Abstract
Recovery from acute hepatitis B virus (HBV) infection requires a broad, vigorous T-cell response, which is enhanced in mice when chemokine receptor 5 (CCR5) is missing. To test the hypothesis that production of a nonfunctional CCR5 (CCR5Δ32 [a functionally null allele containing a 32-bp deletion]) increases the likelihood of recovery from hepatitis B in humans, we studied 526 persons from three cohorts in which one person with HBV persistence was matched to two persons who recovered from an HBV infection. Recovery or persistence was determined prior to availability of lamivudine. We determined genotypes for CCR5Δ32 and for polymorphisms in the CCR5 promoter and in coding regions of the neighboring genes, chemokine receptor 2 (CCR2) and chemokine receptor-like 2 (CCRL2). Allele and haplotype frequencies were compared among the 190 persons with viral recovery and the 336 with persistence by use of conditional logistic regression. CCR5Δ32 reduced the risk of developing a persistent HBV infection by nearly half (odds ratio [OR], 0.53; 95% confidence interval [CI], 0.33 to 0.83; P = 0.006). This association was virtually identical in persons with and without a concomitant human immunodeficiency virus infection. Of the nine individuals who were homozygous for the deletion, eight recovered from infection (OR, 0.25; 95% CI, 0.03 to 1.99; P = 0.19). None of the other neighboring polymorphisms examined were associated with HBV outcome. These data demonstrate a protective effect of CCR5Δ32 in recovery from an HBV infection, provide genetic epidemiological evidence for a role of CCR5 in the immune response to HBV, and suggest a potential therapeutic treatment for patients persistently infected with HBV.
Most adults infected with hepatitis B virus (HBV) recover and develop protective antibodies. However, approximately 5% of adults remain chronically infected with HBV and are at risk for developing end-stage liver disease and hepatocellular carcinoma (20). Hepatitis B recovery occurs more often in individuals who develop a broad and strong T-cell response than in those with weak and narrowly focused responses (28).
CCR5 (chemokine receptor 5) is a CC chemokine receptor expressed by granulocytes, macrophages, immature dendritic cells, CD8+ lymphocytes, and Th1 lymphocytes, and it influences their migration and activation (36). It has also been identified as a coreceptor for human immunodeficiency virus (HIV) entry into host cells (37). The CCR5 gene consists of a single coding exon, and a functionally null allele containing a 32-bp deletion (CCR5Δ32) has been identified in 10 to 15% of Caucasians (21). Individuals heterozygous for the deletion (CCR5Δ32/+) have substantially reduced levels of CCR5 on the cell surface compared to those who are homozygous for the normal CCR5 allele (+/+) (38).
One consequence of CCR5 deficiency in mice is a more robust T-cell response to several infectious agents. For example, CCR5-deficient mice (ccr5−/−) infected with Mycobacterium tuberculosis have more dendritic cells in lymph nodes and increased pulmonary inflammation due to a greater T-cell response than ccr5+/+ mice (2). Similar data have been reported for mouse models of Listeria and lymphocytic choriomeningitis virus infections, as well as for enhanced responses to a melanoma dendritic cell vaccine (25, 26, 39). How CCR5 restrains the immune response is unknown.
Given the importance of the T-cell response in hepatitis B recovery and the increased T-cell response to various antigens observed with ccr5−/− mice, we hypothesized that humans with CCR5Δ32 would be more likely to recover from acute hepatitis B. We tested this hypothesis by genotyping CCR5 in a large cohort of Caucasian HBV-infected individuals who were well-characterized as having either HBV recovery or persistence.
MATERIALS AND METHODS
Study participants.
Since CCR5Δ32 is present only in Caucasians, we studied Caucasian persons who were participating in one of the following ongoing studies: (i) the Multicenter AIDS Cohort Study (MACS), which is a study of 5,622 gay men enrolled in one of four U.S. cities between 1984 and 1985 and between 1987 and 1991 (18), (ii) the Multicenter Hemophilia Cohort Study (MHCS), a prospectively followed cohort of patients with hemophilia, von Willebrand's disease, or a related coagulation disorder from 16 comprehensive hemophilia treatment centers enrolled between 1982 and 1986, as previously described (11), or (iii) the Hemophilia Growth and Development Study (HGDS), which is a continuing study of 333 children and adolescents with hemophilia enrolled between March 1989 and May 1990 (13). The majority of the subjects were from the MACS cohort (80%), with the HGDS and the MHCS each contributing 10%. Informed consent was obtained from all participants.
