Abstract
Our whole-genome microarray studies of Neisseria meningitidis MC58 previously identified a set of 153 genes whose transcription was activated during growth in iron. In this study, Fur-mediated regulation of the iron-activated nspA gene was confirmed, whereas iron-activated regulation of the secY gene was demonstrated to be Fur independent. Analysis of the Fur binding sequences in the nspA gene and an additional iron-activated and Fur-regulated gene identified a hexameric (G/T)ATAAT unit in the operator regions of these genes similar to that observed in Fur- and iron-repressed genes. These studies indicate that the expression of the iron-activated nspA and secY genes in N. meningitidis occur by Fur-dependent and -independent mechanisms, respectively.
It is well established that the iron-responsive regulatory protein Fur functions as a repressor of gene transcription in several microorganisms. In its most basic state, Fur forms a dimer together with divalent cations, such as ferrous iron, and binds to a consensus sequence (the Fur box) that overlaps the promoters of iron-regulated genes to prevent their transcription. Recent studies indicate that in some organisms, Fur may also function as a positive regulator of gene transcription, together with iron (8, 9-12, 24), although the mechanism of iron activation by Fur is not well elucidated. Our recent studies of N. meningitidis group B, using a combination of microarray technology, computational analysis, and in vitro binding studies, revealed that a large number of genes are activated during growth in the presence of iron and that a number of these iron-activated genes have putative Fur-binding sequences to which Fur was demonstrated to bind (17). However, the biological significance of Fur binding to the operator regions of these genes has not been defined, as an N. meningitidis fur mutant had not been constructed at the time of that study.
Of interest within the group of iron-activated genes under the potential control of Fur were candidate genes involved in the virulence potential of N. meningitidis, including the nspA (NMB0663) (1, 21) and secY (NMB0162) (18, 28) genes. Neisserial surface protein A (NspA) is an 18.6-kDa membrane protein of unknown function that was first described to confer protection against meningococcal infection in animal models of infection (1, 21, 22). NspA is highly conserved and expressed by all N. meningitidis strains tested (22, 25). Recent studies indicate that conserved epitopes of the NspA protein confer protection against N. meningitidis serogroup B challenge in a mouse model of meningococcal infection (26). The N. meningitidis secY gene encodes a putative preprotein translocase (SecY) whose homolog in Escherichia coli has been studied extensively; in E. coli, it functions as an essential component of the protein translocation machinery of the cytoplasmic membrane. Sec-dependent protein secretion in the pathogenic Neisseria has been reported (18, 28); however, the function of the N. meningitidis SecY protein is not known, nor has the regulation of the secY gene been examined. In E. coli, the secY gene is located in a gene cluster known as the spc operon, which includes the rplN, rplX, rplE, rpsN, rpsH, rplF, rplR, rpsE, rpmD, and rplO genes, encoding ribosomal proteins (3, 19). While the structure and function of the E. coli Sec system has been well defined, the regulation of genes encompassing the E. coli Sec system are not well understood. In this study, we demonstrate that the iron-activated regulation of the N. meningitidis spc operon is Fur independent. Furthermore, we have definitively demonstrated the role of Fur and iron in the regulation of the N. meningitidis nspA gene and identified a hexameric (G/T)ATAAT unit in the operator region which is present in several additional Fur-regulated genes.
Regulation of the N. meningitidis secY and ribosomal protein genes in response to iron.
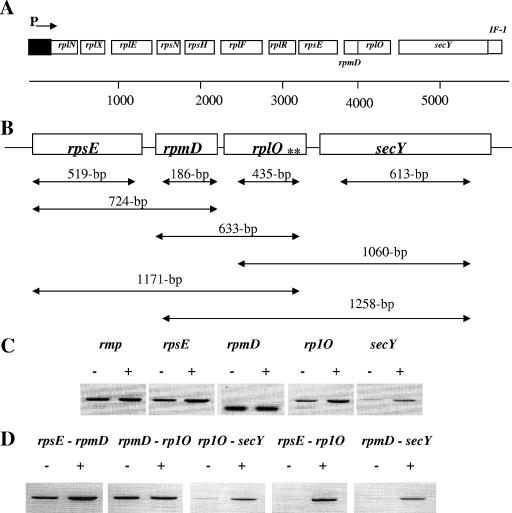
In N. meningitidis MC58, the secY gene is located at the distal region of the putative spc operon, flanking the rplO gene and a transcription initiation factor gene (if-1) within the operon (Fig. 1A). This putative operon is homologous to the previously identified spc operon of E. coli (3). To examine the expression of the ribosomal genes in response to iron and to delineate the operonic nature of this locus, we examined the transcriptional status of the genes by using reverse transcriptase PCR (RT-PCR) with several sets of primer pairs (Fig. 1B and Table 1) and RNA isolated from N. meningitidis MC58 cultures grown for 4 h under iron-depleted and iron-replete conditions. For these studies, N. meningitidis MC58 was grown in chemically defined medium (CDM) under iron-replete (100 μM ferric nitrate) and -depleted (12.5 μM desferal) conditions, as described previously (17). We observed growth restriction of the N. meningitidis MC58 cultures during growth under iron-depleted conditions (data not shown).
