccC and AccA-AccD complexes sufficiently stable for isolation in vitro (18).
E. coli contains only a single species of biotin acceptor protein, the AccB subunit of acetyl coenzyme A (acetyl-CoA) carboxylase (ACC), which is the first enzyme of fatty acid biosynthesis (18, 19). AccB, which is also called biotin carboxyl carrier protein, forms an unstable complex with AccC, the subunit that catalyzes the biotin carboxylase partial reaction of acetyl-CoA carboxylase (Fig. 2B). The AccB-AccC complex was recently shown to consist of an AccC dimer plus four copies of AccB (12). This complex is thought to bind an α2β2 heterotetramer of the AccA and AccD subunits to form active ACC, the enzyme required for production of malonyl-CoA, the key precursor of fatty acid synthesis (18). Since the E. coli ACC subunits are required in a defined stoichiometry, it seems surprising that only the accB and accC genes are cotranscribed. The chromosomal locations of the accA and accD genes are well removed from the locations of the accBC operon and each other (18). The rates of transcription of all four genes are controlled by the cellular growth rate (25), and transcriptional initiation of the accBC operon is autoregulated by accB levels by an unknown mechanism (22, 25).
The accBC gene arrangement first found in E. coli is strikingly conserved among bacteria (22). The two genes have been found to be adjacent in all sequenced proteobacterial genomes, including the smallest and largest such genomes (Pelagibacter ubique and the pseudomonads, respectively). The accBC gene arrangement is also found in gram-positive bacteria, such as the Bacillales and some clostridia. In other clostridia and in the Lactobacillales a fatty acid synthetic gene is located between the accB and accC genes. However, in these organisms all of the ACC subunits, as well as all of the fatty acid synthetic enzymes, are encoded by what appears to be a single long transcript. Therefore, even with an intervening gene, transcription of accB and accC is “hardwired,” with accB being the upstream gene. Moreover, the accBC gene arrangement of E. coli is found in such diverse bacteria as Thermus thermophilus, Chlorobium tepidum, and Chlamydia.
The extremely strong conservation of the accBC gene arrangement coupled with the contrasting highly random genomic locations of accA and accD implies that tight regulation of the AccB:AccC ratio is essential. If this is true, an excess of either AccB or AccC should be detrimental to bacterial physiology. It is known that overproduction of AccB in E. coli shuts down transcription of the accBC operon (22) and also results in derepression of the biotin biosynthetic operon, thereby leading to wasteful synthesis of biotin (10, 17). Would an excess of AccC also be detrimental? Karow and coworkers (23) described complex experiments which suggested that this is the case. In their studies of suppressors of null mutants with mutations in the E. coli htrB gene, a gene later shown to encode an acyltransferase that functions late in lipid A biosynthesis (14), these workers found that overproduction of AccC in E. coli resulted in significant inhibition of fatty acid synthesis, whereas overproduction of both AccB and AccC resulted in no such inhibition (23). A plausible mechanism to explain these results is that excess AccC might inhibit biotinylation of AccB, which in turn would repress the bio operon and inhibit biotin synthesis. This scenario posits that free AccB is the preferred substrate for BirA-catalyzed biotinylation and that excess AccC competes with BirA and ties up a portion of unmodified AccB in an AccB-AccC complex that is a poor substrate for biotinylation (Fig. 1D). We report here that AccC overproduction does indeed block derepression of biotin synthesis at low biotin concentrations and also inhibits biotinylation of AccB. The demonstrated need for precise stoichiometry during production of AccB and AccC provides an excellent rationale for the fact that in the great majority of the extant bacterial genomes the accB and accC genes are adjacent so that they can be cotranscribed to ensure stringent regulation of the ratio of the two proteins.
MATERIALS AND METHODS
Bacterial strains and media.
All strains were derivatives of E. coli K-12. Strain CY1740, a derivative of strain CY486 (17) carrying the lacIq plasmid pMS421, was used in this work. Strain CY1740 carries a chromosomal φ(bioFC-lacZ)501 fusion (4). The medium used in the physiological experiments was medium E supplemented with 0.l% vitamin-free Casamino Acids, 0.4% glucose, and 1 μg/ml thiamine. Cultures were grown at 37°C with vigorous aeration. Glycerol was used instead of glucose for growth of strains containing the PBAD birA plasmid, pCY216 (11). The cultures were grown overnight and then diluted 1:100 into fresh medium having the same composition and grown to the early to mid-log phase before assays were performed. Expression of Ptac-controlled accB and accC genes was induced with 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG), whereas birA expression was induced with 0.2% arabinose.
Plasmid construction.
The sources of the accB and accC genes are shown in Table 1 together with descriptions of the derived plasmids. The vector used was the medium-copy-number Ptac promoter plasmid pKK223-3 (8), although the high-copy-number Plac promoter plasmid pHSG397 (31) was used in some early experiments.
