Abstract
Translation initiation requires the precise positioning of a ribosome at the start codon. The major signals of bacterial mRNA that direct the ribosome to a translational start site are the Shine-Dalgarno (SD) sequence within the untranslated leader and the start codon. Evidence for the presence of many non-SD-led genes in prokaryotes provides a motive for studying additional interactions between ribosomes and mRNA that contribute to translation initiation. A high incidence of adenines has been reported downstream of the start codon for many Escherichia coli genes, and addition of downstream adenine-rich sequences increases expression from several genes in E. coli. Here we describe site-directed mutagenesis of the E. coli aroL, pncB, and cysJ coding sequences that was used to assess the contribution of naturally occurring adenines to in vivo expression and in vitro ribosome binding from mRNAs with different SD-containing untranslated leaders. Base substitutions that decreased the downstream adenines by one or two nucleotides decreased expression significantly from aroL-, pncB-, and cysJ-lacZ fusions; mutations that increased downstream adenines by one or two nucleotides increased expression significantly from aroL- and cysJ-lacZ fusions. Using primer extension inhibition (toeprint) and filter binding assays to measure ribosome binding, the changes in in vivo expression correlated closely with changes in in vitro ribosome binding strength. Our data are consistent with a model in which downstream adenines influence expression through their effects on the mRNA-ribosome association rate and the amount of ternary complex formed. This work provides evidence that adenine-rich sequence motifs might serve as a general enhancer of E. coli translation.
Initiation of bacterial protein synthesis requires selection of an mRNA's translation initiation region (TIR) and initiator tRNA (fMet-tRNAfMet) by the 30S ribosomal subunit aided by the three initiation factors (IF1, IF2, and IF3). Initiation is the rate-limiting step of translation, and the formation of ribosome-initiator tRNA-mRNA ternary complexes is influenced by sequence and structural motifs in and around the mRNA's ribosome binding site (RBS) (33). Features of the mRNA RBS contributing to the efficiency of ribosome binding and translation include the start codon (most frequently AUG in Escherichia coli) and the purine-rich Shine-Dalgarno (SD) sequence, located within the untranslated leader region (UTR), which base pairs with the anti-SD (ASD) sequence near the 3′ end of 16S rRNA (11, 20, 29). In addition to the start codon and SD-ASD interaction, other sequence and structural motifs within mRNA have been suggested to influence ribosome binding and translation, and these motifs include mRNA secondary structure within the TIR (6, 7), specific translation-enhancing sequences upstream (10, 12, 21, 37) and downstream (9, 19, 26, 32) of the start codon, upstream pyrimidine-rich tracts (1, 38, 42), AU-rich sequences within the UTR (14, 15), and downstream A-rich (3) and CA-rich tracts (16). In addition, an increasing number of non-SD-led genes (2) and genes encoding mRNA lacking a 5′-untranslated leader region (leaderless mRNA) (13, 18) are being identified, raising the potential that there are novel sequence and structural motifs within the coding sequence that contribute to the formation of translation initiation complexes.
Statistical analysis of E. coli translational start sites (24, 27) has revealed that the regions downstream of the initiation codon for several genes contain CA- and/or A-rich sequences. In a recent analysis of nucleotide sequences around the boundaries of all open reading frames in E. coli, nucleotide biases were observed immediately downstream of the initiation codon, and the two most frequent second codons, AAA and AAU, were found to enhance translational efficiency (26). In addition to these studies, adenine-rich sequences have been proposed to enhance translation in E. coli, as demonstrated by the increased expression observed for the human gamma interferon and chloramphenicol acetyltransferase (cat) genes after addition of A-rich motifs downstream of the initiation codon (3).
Insertion of CA multimers immediately downstream of the lacZ initiation codon results in an increase in gene expression (16, 30; G. R. Janssen, unpublished data). CA nucleotides, inserted downstream of the start codon for leaderless or SD-leadered mRNAs, provided greater stimulation when they were close to the initiation codon and resulted in dose-dependent increases in expression when increasing numbers of CA repeats were present. Increased expression was also observed after addition of CA repeats to SD-leadered and leaderless neo, kan, and gusA genes, indicating that CA-rich sequences might serve as a general enhancer of expression for leadered and leaderless mRNAs in E. coli (16).
Chen et al. (3) and Martin-Farmer and Janssen (16) demonstrated that addition of adenines and CA repeats, respectively, can dramatically increase gene expression. In the experiments described here we investigated the influence of naturally occurring downstream adenines on in vivo expression and in vitro ribosome binding. The mRNAs encoded by the E. coli aroL, pncB, and cysJ genes include SD-containing untranslated leaders and adenines downstream of their AUG start codons; site-directed mutagenesis was used to decrease and increase the A richness in order to generate putative “down” and “up” mutations, respectively. The putative “down” mutations in aroL, pncB, and cysJ mRNAs decreased both in vivo expression and in vitro ribosome binding, whereas the putative “up” mutations in the aroL and cysJ mRNAs increased both in vivo expression and in vitro ribosome binding. Our data obtained using toeprint, filter binding, and expression assays show that downstream adenines contribute significantly to the rate and amount of ternary complex formed, as well as to the in vivo expression levels of the aroL, pncB, and cysJ genes even in the presence of a canonical SD sequence.
MATERIALS AND METHODS
Bacterial strains.
E. coli DH5α [F− φ80d lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) supE44 λ− thi-1 gyrA relA1] was used as the host strain for all plasmid constructs. E. coli RFS859 (F− thr-1 araC859 leuB6 Δlac74 tsx-274 λ− gyrA111 recA11 relA1 thi-1), a lac deletion strain (28), was used as the host for expression and for the assay of β-galactosidase activity from lacZ reporter genes. E. coli K-12 total genomic DNA was used for isolation of wild-type aroL, cysJ, and pncB gene fragments. E. coli MRE600 (39) was used for isolation of 30S ribosomal subunits.
Reagents and recombinant DNA procedures.
The radiolabeled nucleotides [γ-32P]ATP (6,000 Ci/mmol; 150 mCi/ml) and [α-32P]CTP (3,000 Ci/mmol; 10 mCi/ml) were purchased from Perkin-Elmer. Oligonucleotides were synthesized using a Beckman 1000 M DNA synthesizer or were purchased from commercial suppliers. Restriction endonucleases, T4 DNA ligase, T4 polynucleotide kinase, T4 DNA polymerase, and T7 RNA polymerase were obtained from New England Biolabs. Pfu DNA polymerase was obtained from Stratagene. Avian myeloblastosis virus reverse transcriptase was obtained from Life Sciences, and RNase-free DNase I was purchased from Ambion. All enzymes were used according to the manufacturer's recommendations. Plasmid isolation, E. coli transformation, and other DNA manipulations were carried out by using standard procedures (25).
Mutant construction.
aroL-, cysJ-, and pncB-lacZ fusions were constructed so that they contained either the lacZ untranslated leader or the gene's natural untranslated leader and the first 16 codons of the coding sequence fused to a lacZ reporter gene. Transcription of the lac fusions was provided by the E. coli lac promoter. Plasmids (pBR322 derived) containing the aroL-, cysJ-, and pncB-lacZ fusions were used as templates for site-directed mutagenesis in which one oligonucleotide primer contained the desired mutation(s) (Table 1) and the other primer annealed within the lacZ coding sequence. After amplification, the PCR product was trimmed with appropriate restriction enzymes to facilitate cloning, ultimately producing identical plasmids that varied only by single or double nucleotide mutations within the coding sequence, as shown in Table 1. All DNA regions generated by PCR amplification were verified by DNA sequencing.
