Abstract
Toxin-antitoxin (TA) loci, which were initially characterized as plasmid stabilization agents, have in recent years been detected on the chromosomes of numerous free-living bacteria. Vibrio cholerae, the causative agent of cholera, contains 13 putative TA loci, all of which are clustered within the superintegron on chromosome II. Here we report the characterization of the V. cholerae higBA locus, also known as VCA0391/2. Deletion of higA alone was not possible, consistent with predictions that it encodes an antitoxin, and biochemical analyses confirmed that HigA interacts with HigB. Transient exogenous expression of the toxin HigB dramatically slowed growth of V. cholerae and Escherichia coli and reduced the numbers of CFU by several orders of magnitude. HigB toxicity could be counteracted by simultaneous or delayed production of HigA, although HigA's effect diminished as the delay lengthened. Transcripts from endogenous higBA increased following treatment of V. cholerae with translational inhibitors, presumably due to reduced levels of HigA, which represses the higBA locus. However, no higBA-dependent cell death was observed in response to such stimuli. Thus, at least under the conditions tested, activation of endogenous HigB does not appear to be bactericidal.
Toxin-antitoxin (TA) loci, which were initially characterized as plasmid-borne mediators of plasmid stability (12), have in recent years been identified within the chromosomes of numerous bacterial species (27). These loci typically encode two proteins, including a toxin, which can inhibit cell growth and/or viability, and an antitoxin, encoded upstream of the toxin, which inhibits the activity of the toxin (reviewed in reference 13). Under favorable growth conditions, TA loci produce sufficient antitoxin to bind and inactivate the cognate toxin, and the toxin's effects are blocked. However, under unfavorable conditions, the balance between toxin and antitoxin is perturbed, and the toxin is able to act. Antitoxin insufficiency can develop following loss of a plasmid carrying a TA locus, as antitoxins are less stable than their corresponding toxins; this allows for selection against plasmid loss (38). An altered toxin/antitoxin ratio can also be induced by cell stressors, such as starvation, antibiotics, DNA damage, and oxidative stress, which can prevent antitoxin synthesis and/or activate antitoxin-degrading proteases (6, 8, 16, 23, 34). For chromosomal TA loci, antitoxin depletion results in both an immediate excess of toxin and increased transcription of the TA locus, as antitoxins generally serve as autorepressors of the TA operon.
Although most TA loci have similar genetic organizations and regulatory paradigms, the toxins differ in their modes of action. For example, the toxins CcdB and ParE inhibit DNA replication due to trapping and inactivation of DNA gyrase (9, 20, 26). In contrast, the toxins RelE and MazF inhibit translation, though not via identical means (5, 28, 39, 40), and indirect evidence suggests that Doc inhibits translation as well (17). The mechanisms for the toxins HigB and VapC have not been reported, although sequence and structural analyses may provide some clues (3, 27). Interestingly, however, such analyses of better-characterized toxins have yielded the unexpected findings that toxins with apparently unrelated activities can have similar sequences (e.g., ParE and RelE) (27) or similar structures (e.g., CcdB and MazF) (4).
The cellular role for chromosomally carried TA loci is a subject of controversy. It is clear that exogenous overexpression of toxins in the absence of antitoxins prevents bacterial growth (typically assayed as CFU). Furthermore, TA-dependent cell death has been observed in Escherichia coli following activation of endogenously encoded MazF and YoeB, encoded by two of the species' five known chromosomal TA loci (6, 23, 34). Engelberg-Kulka and colleagues have proposed that the MazF toxin induces programmed cell death, perhaps allowing a subset of cells to be sacrificed to benefit the bacterial population as a whole (1, 11, 22). However, this hypothesis has been contested by Gerdes and colleagues (6, 13, 29), who argued that several toxins, including MazF, have a bacteriostatic, rather than bactericidal, effect that facilitates bacterial adaptation to stress. They reported that transient activation of endogenous TA loci can slow cell metabolism (e.g., protein synthesis) but that cell growth is restored following removal of the toxin-inducing stimulus (5, 7). Analyses of the effects of simultaneous and staggered induction of exogenous TA genes have also been used to investigate this issue; however, the results of these experiments have varied, depending both upon the duration of toxin expression prior to antitoxin induction and upon the media in which the bacteria were cultured (2, 29), and consequently both groups have found support for their respective hypotheses.
Comparative genomic analyses revealed that obligate host-associated organisms are largely free of TA loci, while such loci are present in the vast majority of sequenced free-living bacterial species (27). Based in part on the potential role for TA loci as stress response elements, it was hypothesized that this disparity reflects the different likelihoods that organisms in these two groups will encounter and survive in fluctuating environments. The genome of the diarrheal pathogen Vibrio cholerae, which can thrive in aquatic environments with varying temperature and salinity and can also survive and multiply within the human digestive tract, was found to contain 13 putative TA loci (27, 32). Interestingly, all were located within the superintegron found on ChrII, the smaller of V. cholerae's two chromosomes. The 13 TA loci were associated with attC sites, suggesting that they once were, or may still be, mobile genetic cassettes. Here we characterize VCA0391/2, one of the putative TA loci, which is homologous to higBA of plasmid Rts1 and has also been designated higBAI (27).
The higBA locus was first identified in a temperature-sensitive plasmid (Rts1) of Proteus vulgaris (36); subsequently, 74 similar loci have been detected within the genomes of 31 distinct gram-positive and gram-negative bacterial species (27). To date, no chromosomal HigB and HigA orthologues have been characterized, and only limited information is available regarding the Rts1-encoded proteins. higBA loci differ from other characterized TA loci in that the toxin-encoding gene (higB) lies upstream of the antitoxin-encoding gene (higA); however, as with other TA loci, it appears that the antitoxin represses transcription of the operon. In Rts1, a weak antitoxin-specific promoter not subject to repression by HigA has also been detected (37). There is weak sequence similarity between higB, relE, and parE (3), which provides conflicting clues about the cellular role of HigB. The Rts1-carried locus appears to be responsible for the plasmid's inhibition of host growth at restrictive temperatures (36), but the cellular target(s) for HigB is unknown.
We have analyzed the cellular requirement for higB and higA in V. cholerae, their expression and regulation, and their effects upon the survival of V. cholerae and E. coli. Deletion of higB or higBA did not alter V. cholerae growth, but deletion of higA alone was not possible. Overexpression of HigB inhibited growth of both V. cholerae and E. coli by several orders of magnitude, consistent with its presumed role as a toxin. Coexpression of HigA with HigB, either simultaneously or following initial HigB accumulation, mitigated the effect of HigB; biochemical analyses revealed that HigA interacts with HigB, presumably thereby blocking HigB's toxicity. Conditions that activate endogenous HigB were identified; however, such conditions were not found to cause toxin-dependent cell death.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All V. cholerae strains used here were streptomycin-resistant derivatives of the sequenced clinical isolate N16961 (18). The E. coli strains BI533 (MG1655 Nalr) (19) and BW27784 (22) were used for E. coli toxicity and rescue studies. Cells were cultured in LB broth at 37°C unless otherwise noted. M63 minimal medium (24) was supplemented with 0.2% glucose and 0.1% Casamino Acids. Antibiotics were used at the following concentrations unless otherwise noted: streptomycin, 200 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 20 μg/ml (E. coli) and 5 μg/ml (V. cholerae); and nalidixic acid, 40 μg/ml. Additional antibiotics tested for the ability to induce the higBA locus (noted in text) were all used at concentrations that typically prevent growth of V. cholerae, although in a few cases (e.g., 50 to 100 μg/ml spectinomycin) growth inhibition was not detected during the relatively short assay period. Arabinose (0.02%) and glucose (0.2%) were added to induce and repress PBAD, respectively, and IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM) was used to induce Ptac. Mitomycin C was used at 20 ng/ml.