TABLE 1.
Plasmids and oligonucleotides used in this work
| Plasmid or oligonucleotide | Properties or sequence (5′-3′)a | Reference |
|---|---|---|
| Plasmids | ||
| pKK223-3 | Ptac promoter vector, pBR322 origin, Ampr | 8 |
| pCY701 | pKK223-3 containing accC under Ptac control | This study |
| pCY705 | pKK223-3 containing accB and accC under Ptac control | This study |
| pCY70 | pKK223-3 containing accC under Ptac control | This study |
| pCY708 | pKK223-3 containing accB under Ptac control | This study |
| pAH1 | pKK223-3 containing ′accB under Ptac control | This study |
| pAH2 | pKK223-3 containing ′accB and accC under Ptac control | This study |
| pCY730 | pKK223-3 containing accB K122R and accC under Ptac control | This study |
| pAH15 | pKK223-3 containing accB K122E and accC under Ptac control | This study |
| pAH16 | pKK223-3 containing accB K122M and accC under Ptac control | This study |
| pAH7 | pKK223-3 containing accB K122R under Ptac | This study |
| pAH13 | pKK223-3 containing accB K122E under Ptac control | This study |
| pAH14 | pKK223-3 containing accB K122M under Ptac control | This study |
| pCY216 | birA under PBAD arabinose promoter control in a p15a vector, chloramphenicol resistant | 11 |
| pMS421 | lacIq in a pSC101 origin, spectinomycin resistant | 21 |
| pLS24 | accB chromosomal segment in pGEM5 | 24 |
| pLS83 | accC chromosomal segment in pGEM7 | 24 |
| pCY465 | accB chromosomal segment in pCY462 | 15 |
| pGB7 | accB accC chromosomal segment in pGEM7 | 24 |
| Oligonucleotide primersb | ||
| accB87-1 | GGAATTCCATGGAAGCGCCAGCAGC | |
| accB87-2 | AGAAAGCTTTCTCCATGGACGCGTG | |
| accB-E1 | GCATCGTTGAAGCCATGGAAATGATGAACCAGATCG | |
| accB-E2 | CGATCTGGTTCATCATTTCCATGGCTTCAACGATGC | |
| accB-M1 | CGTTGAAGCCATGATGATGATGAACCAGATCG | |
| accB-M2 | CGATCTGGTTCATCATCATCATGGCTTCAACG | |
| BCCPKR-FA | GTGCATCGTTGAAGCCATGAGAATGATGAACCAGATCGAAG | |
| BCCPKR-RE | CTTCGATCTGGTTCATCATTCTCATGGCTTCAACGATGCAC |
The ′accB gene encodes the last 87 residues of AccB. Any mutational alteration of accB is indicated by the amino acid residue alteration.
Primers accB87-1 (forward) and accB87-2 (reverse) were used to clone a gene encoding the N-terminally truncated protein consisting of the carboxyl 87 residues of AccB. Primers accB-E1 (forward) and accB-E2 (reverse) were used to construct the accB gene encoding the K122E protein. Primers accB-M1 (forward) and accB-M2 (reverse) were used to construct the accB gene encoding the K122M protein. Primers BCCPKR-FA and BCCPKR-RE were used by E. Rhee of our laboratory to construct the accB gene encoding the K122R protein.
Plasmid pCY701 was constructed by digestion of plasmid pLS83 with EcoRI plus NsiI (whose sites lie within the multiple cloning site of the parental plasmid), and the 3,282-bp fragment was ligated to the medium-copy-number tac promoter plasmid pKK223-3 digested with EcoRI and PstI. Plasmid pCY703 was constructed by digesting plasmid pGB7 with HindIII and BssHII and plasmid pCY465 with HindIII and MluI. The 1,836-bp fragment of pGB7 was then ligated to the 2,640-bp fragment of pCY465. The 2,098-bp fragment of pCY703 obtained by digestion with EcoRI plus BglII was then ligated to the 5,014-bp fragment of pCY701 (obtained by digestion with the same enzymes) to obtain pCY705, a tac promoter plasmid carrying accB in its normal chromosomal position upstream of accC. Plasmid pCY708, in which accB was expressed from the tac promoter, was obtained by the same manipulations except that the EcoRI-BglII fragment was the 1,672-bp fragment of pCY465. Plasmid pAH1, which encoded the C-terminal 87-residue biotinylation domain of AccB plus a kanamycin resistance marker, was constructed by PCR amplification using pCY708 as the template with primers incorporating flanking EcoRI and HindIII sites and then cloned into pKK223-3. The fragment containing the accC gene was cut from pCY705 with SstII and HindIII, gel purified, and cloned into pAH1 to generate pAH2. A high-copy-number Plac plasmid carrying accC was constructed by insertion of the XhoI-SacI fragment of pLS83 into pHSG397 cut with the same enzymes. Plasmid pCY703, a ΔaccB derivative of pCY703, was constructed by digestion with KpnI, followed by ligation at a low DNA concentration.
