Abstract
Coxsackievirus B3 (CVB3) is a picornavirus which causes myocarditis and pancreatitis and may play a role in type I diabetes. The viral genome is a single 7,400-nucleotide polyadenylated RNA encoding 11 proteins in a single open reading frame. The 5′ end of the viral genome contains a highly structured nontranslated region (5′NTR) which folds to form an internal ribosome entry site (IRES) as well as structures responsible for genome replication, both of which are critical for virulence. A structural model of the CVB3 5′NTR, generated primarily by comparative sequence analysis and energy minimization, shows seven domains (I to VII). While this model provides a preliminary basis for structural analysis, the model lacks comprehensive experimental validation. Here we provide experimental evidence from chemical modification analysis to determine the structure of the CVB3 5′NTR. Chemical probing results show that the theoretical model for the CVB3 5′NTR is largely, but not completely, supported experimentally. In combination with our chemical probing data, we have used the RNASTRUCTURE algorithm and sequence comparison of 105 enterovirus sequences to provide evidence for novel secondary and tertiary interactions. A comprehensive examination of secondary structure is discussed, along with new evidence for tertiary interactions. These include a loop E motif in domain III and a long-range pairing interaction that links domain II to domain V. The results of our work provide mechanistic insight into key functional elements in the cloverleaf and IRES, thereby establishing a base of structural information from which to interpret experiments with CVB3 and other picornaviruses.
Coxsackievirus B3 (CVB3) is a member of the Enterovirus genus of the family Picornaviridae. As with all picornaviruses, the CVB3 genome is a single-stranded positive-sense RNA that is organized into four sections: a highly structured 5′ nontranslated region (5′NTR), a single open reading frame encoding a polyprotein, a 3′NTR, and a poly(A) tail (48). For CVB3 the 7,400-nucleotide genome is composed of a 5′NTR of approximately 742 bases, a coding region that specifies a 2,185-amino-acid polyprotein, a 98-nucleotide (nt) 3′NTR, and a poly(A) tail (30, 36). Upon entering a permissive host cell, the enterovirus genome first serves as a template for translation, producing the viral polyprotein, and then becomes a template for replication of the minus strand. Both of these functions, as well as the regulatory switch between them, require critical structural elements in the 5′NTR RNA (18, 19, 45, 52). Alteration of structural elements in the 5′NTR severely compromises viral multiplication and also abrogates virulence (6).
The enterovirus cap-independent translation mechanism has been studied extensively, particularly in poliovirus, and serves as a model for all enteroviruses, including CVB3. In this translation mechanism, the 5′NTR contains a cis-acting internal ribosome entry site (IRES) (26, 50, 60) that recruits ribosomes directly to a downstream AUG codon, thereby circumventing canonical cap dependent initiation (8). Elements of the 5′NTR are recognized by cellular translation factors, ribosomes, and other cellular proteins to assemble an initiation complex directly on the downstream AUG (3, 4). For replication, the 5′NTRs of CVB3 and other enteroviruses contain an RNA element known as the cloverleaf, which is required in cis for initiating negative-strand RNA synthesis. These negative strands then serve as templates for production of genomic positive strands (1, 18, 39, 45). Again, RNA-protein complexes, dependent upon structural elements in the 5′NTR RNA, are required for this function. In addition to their direct role in each function, specific RNA structures in the 5′NTR also regulate the conversion from translation to replication on the positive-strand genome (18, 22).
Structural integrity of the 5′NTR is fundamentally important for efficient viral replication and also for virulence. Numerous examples in CVB3 and other picornaviruses prove that mutations in the 5′NTR markedly decrease multiplication efficiency (62), alter cell tropism (54), and attenuate virulence (10, 14, 61). In the best-known example, each of the three attenuated Sabin vaccine strains for poliovirus, considered the prototype picornavirus, contain nucleotide substitutions in domain V of the 5′NTR that are responsible for attenuation (17, 27, 40). These mutants have been shown to multiply poorly in neuronal cells (31), accounting for their decreased neurovirulence and inability to cause poliomyelitis. In CVB3, a cardiovirulent determinant has been identified in domain II of the 5′NTR by characterizing naturally occurring genomes from noncardiovirulent strains (14) and by chimeric studies using echovirus12 and CVB3 (6). Infection studies clearly show reduced multiplication efficiency in cardiomyocytes by noncardiovirulent strains and attenuation of virulence in mice (33). Multiple sequence differences between noncardiovirulent strains and cardiovirulent strains in domain II suggest that virulence depends upon the structure of this domain (33). It is clear that RNA structure in the 5′NTR holds the key to infection and virulence in enteroviruses.