TABLE 1.
DNA sequences of gene fragments, with and without mutations, from the aroL, pncB, and cysJ genes
| Plasmida | Sequence
|
|
|---|---|---|
| Portion of untranslated leaderb | Variable regionc | |
| aroL-lacZ fusions | ||
| pAaroL.WT | 5′...TGGGGAAAACCCACG | ATG ACA CAA CCT CTT TTT CTG...3′ |
| pAaroL.DN1 | 5′...TGGGGAAAACCCACG | ATG ACG CAG CCT CTT TTT CTG...3′ |
| pAaroL.DN1a | 5′...TGGGGAAAACCCACG | ATG ACG CAA CCT CTT TTT CTG...3′ |
| pAaroL.DN1b | 5′...TGGGGAAAACCCACG | ATG ACA CAG CCT CTT TTT CTG...3′ |
| pAaroL.UP1 | 5′...TGGGGAAAACCCACG | ATG ACA CAA CCA CTT TTT CTG...3′ |
| pncB-lacZ fusions | ||
| pApncB.WT | 5′...CAGGATACTGCGCACCT | ATG ACA CAA TTC GCT TCT CCT...3′ |
| pApncB.DN1 | 5′...CAGGATACTGCGCACCT | ATG ACG CAG TTC GCT TCT CCT...3′ |
| pApncB.DN1a | 5′...CAGGATACTGCGCACCT | ATG ACG CAA TTC GCT TCT CCT...3′ |
| pApncB.DN1b | 5′...CAGGATACTGCGCACCT | ATG ACA CAG TTC GCT TCT CCT...3′ |
| pApncB.UP1 | 5′...CAGGATACTGCGCACCT | ATG ACA CAA TTC GCA TCA CCT...3′ |
| cysJ-lacZ fusions | ||
| pAZcysJ.WT | 5′...CAGGAAACAGCC | ATG ACG ACA CAG GTC CCA...3′ |
| pAZcysJ.DN1 | 5′...CAGGAAACAGCC | ATG ACG ACG CAG GTC CCA...3′ |
| pAZcysJ.UP1 | 5′...CAGGAAACAGCC | ATG ACA ACA CAA GTC CCA...3′ |
| pAZcysJ.UP1a | 5′...CAGGAAACAGCC | ATG ACA ACA CAG GTC CCA...3′ |
| pAZcysJ.UP1b | 5′...CAGGAAACAGCC | ATG ACG ACA CAA GTC CCA...3′ |
The pA series of plasmid constructs contained the genes' natural untranslated leader, while the pAZ series contained the lac untranslated leader.
The underlined sequence is the presumed SD sequence.
The ATG codon is the translational start site for the aroL, pncB, and cysJ coding sequences. The amino acid sequence encoded by the variable region for the aroL-lacZ fusions is MTQPPFP; the amino acid sequence encoded by the variable region for the pncB-lacZ fusions is MTQFASP; and the amino acid sequence encoded by the variable region for the cysJ-lacZ fusions is MTTQVP. Boldface type indicates base substitutions in the coding region that create putative “down” or “up” mutations.
β-Galactosidase assays.
Triplicate cultures of plasmid-containing strains were grown in 2× YT (16 g of Difco Bacto tryptone per liter, 10 g of Difco Bacto yeast extract per liter, 10 g of NaCl per liter; pH 7.4) supplemented with 200 μg/ml ampicillin and 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C to an optical density at 600 nm of 0.4 to 0.6 and quick chilled on ice. Triplicate β-galactosidase assays (17) were performed with each of the triplicate cultures.
Primer extension inhibition (toeprinting).
The mRNAs used in primer extension inhibition (toeprint) experiments were generated in vitro with T7 RNA polymerase. DNA templates for T7 transcription were prepared by PCR amplification of miniprep plasmid DNA. Oligonucleotide primers T7lac.lead (5′-GGAATTCTAATACGACTCACTATAGAATTGTGAGCGGATAACAATTTC-3′) and lac.comp2 (5′-ATTAAGTTGGGTAACGCCAG-3′) were used to generate DNA fragments containing the T7 promoter, lac leader, and cysJ sequences (with or without mutations) fused to lacZ from the template plasmids. T7pncB.lead (5′-CTAATACGACTCACTATAGTTCCTGAAGATGTTTATTGTAC-3′) and lac.comp2 were used to generate DNA fragments containing the T7 promoter, pncB leader, and pncB sequences (with or without mutations) fused to lacZ. T7aroL.lead (5′-GGAATTCTAATACGACTCACTATAGATTGAGATTTTCACTTTAAGTGG-3′) and lac.comp2 were used to generate DNA fragments containing the T7 promoter, aroL leader, and aroL sequences (with or with out mutations) fused to lacZ. After gel purification of the resulting PCR-generated fragments, transcription reactions (20-μl mixtures containing 15 mM dithiothreitol, each nucleoside triphosphate at a concentration of 4 mM, 40 mM Tris-HCl [pH 7.9], 20 mM MgCl2, 1 μg template DNA, and 1 μl T7 RNA polymerase [500 U/μl; NEB]) were carried out at 37°C for 1 h. This was followed by incubation with 1 μl of RNase-free DNase I (2 U/μl) for 30 min at 37°C. In vitro-synthesized mRNA was then gel purified on 6% polyacrylamide-7 M urea denaturing gels. After UV shadowing and elution of the mRNA with 0.2% sodium dodecyl sulfate, 0.005 M EDTA, and 0.5 M ammonium acetate, mRNA was extracted with phenol-chloroform-isoamyl alcohol (25:24:1) and precipitated with 0.2 M ammonium acetate and 2.5 volumes of ethanol.
Isolation of E. coli 30S ribosomal subunits and toeprint assays were done as previously described (16). Briefly, 9-μl reaction mixtures containing mRNA (44 nM) with a 32P-labeled primer annealed to the 3′ end, 30S ribosomal subunits, and tRNAfMet (at a 1:10:20 ratio), as well as 1× SB (60 mM NH4Cl, 10 mM Tris-acetate [pH 7.4], 10 mM magnesium acetate, 6 mM β-mercaptoethanol), were incubated at 37°C for 30 min or the times indicated below. The reaction mixtures were placed on ice, and 1 μl of reverse transcriptase (1 U/μl) was added to each mixture; this was followed by incubation at 37°C for 10 min. The reaction mixtures were precipitated with 40 μl of 0.3 M sodium acetate and 2.5 volumes of ethanol for at least 2.5 h at −20°C and electrophoresed on 6% polyacrylamide-7 M urea gels. Dideoxy sequencing reactions mapped toeprint signals to position 16 of mRNA relative to the first base of the start codon (position 1). Toeprint signals were quantified with a Molecular Dynamics PhosphorImager (Storm 800) and were expressed as relative toeprint complexes (RTCs), defined as toeprint pixel value/(toeprint pixel value + full-length pixel value); for aroL, RTC = toeprint pixel value/(toeprint pixel value + full-length pixel value + upstream signal pixel value). RTCs for different mRNAs were compared by using reaction mixtures electrophoresed on the same gel.