FIG. 1.
Regulation of the N. meningitidis rpsE, rpmD, rplO, and secY genes under iron-depleted and iron-replete conditions. (A) Schematic representation of the N. meningitidis MC58 secY gene and genes encoding ribosomal proteins within the spc operon. ORFs within the spc operon are labeled and encode genes for 10 ribosomal proteins [RplN (L14), RplX (L24), RplE (L5), RpsN (S14), RpsH (S8), RplF (L6), RplR (L18), RpsE (S5), RpmD (L30), and RplO (L15)], SecY, and a translation initiation factor (IF-1). The promoter region of the operon is represented as “P,” and the direction of transcription is represented with an arrow, as previously described for the E. coli spc operon (3). The ORFs represented are not drawn to scale. (B) Schematic representation of the DNA fragments amplified during transcription and cotranscription analysis. Each ORF examined in this study is labeled with the corresponding gene; regions amplified are represented with double-headed arrows, and the size of each amplicon is given for each arrow. A previously identified Fur binding sequence in the upstream region of the secY gene (17) is represented by double asterisks (**). (C and D) Transcription (C) and cotranscription (D) analyses of the rpsE, rpmD, rplO, and secY genes. Cultures of N. meningitidis were grown in CDM broth under iron-depleted (−) (12.5 μM desferal) or iron-replete (+) (100 μM ferric nitrate) conditions, and samples were removed at 4 h for RNA preparation. Approximately 250 ng of total RNA was used in each in RT-PCR and was amplified with the primers listed in Table 1.
TABLE 1.
Primers used in this study
| Primera | Sequence (5′→3′)b | Description |
|---|---|---|
| F1 | GAATACAATTCAACCTGCT | Amplifies 435-bp meningococcal rp1O ORF |
| R1 | ATTTCAATCTTACCACCAA | |
| F2 | GTTAAAAGCCTGATTGGTA | Amplifies 186-bp meningococcal rpmD ORF |
| R2 | AAGACTCCACTTTCAACAA | |
| F3 | GTTAACTCCCAAAATGTCTT | Amplifies 519-bp meningococcal rpsE ORF |
| R3 | CAAAACATGAAATTGAAGAA | |
| F4 | CGGGATCCCGTACAGCTCGCTTCTGAAAT | Amplifies 613-bp fragment of the meningococcal secY ORF |
| R4 | CCAAGCTTGGCTATTTGTATCAGCCGAACC | |
| F5 | GAAGATTTTGGATTTGTTCG | Amplifies 339-bp fragment of the meningococcal fur ORF |
| R5 | CACACGCCGTACATATAAAG | |
| F6 | TCAAGCTCTTTAGGTTCTGC | Amplifies 347-bp fragment of the meningococcal nspA ORF |
| R6 | ATGTAGTTGTAGCGGTAGCC | |
| F7 | ATACGGTTGAAGTGGAAT | Amplifies 520-bp fragment of the meningococcal aniA ORF |
| R7 | AACATAAACTTTGTCGAAGA | |
| F8 | CGGGATCCCGCAAGGTTTCATTTAATAAG | Amplifies 111-bp promoter fragment of the meningococcal secY/ribosomal operon |
| R8 | CGGGATCCCGATCTAAGATGGTCTGCATTT | |
| F9 | GCTCTAGAGCAAAATATTGCGATGCAAAA | Amplifies 101-bp promoter fragment of the meningococcal nspA gene |
| R9 | CGGGATCCCGATATTTTGGTTCCTTTATGG | |
| F10 | CGGGATCCCGCTGCTTCTTTATAGTGGAGA | Amplifies 114-bp promoter fragment of the meningococcal rmp gene |
| R10 | CGGGATCCCGCCTCATTAAATTTGTACAGC |
F, forward; R, reverse.
Restriction sites are underlined.