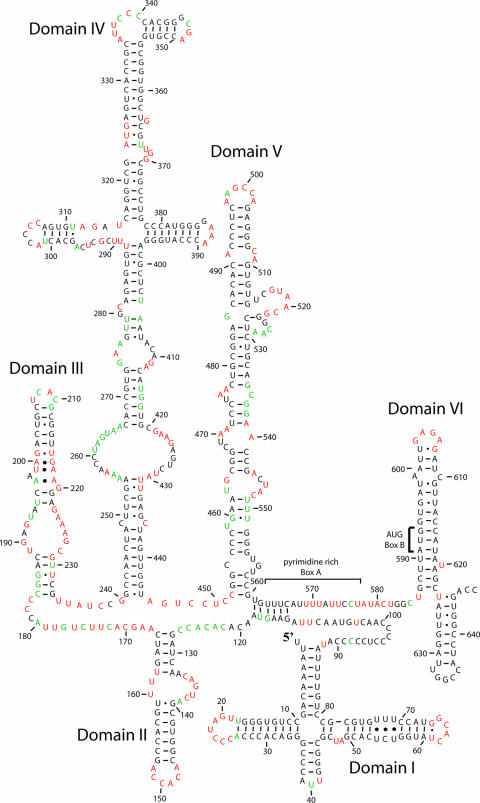
The current structural model for the CVB3 5′NTR (Fig. 1) shows seven secondary structure domains (I to VII) defined by long-range base-pairing interactions. Between these domains are connecting segments that range in length from just 2 nucleotides to over 25 nucleotides. It is generally agreed that domains II to VI house the IRES element (46), although the minimal IRES requires only domains II, IV, and V (11, 21, 46). Domain I is the cloverleaf structure, which contributes to the efficiency of the IRES (55) but is absolutely required for replication functions (2, 45). The structure shown in Fig. 1 was derived from a combination of biochemical studies, energy minimization, and comparative sequence analysis (51, 56, 65). In support of this model, short regions of the molecule have been explored experimentally (41, 47, 56, 58). However, a comprehensive biochemical study of the structure of the 5′NTR RNA in the context of the entire folded molecule has not been completed. We have used chemical modification to analyze the solution structure of domains I to VI of the CVB3 5′NTR in the context of the entire 5′NTR. This analysis provides critical information about both secondary and tertiary interactions within the molecule. The accessibility of each of the four nucleotides to solvent was assessed by probing the molecule with dimethyl sulfate (DMS), 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluenesulfonate (CMCT), and α-keto-β-ethoxybutyraldehyde (kethoxal) (16, 44). We show that much of the theoretical structure of the 5′NTR shown in Fig. 1 is supported experimentally but that there are critical exceptions, particularly in domains II and III, the connecting region between the cloverleaf and domain II, and the connecting region between domains V and VI. Proposals for new secondary and tertiary interactions in the 5′NTR, which include a loop E motif in domain III, an expanded single-stranded region in domain II, and a long-range pairing interaction, are based on the chemical probing analysis, comparative sequence analysis of 105 enterovirus sequences, and folding using the RNASTRUCTURE algorithm (43). Together, these results provide key structural insights into critically important functional regions of the picornaviral 5′NTR.
FIG. 1.
Secondary structure map of the CVB3 5NTR (65). The sequence is that of CVB3/28 (59). The map shows the proposed secondary structures of the seven predicted structural domains (I to VII).
MATERIALS AND METHODS
Viral plasmids.
Plasmids containing cDNA inserts of the CVB3 genome were engineered and kindly provided by Nora Chapman at the Enterovirus Research Laboratory, University of Nebraska Medical Center, Omaha. CVB3/28 (59) contained the 5′NTR derived from cDNAs from a cardiovirulent strain. The original plasmids were full-length infectious clones placed in the pSVN vector (7, 62). For this work, the CVB3 genomic insert was modified by the addition of a T7 promoter sequence just upstream of the 5′ end. We also used a subclone of the CVB3/28 plasmid that was further modified by the addition of a ribozyme sequence between the T7 promoter and the 5′ end. This ribozyme sequence cleaved itself off of the 5′ end to leave the authentic 5′ uridine.
Plasmid DNA was isolated using a Wizard (Promega) miniprep kit according to the procedure provided by the manufacturer. To generate a template for transcription, purified plasmid DNA was digested with an enzyme that leaves a blunt end. Two enzymes were used: EcoRV, which cuts at position 919 of the genome, and Ecl136II, which cuts at position 748.
Viral RNA transcription.
The transcriptions were done in vitro using the Megascript kit (Ambion, Austin, TX) according to the procedure provided by the manufacturer. The reaction mixture included 1 μg of digested template DNA, and the total reaction volume was 20 μl. The transcription reaction mixtures were incubated overnight (12 to 16 h) at 37°C. Following incubation, reaction mixtures were treated with 1 μl of DNase I solution for 30 min at 37°C. The reaction was stopped by adding 115 μl of nuclease-free water and 15 μl of 0.5 M ammonium acetate. RNA was then extracted with phenol-chloroform-isoamyl alcohol (25:24:1), precipitated by incubating at −20° overnight with 300 μl of isopropyl alcohol, pelleted for 20 min at 4°C, washed with 100 μl of 70% ethanol, dried, and resuspended in 24 μl of Tris-EDTA, pH 7.6. RNA was stored frozen at −70° for not more than 4 weeks.
Composite gel electrophoresis.
Folded RNA molecules were analyzed by electrophoresis on nondenaturing agarose-acrylamide composite gels (20). A sample containing 10 μg of RNA transcript was placed in folding buffer (25 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 60 mM KCl) and denatured by heating in an 80°C heat block for 2 min. The sample was allowed to cool slowly in the heat block to 37°C to renature the RNA. The sample was loaded onto a 3% acrylamide-0.5% agarose composite gel in folding buffer and electrophoresed at 150 V for 6 h at 4°C with buffer changes every 2 h. Bands were detected by staining in 0.2% methylene blue, 0.2 M Na-acetate, 0.2 M acetic acid.
RNA structural analysis.
In vitro structural analysis was accomplished by subjecting the folded 5′NTR RNA to chemical modification (16, 44). The nucleotide-modifying agents used were DMS, kethoxal, and CMCT. For each modification reaction, 15 μg of RNA was denatured in either 100 μl of DMS-kethoxal buffer (40 mM K-cacodylate [pH 7.2], 10 mM MgCl2, 50 mM NH4Cl, 0.75 mM dithiothreitol) or 50 μl of CMCT buffer (40 mM K-borate [pH 8.0], 10 mM MgCl2, 50 mM NH4Cl, 0.75 mM dithiothreitol) by incubation at 80°C for 2 min. The reaction mixtures were then slowly cooled to 42°C or lower (over a period of approximately 20 min) to allow the RNA to fold into its native structure and then transferred to 37°C for the modification reactions (16). For each modified RNA, a corresponding control RNA was subjected to all the steps in the protocol without the addition of a modifying agent. For the kethoxal modification, 5 μl of a 1.5 M solution of kethoxal (U.S. Biochemicals) was added to the RNA and incubated at 37°C for 30 min. For the DMS modification, 2 μl of a 20% DMS solution in 95% ethanol was added to the RNA and incubated at 37° for 10 min. For the CMCT modification, 50 μl of a solution containing 42 mg/ml of CMCT dissolved in CMCT buffer was added to the RNA and incubated for 10 min at 37°C. Kethoxal reactions were stopped by adding 50 μl of 150 mM sodium acetate, 250 mM potassium borate, (pH 7.0). DMS reactions were stopped by adding 25 μl of 1 M Tris-HCl (pH 7.5), 1 M β-mercaptoethanol, 0.1 M EDTA, and CMCT reactions were stopped by adding 300 μl of 95% ethanol. Modified RNA was recovered by ethanol precipitation with ammonium acetate.