Plasmid and strain construction.
Constructs for deletion of higB (VCA0391) and higA (VCA0392) were generated using overlap extension PCR. PCR products were cloned into pCVD442 and introduced into the chromosome using standard allelic exchange protocols (10). Primers used for the higB deletion construct were as follows: HigB-A, AGAAAGTGTGGAATCTTCAGC; HigB-B, AAATTCTAATGCCATTGCAACTTGCACTGTCTC; HigB-C, GCATTAGAATTTACTTACTTAGACCCACATAAG; and HigB-D, GGACAATTAAACCGATTTTTG. Primers used for the higA deletion construct were as follows: HigA-A, AGAAAGTGTGGAATCTTCAGC; HigA-B, ATAGTCGTTACCTCAATACTTATGTGGGTCTAA; HigA-C, GGTAACGACTATCAAACGCTTCAAGAGGGACAG; and HigA-D, TTGTTTGAAAGGCATTTTACG.
pBADhigB, which contains the higB open reading frame defined here (see Fig. 2) expressed under the control of the arabinose-inducible promoter PBAD, consists of a PCR product generated with the primers HigBx3 (CGAGCTCGGATGCACAATGAGACAGT) and HigBY (GCTCTAGAAGTCGTTACCTCAATACTTATG), digested with SacI and XbaI, and cloned into pBAD18Kn (15).
FIG. 2.
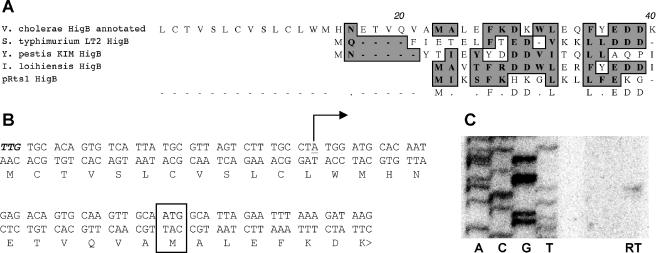
Determination of transcriptional and translational start sites for V. cholerae higB. (A) Alignment of the 5′ ends of annotated V. cholerae HigB and characterized or putative HigBs from Salmonella enterica serovar Typhimurium, Yersinia pestis, Idiomarina loihiensis, and Rts1. Conserved amino acids are shaded. (B) Annotated and experimentally determined start sites for V. cholerae higB and HigB. The annotated translational start is shown in bold italics, the experimentally determined translational start site is boxed, and the experimentally determined transcriptional start site is underlined and marked with a bent arrow. (C) Primer extension analysis of the 5′ end of V. cholerae higB transcripts. The sequence corresponds to the bottom strand shown in panel B. RT, reverse transcription reaction performed with a higB-specific primer. The same start site was also identified using 5′ rapid amplification of cDNA ends (data not shown).
pGZhigA, which contains higA expressed under the control of the IPTG-inducible promoter Ptac, consists of a PCR product generated with HigAX (CGAGCTCCTTACTTAGACCCACATAAGTATTG) and HigAY (GCTCTAGAAAAAACCAACGCATGGTAAA), digested with SacI and XbaI, and cloned into pGZ119EH (25).
To generate pBADHigBHis, the TIGR-annotated higB gene was amplified using HigB-N (TTGTGCACAGTGTCATTATG) and HigB-C2 (ATACTTATGTGGGTCTAAGTAAG) and cloned into pBADTOPO (Invitrogen). The higB-his fragment was then amplified with HigBx3 and 3-His (GCTCTAGATCAATGGTGATGGTGATG), digested with SacI and XbaI, and cloned into pBAD18Kan.
pQE30higBAmyc, which allows for IPTG-induced production of His6HigB and HigA-Myc, consists of higBA amplified with 5B-PQE (CGAGCTCATGGCATTAGAATTTAAAGAT) and 3A-Myc (GCTCTAGATCACAGATCTTCTTCGCTAATCAGTTTCTGTTCTGTACGTGCGCTTTGTTCC), digested with SacI and XbaI, cloned into pBAD18Kan, released with SacI and PstI, and ligated into pQE30 (QIAGEN).
pGZhigAmyc, which encodes a HigA-Myc fusion protein under the control of Ptac, consists of higA amplified with HigAX and 3A-Myc, digested with SacI and XbaI, and ligated into pGZ119EH.
phigBlacZ contains sequences amplified with 5phigB (GAAGATCTGCGTAAAATTGCTTTCTCA) and 3phigB (CCCAAGCTTGTGCATCCATAGGCAAA), digested with BglII and HindIII, and ligated to pCB182N (provided by H. H. Kimsey) to generate a transcriptional reporter fusion for the higB promoter.
phigAlacZ contains sequences amplified with pHigAf (CGGGATCCTGGTTAGAGCAGTTTTACGAGGATG) and phigAr (GCTCTAGATGATGTGATGCCCATTGGTTC), digested with BamHI and XbaI, and ligated to pCB182 (35) to generate a transcriptional reporter fusion for the higA promoter.
All sequences were amplified from N16961 DNA. Plasmid construction was confirmed by DNA sequencing. pCVD442 derivatives were introduced into V. cholerae by conjugation; all other plasmids were introduced into V. cholerae and E. coli by electroporation.
RNA analyses.
RNAs were extracted from bacterial cell pellets using Trizol (Invitrogen) and then treated with DNase I (QIAGEN). For Northern blots, RNAs were run in glyoxal gels (Ambion) and then transferred to Bright Star Plus nylon membranes (Ambion). Blots were hybridized to in vitro-transcribed probes (Strip-EZ RNA; Ambion) in Ultrahyb (Ambion) at 68°C. Templates for probe transcription were linearized derivatives of pCRII-TOPO (Invitrogen) containing higA (nucleotides [nt] 9 to 251 relative to the translational start) or higB (nt −29 to 254). Primer extension analyses were performed on RNAs from wild-type (wt) V. cholerae treated with 10 μg/ml chloramphenicol for 20 min to induce expression of higBA. RNAs were reverse transcribed from the primer Gsp3 (CACCAACCTTTAAGATTTCCTTCA), using MonsterScript (Epicenter Biotechnologies), at 60°C. The sequencing ladder was generated from a Gsp3-5pHigB DNA template, using Gsp3.
Protein purification and analysis.