For RNA and protein isolation, 2.0-ml samples of bacterial cultures were collected at 1-h intervals for up to 6 h. Samples were centrifuged at 13,800 × g for 5 min, the supernatant was aspirated, and the remaining pellet was used for either RNA or protein extraction. Total RNA was isolated with the RNeasy kit (QIAGEN, Valencia, CA) and was DNase I treated to remove residual DNA contamination. The effectiveness of DNA removal was confirmed by PCR amplification of the RNA prior to reverse transcription. These DNA-free RNA samples served as the template for subsequent RT-PCR with the primers listed in Table 1. All RT-PCRs were performed as previously described (17). Briefly, the reaction mixture contained 250 ng of DNA-free RNA, 25 μl of 2× reaction mixture, 100 ng of each primer, 1 μl of RT-Taq mix, and diethyl pyrocarbonate-treated water to a final volume of 50 μl. cDNA synthesis was performed at 50°C for 30 min, followed by predenaturing at 94°C for 2 min. DNA amplification was carried out under the following conditions: 25 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 2 min, and elongation at 72°C for 2 min. RT-PCR analysis demonstrated increased levels of N. meningitidis rpsE, rplO, and secY fragments during growth under iron-replete conditions compared with cultures grown under iron-depleted conditions (Fig. 1C). Interestingly, no significant differences in the expression levels of the rpmD gene during growth of N. meningitidis under iron-depleted and -replete conditions were observed. One likely explanation for the lack of difference in the transcript level of this gene may be due to the relatively small size of the rpmD amplicon, which in turn would have resulted in saturation of DNA amplification under the conditions used.
We next analyzed cotranscription of the rpsE, rpmD, rplO, and secY genes by using specific primers to amplify single DNA fragments spanning the rpsE-rpmD, rpmD-rplO, rplO-secY, rpsE-rplO, and rpmD-secY open reading frames (ORFs) within the operon, as depicted in Fig. 1B. Using several different combinations of these primers, we observed that the rpsE, rpmD, rplO, and secY genes were cotranscribed within the operon and that this was most pronounced in samples obtained from cultures under iron-replete growth conditions (Fig. 1D). As expected, we did not observe differences in the transcript levels of the rmp gene (a gene which is not regulated in response to iron) (17) in N. meningitidis MC58 grown under iron-replete or -depleted conditions (data not shown). Our cotranscription results are in agreement with previous studies of E. coli indicating that transcription of the spc operon initiates from its own promoter and continues into the downstream operon containing additional ribosomal protein genes (3). Likewise, our studies suggest that genes of the spc operon are regulated by a common promoter and that the expression of these genes is transcriptionally activated by growth in the presence of iron. Increased expression of genes encoding ribosomal proteins and secretion proteins in response to iron may contribute to the high-efficiency protein synthesis and subsequent protein transport across the membrane in response to iron. Our results are also in agreement with recent studies by Ducey et al. (13), which suggest that several gonococcal genes, including genes encoding ribosomal proteins, are upregulated under iron-replete growth conditions.
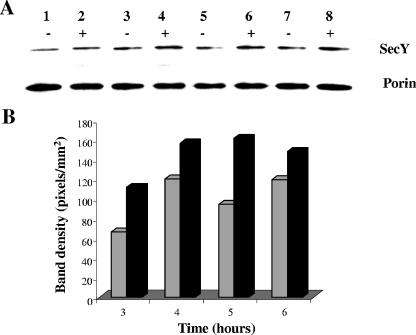
The increased expression of the secY gene under iron-replete conditions was further confirmed at the protein level by Western blot analysis. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of whole-cell lysates was performed as described previously (29) with 10% polyacrylamide gels. Proteins were electrophoretically transferred to nitrocellulose membranes and probed with either anti-SecY serum raised against the N-terminal region of E. coli SecY (1:2,500) (20) or anti-porin B serum (1:2,500) (23). Blots were then incubated with appropriate secondary antibodies conjugated to horseradish peroxidase conjugates (Sigma). Immunoreactive proteins were detected by the ECL+Plus Western blot detection system (Amersham Biosciences, Piscataway, NJ), and the intensity of the immunoreactive bands was quantitated with the Bio-Rad Quantity One program. As shown in Fig. 2A, a modest increase in the level of SecY protein expression was observed under iron-replete growth conditions. The increase in SecY protein in response to growth under iron-replete conditions was maximally observed at 5 h (Fig. 2B).
FIG. 2.