Primer extension.
Sites of chemical modification were identified by reverse transcriptase primer extension analysis (16, 44). To cover the entire 5′NTR, oligonucleotides were designed to prime the extension from different sections of the RNA molecule. Table 1 lists the oligonucleotides used in the extension reactions, their positions on the 5′NTR, and their sequences.
TABLE 1.
Oligonucleotide primers
| Priming position | Sequence (5′→3′) |
|---|---|
| 62 | CAGGCGCACAAAGTACCGT |
| 108 | CGGTGTGTGTTACTTCTAAG |
| 162 | GGTAACAGAAGTGCTTGATC |
| 227 | TGGGCCGGATAACGAACG |
| 240 | AGGTTTTTCGAAGTAGTTGGC |
| 321 | CGTGGGGAATGCGGTGACTCATGG |
| 351 | GCCAACGCAGCCACCGCCAC |
| 382 | GAGCGTCCCATGGGTTTCCCC |
| 463 | TCCGCAGTTAGGATTAGCCGC |
| 557 | TAGGAATAAAATGAAACACGG |
| 627 | AGTCACCGGATGGCCAATCC |
Oligonucleotide primers were labeled on their 5′ ends by using [32P]ATP and T4 polynucleotide kinase. For the labeling reaction, 100 pmol of oligonucleotide DNA was added to 4 μl of 5× forward buffer (350 mM Tris-HCl [pH 7.6], 500 mM KCl, 50 mM MgCl2, 5 mM 2-mercaptoethanol), 100 μCi of [γ-32P]ATP (10 μCi/μl), and 10 units of T4 polynucleotide kinase in a total reaction volume of 20 μl. The reaction mixture was incubated for 40 min at 37°C, and then the kinase enzyme was inactivated by incubation at 60°C for 20 min. After labeling, the reaction mixture was diluted with 30 μl of Tris-EDTA (pH 7.6) to bring the final concentration of the radiolabeled oligonucleotide to 2 pmol/μl.
For the annealing reaction, 1 μg of chemically modified RNA, 2 μl of 5× annealing buffer (250 mM Tris-HCl [pH 8.3], 200 mM KCl) and 1 μl (2 pmol) of 32P-labeled oligonucleotide were placed into a reaction mixture of 10 μl total volume. The reaction mixture was heated to 80°C for 2 min and then cooled slowly to 42°C. Once the reaction mixtures reached 42°C, 2 μl of each annealing mixture was added to 2 μl of 2× extension mix (100 mM Tris-HCl [pH 8.3], 80 mM KCl, 12 mM MgCl2, 4 mM of each deoxynucleoside triphosphate) and 1 unit of avian myeloblastosis virus reverse transcriptase (Life Sciences, St. Petersburg, FL). To generate a sequence ladder, 2 μl of unmodified RNA was added separately to one of the four different termination mixes (50 mM Tris-HCl [pH 8.3], 40 mM KCl, 6 mM MgCl2, 1 mM of each deoxynucleoside triphosphate, and 0.1 mM of one dideoxynucleoside triphosphate [ddATP, ddCTP, ddGTP, or ddTTP]) and 1 unit of avian myeloblastosis virus reverse transcriptase. All the reaction mixtures were incubated at 42°C for 23 min. Reactions were stopped using 2 μl of stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol), and the mixtures were stored at −70°C.
Gel electrophoresis.
Primer extension products were analyzed on 12% sequencing gels. The samples were electrophoresed for either 4 or 6 h at 60-W constant power in 90 mM Tris-borate (pH 8.3), 1 mM EDTA. Images of the gel were captured using a Packard Cyclone phosphorimager.
Sequence comparisons.
Table 2 shows the 105 enterovirus sequences used in the sequence comparisons. Sequences were downloaded from GenBank, National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and analyzed in Vector NTI 9.1. A complete comparative sequence analysis of proposed base pairs is available at http://www.unomaha.edu/biology/tapprich.html. For several domains, specific examples of comparisons that provide strong support for inclusion of a base pair or strong support for removal of a base pair have been included in the figures. These examples are indicative but are not a comprehensive representation of the analysis that was conducted. The complete analysis is available at the website.
TABLE 2.