Filter binding assays.
mRNA used in filter binding assays was synthesized in a 5-μl reaction mixture containing 5 mM dithiothreitol, each nucleoside triphosphate at a concentration of 2 mM, 14 mM MgCl2, 0.5 μl 10× T7 RNA polymerase buffer (NEB), 0.05 μg template DNA, 0.35 μl T7 RNA polymerase (500 U/μl; NEB), and 2.3 μl [α-32P]CTP. Filter binding reaction mixtures (10 μl) containing radiolabeled mRNA (44 nM), 30S ribosomal subunits, and tRNAfMet (at a 1:10:20 ratio), as well as 1× SB, were incubated for various amounts of time at 37°C. The reaction mixtures were then diluted to 500 μl with chilled 1× SB and filtered through nitrocellulose membranes (pore size, 0.45 μm) in a Mini-Fold slot blot manifold (Schleicher and Schuell), and this was followed by washing each well with 3 ml of 1× SB. The membranes were then dried at room temperature, and samples were cross-linked to the membranes by UV light (FB-UVXL-1000; Fisher Scientific). The amount of complex bound to the membrane was determined with a Molecular Dynamics PhosphorImager (Storm 800); standard curves were used to convert pixel values to picomoles, as previously described (4), and the data were expressed in picomoles of mRNA bound.
RESULTS
All base substitutions in the aroL, pncB, and cysJ genes were made via site-directed mutagenesis and maintained the natural amino acid sequence; putative “down” mutations decreased the downstream adenine content, whereas putative “up” mutations increased the downstream adenine content (Table 1). The effects of the mutations on gene expression were assessed by translational fusions of aroL, pncB, and cysJ gene fragments to an E. coli lacZ reporter gene, followed by β-galactosidase assays. Toeprint and filter binding assays were used to compare the rates of ternary complex formation, defined here as the amount of complex formed (ribosomes, tRNA, mRNA) over time, for aroL, pncB, and cysJ mRNAs with and without mutations that altered the downstream adenine content (Table 1). In toeprint assays, variations in a specific RTC might be observed in different assays, but the correlations between the wild-type and mutant binding strengths were consistent and reproducible. These assays were performed in an effort to correlate the in vitro ribosome binding strengths of aroL, pncB, and cysJ mRNAs with the in vivo expression data.
Downstream adenines of aroL.
Transcription of the aroL gene, encoding shikimate kinase II, results in an mRNA with a 124-nucleotide leader containing an optimally spaced (7 to 9 nucleotides), canonical SD sequence (5). Mutations in aroL's coding sequence altering the downstream adenine content are shown in Table 1.
(i) Effect of aroL downstream adenines on in vivo expression.
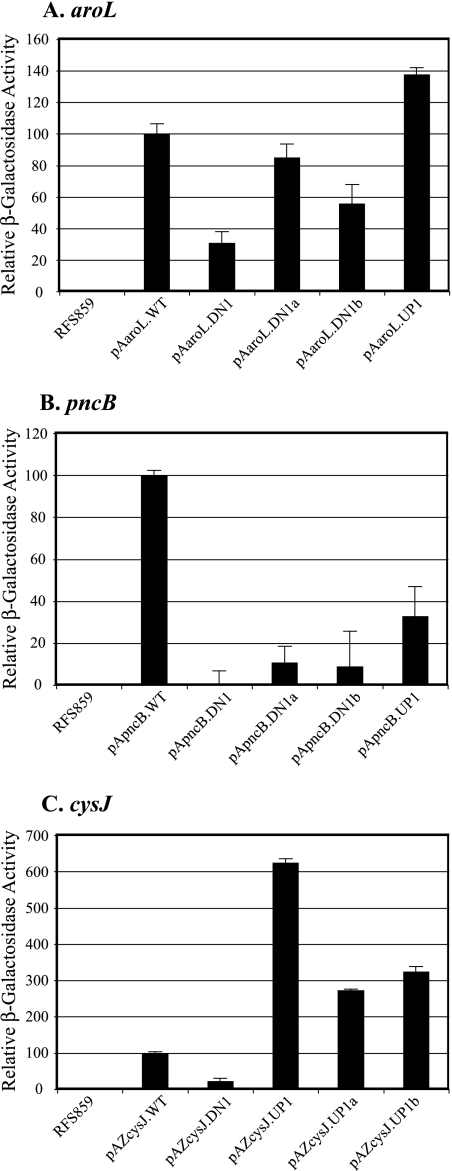
Comparisons of the β-galactosidase activities of cells containing the wild-type (pAaroL.WT) and putative “down” aroL-lacZ constructs revealed that the A→G substitutions in codons 2 and 3 (pAaroL.DN1) decreased lacZ expression 72%; incorporation of the individual substitutions resulted in 15% (pAaroL.DN1a) and 44% (pAaroL.DN1b) decreases in lacZ expression (Fig. 1A). Analysis of the putative “up” aroL-lacZ construct revealed that the U→A substitution at codon 4 (pAaroL.UP1) increased lacZ expression 38% (Fig. 1A). These results indicate that the downstream adenines influence aroL expression significantly, despite the presence of a canonical SD-ASD interaction.
FIG. 1.
Effects of aroL, pncB, and cysJ downstream adenines on expression in vivo. β-Galactosidase activities from aroL-, pncB-, and cysJ-lacZ fusions are compared for cells containing plasmids with and without mutations altering the downstream adenine content (Table 1). (A) Plasmid-free cells and cells containing plasmids with the following aroL-lacZ fusions: pAaroL.WT (100%), pAaroL.DN1 (28%), pAaroL.DN1a (85%), pAaroL.DN1b (56%), and pAaroL.UP1 (138%). (B) Plasmid-free cells and cells containing plasmids with the following pncB-lacZ fusions: pApncB.WT (100%), pApncB.DN1 (0.1%), pApncB.DN1a (11%), pApncB.DN1b (8%), and pApncB.UP1 (33%). (C) Plasmid-free cells and cells containing plasmids with the following cysJ-lacZ fusions: pAZcysJ.WT (100%), pAZcysJ.DN1 (23%), pAZcysJ.Up1 (625%), pAZcysJ.UP1a (274%), and pAZcysJ.UP1b (325%).
(ii) Effect of aroL downstream adenines on in vitro ribosome binding.
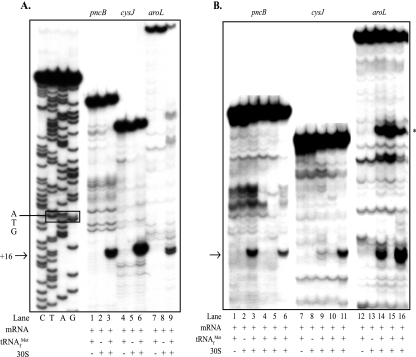
Toeprint assays with aroL-lacZ mRNA and 30S subunits revealed a tRNA-dependent signal corresponding to position 16 relative to the A residue (position 1) of the AUG start codon (Fig. 2A, lanes 7 to 9). Phosphorimage analysis of toeprint signal intensity revealed a 97% reduction for the putative “down” mutant mRNA (Fig. 2B, lane 15) compared to the wild-type mRNA (Fig. 2B, lane 14). Incorporation of the U→A putative “up” mutation at codon 4 increased the toeprint signal 278% (Fig. 2B, lane 16) compared to the wild-type signal (Fig. 2B, lane 14). The toeprint results, together with the aroL-lacZ expression levels, showed that there was a positive correlation between the downstream adenine content, in vitro ribosome binding, and in vivo expression levels.