Log-phase cultures of E. coli XL1-Blue(phigBhis) were grown with 0.2% arabinose for 3 h to induce expression of His6HigB. The tagged protein was affinity purified under native conditions, using Ni-nitrilotriacetic acid (NTA) (QIAGEN) according to the manufacturer's instructions. Purified protein was run in a denaturing polyacrylamide gel, transferred to a polyvinylidene difluoride membrane, and stained with Coomassie blue. The membrane with purified protein was submitted for N-terminal sequencing (Tufts University Core Facility). Similarly, log-phase cultures of XL1-Blue(pQE30higBAmyc) were grown for 3 h with 0.2 mM IPTG to induce expression of His6HigB and HigA-Myc. Proteins were affinity purified as described above and then eluted from the resin in the presence of 250 mM imidazole. Purified proteins were run in denaturing polyacrylamide gels and either stained with colloidal Coomassie blue or transferred to nitrocellulose membranes for Western blotting. Blots were probed with a penta-His antibody (QIAGEN) or anti-Myc antibody (Invitrogen). Detection of HigA-Myc in control purifications and total cell lysates from BW27784(pBADhigB/pGZHigAmyc) was performed in the same manner.
Viability assays.
wt and higBA mutant V. cholerae strains were cultured in LB until an optical density at 600 nm (OD600) of ∼0.7, and then the cultures were divided and either treated with 1 to 10 μg/ml chloramphenicol or 50 μg/ml kanamycin or left untreated. Subsequently, cultures were plated at various time points on LB agar lacking either antibiotic to enumerate CFU. All antibiotic concentrations tested slowed or halted culture growth (measured as OD600).
HigB toxicity assays.
V. cholerae N16961 and N16961 higBA and E. coli BI533 were each transformed with pBAD18Kan and pBADhigB. Transformed strains were grown to exponential phase in LB with kanamycin, 0.2% glucose, and streptomycin (V. cholerae) or nalidixic acid (E. coli). Cultures were then spun down, washed twice with LB, resuspended to an OD600 of ∼0.3 in LB with antibiotics and either 0.02% glucose or 0.02% arabinose, and returned to 37°C. Aliquots were removed every 30 min for assessment of OD600 and for dilution onto selective plates containing 0.2% glucose to enumerate CFU.
HigA activity assays.
The effect of simultaneous production of HigB and HigA was assessed using N16961 higBA(pBADhigB/pGZHigA). The effect of delayed HigA production was assessed using N16961 higBA(pBADhigB/pGZHigAmyc) and E. coli BW27784(pBADhigB/pGZHigAmyc). Strains were grown in LB with antibiotics and 0.2% glucose to exponential phase, spun down, rinsed twice with LB, resuspended to an OD600 of ∼0.3 in LB with antibiotics and either 0.02% glucose or 0.02% arabinose, and returned to 37°C. At time zero or later time points, 1 mM IPTG was added to some cultures to induce expression of HigA. For the assay of simultaneous HigB and HigA induction, aliquots were removed every 30 min for dilution onto selective plates containing 0.2% glucose to enumerate CFU. For the assay of delayed HigA induction, cells were cultured for 30 min in the presence of IPTG prior to being plated on selective plates containing 0.2% glucose.
RESULTS
Generation of V. cholerae lacking higB and higBA.
To study the role of and requirement for the higBA locus in V. cholerae (VCA0391/2) (Fig. 1A), we attempted to generate derivatives of a sequenced clinical isolate, N16961 (18), lacking one or both genes. Mutants containing an in-frame deletion in higB were readily isolated; however, we could not generate an in-frame deletion within higA in wt V. cholerae, even if higA was provided in trans on a plasmid. In contrast, higA deletion mutants could be derived from the N16961 higB mutant. These data are consistent with the hypothesis that HigB is a toxin whose activity can be blocked by an antitoxin, HigA. The growth rates (OD600) and viability (CFU) of wt and higB and higBA mutant V. cholerae strains did not differ notably in either LB or M63 minimal medium (data not shown).
FIG. 1.
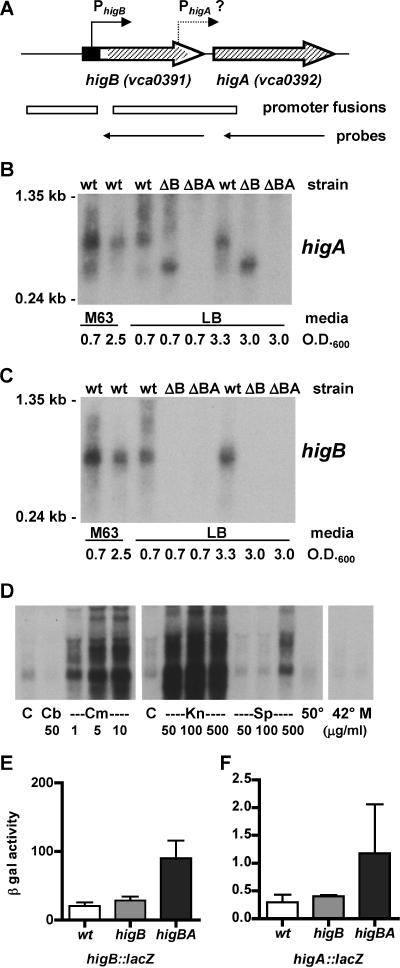
Expression and regulation of V. cholerae higBA. (A) Schematic representation of the higBA region of V. cholerae chromosome II, drawn approximately to scale. Thick arrows show genes, as annotated by TIGR, pointed in the direction of transcription. Bent arrows show the locations of the experimentally determined higBA promoter and the predicted higA promoter. The filled rectangle corresponds to the first 22 aa codons of annotated higB, which are not believed to be translated (see Fig. 2). Hatched areas denote regions deleted from higB and higA. Open rectangles show regions cloned to generate transcriptional reporter fusions in phigBlacZ and phigAlacZ. Thin arrows show regions transcribed in vitro to generate antisense riboprobes. (B) Northern blot of RNAs from wt, higB, and higBA strains of V. cholerae grown in either M63 or LB medium to various densities. The blot was probed with a riboprobe complementary to higA transcripts. (C) Northern blot as described for panel B, but probed with a riboprobe complementary to higB transcripts. (D) Northern blot of RNAs from wt V. cholerae grown in LB medium and treated with a variety of stresses. Cultures were grown at 37°C to an OD600 of ∼0.7 (left and right panels) or 0.5 (middle panel) and then either left at 37°C for an additional 20 min (control [C]) or incubated with carbenicillin (50 μg/ml [Cb]) or chloramphenicol (1 to 10 μg/ml [Cm]) for 20 min, with kanamycin (50 to 500 μg/ml [Kn]) or spectinomycin (50 to 100 μg/ml [Sp]) for 1 h, or with spectinomycin (500 μg/ml) or mitomycin C (20 ng/ml [M]) for 20 min. Heat-treated cells were shifted either to 42°C for 20 min or to 50°C for 10 min. The blot was probed with a riboprobe complementary to higB transcripts. (E) β-Galactosidase activities (Miller units) from a higB::lacZ transcriptional reporter fusion (phigBlacZ) in wt, higB, and higBA strains of V. cholerae. Data are averages for at least three experiments. (F) β-Galactosidase activities (Miller units) from a higA::lacZ transcriptional reporter fusion (phigAlacZ) in wt, ΔhigB, and ΔhigBA strains of V. cholerae. Data are averages for at least three experiments.