Enterovirus sequences
| Accession no. | Length (nt) | Description | Type |
|---|---|---|---|
| AB192877 | 7,425 | Human enterovirus 90 genomic RNA | Enterovirus A |
| AB204852 | 7,408 | Human enterovirus 71 genomic RNA | Enterovirus A |
| AF316321 | 7,411 | Enterovirus 5865/sin/000009 | Enterovirus A |
| AY421760 | 7,398 | Human coxsackievirus A2 strain Fleetwood | Enterovirus A |
| AY421761 | 7,395 | Human coxsackievirus A3 strain Olson | Enterovirus A |
| AY421762 | 7,434 | Human coxsackievirus A4 strain High Point | Enterovirus A |
| AY421763 | 7,400 | Human coxsackievirus A5 strain Swartz | Enterovirus A |
| AY421764 | 7,434 | Human coxsackievirus A6 strain Gdula | Enterovirus A |
| AY421765 | 7,404 | Human coxsackievirus A7 strain Parker | Enterovirus A |
| AY421766 | 7,396 | Human coxsackievirus A8 strain Donovan | Enterovirus A |
| AY421767 | 7,409 | Human coxsackievirus A10 strain Kowalik | Enterovirus A |
| AY421768 | 7,404 | Human coxsackievirus A12 strain Texas-12 | Enterovirus A |
| AY421769 | 7,415 | Human coxsackievirus A14 strain G-14 | Enterovirus A |
| AY790926 | 7,410 | Human coxsackievirus A16 strain shzh00-1 | Enterovirus A |
| CAU05876 | 7,413 | Coxsackievirus A16 G-10 | Enterovirus A |
| NC_001612 | 7,413 | Human enterovirus A | Enterovirus A |
| AF039205 | 7,398 | Coxsackievirus B6 strain Schmitt | Enterovirus B |
| AF081485 | 7,403 | Coxsackievirus B2 strain Ohio | Enterovirus B |
| AF083069 | 7,433 | Echovirus 5 | Enterovirus B |
| AF105342 | 7,398 | Coxsackievirus B6 strain Schmitt | Enterovirus B |
| AF114383 | 7,400 | Coxsackievirus B5 strain Faulkner | Enterovirus B |
| AF114384 | 7,397 | Coxsackievirus B6 strain Schmitt | Enterovirus B |
| AF162711 | 7,440 | Echovirus 30 strain Bastianni | Enterovirus B |
| AF231763 | 7,400 | Coxsackievirus B3 strain 31-1-93 | Enterovirus B |
| AF231764 | 7,400 | Coxsackievirus B3 strain P | Enterovirus B |
| AF231765 | 7,400 | Coxsackievirus B3 strain PD | Enterovirus B |
| AF311939 | 7,397 | Human coxsackievirus B4 strain E2 variant | Enterovirus B |
| AF465516 | 7,427 | Human echovirus 7 strain Wallace | Enterovirus B |
| AF465517 | 7,418 | Human echovirus 6 strain Charles | Enterovirus B |
| AF465518 | 7,435 | Human echovirus 2 strain Cornelis | Enterovirus B |
| AF524866 | 7,453 | Human echovirus 9 strain Barty | Enterovirus B |
| AY302540 | 7,450 | Human echovirus 14 strain Tow | Enterovirus B |
| AY302541 | 7,437 | Human echovirus 15 strain CH 96-51 | Enterovirus B |
| AY302542 | 7,437 | Human echovirus 16 strain Harrington | Enterovirus B |
| AY302543 | 7,416 | Human echovirus 17 strain CHHE-29 | Enterovirus B |
| AY302544 | 7,433 | Human echovirus 19 strain Burke | Enterovirus B |
| AY302545 | 7,435 | Human echovirus 2 strain Cornelis | Enterovirus B |
| AY302546 | 7,394 | Human echovirus 20 strain JV-1 | Enterovirus B |
| AY302547 | 7,426 | Human echovirus 21 strain Farina | Enterovirus B |
| AY302548 | 7,433 | Human echovirus 24 strain DeCamp | Enterovirus B |
| AY302549 | 7,426 | Human echovirus 25 strain JV-4 | Enterovirus B |
| AY302550 | 7,424 | Human echovirus 26 strain Coronel | Enterovirus B |
| AY302551 | 7,412 | Human echovirus 27 strain Bacon | Enterovirus B |
| AY302552 | 7,427 | Human echovirus 29 strain JV-10 | Enterovirus B |
| AY302553 | 7,428 | Human echovirus 3 strain Morrisey | Enterovirus B |
| AY302554 | 7,432 | Human echovirus 31 strain Caldwell | Enterovirus B |
| AY302555 | 7,420 | Human echovirus 32 strain PR10 | Enterovirus B |
| AY302556 | 7,394 | Human echovirus 33 strain Toluca-3 | Enterovirus B |
| AY302557 | 7,394 | Human echovirus 4 strain Pesacek | Enterovirus B |
| AY302558 | 7,418 | Human echovirus 6 strain D'Amori | Enterovirus B |
| AY302559 | 7,427 | Human echovirus 7 strain Wallace | Enterovirus B |
| AY302560 | 7,411 | Enterovirus 69 strain Toluca-1 | Enterovirus B |
| AY429470 | 7,401 | Swine vesicular disease virus strain HK70 | Enterovirus B |
| AY673831 | 7,400 | Human coxsackievirus B3 strain GA | Enterovirus B |
| AY752944 | 7,400 | Human coxsackievirus B3 strain 28 | Enterovirus B |
| AY752945 | 7,400 | Human coxsackievirus B3 strain 0 | Enterovirus B |
| AY752946 | 7,399 | Human coxsackievirus B3 strain 20 | Enterovirus B |
| AY875692 | 7,403 | Human coxsackievirus B5 isolate 2000/CSF/KOR | Enterovirus B |
| CXA1G | 7,389 | Coxsackievirus B1 | Enterovirus B |
| CXA3CG | 7,399 | Coxsackievirus B3 | Enterovirus B |
| CXA3G | 7,396 | Coxsackievirus B3 | Enterovirus B |
| CXA9CG | 7,452 | Coxsackievirus A9 genomic RNA | Enterovirus B |
| CXAB3CG | 7,399 | Coxsackievirus B3 mRNA | Enterovirus B |
| CXB5CGA | 7,402 | Coxsackievirus B5 | Enterovirus B |
| CXU57056 | 7,400 | Coxsackievirus B3 Woodruff variant | Enterovirus B |
| NC_001472 | 7,389 | Human enterovirus B | Enterovirus B |
| PICOXB4 | 7,395 | Coxsackievirus B4 | Enterovirus B |
| AB205396 | 7,460 | Human coxsackievirus A18 genomic RNA | Enterovirus C |
| AF465511 | 7,458 | Human coxsackievirus A13 strain Flores | Enterovirus C |
| AF465512 | 7,441 | Human coxsackievirus A15 strain G-9 | Enterovirus C |
| AF465513 | 7,457 | Human coxsackievirus A18 strain G-13 | Enterovirus C |