FIG. 2.
Effects of aroL, pncB, and cysJ downstream adenines on ribosome binding in vitro: toeprint assays with aroL-, pncB-, and cysJ-lacZ mRNAs and 30S subunits comparing ribosome binding to mRNAs with and without mutations altering the downstream adenine content (Table 1). The arrows indicate the position of the ternary complex-dependent toeprint signals at position 16 relative to the A residue (position 1) of the AUG start codons that result from bound 30S subunits, and the asterisk in panel B indicates the position of an uncharacterized ternary complex-dependent signal in the aroL upstream UTR. Bands observed in the absence of tRNA and ribosomes were artifacts likely due to structures formed within the mRNA. (A) Lanes G, A, T, and C, dideoxy DNA sequence ladder for the pncB template (the ATG initiation triplet is enclosed in a box); lanes 1 to 3, pncB-lacZ wild-type mRNA; lanes 4 to 6, cysJ-lacZ wild-type mRNA; lanes 7 to 9, aroL-lacZ wild-type mRNA. (B) Lanes 1 to 6, pncB-lacZ mRNAs made from the templates pApncB.WT (lanes 1 to 3), pApncB.DN1b (lane 4), pApncB.DN1 (lane 5), and pApncB.UP1 (lane 6); lanes 7 to 11, cysJ-lacZ mRNAs made from the templates pAZcysJ.WT (lanes 7 to 9), pAZcysJ.DN1 (lane 10), and pAZcysJ.UP1 (lane 11); lanes 12 to 16, aroL-lacZ mRNAs made from the templates pAaroL.WT (lanes 12 to 14), pAaroL.DN1 (lane 13), and pAaroL.UP1 (lane 14).
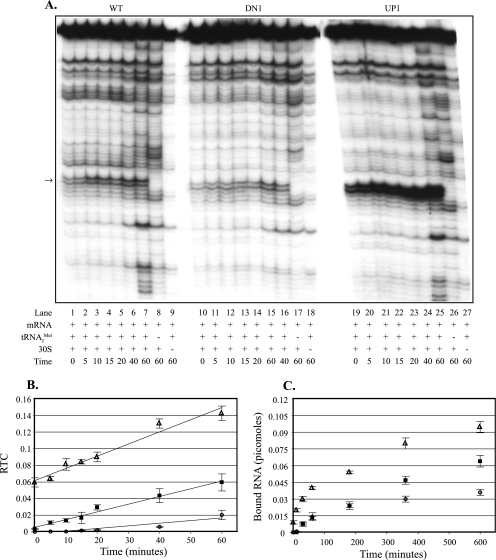
(iii) Effect of aroL downstream adenines on the rate and amount of ternary complex formed.
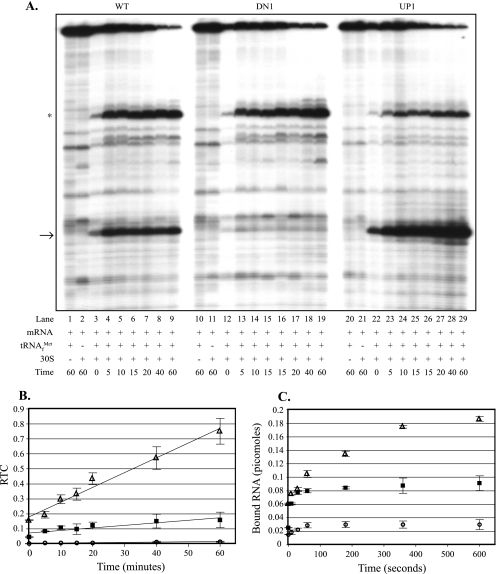
Incorporation of the A→G putative “down” mutations at aroL's second and third codons (Fig. 3A, lanes 10 to 19) resulted in a reduced rate and amount of ternary complex formation compared to the wild-type values (Fig. 3A, lanes 1 to 9), as shown in Fig. 3B. Incorporation of the U→A putative “up” mutation at aroL's fourth codon (Fig. 3A, lanes 20 to 29) resulted in an increased rate and amount of ternary complex formation compared to the wild-type values (Fig. 3A, lanes 1 to 9), as shown in Fig. 3B. The significant amount of toeprint signal at zero time (Fig. 3A and B) reflects the complex formed and cDNA produced during the 10-min incubation with reverse transcriptase. An additional uncharacterized ternary complex signal was observed within aroL's untranslated leader, and its position is indicated in Fig. 3A. Because this signal occurred upstream of the aroL toeprint signal (Fig. 3A), it was included with the full-length signal for phosphorimage quantification of RTC values in Fig. 3B. Calculation of RTC values without the additional band resulted in higher RTC values, but the line slopes, relative to each other, were basically unchanged (data not shown).
FIG. 3.
Effects of aroL downstream adenines on the rate and amount of ternary complex formed: toeprint and filter binding assays comparing ternary complex formation on aroL-lacZ mRNAs with and without mutations altering the downstream adenine content. (A) In toeprint assays, 30S subunits were incubated with tRNAfMet and aroL-lacZ mRNA for different times (in minutes). Lanes 1 to 29 contained aroL-lacZ mRNAs made from the following templates: pAaroL.WT (lanes 1 to 9), pAaroL.DN1 (lanes 10 to 19), and pAaroL.UP1 (lanes 20 to 29). The arrow indicates the position of the ternary complex-dependent toeprint signals at position 16, and the asterisk indicates the position of an uncharacterized ternary complex-dependent signal in the upstream UTR. (B) The RTC formed over time was quantified, and the data (averages from three independent assays) were plotted. ▪, aroL-lacZ wild-type mRNA (pAaroL.WT); ○, aroL-lacZ “down” mutant mRNA (pAaroL.DN1); ▵, aroL-lacZ “up” mutant mRNA (pAaroL.UP1). (C) In filter binding assays, 30S subunits were incubated with tRNAfMet and aroL-lacZ mRNA for different times, and the amounts of bound RNA (averages from three independent assays) were plotted. ▪, aroL-lacZ wild-type mRNA (pAaroL.WT); ○, aroL-lacZ “down” mutant mRNA (pAaroL.DN1); ▵, aroL-lacZ “up” mutant mRNA (pAaroL.UP1). WT, wild type.
In order to obtain a more accurate assessment of complex formation at earlier times, filter binding assays were used. Filter binding assays measure complex formation based on the amount of mRNA bound by ribosomes when preparations are filtered through a nitrocellulose membrane. Incorporation of the A→G putative “down” mutations at aroL's second and third codons reduced the rate and amount of ternary complex formed, whereas the U→A putative “up” mutation at codon 4 increased the amount of ternary complex formed compared to the wild-type amount (Fig. 3C). Ternary complex formation with the “up” mutant occurred with a rate similar to the wild-type rate during the first 60 s of incubation, after which the amount of product formed continued to increase with the “up” mutant but not with the wild type (Fig. 3C). The toeprint and filter binding results provide evidence that downstream adenines enhance the rate and/or amount of ternary complex formation with aroL mRNA.