Expression and regulation of higBA.
Northern blotting was used to assess the expression of higBA in the three strains under a variety of conditions. Low-abundance transcripts of higB and higA were detected in RNAs of wt V. cholerae grown in LB and minimal medium to both log and stationary phase, although expression was somewhat higher in log-phase cultures grown in minimal medium (Fig. 1B and C). Under all conditions tested, the RNA from the higB mutant that hybridized to the higA probe was shorter than that from wt V. cholerae, suggesting that HigB and HigA are produced from a single transcript. A short higA-hybridizing species, which was much less abundant than the predominant species, was also detected in wt V. cholerae. This transcript is probably processed from the longer transcript, as promoter analyses (see below) suggest that a higA-specific promoter has, at best, very weak activity under the conditions assayed.
We also monitored higBA transcript abundance following treatment of cultures with antibiotics, with mitomycin C, and with elevated temperatures, as previous studies have suggested that such stresses can induce expression of TA loci (8, 16, 23). No increase in transcript abundance was detected 20 min after the addition of the ampicillin analog carbenicillin (50 μg/ml) or mitomycin C (20 ng/ml) to cultures or following incubation of cultures at 42°C for 20 min or 55°C for 10 min (Fig. 1D). Exposure of cells to carbenicillin (50 to 250 μg/ml) for 1 h or to rifampin (50 μg/ml) for 20 min also did not increase higB transcript abundance, despite the marked effect of these agents on culture density (data not shown). However, addition of the protein synthesis inhibitors chloramphenicol (1 to 10 μg/ml) and kanamycin (50 to 500 μg/ml) to cultures caused a dramatic increase in higBA transcripts in as little as 20 min (Fig. 1D). The protein synthesis inhibitor spectinomycin also increased higBA transcript abundance, but to a lesser extent, perhaps related to its smaller impact on cell replication. The results obtained with protein synthesis inhibitors are consistent with those from previous studies of TA loci, as reduced production of the presumably unstable antitoxin HigA should release the higBA locus from HigA-mediated autorepression. Surprisingly, however, transcripts larger than those typically observed for the higBA locus were also detected following higBA-inducing treatments. The biological significance of the latter observation is unknown.
To confirm that the V. cholerae higBA locus is autoregulated, we assessed the activity of a higB::lacZ transcriptional reporter fusion (Fig. 1A) in wt and higB and higBA mutant V. cholerae strains (Fig. 1E). No difference was observed between the activities of the reporter in wt and higB mutant V. cholerae, suggesting that, in contrast to regulation of other TA loci (13), HigB does not play a significant role in downregulating its own expression (e.g., as a corepressor). This conclusion is consistent with the similar abundances of higA transcripts detected in wt and higB mutant V. cholerae by Northern blotting (Fig. 1B). However, the activity of the higB reporter was ∼3.5 times greater in the higBA background than in the wt and higB background. Thus, it appears that HigA either directly or indirectly represses expression of the higBA locus, as observed for other antitoxins.
Analysis of the Rts1 higBA locus revealed that it contains a higA-specific promoter (PhigA) as well as the operon promoter (PhigB) (37). Bioinformatic analysis of V. cholerae higBA (BPROM; http://www.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb) suggested that this might also be the case in V. cholerae (data not shown). However, very little β-galactosidase activity was detectable from a higA::lacZ transcriptional reporter fusion containing the potential promoter (Fig. 1A), although this activity was higher than that from a vector control (0.3 to 1.2 Miller units versus <0.1 units) (Fig. 1F and data not shown). These data indicate that if a higA-specific promoter is present, it is active either at very low levels or under different growth conditions from those that activate the higB promoter.
Cloning and 5′-end mapping of V. cholerae higB.
To perform more comprehensive analyses of the role of HigB in the absence of HigA, we attempted to create an inducible higB expression vector, using the arabinose-inducible promoter PBAD in pBAD18Kn (15). Even though glucose, the repressor of PBAD, was used in our attempts to insert higB downstream of pBAD, we were at first unable to obtain clones containing the wt higB sequence. Subsequent sequence analyses comparing the annotated V. cholerae HigB protein to predicted HigB proteins from other organisms (27) revealed minimal homology upstream of amino acid (aa) 23 of the predicted V. cholerae HigB protein, suggesting that higB might be misannotated in the V. cholerae genome and that our cloned fragments could include the higB promoter, PhigB. Primer extension and 5′ rapid amplification of cDNA ends confirmed this theory and indicated that the true transcriptional start site for V. cholerae higB lies 36 nt downstream of TIGR's annotated translational start site (Fig. 2B and C and data not shown). Based on these findings, we hypothesized that the true HigB translational start site is situated at amino acid 23 relative to the annotated start site. Subsequent attempts to generate a new pBADhigB construct lacking the endogenous PhigB promoter were successful. In addition, we created a variant of this construct (pBADHigBHis) predicted to encode HigB with a His6 tag fused to its carboxyl terminus. Affinity purification and amino acid sequencing of this protein (which was found to be nontoxic) confirmed that its amino terminus had the predicted ALEFK sequence; thus, the presumed translational start site (Fig. 2B) is associated with a functional ribosome binding site.
HigB is a potent toxin that prevents growth of both V. cholerae and E. coli.
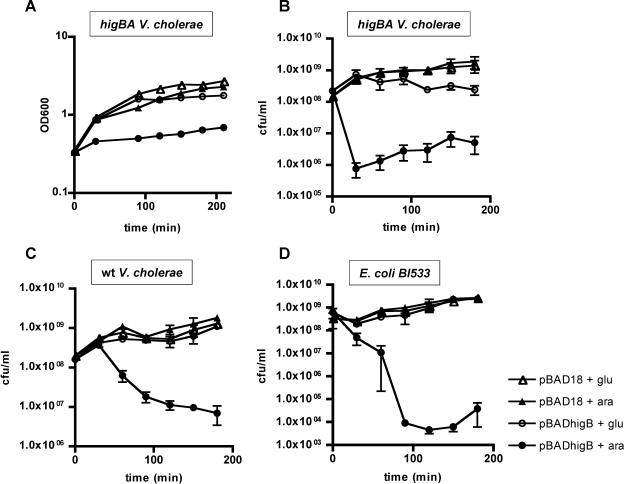
We introduced pBADhigB into wt and higBA mutant V. cholerae strains and into E. coli BI533 and measured if induction of higB expression influenced cell growth and/or survival. As measured by OD600, induction of HigB synthesis in the N16961 higBA strain dramatically slowed the rate of culture growth but did not reduce the culture density below its starting point, perhaps suggesting that HigB did not induce cell lysis (Fig. 3A). However, when we assessed the viability of cells in induced cultures (via quantitation of CFU), we found that production of HigB for as little as 30 min reduced the number of CFU by several orders of magnitude (Fig. 3B). This marked reduction occurred despite the presence of glucose, the repressor of pBADhigB, within the assay plates. Thus, a short period of HigB production in the absence of HigA can dramatically interfere with V. cholerae's ability to replicate. HigB induction also reduced the plating efficiency of N16961(pBADhigB) (Fig. 3C), although the reduction was less rapid, perhaps because of the presence of endogenous HigA. HigB had an even more dramatic effect in E. coli BI533 (Fig. 3D), indicating that HigB's effects are not dependent upon the presence of additional vibrio-specific factors. For all three strains tested, the number of CFU was not reduced to zero; a subpopulation of cells continued to replicate, which is consistent with the slight increase in culture density (Fig. 3A).