| AF465514 | 7,436 | Human coxsackievirus A20 strain IH pool 35 | Enterovirus C |
| AF465515 | 7,405 | Human coxsackievirus A21 strain Kuykendall | Enterovirus C |
| AF499635 | 7,397 | Human coxsackievirus A1 strain Tompkins | Enterovirus C |
| AF499636 | 7,453 | Human coxsackievirus A11 strain Belgium-1 | Enterovirus C |
| AF499637 | 7,458 | Human coxsackievirus A13 strain Flores | Enterovirus C |
| AF499638 | 7,441 | Human coxsackievirus A15 strain G9 | Enterovirus C |
| AF499639 | 7,457 | Human coxsackievirus A17 strain G12 | Enterovirus C |
| AF499640 | 7,458 | Human coxsackievirus A18 strain G13 | Enterovirus C |
| AF499641 | 7,410 | Human coxsackievirus A19 strain 8663 | Enterovirus C |
| AF499642 | 7,436 | Human coxsackievirus A20 strain IH35 | Enterovirus C |
| AF499643 | 7,406 | Human coxsackievirus A22 strain Chulman | Enterovirus C |
| AF546702 | 7,406 | Human coxsackievirus A21 strain Kuykendall | Enterovirus C |
| CXA21CG | 7,401 | Human coxsackievirus A21 (strain Coe) genomic RNA | Enterovirus C |
| CXA24CG | 7,461 | Human coxsackievirus A24 DNA | Enterovirus C |
| NC_001428 | 7,401 | Human enterovirus C | Enterovirus C |
| AY426531 | 7,367 | Human enterovirus 68 strain Fermon | Enterovirus D |
| AB205395 | 7,431 | Poliovirus genomic RNA | Poliovirus |
| AF111953 | 7,445 | Human poliovirus 1 isolate CHN-Hebei/91-2 | Poliovirus |
| AF111961 | 7,444 | Human poliovirus 1 isolate CHN-Guangdong/92-2 | Poliovirus |
| AF111966 | 7,444 | Human poliovirus 1 isolate CHN-Hainan/93-2 | Poliovirus |
| AF405669 | 7,442 | Human poliovirus 1 isolate HAI00003 | Poliovirus |
| AF448782 | 7,439 | Human poliovirus 2 strain EGY88-074 | Poliovirus |
| AY184219 | 7,441 | Human poliovirus 1 strain Sabin 1 | Poliovirus |
| AY184220 | 7,439 | Human poliovirus 2 strain Sabin 2 | Poliovirus |
| AY184221 | 7,432 | Human poliovirus 3 strain Sabin 3 | Poliovirus |
| AY278549 | 7,439 | Human poliovirus 2 isolate P2S/Mog65-3 (20120) | Poliovirus |
| AY278553 | 7,436 | Human poliovirus 1 isolate P1W/Bar65 (19276) | Poliovirus |
| PIPOLS2 | 7,439 | Poliovirus type 2 genome (strain Sabin 2 [P712, Ch, 2ab]) | Poliovirus |
| PIPOLS3 | 7,434 | Poliovirus type 3 mRNA (vaccine strain Sabin 3) | Poliovirus |
| POL2LAN | 7,440 | Poliovirus type 2 (Lansing strain) | Poliovirus |
| POL3L37 | 7,431 | Poliovirus P3/Leon/37 (type 3) | Poliovirus |
| POLIO1B | 7,440 | Human poliovirus 1 Mahoney | Poliovirus |
| POLIOS1 | 7,441 | Human poliovirus strain Sabin 1 | Poliovirus |
| AY876912 | 7,456 | Human enterovirus Ningbo3-02 | Unclassified |
RESULTS
We investigated the structure of the CVB3 5′NTR in solution by using a series of chemical probes. The cardiovirulent strain CVB3/28 was used in the analysis (61). CVB3/28 was constructed by alteration of nucleotide 234 of a noncardiovirulent virus, CVB3/0, from C to T (59), thus converting it to a cardiovirulent strain (61). RNA transcripts were generated through in vitro transcription of linearized plasmid DNA. The plasmids contained the entire viral genomic sequence fused to a T7 promoter. Runoff transcription with T7 RNA polymerase generated a 919-nucleotide RNA that contained the entire 5′NTR (742 nt) as well as 177 nucleotides of coding region when the plasmids were linearized with EcoRV. Transcripts of 748 nucleotides, containing the entire 5′NTR and only 6 nucleotides of coding region, were also generated by linearizing the plasmids with Ecl136II. Probing results from both transcripts are presented, as we found no differences in 5′NTR modification between the 919-nucleotide transcript and the 748-nucleotide transcript. Thus, in our hands, the coding-region RNA between nucleotides 744 and 919 did not have any influence on the structure of the 5′NTR.
Three chemical probes were used to determine the accessibility of each of the four nucleotides to solvent. These chemical probes modify positions on the base that are involved in Watson-Crick base pairing, so only bases that are single stranded and are also accessible to solvent in the folded molecule are reactive. DMS modifies the N1 position of adenosine and the N3 position of cytosine, CMCT modifies the N3 position of uridine, and kethoxal creates a cyclic adduct using the N1 and N2 positions of guanosine (16, 44).
5′NTR conformation.
Folding of the 5′NTR transcripts was accomplished by slow cooling of heat-denatured molecules to a temperature of 37°C in a folding buffer containing 10 mM MgCl2 and 60 mM KCl. To determine whether the 5′NTR transcripts folded into a single conformer, molecules taken through the heat denaturation and slow cooling procedure were analyzed on a 3% acrylamide-0.5% agarose native composite gel that was electrophoresed in folding buffer (25 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 60 mM KCl). Composite gel electrophoresis is very sensitive to RNA conformation (20), and native gels have previously demonstrated conformational heterogeneity in the 5′NTR derived from hepatitis C virus (28). Migration as a single tight band indicated that the molecules folded into conformers of similar overall three-dimensional shape (data not shown).
Chemical probing. (i) Domain I.