Downstream adenines of pncB.
Transcription of the pncB gene, encoding a nicotinic acid phosphoribosyltransferase, results in an mRNA with a 58-nucleotide untranslated leader containing a canonical SD sequence with suboptimal spacing (12 nucleotides) from the start codon (40) and second and third codons identical to those of aroL. Mutations in pncB's coding sequence altering the downstream adenine content are shown in Table 1.
(i) Effect of pncB downstream adenines on in vivo expression.
Comparisons of the β-galactosidase activities of cells containing the wild-type (pApncB.WT) and putative “down” pncB-lacZ constructs revealed that the A→G substitutions in codons 2 and 3 (pApncB.DN1) decreased lacZ expression 99.9%; incorporation of the individual substitutions resulted in 89% (pApncB.DN1a) and 92% (pApncB.DN1b) decreases in lacZ expression (Fig. 1B). Analysis of the putative “up” pncB-lacZ construct revealed that the U→A substitutions in codons 5 and 6 (pApncB.UP1) unexpectedly decreased lacZ expression 67% (Fig. 1B).
To determine if altering the pncB downstream adenine content had a similar effect on expression irrespective of the untranslated leader sequence, analogous pncB-lacZ coding sequence fusions were placed downstream of the E. coli lacZ untranslated leader containing a canonical SD sequence with optimal spacing (7 nucleotides) from the start codon. Putative “down” mutations in codons 2 and 3 of the lac-leadered pncB-lacZ mRNA reduced gene expression 87% relative to the wild-type gene expression (data not shown). These results indicate that the properly spaced canonical SD sequence of the lac leader could not fully compensate for the loss of downstream adenines.
(ii) Effect of pncB downstream adenines on in vitro ribosome binding.
Toeprint assays with pncB-lacZ mRNA and 30S subunits revealed a tRNA-dependent signal corresponding to position 16 relative to the A residue (position 1) of the AUG start codon (Fig. 2A, lanes 1 to 3). pncB-lacZ mRNA containing putative “down” mutations at codon 3 (Fig. 2B, lane 4) or codons 2 and 3 (Fig. 2B, lane 5) nearly eliminated ribosome binding (the toeprint signal was reduced >95%) compared to the wild-type binding (Fig. 2B, lane 3). Incorporation of the putative “up” mutations at codons 5 and 6 resulted in a 62% reduction in toeprint signal intensity (Fig. 2B, lane 6) compared to the wild-type intensity (Fig. 2B, lane 3). The toeprint results, together with the pncB-lacZ expression levels, showed that there was a positive correlation between in vitro ribosome binding and the in vivo expression levels.
(iii) Effect of pncB downstream adenines on the rate and amount of ternary complex formed.
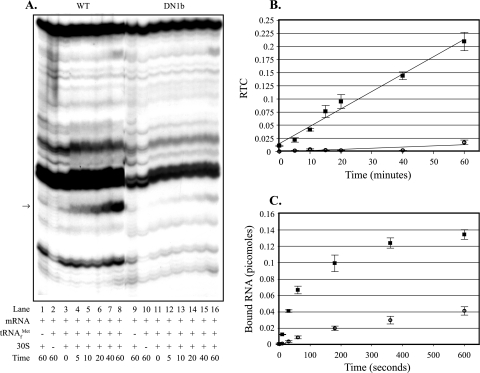
Incorporation of the A→G putative “down” mutation at pncB's third codon (Fig. 4A, lanes 9 to 16) resulted in a dramatically reduced rate and amount of ternary complex formation compared to the wild-type values (Fig. 4A, lanes 1 to 8), as shown in Fig. 4B. The bands obtained in the absence of tRNA and ribosomes are artifacts likely due to structures formed within the mRNA. When preparations were analyzed by filter binding assays, incorporation of the A→G putative “down” mutation at pncB's third codon resulted in a reduced rate and amount of ternary complex formation compared to the wild-type values (Fig. 4C). The toeprint and filter binding results provide evidence that downstream adenines contribute to the rate and amount of ternary complex formation with pncB mRNA.
FIG. 4.
Effects of pncB downstream adenines on the rate and amount of ternary complex formed: toeprint and filter binding assays comparing ternary complex formation on pncB-lacZ mRNAs with and without mutations altering the downstream adenine content. (A) In toeprint assays, 30S subunits were incubated with tRNAfMet and pncB-lacZ mRNA for different times (in minutes). Lanes 1 to 16 contained pncB-lacZ mRNAs made from the following templates: pApncB.WT (lanes 1 to 8) and pApncB.DN1b (lanes 9 to 16). The arrow indicates the position of the ternary complex-dependent toeprint signals. (B) The RTC formed over time was quantified, and the data (averages from three independent assays) were plotted. ▪, pncB-lacZ wild-type mRNA (pApncB.WT); ○, pncB-lacZ “down” mutant mRNA (pApncB.DN1b). (C) In filter binding assays, 30S subunits were incubated with tRNAfMet and pncB-lacZ mRNA for different times, and the amounts of bound RNA (averages from three independent assays) were plotted. ▪, pncB-lacZ wild-type mRNA (pApncB.WT); ○, pncB-lacZ “down” mutant mRNA (pApncB.DN1b). WT, wild type.
Downstream adenines of cysJ.
The E. coli cysJ gene, encoding a NADPH-sulfite reductase flavoprotein component (22), has different second and third codons than aroL and pncB. Cloning a DNA fragment encoding the cysJ untranslated leader and a fragment of cysJ coding sequence into a multicopy plasmid resulted in a dramatic reduction in plasmid copy number (data not shown); therefore, cysJ-lacZ coding sequence fusions were placed downstream of the 38-nucleotide lacZ untranslated leader containing a canonical SD sequence with optimal spacing (7 to 9 nucleotides) from the cysJ start codon. Mutations in cysJ's coding sequence altering the downstream adenine content are shown in Table 1.
(i) Effect of cysJ downstream adenines on in vivo expression.
Comparisons of the β-galactosidase activities of cells containing the wild-type (pAZcysJ.WT) and putative “down” (pAZcysJ.DN1) cysJ-lacZ constructs revealed that the A→G base substitution at the third codon resulted in a 77% decrease in lacZ expression (Fig. 1C). Examination of the putative “up” cysJ-lacZ constructs revealed that the G→A substitutions in codons 2 and 4 (pAZcysJ.UP1) increased lacZ expression 525%; incorporation of the individual substitutions resulted in 174% (pAZcysJ.UP1a) and 225% (pAZcysJ.UP1b) increases in expression (Fig. 1C).
(ii) Effect of cysJ downstream adenines on in vitro ribosome binding.
Toeprint assays with cysJ-lacZ mRNA and 30S subunits revealed a tRNA-dependent signal corresponding to position 16 relative to the A residue (position 1) of the AUG start codon (Fig. 2A, lanes 4 to 6). Phosphorimage analysis of toeprint signal intensity revealed that there was an 86% reduction for the putative “down” mutant (Fig. 2B, lane 10) compared to the wild type (Fig. 2B, lane 9). Incorporation of putative “up” mutations at codons 2 and 4 resulted in a 202% increase in toeprint signal intensity (Fig. 2B, lane 11) compared to the wild-type signal intensity (Fig. 2B, lane 9). The toeprint results, together with the cysJ-lacZ expression levels, showed that there was a strong correlation between the downstream adenine content, in vitro ribosome binding, and in vivo expression levels.