Site-directed mutagenesis of accB was carried out using the Stratagene QuickChange II protocol and the primers shown in Table 1. PCR amplification was carried out using Pfu Turbo DNA polymerase for 16 cycles. The template plasmid DNAs were digested with DpnI, and the PCR product containing the mutated plasmid was used to transform DH5α. Codon K122 of the accB gene of pCY708 was changed to glutamate and methionine codons in order to construct plasmids pAH13 and pAH14, respectively. The mutant accB genes were sequenced to confirm the expected mutations at the Keck Center for Comparative and Functional Genomics at the University of Illinois. Plasmid pCY730, which expressed K122R AccB, was constructed by exchanging the BsiWI-KpnI fragment of plasmid pER73 for the fragment of pCY705. E. Choi-Rhee of our laboratory constructed plasmid pER73 using the primers shown in Table 1. Plasmid pAH7 was constructed from pCY730 by replacement of the accC gene with the SstII-BglII kanamycin resistance cassette of pCY708. The fragment containing the accC gene was cut with SstII and BglI and used to replace the kanamycin resistance gene of plasmids pAH13 and pAH14 in order to construct plasmids pAH15 and pAH16, respectively. Two plasmids, pAH8 and pAH9, which encoded a derivative of the 87-residue C-terminal fragment of AccB in which the biotinylated lysine was replaced with arginine, were constructed by cloning the NdeI-BsiWI fragment of plasmid pAH1 into pAH7 and pCY730, respectively.
Assays.
β-Galactosidase activity was determined as described by Miller (27) following disruption of the cells by sodium dodecyl sulfate-chloroform treatment. Protein biotinylation was measured by pipetting 100-μl samples of cultures grown on [8,9-3H]biotin (1 μCi/ml) at the required biotin concentrations onto 2.5-cm disks of Whatman 3MM filter paper previously soaked with 50 μl of 0.5 mM nonradioactive biotin (11, 17). The filter disks were washed twice with 10% trichloroacetic acid and then twice with 5% trichloroacetic acid. The radioactivity was determined by scintillation counting with an LS6500 multipurpose scintillation counter (Beckman Coulter). The counting efficiency was obtained by quantitative elution of the labeled proteins from the filter disks by boiling the disks for 15 min in 0.1 M NaOH (17).
RESULTS
Overproduction of AccC results in down-regulation of bio operon transcription.
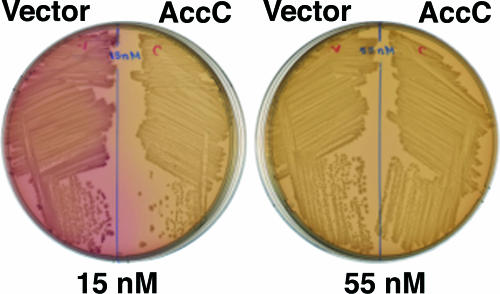
We first assayed expression of the biotin biosynthetic operon using the bio-lac fusion strain BM2661 of Barker and Campbell (4) or an essentially identical strain, strain CY486, constructed by Cronan (17). In these strains the lacZ and lacY genes are fused to the rightward bio promoter (the promoter of bioBFCD), resulting a lactose-positive phenotype when bio operon transcription is derepressed and a lactose-negative phenotype when the operon is repressed. These strains also carry a deletion of the chromosomal lactose operon and are biotin auxotrophs due to insertion of the lactose utilization genes into bioF. These strains form bright red colonies when they are spread on MacConkey agar plates due to lactose utilization resulting from derepression of bio operon transcription by the low concentration of biotin present in the medium (15 nM according to data provided by the manufacturer). However, upon plating on MacConkey agar supplemented with additional biotin (40 nM or a higher concentration) white colonies are formed, indicating that there is repression of bio operon transcription by biotin (4). In preliminary tests we found that introduction of a plasmid that expressed high levels of AccC resulted in white colonies on MacConkey plates that were not supplemented with additional biotin, indicating that there was inhibition of the depression of bio operon transcription normally seen on this medium (Fig. 3). Three different accC plasmids gave this result. These plasmids were (i) a medium-copy-number tac promoter plasmid (see below), (ii) a very-high-copy-number lac promoter plasmid (similar to the plasmid used by Karow et al. [23]) (data not shown), and (iii) a low- to medium-copy number plasmid in which accC was expressed from a plasmid tet promoter (data not shown). In contrast, an accC plasmid derived from pCY465, a plasmid expected to express AccC at a level similar to the level expressed by the chromosomal gene (15), failed to block depression. Hence, significant overexpression of AccC was needed to observe inhibition of bio operon transcription. It should be noted that accB, accC, and birA are all essential for growth of E. coli.