To investigate our hypothesis, a nested case-control design was used. In this design, each individual from one of the above-described cohorts who had a persistent hepatitis B infection was matched to two persons from the same cohort who recovered from an HBV infection but were otherwise similar with regard to nongenetic factors. Matching criteria included geographic location and factors that have been associated with HBV recovery, including age within 10 years, gender, and HIV type 1 (HIV-1) status (16). In this study, for 44 persistently infected persons, only one match was available. There were no persistently infected persons included who did not have at least one match.
Subjects were considered persistently infected with HBV if their serum or plasma tested positive for hepatitis B surface antigen (HBsAg) at two visits separated by a minimum of 6 months. Testing for antibodies against hepatitis B core antigen (anti-HBc) and HBsAg (anti-HBs) was performed as needed to exclude primary HBV infection. Individuals with HBV recovery were positive for anti-HBc and anti-HBs without the presence of HBsAg at two time points separated by a minimum of 6 months. All initial tests were performed on serum at the time of entry into the cohort study. HBV status of HIV-positive subjects was determined before antiretroviral therapy (including lamivudine) was available.
Serologic testing.
All serum specimens were stored at −70°C prior to testing. HIV-1 antibody testing was done by enzyme immunoassay with reactive results confirmed as positive by Western blotting (11, 18). HBsAg, anti-HBs, and anti-HBc testing was done by using commercially available kits according to the manufacturer's specifications (AUSZYME, AUSAB, and CORZYME, respectively; Abbott Laboratories, Abbott Park, IL).
DNA extraction and genotyping.
For each individual, Epstein-Barr virus-transformed cell lines were established, and genomic DNA was extracted from these cell lines by use of phenol-chloroform.
CCR5Δ32 genotyping was performed using PCR amplification of genomic DNA with primers CCR-5A, 5′-AGGTCTTCATTACACCTGCAGC-3′, and CCR-5B, 5′-CCTCTCATTTCGACACCGAAGC-3′. The PCR products were separated by electrophoresis in a 3% agarose gel (169 bp for the wild-type allele and 137 bp for the 32-bp-deletion allele). CCR5 promoter polymorphism 59029 (rs1799987, CCR5-2459) was determined by restriction fragment length polymorphism as previously described (3).
To determine if the association with CCR5Δ32 was due to linkage disequilibrium with a neighboring gene, we also genotyped polymorphisms leading to nonsynonymous changes in adjacent genes CCR2 (chemokine receptor 2) and CCRL2 (chemokine receptor-like 2). The CCR2-64I polymorphism (rs1799864) was defined using the Assay-by-Design service from Applied Biosystems under conditions recommended by the manufacturer (forward primer, 5′-TCTTTGGTTTTGTGGGCAACATG-3′; reverse primer, 5′-AGGTAAATGTCAGTCAAGCACTTCA-3′; probe, 5′-TGGTCA[G]TCCTCATC-3′). Three CCRL2 polymorphisms, T500A (rs3204849), G502A (rs6441977), and A727G (rs3204850), for which the minor allele at each position had a frequency of >5%, were genotyped using an AcycloPrime-FP single-nucleotide polymorphism (SNP) detection assay (Perkin Elmer, Boston, MA), a single-base extension method performed according to the manufacturer's specifications (15). In this method, a 200-bp fragment containing the SNP of interest is amplified (primers available upon request) with cycling conditions of 95°C for 10 min, 35 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 60 s, and then a final extension step of 72°C for 10 min. After amplification, the excess primer and deoxynucleoside triphosphates were degraded with shrimp alkaline phosphatase and exonuclease I used according to the manufacturer's specifications. These enzymes are heat inactivated, allowing for the final step, which adds one of two fluorescent terminators, representing the alleles present at the SNP of interest, to a primer ending immediately upstream of the SNP site (extension primer). The cycling protocol for the single base extension step is 95°C for 2 min followed by 15 cycles of 95°C for 15 s and 55°C for 30 s. The results are identified by the amount of fluorescence polarization of each allele as determined by a Victor2V instrument (Perkin Elmer, Boston, MA). In order to determine the accuracy of this method, we verified the results with direct sequencing in 15 samples for each SNP and found no errors.
All polymorphisms in this study were in Hardy-Weinberg equilibrium, which was assessed for each SNP/deletion by using the chi-square test with 1 degree of freedom.
Statistical analysis.