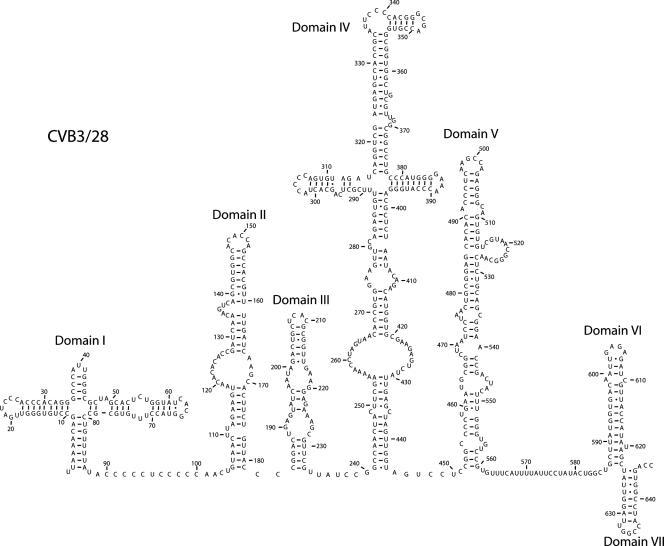
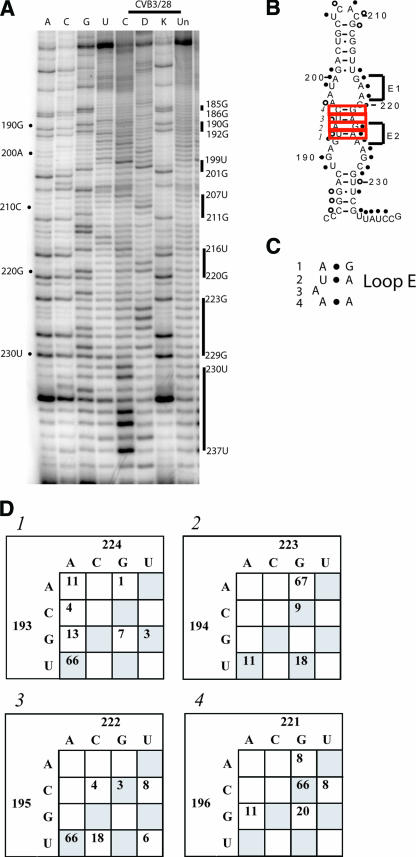
The cloverleaf structure of domain I is well supported by comparative sequence analysis, and the 3′ stem-loop, known as stem-loop d, has been analyzed by nuclear magnetic resonance (NMR) (12, 23, 47). Our probing results provide solid experimental support for the cloverleaf structure. Figure 2 shows an example of the primer extension results for domain I (Fig. 2A) and the connecting region between domain I and domain II (Fig. 2A and B), as well as a summary of the modifications on the predicted secondary structure map (Fig. 2C). Comparative sequence analysis for selected positions is shown in Fig. 2D.
FIG. 2.
Chemical probing results for domain I (cloverleaf). A. A 12% sequencing gel showing primer extension analysis of modified and unmodified CVB3 RNA. Labels on the left indicate nucleotide positions according to the sequencing tracks (lanes A, C, G, and U), and labels on the right identify positions that are modified. Lane Un, unmodified; lane K, kethoxal; lane D, DMS; lane C, CMCT. B. A 12% sequencing gel to show detailed results for the connecting region between domain I and domain II. Labels on the right indicate nucleotide positions according to the sequencing tracks (lanes U, G, C, and A), and labels on the left identify positions that are modified. Lane Un, unmodified; lane K, kethoxal; lane D, DMS; lane C, CMCT. C. Predicted secondary structure map of domain I, showing modified positions. Filled circles identify strongly modified positions; open circles identify moderately modified positions. Red boxes indicate examples of pairs that are not supported by the comparative sequence analysis results shown in panel D. Green boxes indicate examples of pairs that are supported by phylogentic analysis in panel D. D. Analysis of representative paired positions, showing the number of occurrences of nucleotide identities among 105 enterovirus sequences.
The basal stem of the cloverleaf (stem a) and all but the final pair of the stem of stem-loop b are protected from chemical modification as expected for base-paired stem regions. Comparative sequence analysis gives strong support to both of these stems, with eight of the nine pairs in stem a completely conserved and eight of the nine pairs in stem b showing many examples of compensatory changes (Fig. 2D). One exception is the proposed pair of 11C-33G, which shows 19 examples where the pair is a mismatch (Fig. 2D). Even so, our probing results do not show exposure of either 11C or 33G. The loop region of stem-loop b is accessible, beginning with a CMCT modification at position 19U. Bases 19U, 20G, 21A, 22U, 23C, 24C, and 25C form a hairpin loop in the cloverleaf, and all are exposed to solvent. The stem region of stem-loop c is protected, as are positions 39A and 40U in the loop region. Some results indicate a very light modification at 40U, but this is variable. The folding of the cloverleaf appears to protect these positions. Base 41U in the loop region consistently displayed accessibility. The pair 38C-42G is not supported by comparative sequence analysis, showing a change to C-U in 19 poliovirus sequences and suggesting there is a 5-base hairpin loop rather than a 3-base hairpin loop (Fig. 2D). But again, our probing results suggest that these bases are involved in structure in CVB3.