FIG. 3.
Repression of bio operon transcription upon overproduction of AccC. The plates are MacConkey agar plates containing 100 μM IPTG, and the biotin concentrations are indicated below the plates. The red color is due to depression of the bioBFCD promoter that drives expression of the lacZ and lacY genes (4). The host was strain CY1740, and the accC and vector plasmids were pCY701 and pKK223-3, respectively. Essentially identical results were obtained with strains BM2661 and CY486.
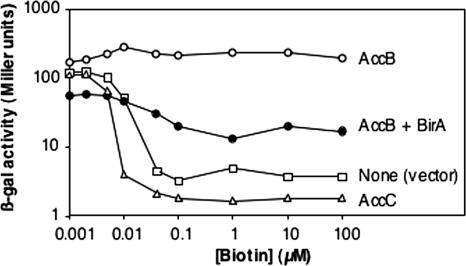
The effects of AccC overproduction on bio operon transcription were quantitated by assaying β-galactosidase activity in strain CY1740 (Fig. 4). The presence of high levels of LacI in this strain allowed the effects of induction of the tac promoter-controlled acc genes to be tested. Derivatives of strain CY1740 carrying either the tac vector plasmid or plasmids that encoded AccC, AccB, or both AccB and BirA (on separate compatible plasmids) were constructed. The strain carrying the vector plasmid showed the expected decreases in bio operon transcription with increasing biotin concentrations first reported by Barker and Campbell (4) and subsequently confirmed in our laboratory (17). The most responsive range of biotin concentrations is about 4 to 40 nM (4, 17), as shown by the slope of the curve for the vector-containing strain (Fig. 4). As expected from previous work, addition of biotin at a concentration of 40 nM resulted in almost maximal repression of bio operon transcription in the vector-containing strain, whereas addition of 10 nM biotin resulted in only slight repression (Fig. 4). In contrast, the slope for the strain overproducing AccC was much steeper, and at 10 nM biotin the strain overproducing AccC showed almost full repression (Fig. 4). This down-regulation, which increased with the biotin concentration up to a concentration of about 100 μM, was dependent on induction of AccC overproduction since in the absence of inducer the strain had an induction profile similar to that of the vector-containing control strain (Fig. 4). Derepression due to AccB overproduction was partially offset by overproduction of BirA (Fig. 4), as expected from previous work (17).
FIG. 4.
Expression with various biotin concentrations of the chromosomal φ(bioFC-lacZ)501 fusion of derivatives of strain CY1740 carrying a plasmid encoding AccB (○) or AccC (▵) or the vector pKK223-3 (□). A derivative carrying two compatible plasmids, one encoding AccB and the other encoding BirA (•), was also assayed. The accC, accB, and birA plasmids were pCY701, pCY708, and pCY216, respectively. Note that both axes have log scales. The experiment was repeated twice in its entirety, and in addition, the vector and AccC overproduction cultures were compared two more times. The results were essentially identical to those shown. β-gal, β-galactosidase.
Overproduction of both AccB and AccC relieves the down-regulation observed with overproduction of AccC alone.
A plausible mechanism for the down-regulation observed when AccC is overproduced is that AccC forms a complex with the AccB produced by the chromosomal accB gene and the complex is a poorer substrate for BirA-catalyzed biotinylation than free AccB is. Thus, high intracellular levels of BirA liganded to biotinoyl-5′-AMP accumulate, which result in increased occupancy of the bio operator and in greater repression of biotin synthesis (given sufficiently high intracellular biotin levels) (Fig. 1D). If this is true, overexpression of AccB together with overexpression of AccC should eliminate the inhibition observed when AccC is overproduced. Therefore, we constructed a series of plasmids based on the tac promoter vector that coexpressed both AccB and AccC or expressed only AccB using the native ribosome binding sites. Derivatives of strain CY1740 carrying these plasmids were then tested on MacConkey plates containing various concentrations of biotin. In studies done in parallel with the AccC plasmid and the empty vector we found that the strain carrying the accBC plasmid produced red colonies on unsupplemented MacConkey medium (containing15 nM biotin), indicating that coordinated overproduction of AccB and AccC relieved the inhibitory effects seen when AccC alone was overproduced (data not shown). As expected from previous work (17), overproduction of AccB resulted in derepression of bio operon transcription (red colonies) with both 15 nM and 55 nM biotin and pink colonies with 1 μM biotin due to consumption of the BirA-bound biotinoyl-5′-AMP and concomitant freeing of the bio operator (data not shown). These results were supported by β-galactosidase assays that showed that the effects of AccC overproduction were completely abolished when AccB was overproduced simultaneously with AccC (Fig. 5). Indeed, overproduction of both AccB and AccC resulted in derepression of bio operon transcription similar to the derepression seen when only AccB was overexpressed (Fig. 4).