Allele frequencies for CCR5Δ32 were calculated and compared between those persons with viral recovery and persistence by use of conditional logistic regression (SAS version 10; SAS, Cary, NC). An odds ratio (OR) of >1 was associated with persistence and <1 with recovery. Haplotypes were inferred from the genotypes in the study population and were constructed using a program designed for population-based studies, PHASE v 2.0 (http://www.stat.washington.edu/stephens/software.html) (30). A P value of <0.05 was considered significant in all analyses, and any genotype that met this criterion was stratified by HIV status to exclude an HIV association.
RESULTS
This study included 526 Caucasian subjects who had been infected with HBV, of whom 190 were chronically infected and were matched to 336 individuals who had recovered from the infection. No significant differences were detected between cases and controls in terms of nongenetic factors, with a median age of 31.5, 100% male, and 69% HIV infected.
CCR5Δ32 is associated with recovery from HBV infection.
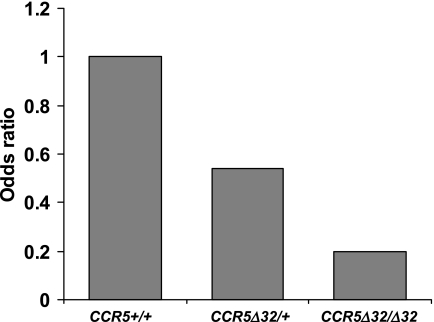
The overall allele frequency of CCR5Δ32 for all study subjects was 11%, which is consistent with its known population frequency (7). CCR5Δ32 was more frequent in those subjects who recovered from infection than in those with a persistent HBV infection (Table 1) . In fact, based on an allelic analysis, CCR5Δ32 reduced the risk of developing a persistent hepatitis B infection by nearly half (OR, 0.53; 95% confidence interval [CI], 0.33 to 0.83; P = 0.006). In addition to matching cases and controls by HIV status, we stratified the analyses by HIV status to rule out any possibility that HIV infection could confound our analysis; notably, CCR5Δ32 showed virtually identical levels of protection in the HIV-negative pairs and in the HIV-positive pairs (OR, 0.49 and 0.54, respectively). Of the nine individuals who were homozygous for the deletion (CCR5Δ32/Δ32), eight recovered from infection, suggesting strong protection against HBV persistence in the absence of functional CCR5 molecules altogether (OR, 0.25; 95% CI, 0.03 to 1.99; P = 0.19). Thus, CCR5Δ32 appears to have a codominant effect on recovery in that the likelihood of recovery among heterozygotes was intermediate compared to the likelihoods for the CCR5Δ32/Δ32 and CCR5+/+ genotypes (Fig. 1). Viral persistence was more common among individuals with the CCR5+/+ genotype (OR, 1.95; 95% CI, 1.17 to 3.23; P = 0.009) than among individuals with at least one copy of CCR5Δ32.
TABLE 1.
Distribution of CCR5Δ32 and CCR5−2459G promoter variant, stratified by HBV recovery and persistence
| Genotype | % of alleles or subjects with genotypea
|
ORb | 95% CI | P | |
|---|---|---|---|---|---|
| HBV recovery (n = 336) | HBV persistence (n = 190) | ||||
| CCR5Δ32 | 13.0* | 7.4* | 0.53 | 0.33-0.83 | 0.006 |
| CCR5+/+ | 76.4** | 85.7** | 1.95 | 1.18-3.23 | 0.009 |
| −2459G | 42.0* | 45.4* | 1.14 | 0.88-1.48 | 0.34 |
| −2459GG | 19.1** | 22.0** | 1.23 | 0.77-1.96 | 0.38 |
*, values represent percentages of alleles (chromosomes); **, values represent percentages of subjects with genotype.
An odds ratio of >1 is associated with viral persistence, and an odds ratio of <1 is associated with viral recovery.
FIG. 1.
Graphic representation of odds ratios for recovery from HBV infection based on different CCR5 genotypes. Odds ratios were calculated using a model in which the CCR5+/+ genotype (homozygous wild type) was the reference group (set to an odds ratio of 1) and the genotypes CCR5Δ32/+ (OR, 0.54; 95% CI, 0.32 to 0.90) and CCR5Δ32/Δ32 (OR, 0.20; 95% CI, 0.03 to 1.61) were separate terms. Odds ratios of <1 favor viral recovery.