(iii) Effect of cysJ downstream adenines on the rate and amount of ternary complex formation.
Incorporation of the A→G putative “down” mutation at cysJ's third codon (Fig. 5A, lanes 10 to 18) resulted in a reduced rate and amount of ternary complex formation compared to the wild-type values (Fig. 5A, lanes 1 to 9), as shown in Fig. 5B. Incorporation of the G→A putative “up” mutations at cysJ's second and fourth codons (Fig. 5A, lanes 19 to 27) resulted in an increased rate and amount of ternary complex formation compared to the wild-type values (Fig. 5A, lanes 1 to 9), as shown in Fig. 5B. Phosphorimage analysis of the full-length, runoff signal yielded consistently high pixel values, resulting in low RTC values; however, the rate comparisons for mutant and wild-type mRNAs still correlated well with the results visualized by autoradiography. When preparations were analyzed by filter binding assays, incorporation of the A→G putative “down” mutation at cysJ's third codon resulted in a rate and amount of ternary complex formation similar to the rate and amount obtained with wild-type mRNA during the first 3 min of incubation; after 3 min, the rate and amount of ternary complex formed were reduced compared to the wild-type values (Fig. 5C). Incorporation of the G→A putative “up” mutations at cysJ's second and fourth codons resulted in an increased rate and amount of ternary complex formation compared to the wild-type values, with the most significant difference in the rate occurring in the initial 3 min of binding (Fig. 5C). The toeprint and filter binding results provided evidence that downstream adenines enhance the rate and amount of ternary complex formed with cysJ mRNA.
FIG. 5.
Effects of cysJ downstream adenines on the rate and amount of ternary complex formed: toeprint and filter binding assays comparing ternary complex formation on cysJ-lacZ mRNAs with and without mutations altering the downstream adenine content. (A) In toeprint assays, 30S subunits were incubated with tRNAfMet and cysJ-lacZ mRNA for different times (in minutes). Lanes 1 to 27 contained cysJ-lacZ mRNAs made from the following templates: pAZcysJ.WT (lanes 1 to 9), pAZcysJ.DN1 (lanes 10 to 18), and pAZcysJ.UP1 (lanes 19 to 27). The arrow indicates the position of the ternary complex-dependent toeprint signals. (B) The RTC formed over time was quantified, and the data (averages from three independent assays) were plotted. ▪, cysJ-lacZ wild-type mRNA (pAZcysJ.WT); ○, cysJ-lacZ “down” mutant mRNA (pAZcysJ.DN1); ▵, cysJ-lacZ “up” mutant mRNA (pAZcysJ.UP1). (C) In filter binding assays, 30S subunits were incubated with tRNAfMet and cysJ-lacZ mRNA for different times, and the amounts of bound RNA (averages from three independent assays) were plotted. ▪, cysJ-lacZ wild-type mRNA (pAZcysJ.WT); ○, cysJ-lacZ “down” mutant mRNA (pAZcysJ.DN1); ▵, cysJ-lacZ “up” mutant mRNA (pAZcysJ.UP1). WT, wild type.
DISCUSSION
The correlation of adenine-rich regions with efficient translation in E. coli was first noted by Dreyfus (8), and in later studies workers reported stimulatory effects on translation due to the addition of adenines (3, 16, 30). We report here that adenines occurring naturally downstream of the initiation codon can contribute significantly to ribosome binding and expression in E. coli. Our data show that downstream adenines can have major effects on expression even in the presence of canonical SD sequences. Demonstration that downstream nucleotides can function as major effectors of expression is especially interesting in light of a recent report (2) that non-SD-led genes, including leaderless mRNAs, are as common as SD-led genes in prokaryotes. The absence of an SD sequence requires that other sequence or structural features of the mRNA help direct the ribosome to the correct initiation site. Based on work described here, the position and number of downstream adenines may contribute to the ribosome binding strength of mRNAs with, and possibly without, canonical SD sequences.
In our experiments we investigated the influence of naturally occurring downstream adenines on in vitro ribosome binding and in vivo expression for the E. coli aroL, pncB, and cysJ mRNAs. Compared to the data for the wild-type mRNAs, the putative “down” mutations significantly decreased in vitro ribosome binding and in vivo expression; putative “up” mutations in aroL and cysJ significantly increased in vitro ribosome binding and in vivo expression. These data are consistent with previous reports which showed that A-rich sequences added immediately downstream of the start codon stimulated expression from several natural and heterologous genes in E. coli (3, 16). While the pncB “down” mutations demonstrated clearly that adenines in codons 2 and 3 are important for expression, it is not clear why the putative “up” mutations in codons 5 and 6 did not increase expression; two possible explanations are that the substitutions were too far from the start codon (i.e., compared to codons 2 and 4 for cysJ and aroL “up” mutations) and that the mutations resulted in a secondary structure that reduced access of a ribosome to the initiation region.
Downstream adenines affect the rate and amount of ternary complex formed.
Using toeprint and filter binding assays as two independent approaches to estimate ribosome binding, we observed a strong correlation between the downstream adenine content, in vivo expression, and the rate and amount of in vitro ribosome binding, suggesting strongly that the variations in expression were mediated through the observed effects on ribosome binding. Toeprint assays also revealed that aroL, pncB, and cysJ ternary complexes, once formed, did not dissociate in the presence of a competitor mRNA (data not shown), in agreement with a previous report which showed that ternary complexes are stable and irreversible in the presence of competitor mRNA (31). Therefore, downstream adenines appear to influence expression through their effects on mRNA-ribosome association and not by preventing ternary complex dissociation.
Possible contributions of downstream adenines to ribosome binding and expression.
Comparison of the codons resulting from aroL, pncB, and cysJ mutagenesis to an E. coli genomic codon usage table (GenBank) suggested that the effects on expression were not due to the introduction of rare codons or to low tRNA availability. Also, conservation of the wild-type amino acid sequence ensured that expression differences from the mutant constructs were not the result of an altered peptide sequence. Furthermore, the strong correlation between in vitro ribosome binding and in vivo expression, for both the wild type and mutants, suggested that the downstream adenines exerted their effect at the initiation stage rather than the elongation stage.
It is possible that the adenine effects seen here were due to the structure within the TIR. It has been reported that the secondary structure within the TIR may inhibit expression, whereas a more open TIR with less structure allows increased expression (6, 7). In an effort to explore the possible contribution of structure to our results, various lengths of the cysJ-, pncB-, and aroL-lacZ mRNAs, with or without mutations, were subjected to analysis with MFOLD [version 5.a (17:13); M. Zuker, Rensselaer Polytechnic Institute, Troy, NY (http://bioweb.pasteur.fr/cgi-bin/seqanal/mfold.pl.)] for prediction of mRNA secondary structures by energy minimization. While slight variations in the predicted structures resulted from the downstream base substitutions, a general correlation of more closed or open structures with corresponding decreases or increases in expression and ribosome binding was not observed (data not shown). In addition, a comparison of the signals resulting from reverse transcriptase pausing or terminating at inhibitory secondary structures during toeprint assays did not suggest significant differences in structure between wild-type and mutant mRNAs (Fig. 3 to 5). Another possibility involving structure is the formation of adenine-rich pseudoknots, described previously as high-affinity RNA ligands to 30S subunits and ribosomal protein S1 (23). While a contribution by downstream adenines to a stimulatory secondary structure has not been discounted, we do not have any data to support this model.