FIG. 5.
Expression with various biotin concentrations of the chromosomal φ(bioFC-lacZ)501 fusion of derivatives of strain CY1740 carrying a plasmid encoding AccBC (▪ and □) or AccC (• and ○). The solid symbols and plus signs indicate induction by addition of 0.1 mM IPTG, whereas the open symbols and minus signs indicate uninduced cultures. The accC and accBC plasmids were pCY701 and pCY705, respectively. The experiment was repeated twice in its entirety, and in addition, the AccBC and AccC overproduction cultures were compared two more times. The results were essentially identical to those shown. β-gal, β-galactosidase.
AccB-AccC interaction is required for AccB overproduction to counter the down-regulation resulting from AccC overproduction.
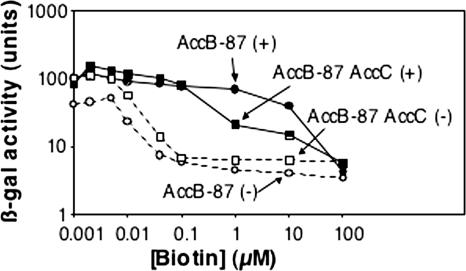
The reversal of the inhibitory effects of AccC overproduction by coupled overproduction with AccB indicated that when there was no longer an excess of AccC, AccB became biotinylated and was assembled into AccB-AccC complexes. If this is true, a mutant AccB protein that is unable to bind AccC should not reverse the down-regulation seen when AccC is overproduced. This is because excess AccC would remain free to interact with the AccB encoded by the chromosome. However, due to the regulatory circuitry, interpretation of this experiment is complicated by the derepression of bio operon transcription resulting from overproduction of a biotin acceptor protein. Hence, we expected to observe levels of repression intermediate between those seen when the mutant AccB alone was overexpressed and those seen when the mutant AccC alone was overexpressed, and perhaps the intermediate levels would be seen only at high biotin concentrations, when much of the AccB would have become biotinylated (and hence neutralized). The mutant AccB used was a protein comprising the carboxyl-terminal 87 residues of AccB. This protein is unable to bind AccC (1, 12). Coupled overexpression of AccC with the truncated AccB protein resulted in increased repression compared to that of a strain that overexpressed only the truncated AccB protein (Fig. 6) or a strain that overproduced both the native AccB and AccC proteins (Fig. 5). However, as expected, these different effects were seen only at high biotin concentrations; the effects at lower concentrations were masked by biotin acceptor protein overproduction. Similar results were obtained using a plasmid that encoded wild-type AccC plus a mutant AccB in which a stretch of three N-terminal residues required to form the metastable AccB-AccC complex (12) was deleted (data not shown). However, this construct showed greater repression at low biotin concentrations than the C-terminal 87-residue construct showed, probably because the full-length AccB protein is a poorer BirA substrate (it has a higher Km) than the 87-residue protein (28).
FIG. 6.
Expression with various biotin concentrations of the chromosomal φ(bioFC-lacZ)501 fusion of derivatives of strain CY1740 carrying plasmid pAH1 encoding the carboxyl 87 residues of AccB (AccB-87) (• and ○) or plasmid pAH2 encoding AccB-87 plus AccC (▪ and □). The solid symbols and plus signs indicate induction by addition of 0.1 mM IPTG, whereas the open symbols and minus signs indicate uninduced cultures. The experiment was repeated twice, and the results were essentially identical to those shown. β-gal, β-galactosidase.
In order to remove the complication of derepression of bio operon transcription by increased biotin acceptor protein levels, we tried to construct a mutant AccB that bound AccC but was not biotinylated. Our hypothesis was that upon coexpression with AccC this mutant AccB protein would interact with and thereby neutralize the overproduced AccC, thus relieving the repression seen when AccC was overproduced alone. The obvious approach to construct an AccB protein that could not be biotinylated was to substitute another residue for the lysine residue (K122) to which biotin is attached. However, because there is not a good mimic of lysine among the natural amino acids, we constructed accBC plasmids encoding three different K122 substitutions, K122R, K122M, and K122E, in the hope that one or more of the proteins would retain the ability to fold normally and efficiently bind AccC. The K122R substitution would preserve a positive charge like that of lysine and an aliphatic chain (albeit a longer chain). K122E would provide an aliphatic chain (albeit a shorter chain) plus a charge (albeit the opposite charge), whereas K122M would provide an uncharged mimic of the lysine aliphatic chain (although shorter and kinked due to the thioether bonds). Coexpression of AccB K122R with AccC almost fully relieved the down-regulation of bio operon transcription seen when AccC was overexpressed alone; almost full derepression was seen with 10 nM biotin (Fig. 7). In contrast, the other two K122 mutant proteins, K122M and K122E, only weakly relieved down-regulation when they were overexpressed along with AccC, suggesting that these proteins bound AccC poorly. Relief of down-regulation by AccC overexpression required interaction of the mutant AccB with AccC because AccC coexpression with a K122R derivative of a protein consisting of the carboxyl 87 residues of AccB resulted in levels of repression very similar to the level seen when AccC was overproduced alone (data not shown). Previous work showed that the K122R AccB protein cannot be biotinylated, as would be expected from the very different pKa values of the lysine and arginine side chains (15). Consistent with this result, we found that high-level expression of all three K122 mutant AccB proteins in the absence of coupled AccC expression did not derepress bio operon transcription (data not shown).