The A−2459G CCR5 promoter variant (−2459G) has been associated with slower AIDS progression (5) and may decrease CCR5 transcription (23). Thus, we tested whether this promoter variant was also associated with HBV recovery. In our study sample, the −2459G variant was in complete linkage disequilibrium with the CCR5+ allele and thus was never on the same haplotype as CCR5Δ32. Univariate analysis did not demonstrate a difference in allele frequency of the promoter variant between those subjects who recovered from HBV infection and those who did not (Table 1). Similarly, homozygosity for either the variant or the wild type was not associated with HBV outcome. A multivariate model that included CCR5Δ32 and −2459G as covariables in the analysis did not reveal any association with the promoter variant, indicating that the strong association of CCR5Δ32 with HBV recovery did not mask a CCR5 promoter association.
Polymorphisms in genes neighboring CCR5Δ32 are not associated with HBV outcome.
In order to ascertain whether the association between HBV recovery and CCR5Δ32 could be due to a neighboring locus in strong linkage disequilibrium with CCR5Δ32, nonsynonymous SNPs within the two neighboring genes, CCR2 and CCRL2, were typed in our cohort. A previously identified polymorphism in CCR2, which involves an amino acid change from valine to isoleucine at position 64 (CCR2-64I), has been associated with slower progression to AIDS, suggesting an effect on CCR2 function (29), and was therefore genotyped in our HBV samples. Univariate analysis of CCR2-64I revealed a trend with HBV recovery, although it was not significant (OR, 0.72; 95% CI, 0.48 to 1.07; P = 0.11) (Table 2) . CCR2-64I was always present on a CCR5+ haplotype, similarly to results from previous studies (29), and thus cannot explain the association of HBV recovery with CCR5Δ32. To determine whether the strong association of CCR5Δ32 with viral clearance masked an association with CCR2-64I, we constructed a multivariate model with CCR5Δ32 and CCR2-64I as covariables. The weak association with CCR2-64I was not strengthened under this model, nor did it alter the CCR5Δ32 association.
TABLE 2.
Percentages of alleles with indicated CCR2 or CCRL2 polymorphisms and percentages of indicated CCRL2-CCR5 haplotypes in subjects with HBV recovery and persistence
| Genotype or haplotype | % of alleles or subjects with haplotype or genotypea
|
ORb | 95% CI | P | |
|---|---|---|---|---|---|
| HBV recovery | HBV persistence | ||||
| Genotype | |||||
| CCR2-64I | 11.1* | 8.5* | 0.72 | 0.48-1.07 | 0.11 |
| CCRL2-500T | 62.8* | 55.8* | 0.74 | 0.54-1.00 | 0.05 |
| CCRL2-502A | 18.8* | 11.2* | 0.50 | 0.32-0.80 | 0.003 |
| CCRL2−727G | 8.2* | 6.9* | 0.80 | 0.45-1.42 | 0.45 |
| CCRL2-CCR5 haplotype | |||||
| 500T-WTc | 48.3** | 48.8** | 1.00 | 0.73-1.38 | 0.98 |
| 500T-Δ32 | 14.2** | 7.3** | 0.44 | 0.25-0.78 | 0.005 |
| 502A-WT | 5.8** | 4.4** | 0.70 | 0.33-1.49 | 0.36 |
| 502A-Δ32 | 13.6** | 7.3** | 0.47 | 0.26-0.84 | 0.01 |
*, values represent percentages of alleles; **, values represent percentages of subjects with haplotype. For CCRL2 polymorphisms, there were 244 subjects with HBV recovery and 138 with HBV persistence, since these polymorphisms were tested only in a subset of subjects. For CCR2, there were 336 subjects with viral recovery and 190 with viral persistence.
An odds ratio of >1 is associated with viral persistence, and an odds ratio of <1 is associated with viral recovery.
WT, wild-type CCR5.
The three nonsynonymous coding region polymorphisms in CCRL2 that had frequencies of >5% (T500A, G502A, and A727G) were genotyped in a subset of subjects (n = 382). In univariate analysis, the 500T and 502A alleles were associated with HBV recovery (OR, 0.74; 95% CI, 0.54 to 1.00; P = 0.05 and OR, 0.50; 95% CI, 0.32 to 0.80; P = 0.003, respectively) (Table 2). However, haplotype analyses indicated that these associations were due to linkage disequilibrium with CCR5Δ32. Haplotypes containing either the 500T or the 502A variant without CCR5Δ32 were not associated with HBV recovery. Neither the wild type nor the variant of CCRL2 at −727 was associated with HBV outcomes. Stratification by HIV status revealed no difference between the HIV-positive and HIV-negative subjects in these analyses.