Modifications at positions 48U and 49A confirm the bulged loop in stem d. The RNA then closes for the remainder of the stem region. Bases 54 to 56 in this stem are predicted to be available for modification as part of an internal loop opposite positions 71 to 73. However, NMR analysis indicates that these bases participate in noncanonical pyrimidine-pyrimidine pairs (47), creating a continuous stem. Our probing results confirm the paired arrangement, as none of the pyrimidines are accessible. Comparative sequence analysis shows that 54U changes to A 37 times, which creates an A-U pair with 73U in every case. The 55C-72U pair is completely conserved. The 56U-71U pyrimidine-pyrimidine pair changes to 56C-71C 18 times and so is either U-U or C-C in 104 of 105 sequences. There is only one occurrence of C-U. The G-U pair at the end of stem-loop d is modified, showing a light hit at 61U and a strong hit at 66G. The proposed tetraloop at the end of stem loop d is completely exposed, with modifications between 62C and 65G. The 5′-CACG-3′ tetraloop sequence found in CVB3/28 is not one of the canonical tetraloop motifs (12). Interestingly, 49 of the 105 enterovirus sequences have 62U rather than 62C, which creates a canonical 5′-UNCG-3′ tetraloop, and NMR results show that the 5′-CACG-3′ sequence present in 53 of the 105 enterovirus strains adopts a fold very similar to a 5′-UNCG-3′ tetraloop (12). In fact, the NMR results for these two sequences have helped to expand the tetraloop motif to include 5′-CACG-3′ (12).
The long pyrimidine-rich connecting region between domain I and domain II is protected from our chemical probes, aside from strong modifications at 88U and 104U and a weak modification at 92C (Fig. 2B). We have observed this protection in multiple analyses and using two different oligonucleotide primers. At 16 nucleotides in length, this connecting region is the longest inaccessible stretch of bases in the entire 5′NTR. We suggest that the connecting region between domains I and II is folding into a unique structure that is involved in interactions with other regions of the molecule. Furthermore, as outlined in the section “Domain II” below, we propose that the connecting region between domain I and domain II extends much further than the present model suggests and terminates with a long-range pairing interaction.
(ii) Domain II.
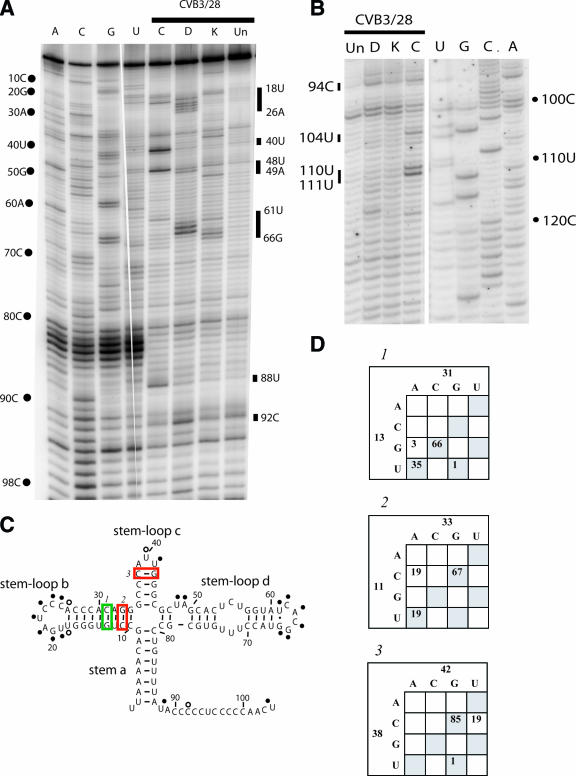
Domain II is a critical region of the enterovirus 5′NTR. Studies of both natural isolates and laboratory strains have shown that the sequence of domain II is a determinant for virulence in CVB3 (6, 14, 15, 33) and poliovirus (10). From our chemical probing results, it is clear that the structure of domain II is extremely complex. Figure 3 shows examples of the primer extension results (Fig. 3A) and also shows the modifications on the predicted secondary structure map (Fig. 3B). In the basal stem region of domain II there are several unusual probing results. In the lower stem (nt 105-117:171-181), the 5′ strand shows protections that would be expected for a stem region, with modification only at the bulged uridines at positions 110 and 111. However, the 3′ strand of this same stem region is largely exposed, showing prominent hits at 5 of the 10 positions and light hits at other 5. Given the variable intensity of the modifications in the 3′ strand and the protection of the 5′ strand, we suggest that this stem region is a dynamic element in the molecule, with the 5′ partner consistently involved in either secondary or tertiary interaction and with the 3′ partner adopting an unstructured state. Comparative sequence analysis also suggests that several positions do not conserve pairing. For example, 106U-180A is a mismatch more often than it is complementary, and 107A-179U is a mismatch in 21 cases (Fig. 3C).
FIG. 3.
Chemical probing results for domain II. A. A 12% sequencing gel showing primer extension analysis of modified and unmodified CVB3 RNA. Labels on the right indicate nucleotide positions according to the sequencing tracks (lanes U, G, C, and A), and labels on the left identify positions that are modified. Lane Un, unmodified; lane K, kethoxal; lane D, DMS; lane C, CMCT. B. Predicted secondary structure map of domain II, showing modified positions. Filled circles identify strongly modified positions; open circles identify moderately modified positions. Red boxes indicate examples of pairs that are not supported by the comparative sequence analysis results shown in panel C. Green boxes indicate examples of pairs that are supported by phylogentic analysis in panel C. C. Analysis of representative paired positions, showing the number of occurrences of nucleotide identities among 105 enterovirus sequences.