FIG. 3.
Effects of HigB production on culture density and CFU of V. cholerae and E. coli. Strains contained either pBAD18 (triangles) or pBADhigB (circles). All cultures were grown in LB medium plus 0.2% glucose, washed, and resuspended in either LB plus 0.02% glucose (open symbols) or LB plus 0.02% arabinose (filled symbols) at time zero. Culture densities and/or CFU were enumerated at subsequent time points. (A) OD600 for V. cholerae higBA strain. (B) CFU of V. cholerae higBA strain. (C) CFU of wt V. cholerae. (D) CFU of E. coli BI533.
HigA prevents the toxicity of HigB.
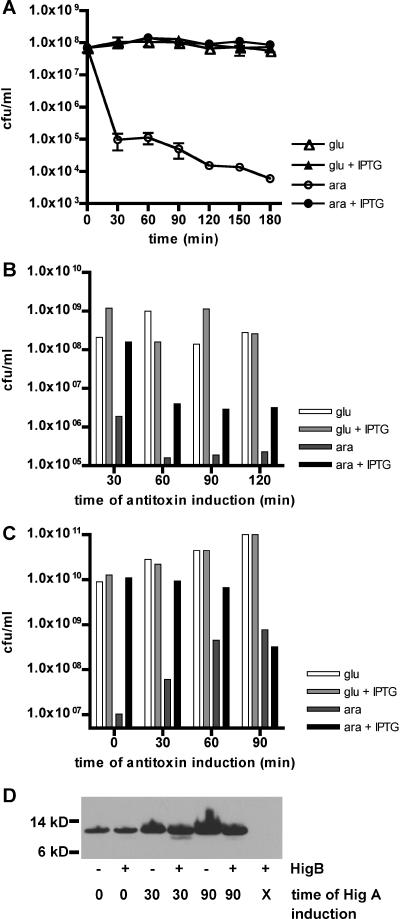
To assess directly whether HigA could block the toxicity of HigB, we cotransformed the N16961 higBA strain with pBADhigB and pGZhigA, a plasmid containing higA under the control of the IPTG-inducible promoter Ptac. CFU were then quantitated for cultures grown in glucose, with or without IPTG, and in arabinose, with or without IPTG. Production of HigA had no effect on the viability of cells grown in glucose (higB repressed); however, it increased the viability of cells cultured with arabinose (higB induced) by several orders of magnitude, restoring CFU to the level observed for cells with repressed higB (Fig. 4A). Thus, V. cholerae HigA appears to function as an antitoxin that can prevent the growth-inhibitory effect of HigB.
FIG. 4.
Effects of HigA production coincident with or subsequent to induction of HigB in V. cholerae and E. coli. All cultures were grown in LB medium plus 0.2% glucose, washed, and resuspended in either LB plus 0.2% glucose or LB plus 0.02% arabinose at time zero. At either time zero (A) or subsequent time points (B, C, and D), IPTG was added to some cultures to induce production of HigA (A) or HigA-Myc (B, C, and D). After further incubation, cells were plated to enumerate CFU (A, B, and C) or harvested for protein isolation and Western blotting. Results of representative experiments are shown in each panel. (A) CFU of N16961 ΔhigBA(pBADhigB/pGZHigA). (B) CFU of N16961 ΔhigBA(pBADhigB/pGZHigAmyc). (C) CFU of E. coli BW27784(pBADhigB/pGZHigAmyc). (D) Western blot of E. coli BW27784(pBADhigB/pGZHigAmyc) probed with anti-Myc antibody. Lane X, no addition of IPTG to induce HigA-Myc production.
Studies of additional TA loci have suggested that the growth-inhibitory effect of some toxins (e.g., RelE) can be bacteriostatic rather than bactericidal (29). For such toxins, it is possible to reverse the toxin's effects by subsequent (rather than simultaneous) production of antitoxin. We performed analogous “rescue” experiments with both the higBA mutant V. cholerae strain and E. coli BW27784 (22). Both strains were cotransformed with pBADhigB and pGZhigAmyc. The latter plasmid, which was shown to have antitoxin activity in experiments like those shown in Fig. 4A (data not shown), encodes an epitope-tagged version of HigA, thus permitting biochemical as well as functional detection of HigA.
For the rescue experiments, cells were initially cultured either in glucose (no HigB) or in arabinose (with HigB) for various amounts of time, followed by an additional 30 min of growth in the presence or absence of IPTG (i.e., with or without HigA). CFU were then enumerated on plates containing glucose and lacking IPTG. In the V. cholerae higBA background, induction of higB for 30 min followed by induction of higA resulted in little or no reduction of CFU relative to that for cultures in which toxin was not induced (Fig. 4B, 30 min). In contrast, a 30-min induction of toxin with no subsequent antitoxin induction resulted in a >100-fold drop in CFU (Fig. 3B). Longer delays (60 to 120 min) prior to higA induction resulted in incomplete rescue of culture viability; still, even when higA was induced 120 min after induction of higB, this increased the number of CFU ∼10-fold relative to that for a parallel culture in which higB was induced but higA was not (Fig. 4B, arabinose with or without IPTG, 120 min). Similar results were obtained from experiments performed with E. coli, although both toxicity and rescue were somewhat less effective. Complete blockage of HigB toxicity in E. coli was obtained only with simultaneous induction of HigB and HigA production, and no rescue was observed if HigA synthesis was induced 90 min after that of HigB, but some rescue was observed at intermediate time points (Fig. 4C). Overall, it appears that the effects of short-term (∼30 min) production of HigB can effectively be countered by HigA induction but that those from longer (>60 min) HigB exposure are generally, though not entirely, irreversible.