FIG. 7.
Expression of the chromosomal φ(bioFC-lacZ)501 fusion of derivatives of strain CY1740 carrying plasmids encoding AccC plus mutant AccB proteins. The AccB K122R protein (•), mutant AccB K122E protein (□), and mutant AccB K122M protein (▴) were encoded by plasmids pAH7, pAH13, and pAH14, respectively. The experiment was repeated twice, and the results were essentially identical to those shown. β-gal, β-galactosidase.
Overproduction of AccC inhibits biotinylation of AccB.
The bio operon expression data indicate that inhibition results from interaction of excess AccC with AccB to form a complex that is a poor (compared to free AccB) BirA substrate. Therefore, a strain overproducing AccC should contain less biotinylated AccB than a wild-type strain contains. Indeed, this was the case. At biotin concentrations less than about 10 nM, AccB (encoded by the chromosomal gene) was 50 to 75% underbiotinylated when AccC was overproduced (Table 2). This deficiency in biotinylation was reversed when there was simultaneous overproduction of BirA, indicating that AccC and BirA compete for AccB, as expected from the bio operon expression results. Increased biotin concentrations also partially relieved underbiotinylation, presumably by increasing the activity of BirA (Table 2). Finally, expression of K122R AccB together with AccC relieved underbiotinylation of the chromosomally expressed AccB (Table 2).
TABLE 2.
Inhibition of AccB biotinylation by AccC overproduction and reversal of this inhibition by BirA overproductiona
| Biotin concn (nM) | Biotinylated AccB concn (pmol/109 cells) with:
|
|||
|---|---|---|---|---|
| Vector | AccC | AccC + BirA | AccC + AccB (K122R) | |
| 1 | 2.5 | 1.1 | 4.1 | 10.5 |
| 2 | 3.6 | 1.2 | 3.7 | 9.8 |
| 5 | 6.1 | 4.1 | 7.2 | 23.1 |
| 10 | 13.1 | 4.0 | 16.5 | 18.7 |
| 40 | 13.8 | 11.6 | 17.4 | 17.1 |
| 100 | 17.5 | 12.9 | 18.2 | 21.4 |
Cultures of strain CY1740 carrying plasmids encoding AccC (pCY701), AccC plus BirA (pCY701 and pCY216), AccB K122 plus AccC (pCY730), or the vector (pKK223-3) were used for AccC plasmid construction. The strains were grown in the presence of 3H-labeled biotin at the concentrations indicated, and attachment to AccB (the sole biotinylated protein of E. coli) was measured as described in Materials and Methods.
DISCUSSION
The transcriptional coupling of the E. coli accB and accC genes can now be rationalized by the fact that overproduction of either AccB or AccC without the other disrupts regulation of the bio operon, leading to either overproduction or underproduction of biotin. Moreover, Karow et al. (23) reported that overproduction of either protein alone results in appreciable inhibition of the rate of fatty acid synthesis. This effect cannot be attributed to disturbed biotin production because these workers used a medium that contained biotin. However, in the absence of an exogenous source of biotin, the decreased biotinylation of AccB seen when AccC is overproduced has obvious physiological consequences. It should also be noted that overproduction of biotin is metabolically expensive despite the low levels of this vitamin required by E. coli. This is because the pathway requires expenditure of at least seven ATP equivalents per biotin molecule and six different enzymes, one of which, BioB, appears to be degraded often as a consequence of its catalytic cycle (13).
Our data suggest that free AccB is the preferred substrate for BirA-catalyzed biotinylation in vivo and that biotinylated AccB is the form normally incorporated into the AccB-AccC complex. Thus, in vivo it seems that AccC and BirA compete for unbiotinylated AccB, as shown in Table 2, and the system is set such that BirA wins the competition. The bifunctional nature of BirA precludes unambiguous interpretation of bio operon regulation experiments in which BirA and AccC are both overexpressed. Interpretable experiments could be done using a BirA mutant protein that was completely defective in operator binding but that retained full biotinylation activity. Unfortunately, no such mutant is known. Indeed, removal of the N-terminal DNA binding domain from BirA severely compromises the ability of the protein to bind biotin and biotinoyl-5′-AMP (34). It should be noted that our laboratory previously reported that unbiotinylated AccB complexed with AccC was a substrate for BirA-catalyzed biotinylation in vitro (12). However, the effect was modest, and due to the metastable nature of the isolated complex we cannot exclude the possibility that unbiotinylated AccB dissociated from the complex became biotinylated and then reformed a complex. Our interpretation that free AccB is the preferred substrate for BirA-catalyzed biotinylation in vivo is consistent with the finding that fusion proteins having the 87-residue AccB biotin domain as the downstream partner compete well with the full-length AccB protein for biotinylation in vitro (10, 24).