Expression of the N. meningitidis SecY protein in response to growth under iron-depleted and -replete conditions. (A) Western blot analysis. Approximately 1.5 mg of total cell lysate was prepared from N. meningitidis MC58 grown in CDM broth under iron-depleted (−) and -replete (+) conditions and loaded onto sodium dodecyl sulfate-polyacrylamide gels. Proteins were transferred to membranes and probed with either E. coli SecY antiserum or N. meningitidis porin B antiserum (positive control for protein loading). Lanes 1 and 2, 3 and 4, 5 and 6, and 7 and 8 are samples obtained at 3, 4, 5, and 6 h, respectively. (B) Quantitative analysis. Densities of the immunoreactive bands were quantitated with the Bio-Rad Quantity One program (P < 0.001). Samples were obtained from cultures grown under iron-depleted (gray bars) and iron-replete (black bars) growth conditions. The results are representative of three independent experiments.
Role of Fur in the regulation of the N. meningitidis secY and nspA genes.
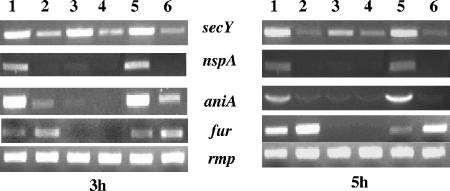
To investigate the role of Fur in the iron-activated expression of the secY and nspA genes, we utilized a N. meningitidis fur deletion mutant (MC-Fko′) and a fur-complemented mutant (MC-Fko′-C), which we constructed with the plasmid constructs pGemFkoB:Km and pSLFur-C1, respectively, according to the method of Delany et al. (6). Positive transformants were confirmed by PCR, sequencing, and Western blot analysis with anti-Fur antibody (data not shown). To analyze the expression of iron-regulated secY and nspA genes by Fur, total RNA was isolated from wild-type, fur mutant, and fur-complemented mutant strains after 3 and 5 h of growth in CDM under iron-replete and -depleted conditions, and RT-PCR analyses were performed as described above. All three strains (wild type, fur mutant, and fur-complemented mutant) grew to similar levels under iron-replete and -depleted conditions, with a similar level of growth restriction upon growth under iron-deplete conditions (data not shown). As expected, an increased level of the secY transcript was observed in the wild-type (Fig. 3, lanes 1 and 2) and fur-complemented mutant strains (Fig. 3, lanes 5 and 6) under iron-replete conditions. Notably, an increased level of the secY transcript was also observed in the fur mutant grown under iron-replete conditions compared to iron-depleted conditions, similar to that of the wild type and complemented fur mutant strain under similar growth conditions. Similarly, our Western blot analysis of total cell lysates obtained from the wild-type, fur mutant, and complemented fur mutant strains also indicated that expression of secY is transcriptionally activated by iron and is not regulated by Fur (data not shown). Unlike the secY gene, transcription of the nspA gene is iron activated in a Fur-dependent manner, as determined by the expression of the nspA gene in both the wild-type and fur complemented strain but not in the fur mutant strain (Fig. 3, lanes 3 and 4). As expected, we did not observe differences in the transcript levels of the rmp gene (17) in any of the three strains grown under iron-replete or -depleted conditions. Likewise, the iron-repressed and Fur-regulated meningococcal fur gene (6) and iron-activated and Fur-regulated aniA gene (7) were confirmed to be regulated by Fur and iron. These results indicate that both the secY and nspA genes are regulated in response to growth with iron but that only the nspA gene is regulated by Fur.
FIG. 3.
Expression of the N. meningitidis secY and nspA genes in response to iron and Fur. Transcriptional analysis of the secY and nspA genes from RNA isolated from N. meningitidis MC58 (lanes 1 and 2), N. meningitidis Fko′ (lanes 3 and 4), and N. meningitidis Fko′-C (lanes 5 and 6) by RT-PCR is shown. Meningococcal rmp, fur, and aniA genes were utilized as controls. Lanes 1, 3, and 5 correspond to samples obtained from cultures grown under iron-replete conditions, and lanes 2, 4, and 6 correspond to samples obtained from cultures grown under iron-depleted conditions. The time at which samples were examined is indicated below each panel.
Binding studies of meningococcal Fur to operator sequences of the iron-activated nspA and secY genes.