Since several toxins have been shown to either directly or indirectly interfere with protein synthesis (5, 7), we explored the possibility that HigA's apparent inability to completely prevent HigB-dependent toxicity after delayed induction might actually reflect a failure in HigA synthesis after substantial exposure to toxin. To do this, we compared the levels of HigA-Myc generated in the presence versus the absence of toxin. Cells were harvested for Western analysis 60 min after induction of HigA-Myc production. Analyses were initially attempted using cell lysates from N16961 higBA(pBADhigB/pGZhigAmyc); surprisingly, we were never able to detect HigA-Myc in these lysates, despite clear functional evidence that HigA is produced (Fig. 4B). We hypothesize that protein processing removes or modifies the Myc epitope tag in V. cholerae cultures. However, HigA-Myc was readily detected in E. coli transformed with the above plasmids, both in the presence and in the absence of HigB (Fig. 4D). Antitoxin was apparent even when its synthesis was induced 90 min after induction of HigB, at which point no amelioration of toxicity was observed; antitoxin levels per ml of culture were slightly reduced at this time point relative to levels in cultures lacking toxin. Thus, the absence of rescue when antitoxin induction lags sufficiently behind toxin induction is not simply due to a failure of antitoxin production. However, it is possible that the quantity of HigA produced following late induction is insufficient to inactivate the HigB already accumulated. Alternatively, it may be that after 90 min of HigB production, downstream pathways have been altered to such an extent that even complete inactivation of HigB by HigA cannot restore normal cellular functions.
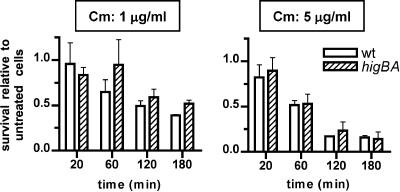
We also assessed whether induction of the endogenous higBA locus is linked to the death of V. cholerae cells. Both wt and higBA cells were treated with agents shown to induce higBA transcript accumulation (Fig. 1D), which presumably reflects inactivation of the repressor/antitoxin HigA and a corresponding activation of HigB. Exposure of these strains to chloramphenicol (1 to 10 μg/ml) or kanamycin (50 μg/ml) slowed or halted growth of both strains; however, when cells were enumerated (CFU) following up to 3 h of antibiotic treatment, no significant differences were detected between survival rates for wt and higBA mutant V. cholerae (Fig. 5 and data not shown). Thus, it does not appear that transient activation of endogenous HigB promotes programmed cell death, although it is possible that a HigB-dependent loss of viability might be detected following lengthier periods of antibiotic treatment. For these experiments, it is not possible to assess whether toxin activation inhibits cell growth, as the antibiotics used to induce HigB activity independently have such an effect.
FIG. 5.
Survival of wt and higBA mutant V. cholerae cells following exposure to chloramphenicol. Cultures of wt (open bars) and higBA mutant (hatched bars) V. cholerae were grown to an OD600 of ∼0.7, split, and either treated with 1 or 5 μg/ml chloramphenicol (Cm) or left untreated. After various times, cultures were plated on LB agar without chloramphenicol to enumerate CFU. Survival represents the number of CFU in treated cultures relative to the number of cells in untreated cultures at the same time point. Each bar is the average value for two to four experiments.
HigB and HigA interact in vivo.
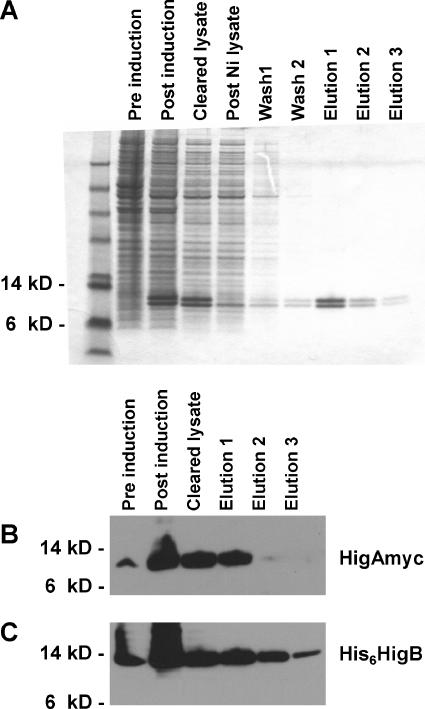
It is generally believed that antitoxins block the activities of their cognate toxins by binding to them and thereby inhibiting their interactions with other cellular factors. To determine whether V. cholerae HigB and HigA interact, we coexpressed epitope-tagged versions of each protein and assessed whether they could be copurified. Production of the tagged proteins was controlled by a single promoter in the expression construct pQE30HigBAmyc, allowing IPTG-inducible expression of balanced amounts of toxin and antitoxin. His6HigB was affinity purified under native conditions, using Ni-NTA resin; Coomassie blue staining of the protein eluted from the resin revealed a closely migrating doublet of between 10 and 15 kDa, consistent with the sizes of His6HigB (110 aa) and HigA-Myc (115 aa) (Fig. 6A). Western blotting of purified samples confirmed that both His6HigB and HigA-Myc had been purified (Fig. 6B and C). In contrast, no HigA-Myc was detected in samples purified in parallel from E. coli expressing HigB and HigA from pBADHigB (untagged protein) and pGZHigAmyc (data not shown), suggesting that HigA-Myc does not associate nonspecifically with the Ni-NTA resin but was instead purified due to its interaction with HigB. Thus, it appears likely that HigA's inhibition of HigB toxicity is, as with other antitoxins, dependent upon direct interaction between the two proteins.
FIG. 6.
Affinity purification of His6HigB and associated HigA-Myc. His6HigB and HigA-Myc were coexpressed from pQE30HigBAmyc in E. coli XL1Blue, and His6HigB was affinity purified from cell extracts, using Ni-NTA resin under nondenaturing conditions. Cell extracts and subsequent fractions from the purification were run in denaturing polyacrylamide gels and then analyzed using either Coomassie blue staining or Western blotting. (A) Proteins from various purification stages stained with colloidal Coomassie blue. (B) Western blot of pre- and postpurification fractions probed with anti-Myc antibody. (C) Western blot of pre- and postpurification fractions probed with anti-His6 antibody.
DISCUSSION
We have characterized the V. cholerae higBA locus, the first chromosomally carried locus of this class to be studied. We found notable similarities between V. cholerae higBA, Rts1 higBA, and several characterized TA loci carried on the E. coli chromosome. Overexpression of the toxin (HigB) blocks increases in culture density and rapidly reduces CFU in a culture by several orders of magnitude. This toxicity is preventable by simultaneous expression of the antitoxin (HigA). Toxicity can also be reduced by delayed induction of the antitoxin; HigA's effect is noticeable when it is induced up to 120 min after induction of HigB, although delays significantly reduce its efficacy (Fig. 4B). Endogenous higB and higA are not usually differentially expressed, as they are largely, if not entirely, controlled by a shared promoter. This promoter is repressed by HigA (Fig. 1E). Although toxins have been reported to act as corepressors for other TA loci, including Rts1 higBA, there is no indication that V. cholerae HigB functions as a corepressor; the activities of a higB::lacZ transcriptional reporter fusion did not differ in wt and higB V. cholerae strains. This result also suggests that stabilization of HigA by HigB is not a key factor in maintaining adequate antitoxin levels.