DISCUSSION
These data demonstrate a protective effect of CCR5Δ32 in recovery from an HBV infection, which is unlikely to involve neighboring loci in linkage disequilibrium with CCR5. We found that individuals who have at least one copy of the gene encoding a nonfunctional receptor (CCR5Δ32) are twice as likely to recover from hepatitis B, and the protective effect appears to be codominant. Based on data from ccr5−/− mouse models, there are at least two potential explanations for these observations. First, several studies demonstrate that CCR5-deficient mice have increased CD4+ and CD8+ T-cell responses to a variety of antigens and to a dendritic cell vaccine (25, 26, 39). Such findings suggest that CCR5 may behave as a negative regulator of T cells in an immune response. Data from a cohort of hemophiliacs support this increase in T-cell responses with CCR5 deficiency because CCR5Δ32 homozygous individuals had a 30% increase in lymphocytes compared to CCR5+/+ individuals (27). Within the hepatitis C-infected subgroup of these hemophiliacs, the CCR5Δ32 homozygous individuals had a 117% increase in alanine aminotransferase, a marker of liver inflammation, compared to levels for those without CCR5Δ32, suggesting a greater T-cell-dependent inflammatory response among individuals with low/no CCR5. Thus, CCR5-mediated attenuation of the immune response may increase the risk of HBV persistence. Second, based on a concanavalin A (ConA)-induced fulminant hepatitis murine model, which is a model of T-cell-mediated hepatitis, CCR5 deficiency prevents hepatic natural killer T (NKT) cell apoptosis and upregulates NKT cell function (1), phenomena that would favor recovery from HBV infection since NK and NKT cells are important in controlling HBV replication in HBV transgenic mouse models (17). A second study of ConA-induced hepatitis demonstrated that CCL5, which is the ligand for CCR5 and is increased in CCR5 deficiency, enhanced recruitment of chemokine receptor 1 (CCR1)-expressing CD4+ T cells, NKT cells, and macrophages into the liver (24). These CCR1-expressing CD4+ T cells produce gamma interferon during ConA-induced hepatitis in normal mice, and if this occurs during hepatitis B infection, it would also increase the likelihood of recovery from hepatitis B (12). Our data also suggest that there is a gene dosage effect, since one copy of CCR5Δ32 was intermediate in protective effect compared to zero and two copies; however, mice heterozygous for the ccr5 deletion have not been studied to confirm this.
Given these seemingly functional benefits of missing CCR5, its absence may have a dual protective effect against HIV: decreased viral entry into host cells and increased innate/acquired immunity against the virus. The latter effect may be operative against a number of viral infections, including HBV. On the other hand, increased susceptibility to West Nile virus both in mice that are CCR5 deficient and in humans who have the CCR5Δ32 allele has also been reported previously (9, 10). Thus, CCR5 deficiency appears to impact the response to a variety of infections in ways that are not necessarily easy to predict, possibly due to differences in pathogenic mechanisms employed by different infectious organisms. Given the effects of CCR5 on such a broad range of pathogens, perhaps the force that presumably drove CCR5Δ32 to its present frequency in Caucasians was actually a diverse group of infectious diseases rather than a single, deadly pathogen.
CCR5 has been examined in one other study, involving an HBV-infected Korean cohort that enrolled 377 individuals, of whom 138 spontaneously recovered from infection (6). CCR5Δ32 was absent from this cohort, but an association between the promoter variant CCR5−2459G, which might confer lower promoter activity (23), and HBV recovery was reported. This finding is consistent with a protective effect of lower CCR5 levels, as is suggested in our study, although we did not observe a protective effect of this variant in our study.
HIV coinfection in approximately 70% of our subjects was carefully considered in our study, since CCR5Δ32 is known to protect against AIDS progression. We conclude that the effect of CCR5Δ32 on HIV infection cannot explain or confound the effect of this allele on HBV clearance reported herein, for several definitive reasons: (i) stratification of the analysis by HIV status demonstrated an identical protective association of CCR5Δ32 in HIV-negative and -positive subjects, (ii) those with viral recovery and persistence were matched on HIV status in our study, and (iii) HBV infection occurred prior to HIV infection in nearly all cases, so the outcome of the viral hepatitis infection was determined prior to acquiring HIV (19). For those cases in which the HIV infection did occur first, it is unlikely that immunosuppression from HIV played a significant role in the results since the HBV status was determined at study entry, which is prior to profound HIV-induced immunosuppression. Also, the status of the HBV infection was established prior to the availability of oral antiretroviral agents with activity against both HIV and HBV; thus, none of the individuals had been treated for HIV or HBV at the time the HBV infection status was determined.