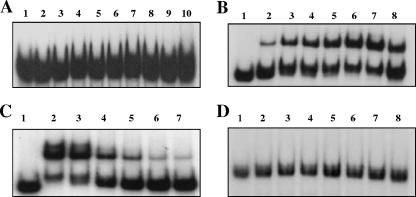
Previously, we demonstrated by electrophoretic mobility shift assay (EMSA) analysis that meningococcal Fur (17) was able to bind to the putative Fur binding sequences in the upstream regions of the iron-activated secY and nspA genes (30). Briefly, the promoter fragments for EMSA were PCR amplified from N. meningitidis MC58 with the oligonucleotides shown in Table 1. DNA fragments were restriction digested, gel purified, and end labeled by Klenow fragment with [α-32P]dATP and a nucleoside triphosphate mixture (dCTP, dTTP, and dGTP). The reaction mixture was purified with MicroSpin G-25 columns (Amersham Pharmacia Biotech, Piscataway, NJ) to remove unincorporated nucleotides. Labeled DNA probes were then incubated with N. meningitidis Fur protein in binding buffer (100 mM Tris-Cl, pH 7.2-25 mM MgCl2-200 mM KCl-0.625 mM MnCl2-10 mM dithiothreitol [DTT]-50% glycerol-3.0 μg poly(dI-dC)-5.0 μg bovine serum albumin [BSA]) at room temperature for 20 min and electrophoresed on a native 6% polyacrylamide gel to separate different complexes. Following electrophoresis, the gels were transferred to filter paper and dried for 1 h at 80°C, and radioactive bands were visualized by autoradiography. Our previous analysis revealed that the putative Fur box to which Fur binding was observed was located in the upstream region of the secY gene and overlapped the ORF of the rplO gene (Fig. 1B). Interestingly, our in silico analysis of the annotated N. meningitidis MC58 genome failed to identify any promoter-like sequences within this short intergenic region (12 bp) between the rplO and secY genes. In E. coli, transcription of genes within the spc operon is initiated from the spc promoter (3), and we postulated that most likely the transcription of N. meningitidis secY/ribosomal operon genes is initiated in a similar fashion. To clarify the discrepancy between the ability of Fur to bind to the putative operator region of the secY gene and the lack of the involvement of Fur in the regulation of the secY gene, we scanned the promoter region of the N. meningitidis secY/ribosomal operon, using a 19-bp E. coli palindromic Fur binding sequence (2, 9, 10) or a 21-bp Neisseria consensus sequence that has ∼80% homology with the E. coli consensus (16), and identified a putative Fur binding sequence in this region (see Fig. 5B). EMSA analysis was then performed (17) to examine the direct interaction of Fur with the in silico-identified Fur box sequence within the putative promoter region of the secY/ribosomal operon. As shown in Fig. 4A, we did not observe a shift when the Fur protein was used with the in silico-identified Fur box sequence within the putative promoter region of the secY/ribosomal operon, even with a relatively high concentration (400 nM) of Fur.
FIG. 5.
DNase I footprinting analysis of N. meningitidis Fur with the nspA operator probe. (A) Footprinting analysis. The plus strand of the nspA promoter fragment was labeled and incubated with increasing concentrations of N. meningitidis Fur prior to digestion with DNase I. Lane 1, no Fur; lane 2, 150 nM Fur; lane 3, 460 nM Fur; lane 4, 1.22 μM Fur. DNA standards (GATC) are shown on the right. The region protected by Fur is indicated by the dark line. (B) Schematic representation of the operator elements of the iron-activated nspA, aniA, and secY genes and the iron-repressed fur gene. DNA sequences of the iron-activated and Fur-regulated nspA and aniA genes, the iron-repressed and Fur-regulated fur gene, and the iron-activated secY gene operators were amplified for in vitro binding studies using the primers listed in Table 1. Boxed nucleotides represent the in silico-identified Fur box. The previously reported hexameric repeat 5′-NATW(A/T)AT-3′ (17) is represented in boldface type. In the case of the nspA gene, the extent of the footprinted region is represented by a double-headed arrow.
FIG. 4.
EMSA analysis of meningococcal Fur binding to meningococcal secY and nspA operator DNA probes. DNA probes used are as follows: A, secY; B and C, nspA; D, rmp. One nanogram of 32P-labeled DNA fragments was incubated with increasing concentrations of N. meningitidis Fur protein. (A) Lane 1, no Fur; lanes 2 to 10, 80 nM, 120 nM, 140 nM, 160 nM, 180 nM, 200 nM, 240 nM, 300 nM, and 400 nM Fur, respectively. (B and D) Lane 1, no Fur; lanes 2 to 8, 160 nM, 320 nM, 400 nM, 480 nM, 560 nM, 720 nM, and 800 nM Fur, respectively. (C) EMSA analysis of 32P-labeled nspA DNA after incubation with meningococcal Fur and an excess of cold probe (unlabeled operator DNA fragments). Lane 1, no Fur and no cold probe; lane 2, 800 nM Fur and no cold probe; lanes 3 to 7 800 nM Fur and 50-fold, 100-fold, 200-fold, 300-fold, and 400-fold cold probe, respectively.