The expression and/or activity of some TA loci can be altered by numerous stimuli, typically stresses, such as amino acid and glucose starvation, exposure to various classes of antibiotics, oxidative stress, elevated temperature, and exposure to DNA-damaging agents (6, 8, 16, 23, 33, 34). This does not appear to be the case for the V. cholerae higBA genes, which instead appear to be regulated by a more limited set of stimuli. Carbenicillin, rifampin, mitomycin C (sufficient to activate RecA) (31), and growth at either 42 or 50°C had no effect on higBA transcript abundance, suggesting that these treatments do not alter the level of HigA or its ability to repress the higBA promoter. However, increased higBA transcript levels were detected following exposure of V. cholerae to the protein synthesis inhibitors chloramphenicol, kanamycin, and spectinomycin. Presumably, HigB is more stable than HigA (as seen with other toxin-antitoxin pairs), and consequently, any stimuli that nonspecifically block translation can preferentially reduce antitoxin levels and alleviate repression of the TA locus. Thus, we predict that amino acid starvation, previously shown to promote transcription of relBE and perhaps mazEF (7, 8; contradicted by reference 1), would also increase expression of higBA and that all of the activating stimuli would influence expression of the other TA loci contained within V. cholerae's superintegron as well. Due to the altered balance between toxin and antitoxin levels, such stimuli should also promote toxin activity.
The biological role of chromosomally carried TA loci is the subject of debate. Engelberg-Kulka and colleagues have proposed that TA loci induce programmed cell death (11, 23), while Gerdes and others favor the idea that these loci merely retard cell growth until favorable growth conditions are restored (4, 13, 29). These conflicting ideas may, in part, reflect genuine differences in the activities of TA-encoded toxins, i.e., some may act more as killers than others. For example, mazEF-dependent cell death in E. coli has been reported in response to numerous stimuli (14, 16, 23, 33, 34), whereas induction of relBE (and in certain cases mazEF) has no effect on cell viability (5, 7). The conflicting conclusions also appear to reflect differences between the conditions (e.g., time or culture media) under which the effects of toxin production were assessed.
One approach used to address the question of whether TA-encoded toxins are bacteriostatic or bactericidal has been to monitor whether the effects of toxin production (typically exogenously expressed) can be reversed by subsequent production (again exogenous) of antitoxin. If delayed expression of antitoxin is sufficient to restore culture viability, then it is argued that the effect of the toxin is bacteriostatic; when no rescue is observed, it is argued that the toxin is bactericidal. We performed analogous experiments using V. cholerae HigB and HigA and observed that delayed production of HigA following production of HigB could reduce or eliminate HigB-dependent toxicity (Fig. 4B and C). However, after sufficient delay, the effect of HigA was minimal to undetectable. Thus, it is clear that whether HigB is judged to be bacteriostatic versus bactericidal based on this approach depends largely upon the conditions and times assayed: the effects of short exposure to HigB are reversible, while those of longer exposures largely are not, as has been observed for MazF (2, 29).
A pitfall of “rescue” experiments performed to date is that in most cases investigators have not reported whether antitoxin is actually produced in response to inducing stimuli. Since several toxins have been shown to interfere with protein translation (e.g., RelE and MazF/ChpAK), it is easy to imagine that after a certain period of toxin induction, cells may no longer be capable of synthesizing antitoxin. Under such conditions, it cannot be argued that the antitoxin is ineffective at countering the toxin; instead, it is simply absent. To address this concern in our own rescue experiments, we monitored the accumulation of epitope-tagged antitoxin (HigA-Myc) following its induction at various time points relative to higB induction. We found that even after 90 min of HigB production, cells could be induced to synthesize a significant quantity of HigA; thus, HigB does not appear to have a dramatic global effect on gene expression. HigA accumulation was slightly lower per ml of culture in cells expressing HigB than in cells with repressed HigB expression; however, this may reflect the slower growth of cells expressing toxin. These data strongly suggest that the absence of rescue following 90 min of toxin production is not simply due to a failure to synthesize antitoxin, suggesting that exogenously produced HigB can be bactericidal. However, it is also possible that extended toxin induction does not initiate a new cellular program (e.g., programmed cell death); an alternative explanation might be that the antitoxin produced is simply insufficient to inactivate the toxin that has already accumulated.
We also assessed whether conditions that activate endogenous higBA (e.g., protein synthesis inhibitors) promote toxin-dependent cell death. No significant difference was observed between survival rates of wt and higBA V. cholerae cells following exposure to either chloramphenicol or kanamycin. Furthermore, chloramphenicol treatment had an extremely limited effect on the viability of wild-type cells, despite having a dramatic effect on cell replication (Fig. 5 and data not shown). Because such treatment clearly activates higBA, and probably activates additional TA loci within the V. cholerae superintegron, our data suggest that expression of HigB (and potentially other toxins as well) at endogenous levels for several hours is not bactericidal.
The function of the higBA locus in V. cholerae remains to be determined. One possibility is that toxin activation (presumably in response to unfavorable growth conditions) inhibits bacterial growth until more auspicious growth conditions are restored. Interestingly, it has been observed that TA loci are activated in persister cells, which neither grow nor die in the presence of bactericidal agents (21). However, it is still unclear how, following toxin activation, cells can regenerate a growth-favoring TA balance, particularly if the accumulated toxin inhibits gene expression, as demonstrated for several toxins. The dynamics of toxin inactivation poststress have not been explored thoroughly. It may be that antitoxin-independent processes for inactivation remain to be discovered or that antitoxin production can be uncoupled from toxin production (e.g., via activation of PhigA). Stochastic processes may also play a role (4); however, it is difficult to see how these alone could allow for complete maintenance of culture viability. More understanding of this cycle may develop as natural conditions that promote toxin activation are identified.
Another possible role for chromosomal TA loci is promotion of genomic stability. For example, V. cholerae higBA might protect against loss either of the V. cholerae second chromosome or, as proposed by Rowe-Magnus et al., of the V. cholerae superintegron (32). Segregation and partitioning mechanisms for chromosome II appear to be quite reliable, and hence chromosome loss is a very rare event; however, it is interesting that cells induced to lose chromosome II (and thus presumably containing an excess of residual HigB relative to HigA) share some striking morphological characteristics with cells that overexpress HigB (Y. Yamaichi and M. K. Waldor, unpublished observations). Thus, HigB may aid in selecting against V. cholerae cells containing only chromosome I in the rare cases where such cells develop. In contrast to the overall stability of chromosome II, it is clear that the V. cholerae superintegron is extremely plastic (30). It contains numerous repeat sequences that might be expected to lead to frequent excision events; however, as with a loss of the complete chromosome, loss of a TA-encoding region would cause irreversible activation of the toxin due to the absence of the antitoxin-encoding gene. Such activation may provide strong counterselection against deletion of TA loci and, presumably, against deletion of adjacent loci that might be lost simultaneously. Consequently, although TA loci might initially be seen as “selfish” genes, they may also provide benefits for the organism in which they reside.
ADDENDUM IN PROOF
While this paper was under review, Christensen-Dalsgaard and Gerdes (M. Christensen-Dalsgaard and K. Gerdes, Mol. Microbiol. 62:397-411, 2006) reported the mechanism of action of HigB.