Further work is necessary to determine if the AccB-AccC complex formed when AccC is overproduced has the normal 2:1 stoichiometry or a different stoichiometry. However, if AccC overproduction results in aberrant complexes, these complexes may be less stable than the normal complex and thus very difficult to isolate. It would be interesting to determine if the effects of AccC overproduction seen in E. coli are also seen in the distantly related bacterium Bacillus subtilis. B. subtilis BirA regulates transcription of its bio operon (7), and the accB and accC genes are cotranscribed (26). It would also be interesting to determine if and how stoichiometric production of AccB and AccC occurs in cyanobacteria, where the accB and accC genes are unlinked (20).
Acknowledgments
We thank E. Choi-Rhee for construction of the K122R mutant.
This work was supported by NIH grant AI15650 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 20 October 2006.
REFERENCES
- 1.Alberts, A. W., A. M. Nervi, and P. R. Vagelos. 1969. Acetyl CoA carboxylase. II. Demonstration of biotin-protein and biotin carboxylase subunits. Proc. Natl. Acad. Sci. USA 63:1319-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker, D. F., and A. M. Campbell. 1981. The birA gene of Escherichia coli encodes a biotin holoenzyme synthetase. J. Mol. Biol. 146:451-467. [DOI] [PubMed] [Google Scholar]
- 3.Barker, D. F., and A. M. Campbell. 1981. Genetic and biochemical characterization of the birA gene and its product: evidence for a direct role of biotin holoenzyme synthetase in repression of the biotin operon in Escherichia coli. J. Mol. Biol. 146:469-492. [DOI] [PubMed] [Google Scholar]
- 4.Barker, D. F., and A. M. Campbell. 1980. Use of bio-lac fusion strains to study regulation of biotin biosynthesis in Escherichia coli. J. Bacteriol. 143:789-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckett, D. 2005. The Escherichia coli biotin regulatory system: a transcriptional switch. J. Nutr. Biochem. 16:411-415. [DOI] [PubMed] [Google Scholar]
- 6.Beckett, D. 2005. Multilevel regulation of protein-protein interactions in biological circuitry. Phys. Biol. 2:S67-S73. [DOI] [PubMed] [Google Scholar]
- 7.Bower, S., J. Perkins, R. R. Yocum, P. Serror, A. Sorokin, P. Rahaim, C. L. Howitt, N. Prasad, S. D. Ehrlich, and J. Pero. 1995. Cloning and characterization of the Bacillus subtilis birA gene encoding a repressor of the biotin operon. J. Bacteriol. 177:2572-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosius, J. 1988. Expression vectors employing lambda-, trp-, lac-, and lpp-derived promoters. Bio/Technology 10:205-225. [DOI] [PubMed] [Google Scholar]
- 9.Chapman-Smith, A., and J. E. Cronan, Jr. 1999. The enzymatic biotinylation of proteins: a post-translational modification of exceptional specificity. Trends Biochem. Sci. 24:359-363. [DOI] [PubMed] [Google Scholar]
- 10.Chapman-Smith, A., T. W. Morris, J. C. Wallace, and J. E. Cronan, Jr. 1999. Molecular recognition in a post-translational modification of exceptional specificity. Mutants of the biotinylated domain of acetyl-CoA carboxylase defective in recognition by biotin protein ligase. J. Biol. Chem. 274:1449-1457. [DOI] [PubMed] [Google Scholar]
- 11.Chapman-Smith, A., D. L. Turner, J. E. Cronan, Jr., T. W. Morris, and J. C. Wallace. 1994. Expression, biotinylation and purification of a biotin-domain peptide from the biotin carboxy carrier protein of Escherichia coli acetyl-CoA carboxylase. Biochem. J. 302:881-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi-Rhee, E., and J. E. Cronan. 2003. The biotin carboxylase-biotin carboxyl carrier protein complex of Escherichia coli acetyl-CoA carboxylase. J. Biol. Chem. 278:30806-30812. [DOI] [PubMed] [Google Scholar]
- 13.Choi-Rhee, E., and J. E. Cronan. 2005. Biotin synthase is catalytic in vivo, but catalysis engenders destruction of the protein. Chem. Biol. 12:461-468. [DOI] [PubMed] [Google Scholar]
- 14.Clementz, T., J. J. Bednarski, and C. R. Raetz. 1996. Function of the htrB high temperature requirement gene of Escherichia coli in the acylation of lipid A: HtrB catalyzed incorporation of laurate. J. Biol. Chem. 271:12095-12102. [DOI] [PubMed] [Google Scholar]