Our analysis of the ability of meningococcal Fur to bind the promoter region encompassing the in silico-predicted Fur box of the nspA gene revealed a shift in the mobility of nspA probe with increasing concentrations of Fur (Fig. 4B). Although the amount of the shifted nspA probe increased with concentrations of Fur, a considerable amount of free probe was observed even at the highest concentration of Fur protein (800 nM) used. This could be attributed to the presence of primer dimer contamination from PCR amplification in the nspA probe. The specificity of N. meningitidis Fur binding to the iron-activated and Fur-regulated nspA gene was confirmed by cold competition experiments using an excess (25- to 400-fold) of unlabeled probe (containing the same sequence as the labeled probe). As shown in Fig. 4C, a gradual loss of each of the complexes was observed with a corresponding increase of unlabeled probes. As expected, no shift was observed when the rmp promoter probe was used, even at the highest concentration of Fur (800 nM) used (Fig. 4D). Taken together, our in vitro binding studies conclusively demonstrate the ability of N. meningitidis Fur to interact with nspA but not the operator region of the secY gene.
Mapping of Fur binding sequences in the promoter/operator of the iron-activated nspA gene.
Having demonstrated the ability of N. meningitidis Fur to interact with the 101-bp nspA promoter region by EMSA analysis, we next sought to map the promoter region of nspA by DNase I protection studies, as described previously (15). Briefly, DNA fragments encompassing the putative Fur box of the nspA gene were PCR amplified from N. meningitidis by colony PCR using specific primers (Table 1) and cloned into pBCSK (Stratagene, La Jolla, CA) to generate the recombinant plasmid pBCSK.nspA. Labeling of the DNA probes for footprinting was accomplished by using M13 forward (5′ end-labeled) and reverse (unlabeled) primers in a PCR and purified on a G-25 spin column (Amersham Biosciences, Piscataway, NJ). Approximately 40,000 cpm of labeled probe was used in each reaction. Protein-DNA complex was formed in 100 μl of footprinting buffer containing 10 mM Tris-HCl (pH 8.0), 100 mM KCl, 2 mM MgCl2, 1 mM CaCl2, 100 μM MnCl2, 1 mM DTT, 10% glycerol, 1 μg of sonicated salmon sperm DNA, and 5 μg of acetylated BSA and was incubated for 30 min at room temperature. DNase I (0.1 ng) treatment was carried out for 2 min at 37°C. The reaction was stopped by the addition of 100 μl of stop buffer (0.1 M EDTA, pH 8.0-0.6 M Na acetate-20 μg/ml sonicated salmon sperm DNA). DNA samples were then extracted with phenol-chloroform, ethanol precipitated, and resuspended in 4 μl of sequencing buffer. After denaturation at 95°C for 2 min, samples were subjected to electrophoresis on 6% urea-polyacrylamide gels at 2,000 V, dried, and autoradiographed.
As shown in Fig. 5, in the absence of any added Fur protein, digestion of labeled fragments produced a relatively uniform distribution of DNA fragments (lane 1). A 32-bp region of protection was seen when the nspA promoter region was used with 150 and 460 nM concentrations of meningococcal Fur (lanes 2 and 3). When a higher concentration of Fur was used, an extended region of protection was observed (Fig. 5, lane 4). Further analysis of the Fur binding sequences in the operator regions of the N. meningitidis iron-repressed fur gene and iron-activated aniA (data not shown) and nspA genes by DNase I protection identified a hexameric 5′-(G/T)ATAAT-3′ unit, which was absent in the predicted Fur binding sequence of the secY/ribosomal operon (Fig. 5B). These results are in agreement with previous studies in which we used in silico analysis to define the N. meningitidis Fur box consensus sequence. This highly conserved NATWAT motif (17) was also identified in the operator regions of both iron-activated and iron-repressed Fur-dependent genes, as defined by DNase I protection analysis (Fig. 5B).
Interestingly, our DNase I footprinting analysis indicated that the nspA Fur binding sequence overlapped the −10 region of the N. meningitidis iron-activated nspA gene according to mapping of an nspA gene homolog in N. gonorrhoeae (27). This phenomenon is commonly described for genes that are repressed by this global regulator. Further studies are warranted to fine tune the molecular mechanism of nspA gene activation by Fur.