Acknowledgments
We are grateful to D. Lazinski and S. McLeod for critical readings of the manuscript, to K. Gerdes for helpful discussions and sharing of unpublished results, and to the NEMC GRASP Center for preparation of plates and media.
This work was funded by NIH grants AI42347, AI59698, and P30DK-34928 to the NEMC GRASP Digestive Center and by the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 3 November 2006.
REFERENCES
- 1.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amitai, S., Y. Yassin, and H. Engelberg-Kulka. 2004. MazF-mediated cell death in Escherichia coli: a point of no return. J. Bacteriol. 186:8295-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anantharaman, V., and L. Aravind. 2003. New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol. 4:R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buts, L., J. Lah, M. H. Dao-Thi, L. Wyns, and R. Loris. 2005. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 30:672-679. [DOI] [PubMed] [Google Scholar]
- 5.Christensen, S. K., and K. Gerdes. 2003. RelE toxins from bacteria and archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 48:1389-1400. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, S. K., G. Maenhaut-Michel, N. Mine, S. Gottesman, K. Gerdes, and L. Van Melderen. 2004. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol. Microbiol. 51:1705-1717. [DOI] [PubMed] [Google Scholar]
- 7.Christensen, S. K., K. Pedersen, F. G. Hansen, and K. Gerdes. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332:809-819. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, S. K., M. Mikkelsen, K. Pedersen, and K. Gerdes. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 98:14328-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Critchlow, S. E., M. H. O'Dea, A. J. Howells, M. Couturier, M. Gellert, and A. Maxwell. 1997. The interaction of the F plasmid killer protein, CcdB, with DNA gyrase: induction of DNA cleavage and blocking of transcription. J. Mol. Biol. 273:826-839. [DOI] [PubMed] [Google Scholar]
- 10.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelberg-Kulka, H., R. Hazan, and S. Amitai. 2005. mazEF: a chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J. Cell Sci. 118:4327-4332. [DOI] [PubMed] [Google Scholar]
- 12.Gerdes, K., P. B. Rasmussen, and S. Molin. 1986. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 83:3116-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerdes, K., S. K. Christensen, and A. Løbner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 14.Godoy, V. G., D. F. Jarosz, F. L. Walker, L. A. Simmons, and G. C. Walker. 2006. Y-family DNA polymerases respond to DNA damage-independent inhibition of replication fork progression. EMBO J. 25:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hazan, R., B. Sat, and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 186:3663-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hazan, R., B. Sat, M. Reches, and H. Engelberg-Kulka. 2001. Postsegregational killing mediated by the P1 phage “addiction module” phd-doc requires the Escherichia coli programmed cell death system mazEF. J. Bacteriol. 183:2046-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hochhut, B., J. Marrero, and M. K. Waldor. 2000. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J. Bacteriol. 182:2043-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, Y., J. Pogliano, D. R. Helinski, and I. Konieczny. 2002. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 44:971-979. [DOI] [PubMed] [Google Scholar]
- 21.Keren, I., D. Shah, A. Spoering, N. Kaldalu, and K. Lewis. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khlebnikov, A., K. A. Datsenko, T. Skaug, B. L. Wanner, and J. D. Keasling. 2001. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology 147:3241-3247. [DOI] [PubMed] [Google Scholar]
- 23.Kolodkin-Gal, I., and H. Engelberg-Kulka. 2006. Induction of Escherichia coli chromosomal mazEF by stressful conditions causes an irreversible loss of viability. J. Bacteriol. 188:3420-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lech, K., and R. Brent. 1995. E. coli, plasmids, and bacteriophages, p. 1.0.3-1.15.8. In F. M. Ausubel, R. Brent, R. E. Kingston, D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley and Sons, Inc., New York, NY.
- 25.Lessl, M., D. Balzer, R. Lurz, V. L. Waters, D. G. Guiney, and E. Lanka. 1992. Dissection of IncP conjugative plasmid transfer: definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J. Bacteriol. 174:2493-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maki, S., S. Takiguchi, T. Miki, and T. Horiuchi. 1992. Modulation of DNA supercoiling activity of Escherichia coli DNA gyrase by F plasmid proteins. Antagonistic actions of LetA (CcdA) and LetD (CcdB) proteins. J. Biol. Chem. 267:12244-12251. [PubMed] [Google Scholar]
- 27.Pandey, D. P., and K. Gerdes. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33:966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pedersen, K., A. V. Zavialov, M. Y. Pavlov, J. Elf, K. Gerdes, and M. Ehrenberg. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112:131-140. [DOI] [PubMed] [Google Scholar]
- 29.Pedersen, K., S. K. Christensen, and K. Gerdes. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45:501-510. [DOI] [PubMed] [Google Scholar]
- 30.Purins, L., C. A. Clark, P. Kaewrakon, T. Focareta, and P. A. Manning. 2000. The Vibrio cholerae O1 chromosomal integron. Microbiology 146:2605-2612. [DOI] [PubMed] [Google Scholar]
- 31.Quinones, M., B. M. Davis, and M. K. Waldor. 2006. Activation of the Vibrio cholerae SOS response is not required for intestinal cholera toxin production or colonization. Infect. Immun. 74:927-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rowe-Magnus, D. A., A. M. Guerout, L. Biskri, P. Bouige, and D. Mazel. 2003. Comparative analysis of superintegrons: engineering extensive genetic diversity in the Vibrionaceae. Genome Res. 13:428-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sat, B., M. Reches, and H. Engelberg-Kulka. 2003. The Escherichia coli mazEF suicide module mediates thymineless death. J. Bacteriol. 185:1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sat, B., R. Hazan, T. Fisher, H. Khaner, G. Glaser, and H. Engelberg-Kulka. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 183:2041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider, K., and C. F. Beck. 1986. Promoter-probe vectors for the analysis of divergently arranged promoters. Gene 42:37-48. [DOI] [PubMed] [Google Scholar]
- 36.Tian, Q. B., M. Ohnishi, A. Tabuchi, and Y. Terawaki. 1996. A new plasmid-encoded proteic killer gene system: cloning, sequencing, and analyzing hig locus of plasmid Rts1. Biochem. Biophys. Res. Commun. 220:280-284. [DOI] [PubMed] [Google Scholar]
- 37.Tian, Q. B., T. Hayashi, T. Murata, and Y. Terawaki. 1996. Gene product identification and promoter analysis of hig locus of plasmid Rts1. Biochem. Biophys. Res. Commun. 225:679-684. [DOI] [PubMed] [Google Scholar]
- 38.Van Melderen, L., P. Bernard, and M. Couturier. 1994. Lon-dependent proteolysis of CcdA is the key control for activation of CcdB in plasmid-free segregant bacteria. Mol. Microbiol. 11:1151-1157. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, Y., J. Zhang, H. Hara, I. Kato, and M. Inouye. 2005. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 280:3143-3150. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, Y., J. Zhang, K. P. Hoeflich, M. Ikura, G. Qing, and M. Inouye. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12:913-923. [DOI] [PubMed] [Google Scholar]