It is also possible that downstream adenines, perhaps in association with pyrimidines (16), provide a region within the RBS that is recognized specifically by ribosome-associated proteins that promote initiation complex formation. For example, the 30S subunit protein S1 is a strong RNA binding protein reported to concentrate mRNA near the ribosome decoding site (1, 35, 36). Previous studies revealed the binding of S1 to various nucleotide motifs within the untranslated leader, including an AU-rich omega sequence (1, 10, 14, 15), a CAU-rich omega-like sequence (38), and a (CAA)n repeat (38). Based on these data it is conceivable that S1 could bind downstream A-rich regions for more efficient delivery of mRNA to the ribosome decoding site; however, there has been no direct evidence that S1 binds specific nucleotide motifs downstream of the start codon. Interaction between downstream A-rich regions and 16S rRNA might also contribute to mRNA-ribosome association. In possible support of this, poly(A) RNA formed cross-links to 16S rRNA nucleotides 1394 to 1399, located adjacent to the ribosomal P-site (34).
Crystal structures of ribosome-mRNA complexes provide evidence that mRNA positions −3 to 10 (with the A residue of the AUG start codon as position 1) are wrapped closely around the neck of the 30S subunit, with most ribosome contacts involving the mRNA backbone rather than its bases (41). However, these crystal structures represent stable complexes and provide little information about interactions occurring during ribosome loading that lead to a stable complex.
Recent experiments in our lab also suggested that there is contact between a natural adenine-rich sequence of a leaderless mRNA and proteins associated with E. coli 30S subunits. In this work, leaderless mRNA encoded by the cI gene of bacteriophage lambda, with a photoactivatable 4-thiouridine in the AUG start codon, was cross-linked to 30S subunit proteins (J. E. Brock and G. R. Janssen, unpublished data). However, cross-linking was inhibited by a competitor cI mRNA that lacked a start codon but contained the natural adenine-rich sequence immediately downstream of the start codon. Also, the same cI competitor mRNA inhibited ribosome binding to wild-type cI mRNA in toeprint assays, suggesting that the adenines contribute to an interaction between mRNA and components of the ribosome that facilitate ternary complex formation. Additional support for downstream adenine stimulation of leaderless mRNA translation in E. coli was provided by a report (16) which showed that addition of CA multimers increased expression from leaderless lacZ, gusA, and neo mRNAs.
Implications and application of downstream adenines for expression.
Although the mechanistic details of adenine-enhanced ribosome binding and expression remain to be determined, the ability to stimulate translation from leaderless mRNA (16) indicates that an untranslated leader or SD-ASD interaction is not required. Even in the presence of a canonical SD-ASD interaction, downstream adenines significantly influenced ribosome binding and expression from aroL, pncB, and cysJ mRNAs. The first five codons of coding sequence are protected by a ribosome during formation of an initiation complex and thereby represent potential contacts that could contribute to an mRNA's ribosome binding strength; however, constraints imposed by the encoded amino acid sequence have made it difficult to consider sequence-specific translation enhancers in this region of the mRNA. Genetic code degeneracy and “wobble position” variability, however, could allow a bias toward adenines within the first few codons while imposing minimal constraints on the encoded amino acids. Downstream adenines, possibly in conjunction with the start codon or other features of the mRNA, might present a recognition motif (sequence and/or structural) to a component of the translational machinery and contribute importantly to the ribosome binding strength, with the number and position of adenines allowing “fine-tuning” of expression.
If downstream adenines merely provide an open structure for access of the start codon to ribosomes, then one might expect adenines to stimulate expression in a variety of translation systems; however, addition of downstream adenines to a neo reporter gene did not increase expression in the G+C-rich, gram-positive organism Streptomyces lividans (J. M. Day and G. R. Janssen, unpublished data). The possibility that adenine stimulation might not occur in G+C-rich organisms supports the hypothesis that the enhanced ribosome binding and expression are at least partially based on sequence and suggests that adenine stimulation of translation might be limited to E. coli and related organisms. Using cross-linking techniques to identify downstream adenine-ribosome interactions, along with further structural studies, may provide evidence that distinguishes between sequence and structural components of adenine stimulation.
Characterization of the adenine effect might also allow simple engineering of expression levels, up or down, by increasing or decreasing the number of downstream adenines. Identification of mRNA features contributing to ribosome binding and translation, from coding sequences with or without SD sequences (2), is essential for understanding these important stages of gene expression in E. coli and other organisms.
Acknowledgments
This work was supported by grant GM065120 from the National Institutes of Health. J.E.B. thanks the Miami University Graduate School for a Graduate Student Achievement Award and the Center for Bioinformatics and Functional Genomics for a summer research fellowship.
We especially thank Eileen Bridge for helpful comments on the manuscript and Holly Rovito and Michael Day for inspiring discussions and assistance with assay development.
Footnotes
Published ahead of print on 3 November 2006.
REFERENCES
- 1.Boni, I. V., D. M. Isaeva, M. L. Musychenko, and N. V. Tzareva. 1991. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 19:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, B., S. Halgamuge, and S. Tang. 2006. Analysis of SD sequences in completed microbial genomes: non-SD-led genes are as common as SD-led genes. Gene 373:90-99. [DOI] [PubMed] [Google Scholar]
- 3.Chen, H., L. Pomeroy-Cloney, M. Bjerknes, J. Tam, and E. Jay. 1994. The influence of adenine rich motifs in the 3′ portion of the ribosome binding site on human IFN-γ gene expression in Escherichia coli. J. Mol. Biol. 240:20-27. [DOI] [PubMed] [Google Scholar]
- 4.Day, M. J., and G. R. Janssen. 2004. Isolation and characterization of ribosomes and translational initiation factors from the gram-positive soil bacterium Streptomyces lividans. J. Bacteriol. 186:6864-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeFeyter, R. C., B. E. Davidson, and J. Pittard. 1986. Nucleotide sequence of the transcription unit containing the aroL and aroM genes from Escherichia coli K-12. J. Bacteriol. 165:233-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Smit, M. H., and J. van Duin. 1990. Control of prokaryotic translational initiation by mRNA secondary structure. Proc. Nucleic Acid Res. Mol. Biol. 38:1-35. [DOI] [PubMed] [Google Scholar]
- 7.de Smit, M. H., and J. van Duin. 1990. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc. Natl. Acad. Sci. USA 87:7668-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dreyfus, M. 1988. What constitutes the signal for the initiation of protein synthesis on Escherichia coli mRNAs? J. Mol. Biol. 204:79-94. [DOI] [PubMed] [Google Scholar]
- 9.Etchegaray, J. P., and M. Inouye. 1999. Translational enhancement by an element downstream of the initiation codon in Escherichia coli. J. Biol. Chem. 274:10079-10085. [DOI] [PubMed] [Google Scholar]
- 10.Gallie, D. R., D. E. Sleat, J. W. Wyatts, P. C. Turner, and T. M. A. Wilson. 1987. The 5 prime leader sequence of tobacco mosaic virus RNA enhances the expression of foreign gene transcripts in vitro and in vivo. Nucleic Acids Res. 15:3257-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gualerzi, C. O., L. Brandi, E. Caserta, A. La Teana, R. Spurio, J. Tomsic, and C. L. Pon. 2000. Translation initiation in bacteria, p. 477-494. In R. A. Garrett, S. R. Douthwaite, A. Liljas, A. T. Matheson, P. B. Moore, and H. F. Noller (ed.), The ribosome: structure, function, antibiotics, and cellular interactions. ASM Press, Washington, DC.