Even though CCR5Δ32 is associated with recovery from HBV infection, the majority of people who recovered from infection in this study did not have this deletion. This is expected since recovery from hepatitis B is certainly polygenic, so CCR5Δ32 is one of several genes involved in HBV pathogenesis. Although other relevant genes have been identified previously (4, 14, 31-35), this deletion is important since it is present in 13% of people who recovered and it has one of the strongest odds ratios published to date.
The fact that our findings support data from mouse models showing an increased T-cell response to other infectious agents with CCR5 deficiency is particularly intriguing because it offers a potential means to enhance the T-cell response to HBV by blocking CCR5. Current therapeutic options (nucleoside [or nucleotide] analogues) to treat chronic hepatitis B led to a therapeutic response in only a minority of treated individuals (8, 22). Thus, simultaneous administration of a CCR5 blocking agent, which is already being developed for HIV infection, along with an anti-HBV agent may increase HBV clearance through augmentation of the T-cell response to HBV antigens.
Acknowledgments
This work was supported by NIH grant DA00441. C.L.T. was additionally supported in part by the Investigators in the Pathogenesis of Infectious Diseases Award from the Burroughs Wellcome Fund. The MACS (http://www.statepi.jhsph.edu/macs/macs.html) is funded by the National Institute of Allergy and Infectious Diseases, with additional supplemental funding from the National Cancer Institute and the National Heart, Lung, and Blood Institute (UO1-AI-35042, 5-MO1-RR-00722 [GCRC], UO1-AI-35043, UO1-AI-37984, UO1-AI-35039, UO1-AI-35040, UO1-AI-37613, and UO1-AI-35041). The MHCS is supported by National Cancer Institute contract N01-CP-33002 along with the Research Triangle Institute. The HGDS is supported by the Bureau of Maternal and Child Health and Resources Development (MCJ-060570), the National Institute of Child Health and Human Development (NO1-HD-4-3200), the Centers for Disease Control and Prevention, and the National Institute of Mental Health. Additional support has been provided by grants from the National Center for Research Resources of the National Institutes of Health to the New York Hospital-Cornell Medical Center Clinical Research Center (MO1-RR06020), the Mount Sinai General Clinical Research Center, New York (MO1-RR00071), the University of Iowa Clinical Research Center (MO1-RR00059), and the University of Texas Health Science Center, Houston (MO1-RR02558 and R01-HD-4-1224). This project has been supported in part by the Intramural Research Program and by contract no. NO1-CO-12400 from the National Cancer Institute, National Institutes of Health.
Data in the manuscript were collected by the MACS, with centers (names of principal investigators are in parentheses) at The Johns Hopkins University Bloomberg School of Public Health (Joseph B. Margolick and Lisa Jacobson), Howard Brown Health Center and Northwestern University Medical School (John Phair), University of California, Los Angeles (Roger Detels), and the University of Pittsburgh (Charles Rinaldo).
The authors thank Abbott Laboratories for donating HBsAg and anti-HBs kits and all cohort participants for making this study possible.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, and mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. government.