Conclusions.
In the present study, we examined two N. meningitidis iron-activated genes identified by microarray analysis to definitively define the role of Fur in the expression of these genes. While our previous studies determined by EMSA analysis that meningococcal Fur was able to bind to the putative Fur binding sequences in the upstream regions of the iron-activated secY gene (17), in the present study we demonstrated that genes within the secY operon are cotranscribed and that the previously identified putative Fur box is actually within an ORF. We found that Fur did not bind to the promoter region of the N. meningitidis secY/ribosomal operon, nor was Fur required for expression of the N. meningitidis secY gene. In E. coli, the preprotein translocase SecY functions together with SecE and SecG in protein secretion across the cytoplasmic membrane via the SecYEG pathway (4, 14). Similar to what we observed for the meningococcal secY gene, we also found that the transcription of the meningococcal secE and secG genes was increased during growth under iron-replete conditions and was also Fur independent (data not shown), suggesting that expression of entire SecYEG complex is controlled by iron and yet is Fur independent. Although these genes were not identified in a very recently published study on the N. meningitidis fur and iron regulon (5), this could be attributed to variations in experimental design, i.e., the media used and time points chosen for analysis of gene expression. Our results, however, are supported by recent studies of N. gonorrhoeae wherein expression of the protein secretion secY and secE genes and several ribosomal genes were reported to be increased during growth under iron-replete conditions (13), although the role of Fur in the regulation of these genes was not examined.
An additional point regarding in silico predictions and EMSA analysis for the identification of putative Fur binding regions and correlation with biological significance deserves attention. Although these analyses serve as an excellent tool for defining the interaction between protein and DNA, further experiments are warranted to conclusively demonstrate the in vivo relevance of such interactions. In this study, using an N. meningitidis fur mutant, we confirmed the Fur-dependent expression of the N. meningitidis nspA gene, in contrast to the regulation of the secY gene, which was demonstrated to be Fur independent. Furthermore, DNase I footprinting analysis with nspA and several additional Fur-regulated genes confirmed that the hexameric 5′-NAT(A/T)AT-3′ unit is important for N. meningitidis Fur binding and potentially for transcriptional regulation of these genes (17). However, our results further suggest that the presence of a consensus Fur box sequence itself may not be sufficient in determining whether Fur binding results in repression versus activation of gene transcription.
Acknowledgments
We thank Vincenzo Scarlato for the pGemFkoB:Km and pSLFur-C1 plasmid constructs and Tim Yahr and Ann Flower for the SecY antibody.
This study was supported by a grant to C. A. Genco from NIH-NIAID (AI048611).
Footnotes
Published ahead of print on 3 November 2006.
REFERENCES
- 1.Cadieux, N., M. Plante, C. R. Rioux, J. Hamel, B. R. Brodeur, and D. Martin. 1999. Bactericidal and cross-protective activities of a monoclonal antibody directed against Neisseria meningitidis NspA outer membrane protein. Infect. Immun. 67:4955-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calderwood, S. B., and J. J. Mekalanos. 1988. Confirmation of the Fur operator site by insertion of a synthetic oligonucleotide into an operon fusion plasmid. J. Bacteriol. 170:1015-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerretti, D. P., D. Dean, G. R. Davis, D. M. Bedwell, and M. Nomura. 1983. The spc ribosomal protein operon of Escherichia coli: sequence and cotranscription of the ribosomal protein genes and a protein export gene. Nucleic Acids Res. 11:2599-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collinson, I. 2005. The structure of the bacterial protein translocation complex SecYEG. Biochem. Soc. Trans. 33:1225-1230. [DOI] [PubMed] [Google Scholar]
- 5.Delany, I., R. Grifantini, E. Bartolini, R. Rappuoli, and V. Scarlato. 2006. Effect of Neisseria meningitidis fur mutations on global control of gene transcription. J. Bacteriol. 188:2483-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delany, I., R. Ieva, C. Alaimo, R. Rappuoli, and V. Scarlato. 2003. The iron-responsive regulator fur is transcriptionally autoregulated and not essential in Neisseria meningitidis. J. Bacteriol. 185:6032-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delany, I., R. Rappuoli, and V. Scarlato. 2004. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol. Microbiol. 52:1081-1090. [DOI] [PubMed] [Google Scholar]