- 15.Cronan, J. E., Jr. 2001. The biotinyl domain of Escherichia coli acetyl-CoA carboxylase. Evidence that the “thumb” structure is essential and that the domain functions is a dimer. J. Biol. Chem. 276:37355-37364. [DOI] [PubMed] [Google Scholar]
- 16.Cronan, J. E., Jr. 1989. The E. coli bio operon: transcriptional repression by an essential protein modification enzyme. Cell 58:427-429. [DOI] [PubMed] [Google Scholar]
- 17.Cronan, J. E., Jr. 1988. Expression of the biotin biosynthetic operon of Escherichia coli is regulated by the rate of protein biotination. J. Biol. Chem. 263:10332-10336. [PubMed] [Google Scholar]
- 18.Cronan, J. E., Jr., and G. L. Waldrop. 2002. Multi-subunit acetyl-CoA carboxylases. Prog. Lipid Res. 41:407-435. [DOI] [PubMed] [Google Scholar]
- 19.Fall, R. R., A. W. Alberts, and P. R. Vagelos. 1975. Analysis of bacterial biotin-proteins. Biochim. Biophys. Acta 379:496-503. [DOI] [PubMed] [Google Scholar]
- 20.Gornicki, P., L. A. Scappino, and R. Haselkorn. 1993. Genes for two subunits of acetyl coenzyme A carboxylase of Anabaena sp. strain PCC 7120: biotin carboxylase and biotin carboxyl carrier protein. J. Bacteriol. 175:5268-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grana, D., T. Gardella, and M. M. Susskind. 1988. The effects of mutations in the ant promoter of phage P22 depend on context. Genetics 120:319-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James, E. S., and J. E. Cronan. 2004. Expression of two Escherichia coli acetyl-CoA carboxylase subunits is autoregulated. J. Biol. Chem. 279:2520-2527. [DOI] [PubMed] [Google Scholar]
- 23.Karow, M., O. Fayet, and C. Georgopoulos. 1992. The lethal phenotype caused by null mutations in the Escherichia coli htrB gene is suppressed by mutations in the accBC operon, encoding two subunits of acetyl coenzyme A carboxylase. J. Bacteriol. 174:7407-7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, S. J., and J. E. Cronan, Jr. 1992. The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J. Biol. Chem. 267:855-863. [PubMed] [Google Scholar]
- 25.Li, S. J., and J. E. Cronan, Jr. 1993. Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step of lipid biosynthesis. J. Bacteriol. 175:332-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marini, P., S. J. Li, D. Gardiol, J. E. Cronan, Jr., and D. de Mendoza. 1995. The genes encoding the biotin carboxyl carrier protein and biotin carboxylase subunits of Bacillus subtilis acetyl coenzyme A carboxylase, the first enzyme of fatty acid synthesis. J. Bacteriol. 177:7003-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics, 1st ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Nenortas, E., and D. Beckett. 1996. Purification and characterization of intact and truncated forms of the Escherichia coli biotin carboxyl carrier subunit of acetyl-CoA carboxylase. J. Biol. Chem. 271:7559-7567. [DOI] [PubMed] [Google Scholar]
- 29.Otsuka, A., and J. Abelson. 1978. The regulatory region of the biotin operon in Escherichia coli. Nature 276:689-694. [DOI] [PubMed] [Google Scholar]
- 30.Streaker, E. D., and D. Beckett. 2006. The biotin regulatory system: kinetic control of a transcriptional switch. Biochemistry 45:6417-6425. [DOI] [PubMed] [Google Scholar]
- 31.Takeshita, S., M. Sato, M. Toba, W. Masahashi, and T. Hashimoto-Gotoh. 1987. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene 61:63-74. [DOI] [PubMed] [Google Scholar]
- 32.Weaver, L. H., K. Kwon, D. Beckett, and B. W. Matthews. 2001. Competing protein:protein interactions are proposed to control the biological switch of the E coli biotin repressor. Protein Sci. 10:2618-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu, Y., and D. Beckett. 1997. Biotinyl-5′-adenylate synthesis catalyzed by Escherichia coli repressor of biotin biosynthesis. Methods Enzymol. 279:405-421. [DOI] [PubMed] [Google Scholar]
- 34.Xu, Y., and D. Beckett. 1996. Evidence for interdomain interaction in the Escherichia coli repressor of biotin biosynthesis from studies of an N-terminal domain deletion mutant. Biochemistry 35:1783-1792. [DOI] [PubMed] [Google Scholar]