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Ajuebor, M. N., A. I. Aspinall, F. Zhou, T. Le, Y. Yang, S. J. Urbanski, et al. 2005. Lack of chemokine receptor CCR5 promotes murine fulminant liver failure by preventing the apoptosis of activated CD1d-restricted NKT cells. J. Immunol. 174:8027-8037. [DOI] [PubMed] [Google Scholar]
- 2.Algood, H. M., and J. L. Flynn. 2004. CCR5-deficient mice control Mycobacterium tuberculosis infection despite increased pulmonary lymphocytic infiltration. J. Immunol. 173:3287-3296. [DOI] [PubMed] [Google Scholar]
- 3.An, P., M. P. Martin, G. W. Nelson, M. Carrington, M. W. Smith, K. Gong, et al. 2000. Influence of CCR5 promoter haplotypes on AIDS progression in African-Americans. AIDS 14:2117-2122. [DOI] [PubMed] [Google Scholar]
- 4.Bellamy, R., C. Ruwende, T. Corrah, K. P. McAdam, M. Thursz, H. C. Whittle, et al. 1999. Tuberculosis and chronic hepatitis B virus infection in Africans and variation in the vitamin D receptor gene. J. Infect. Dis. 179:721-724. [DOI] [PubMed] [Google Scholar]
- 5.Carrington, M., M. Dean, M. P. Martin, and S. J. O'Brien. 1999. Genetics of HIV-1 infection: chemokine receptor CCR5 polymorphism and its consequences. Hum. Mol. Genet. 8:1939-1945. [DOI] [PubMed] [Google Scholar]
- 6.Chang, H. Y., S. H. Ahn, D. Y. Kim, J. S. Shin, Y. S. Kim, S. P. Hong, et al. 2005. Association between CCR5 promoter polymorphisms and hepatitis B virus infection. Korean J. Hepatol. 11:116-124. (In Korean.) [PubMed] [Google Scholar]
- 7.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, M. W. Smith, R. G. J. Allikmets, et al. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science 273:1856-1862. [DOI] [PubMed] [Google Scholar]
- 8.Dienstag, J. L., E. R. Schiff, T. L. Wright, R. P. Perrillo, H. W. Hann, Z. Goodman, et al. 1999. Lamivudine as initial treatment for chronic hepatitis B in the United States. N. Engl. J. Med. 341:1256-1263. [DOI] [PubMed] [Google Scholar]
- 9.Glass, W. G., J. K. Lim, R. Cholera, A. G. Pletnev, J. L. Gao, and P. M. Murphy. 2005. Chemokine receptor CCR5 promotes leukocyte trafficking to the brain and survival in West Nile virus infection. J. Exp. Med. 202:1087-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glass, W. G., D. H. McDermott, J. K. Lim, S. Lekhong, S. F. Yu, W. A. Frank, et al. 2006. CCR5 deficiency increases risk of symptomatic West Nile virus infection. J. Exp. Med. 203:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goedert, J. J., C. M. Kessler, L. M. Aledort, et al. 1989. A prospective study of human immunodeficiency virus type 1 infection and the development of AIDS in subjects with hemophilia. N. Engl. J. Med. 321:1141-1148. [DOI] [PubMed] [Google Scholar]
- 12.Guidotti, L. G., and F. V. Chisari. 2001. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 19:65-91. [DOI] [PubMed] [Google Scholar]
- 13.Hilgartner, M. W., S. M. Donfield, A. Willoughby, C. F. Contant, Jr., B. L. Evatt, E. D. Gomperts, et al. 1993. Hemophilia growth and development study. Design, methods, and entry data. Am. J. Pediatr. Hematol. Oncol. 15:208-218. [DOI] [PubMed] [Google Scholar]
- 14.Hohler, T., A. Kruger, G. Gerken, P. M. Schneider, K. H. Meyer zum Buschenefelde, and C. Rittner. 1998. A tumor necrosis factor-alpha (TNF-alpha) promoter polymorphism is associated with chronic hepatitis B infection. Clin. Exp. Immunol. 111:579-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu, T. M., X. Chen, S. Duan, R. D. Miller, and P. Y. Kwok. 2001. Universal SNP genotyping assay with fluorescence polarization detection. BioTechniques 31:560-564. [DOI] [PubMed] [Google Scholar]
- 16.Hyams, K. 1995. Risks of chronicity following acute hepatitis B virus infection: a review. Clin. Infect. Dis. 20:992-1000. [DOI] [PubMed] [Google Scholar]
- 17.Kakimi, K., L. G. Guidotti, Y. Koezuka, and F. V. Chisari. 2000. Natural killer T cell activation inhibits hepatitis B virus replication in vivo. J. Exp. Med. 192:921-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaslow, R. A., D. G. Ostrow, R. Detels, J. P. Phair, B. F. Polk, and C. R. Rinaldo. 1987. The Multicenter AIDS Cohort Study: rationale, organization, and selected characteristics of the participants. Am. J. Epidemiol. 126:310-318. [DOI] [PubMed] [Google Scholar]