- 12.Ivanov, I. G., R. Alexandrova, B. Dragulev, D. Leclerc, A. Saraffova, V. Maximova, and M. G. Abouhaidar. 1992. Efficiency of the 5′-terminal sequence (omega) of tobacco mosaic virus RNA for the initiation of eukaryotic gene translation in Escherichia coli. Eur. J. Biochem. 209:151-156. [DOI] [PubMed] [Google Scholar]
- 13.Janssen, G. R. 1993. Eubacterial, archaeabacterial, and eukaryotic genes that encode leaderless mRNA, p. 59-67. In R. Baltz, G. D. Hegeman, and P. L. Skatrud (ed.), Industrial microorganisms: basic and applied molecular genetics. American Society for Microbiology, Washington, DC.
- 14.Komarova, A. V., L. S. Tchufistova, E. V. Supina, and I. V. Boni. 2002. Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA 8:1137-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komarova, A. V., L. S. Tchufistova, M. Dreyfus, and I. V. Boni. 2005. AU-rich sequences within 5′ untranslated leaders enhance translation and stabilize mRNA in Escherichia coli. J. Bacteriol. 187:1344-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin-Farmer, J. A., and G. R. Janssen. 1999. A downstream CA repeat sequence increases translation from leadered and unleadered mRNA in Escherichia coli. Mol. Microbiol. 31:1025-1038. [DOI] [PubMed] [Google Scholar]
- 17.Miller, J. (ed.). 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Moll, I., S. Grill, C. O. Gualerzi, and U. Blasi. 2002. Leaderless mRNAs in bacteria: surprises in ribosomal recruitment and translational control. Mol. Microbiol. 43:239-246. [DOI] [PubMed] [Google Scholar]
- 19.O'Conner, M., T. Asai, C. L. Squires, and A. E. Dahlberg. 1999. Enhancement of translation by the downstream box does not involve base pairing of mRNA with the penultimate stem sequence of 16S rRNA. Proc. Natl. Acad. Sci. USA 96:8973-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Donnell, S. M., and G. R. Janssen. 2001. The initiation codon affects ribosome binding and translational efficiency in Escherichia coli of cI mRNA with or without the 5′ untranslated leader. J. Bacteriol. 183:1277-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olins, P. O., and H. S. Rangwala. 1989. A novel sequence element derived from bacteriophage T7 mRNA acts as an enhancer of translation of the lacZ gene in E. coli. J. Mol. Biol. 264:16973-16976. [PubMed] [Google Scholar]
- 22.Ostrowski, J., and N. M. Kredich. 1989. Molecular characterization of the cysJIH promoters of Salmonella typhimurium and Escherichia coli: regulation by cysB protein and N-acetyl-l-serine. J. Bacteriol. 171:130-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ringquist, S., T. Jones, E. E. Snyder, T. Gibson, I. Boni, and L. Gold. 1995. High-affinity RNA ligands to Escherichia coli ribosomes and ribosomal protein S1: comparison of natural and unnatural binding sites. Biochemistry 34:3640-3648. [DOI] [PubMed] [Google Scholar]
- 24.Rudd, K. E., and T. D. Schneider. 1992. Compilation of E. coli ribosome binding sites, p. 17.19-17.46. In J. Miller (ed.), A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Sato, T., M. Terabe, H. Watanabe, T. Gojobori, C. Hori-Takemoto, and K. Miura. 2001. Codon and base biases after the initiation codon of the open reading frames in the Escherichia coli genome and their influence on the translation efficiency. J. Biochem. 129:851-860. [DOI] [PubMed] [Google Scholar]
- 27.Scherer, G. F. E., M. D. Walkinshaw, S. Arnott, and D. J. Morr. 1980. The ribosome-binding sites recognized by E. coli ribosomes have regions with signal character in both the leader and protein coding segments. Nucleic Acids Res. 8:3895-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schleif, R. 1972. Fine-structure deletion map of the Escherichia coli l-arabinose operon. Proc. Natl. Acad. Sci. USA 11:3479-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shine, J., and L. Dalgarno. 1974. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 71:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slovek, L. E. 1999. Ribosomal binding and enhanced translation of CA-containing mRNA in Escherichia coli. M.S. thesis. Miami University, Oxford, OH.
- 31.Spedding, G., T. Gluick, and D. Draper. 1993. Ribosome initiation complex formation with the pseudoknotted α operon messenger RNA. J. Mol. Biol. 229:609-622. [DOI] [PubMed] [Google Scholar]
- 32.Sprengart, M. L., H. P. Farscher, and E. Fuchs. 1990. The initiation of translation in E. coli: apparent base pairing between the 16S rRNA and the downstream sequences of the mRNA. Nucleic Acids Res. 18:1719-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steitz, J. A. 1969. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature 224:957-964. [DOI] [PubMed] [Google Scholar]
- 34.Stiege, W., K. Stade, D. Schuller, and R. Brimacombe. 1988. Covalent cross-linking of poly (A) to Escherichia coli ribosomes, and localization of the cross-link site within the 16S rRNA. Nucleic Acids Res. 16:2369-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian, A. R. 1984. Structure and function of the largest Escherichia coli ribosomal protein. Trends Biochem. Sci. 9:491-494. [Google Scholar]
- 36.Suryanarayana, T., and A. R. Subramanian. 1983. An essential function of ribosomal protein S1 in messenger ribonucleic acid translation. Biochemistry 22:2715-2719. [DOI] [PubMed] [Google Scholar]
- 37.Thanaraj, T. A., and M. W. Pandit. 1989. An additional ribosome binding site on mRNA of highly expressed genes and a bifunctional site on the colicin fragment of 16S rRNA from E. coli: important determinants of the efficiency of translation initiation. Nucleic Acids Res. 18:2973-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tzareva, N. V., V. I. Makhno, and I. V. Boni. 1994. Ribosome-messenger recognition in the absence of the Shine-Dalgarno interactions. FEBS Lett. 337:189-194. [DOI] [PubMed] [Google Scholar]
- 39.Wade, H. E., and H. K. Robinson. 1966. Magnesium ion-independent ribonucleic acid depolymerases in bacteria. Biochem. J. 101:467-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wubbolts, M. G., P. Terpstra, J. B. van Beilen, J. Kingma, H. A. R. Meesters, and B. Witholt. 1990. Variation of cofactor levels in Escherichia coli. J. Biol. Chem. 265:17665-17672. [PubMed] [Google Scholar]
- 41.Yusupova, G. Z., M. M. Yusupova, J. H. D. Cate, and H. F. Noller. 2001. The path of messenger RNA through the ribosome. Cell 106:233-241. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, J., and M. P. Deutscher. 1992. A uridine rich sequence required for translation of prokaryotic mRNA. Proc. Natl. Acad. Sci. USA 89:2605-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]