- 8.Delany, I., G. Spohn, R. Rappuoli, and V. Scarlato. 2001. The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol. Microbiol. 42:1297-1309. [DOI] [PubMed] [Google Scholar]
- 9.de Lorenzo, V., F. Giovannini, M. Herrero, and J. B. Neilands. 1988. Metal ion regulation of gene expression. Fur repressor-operator interaction at the promoter region of the aerobactin system of pColV-K30. J. Mol. Biol. 203:875-884. [DOI] [PubMed] [Google Scholar]
- 10.de Lorenzo, V., S. Wee, M. Herrero, and J. B. Neilands. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubrac, S., and D. Touati. 2000. Fur-positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J. Bacteriol. 182:3802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubrac, S., and D. Touati. 2002. Fur-mediated transcriptional and post-transcriptional regulation of FeSOD expression in Escherichia coli. Microbiology 148:147-156. [DOI] [PubMed] [Google Scholar]
- 13.Ducey, T. F., M. B. Carson, J. Orvis, A. P. Stintzi, and D. W. Dyer. 2005. Identification of the iron-responsive genes of Neisseria gonorrhoeae by microarray analysis in defined medium. J. Bacteriol. 187:4865-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duong, F., and W. Wickner. 1997. Distinct catalytic roles of the SecYE, SecG and SecDFC subunits of preprotein translocase holoenzyme. EMBO J. 16:2756-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1998. Binding of the fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 283:537-547. [DOI] [PubMed] [Google Scholar]
- 16.Genco, C. A., and P. J. Desai. 1996. Iron acquisition in the pathogenic Neisseria. Trends Microbiol. 4:179-184. [DOI] [PubMed] [Google Scholar]
- 17.Grifantini, R., S. Sebastian, E. Frigimelica, M. Draghi, E. Bartolini, A. Muzzi, R. Rappuoli, G. Grandi, and C. A. Genco. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. USA 100:9542-9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halter, R., J. Pohlner, and T. F. Meyer. 1984. IgA protease of Neisseria gonorrhoeae: isolation and characterization of the gene and its extracellular product. EMBO J. 3:1595-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henkin, T. M., S. H. Moon, L. C. Mattheakis, and M. Nomura. 1989. Cloning and analysis of the spc ribosomal protein operon of Bacillus subtilis: comparison with the spc operon of Escherichia coli. Nucleic Acids Res. 17:7469-7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joly, J. C., M. R. Leonard, and W. T. Wickner. 1994. Subunit dynamics in Escherichia coli preprotein translocase. Proc. Natl. Acad. Sci. USA 91:4703-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin, D., B. R. Brodeur, J. Hamel, F. Couture, U. de Alwis, Z. Lian, S. Martin, D. Andrews, and R. W. Ellis. 2000. Candidate Neisseria meningitidis NspA vaccine. J. Biotechnol. 83:27-31. [DOI] [PubMed] [Google Scholar]
- 22.Martin, D., N. Cadieux, J. Hamel, and B. R. Brodeur. 1997. Highly conserved Neisseria meningitidis surface protein confers protection against experimental infection. J Exp. Med. 185:1173-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massari, P., Y. Ho, and L. M. Wetzler. 2000. Neisseria meningitidis porin PorB interacts with mitochondria and protects cells from apoptosis. Proc. Natl. Acad. Sci. USA 97:9070-9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masse, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moe, G. R., P. Zuno-Mitchell, S. N. Hammond, and D. M. Granoff. 2002. Sequential immunization with vesicles prepared from heterologous Neisseria meningitidis strains elicits broadly protective serum antibodies to group B strains. Infect. Immun. 70:6021-6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norheim, G., E. Arne Hoiby, D. A. Caugant, E. Namork, T. Tangen, E. Fritzsonn, and E. Rosenqvist. 2004. Immunogenicity and bactericidal activity in mice of an outer membrane protein vesicle vaccine against Neisseria meningitidis serogroup A disease. Vaccine 22:2171-2180. [DOI] [PubMed] [Google Scholar]
- 27.Plante, M., N. Cadieux, C. R. Rioux, J. Hamel, B. R. Brodeur, and D. Martin. 1999. Antigenic and molecular conservation of the gonococcal NspA protein. Infect. Immun. 67:2855-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pohlner, J., R. Halter, and T. F. Meyer. 1987. Neisseria gonorrhoeae IgA protease. Secretion and implications for pathogenesis. Antonie Leeuwenhoek 53:479-484. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Sebastian, S., S. Agarwal, J. R. Murphy, and C. A. Genco. 2002. The gonococcal fur regulon: identification of additional genes involved in major catabolic, recombination, and secretory pathways. J. Bacteriol. 184:3965-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]