- 19.Kingsley, L. A., C. R. Rinaldo, Jr., D. W. Lyter, R. O. Valdiserri, S. H. Belle, and M. Ho. 1990. Sexual transmission efficiency of hepatitis B virus and human immunodeficiency virus among homosexual men. JAMA 264:230-234. [PubMed] [Google Scholar]
- 20.Lee, W. M. 1997. Hepatitis B virus infection. N. Engl. J. Med. 337:1733-1745. [DOI] [PubMed] [Google Scholar]
- 21.Liu, R., W. A. Paxton, S. Choe, D. Ceradini, S. R. Martin, R. Horuk, et al. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367-377. [DOI] [PubMed] [Google Scholar]
- 22.Marcellin, P., T. T. Chang, S. G. Lim, M. J. Tong, W. Sievert, M. L. Shiffman, et al. 2003. Adefovir dipivoxil for the treatment of hepatitis B e antigen-positive chronic hepatitis B. N. Engl. J. Med. 348:808-816. [DOI] [PubMed] [Google Scholar]
- 23.McDermott, D. H., P. A. Zimmerman, F. Guignard, C. A. Kleeberger, S. F. Leitman, P. M. Murphy, et al. 1998. CCR5 promoter polymorphism and HIV-1 disease progression. Lancet 352:866-870. [DOI] [PubMed] [Google Scholar]
- 24.Moreno, C., T. Gustot, C. Nicaise, E. Quertinmont, N. Nagy, M. Parmentier, et al. 2005. CCR5 deficiency exacerbates T-cell-mediated hepatitis in mice. Hepatology 42:854-862. [DOI] [PubMed] [Google Scholar]
- 25.Nansen, A., J. P. Christensen, S. O. Andreasen, C. Bartholdy, J. E. Christensen, and A. R. Thomsen. 2002. The role of CC chemokine receptor 5 in antiviral immunity. Blood 99:1237-1245. [DOI] [PubMed] [Google Scholar]
- 26.Ng-Cashin, J., J. J. Kuhns, S. E. Burkett, J. D. Powderly, R. R. Craven, H. W. van Deventer, et al. 2003. Host absence of CCR5 potentiates dendritic cell vaccination. J. Immunol. 170:4201-4208. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen, G. T., M. Carrington, J. A. Beeler, M. Dean, L. M. Aledort, P. M. Blatt, et al. 1999. Phenotypic expressions of CCR5-delta32/delta32 homozygosity. J. Acquir. Immune Defic. Syndr. 22:75-82. [DOI] [PubMed] [Google Scholar]
- 28.Rehermann, B., D. Lau, J. H. Hoofnagle, and F. V. Chisari. 1996. Cytotoxic T lymphocyte responsiveness after resolution of chronic hepatitis B virus infection. J. Clin. Investig. 97:1655-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, M. W., M. Dean, M. Carrington, C. Winkler, G. A. Huttley, D. A. Lomb, et al. 1997. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Science 277:959-965. [DOI] [PubMed] [Google Scholar]
- 30.Stephens, M., N. J. Smith, and P. Donnelly. 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68:978-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thio, C. L., T. Mosbruger, J. Astemborski, S. Greer, G. D. Kirk, S. J. O'Brien, et al. 2005. Mannose binding lectin genotypes influence recovery from hepatitis B virus infection. J. Virol. 79:9192-9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thio, C. L., T. L. Mosbruger, R. A. Kaslow, C. L. Karp, S. A. Strathdee, D. Vlahov, et al. 2004. Cytotoxic T-lymphocyte antigen 4 gene and recovery from hepatitis B virus infection. J. Virol. 78:11258-11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thio, C. L., D. L. Thomas, and M. Carrington. 2000. Chronic viral hepatitis and the human genome. Hepatology 31:819-827. [DOI] [PubMed] [Google Scholar]
- 34.Thio, C. L., D. L. Thomas, P. Karacki, X. Gao, D. Marti, R. A. Kaslow, et al. 2003. Comprehensive analysis of class I and class II HLA antigens and chronic hepatitis B virus infection. J. Virol. 77:12083-12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thursz, M., H. C. Thomas, B. M. Greenwood, and A. V. S. Hill. 1997. Heterozygote advantage for HLA class-II type in hepatitis B virus infection. Nat. Genet. 17:11-12. [DOI] [PubMed] [Google Scholar]
- 36.Wong, M. M., and E. N. Fish. 2003. Chemokines: attractive mediators of the immune response. Semin. Immunol. 15:5-14. [DOI] [PubMed] [Google Scholar]
- 37.Wu, L., G. LaRosa, N. Kassam, C. J. Gordon, H. Heath, N. Ruffing, et al. 1997. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J. Exp. Med. 186:1373-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, L., W. A. Paxton, N. Kassam, N. Ruffing, J. B. Rottman, N. Sullivan, et al. 1997. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J. Exp. Med. 185:1681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou, Y., T. Kurihara, R. P. Ryseck, Y. Yang, C. Ryan, J. Loy, et al. 1998. Impaired macrophage function and enhanced T-cell-dependent immune response in mice lacking CCR5, the mouse homologue of the major HIV-1 coreceptor. J. Immunol. 160:4018-4025. [PubMed] [Google Scholar]