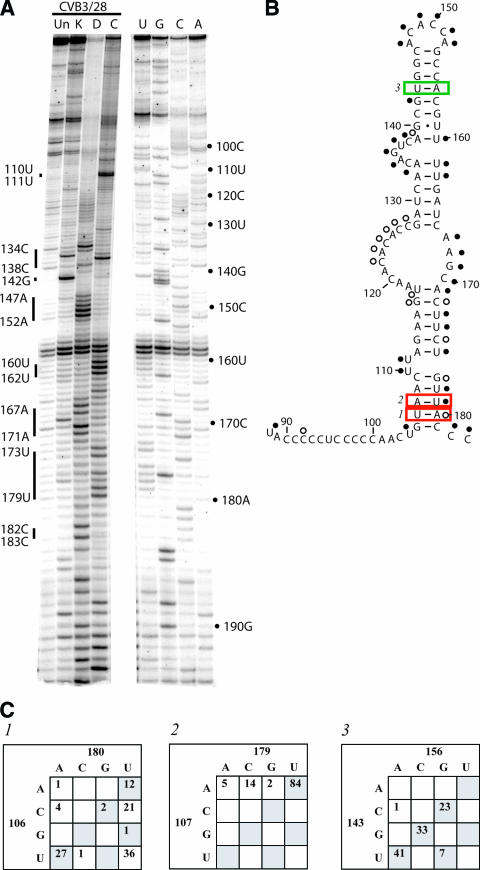
The sequence 5′-GAAGUA-3′ from position 113 to 118 is completely complementary to 5′-UGUUUC-3′ from position 561 to 566. The latter sequence is in the connecting region between domain V and domain VI. It is also protected from chemical modification in our probing analysis (see Fig. 7A). Comparative sequence analysis shows that the stem of nt 113-117:171-175 contains a total of 66 mismatches, while the proposed stem of nt 113-117:562-566 contains a total of 10 mismatches. In the most compelling example, the 117-171 pair is a mismatch 52 times while the 117-562 pair is a mismatch only 5 times. Furthermore, the 117-562 pair has mutated to a compensatory pair 32 times. We propose that a long-range pairing interaction forms between the domain II and domain V sequences and that this is one structure formed by the 5′ strand of domain II.
FIG. 7.
Chemical probing results for domain VI. A. A 12% sequencing gel showing primer extension analysis of modified and unmodified CVB3 RNA. Labels on the right indicate nucleotide positions according to the sequencing tracks (lanes U and G), and labels on the left identify positions that are modified. Results are shown for CVB3/28. Lane Un, unmodified; lane K, kethoxal; lane D, DMS; lane C, CMCT. B. Predicted secondary structure map of domain VI, showing modified positions. Filled circles identify strongly modified positions; open circles identify moderately modified positions. The pyrimidine-rich sequence (box A) and the AUG (box B) are indicated on the map.
The large internal loop involving positions 118 to 127 and 167 to 170 also shows interesting modification results. The 3′ strand of the loop is completely accessible, as might be expected. However, the 5′ strand is largely protected from modification. In the lower part of the loop, the nucleotides are completely protected, indicating that they are involved in secondary or tertiary interaction. In the upper part of the loop, we see very light hits in some experiments and no hits in others. Clearly, this long stretch of bases is participating in structural interactions, but strict base pairing is unlikely as a result of sequence variability. Among enterovirus sequences, the 5′ strand varies from 8 to 15 bases in length and the sequence is extremely variable. There is a strong bias on this strand for A and C, but all bases are represented.
In contrast to the basal stem, the apical stem-loop of domain II produces a modification pattern that is largely consistent with the predicted secondary structure. Strong modifications at 134C, 135G, 136U, and 137C confirm the predicted bulge loop. On the opposite side of the bulge, we see accessibility of a string of uridines from position 160 to 162. Comparative sequence analysis also suggests these uridines are not paired. The proposed 132A-162U pair is a mismatch 50 times, and the proposed 133A-161U pair is a mismatch more often than it is a canonical pair. Interestingly, neither 132A nor 133A is modified. These bases must be involved in other interactions. The stem leading up to the hairpin loop is protected at all seven paired positions except 142G, and the nucleotides in the loop region (position 147 to 152) are all exposed to solvent. Comparative sequence support for the apical stem is convincing, with a large number of compensatory substitutions at most positions (an example is shown in Fig. 3C). The connecting region between domain II and domain III consists of adjacent cytidines that are conserved in all 105 enterovirus sequences. Both of these positions are accessible to chemical probes.
(iii) Domain III.
Deletion analysis with poliovirus has proven that domain III is dispensable for both IRES function and viral replication (11, 21, 46). The predicted secondary structure of domain III shows numerous internal loops separated by only a few base pairs. Our probing results suggest that the majority of the domain is not stably involved in Watson-Crick base pairing. An example of the primer extension results for domain III and a map showing the modifications on a predicted secondary structure map is shown in Fig. 4A and B.
FIG. 4.
Chemical probing results for domain III. A. A 12% sequencing gel showing primer extension analysis of modified and unmodified CVB3 RNA. Labels on the left indicate nucleotide positions according to the sequencing tracks (lanes A, C, G, and U), and labels on the right identify positions that are modified. Lane Un, unmodified; lane K, kethoxal; lane D, DMS; lane C, CMCT. B. Predicted secondary structure map of domain III, showing modified positions. Filled circles identify strongly modified positions; open circles identify moderately modified positions. The two potential loop E motifs are indicated by E1 and E2. Red boxes indicate examples of pairs that are not supported by the comparative sequence analysis results shown in panel D. Green boxes indicate examples of pairs that are supported by phylogentic analysis in panel D. C. Diagram of the loop E motif, showing the noncanonical base pairs and the bulged nucleotide. Elements of the loop E motif are numbered: 1, sheared A-G pair; 2, trans-Hoogsteen U-A; 3, bulged A; 4, trans (locally parallel)-Hoogsteen-Hoogsteen A-A. D. Analysis of representative paired positions, showing the number of occurrences of nucleotide identities among 105 enterovirus sequences.
An argument can be made for the presence of the basal stem from chemical probing results, since most of the bases proposed to be paired, such as 184C, 185G, 186G, 187A, 188C, 230U, and 232C, are either protected or partially protected. The comparative sequence analysis lends support for this stem. The first two base pairs of the stem (184C-233G and 185G-232C) are completely conserved, and the third pair (186G-231U) changes to G-C 49 times. Interestingly, 187A and 188C are missing in 38 enterovirus sequences, which shortens the basal stem from five pairs to three pairs.
Through the series of three internal loops above the basal stem nearly every base is accessible, including most of the proposed pairs that separate the loops. Our probing results do not support the proposed Watson-Crick pairs, and neither does comparative sequence analysis. Sequence comparisons for the four positions separating the large internal loops (193U-196C and 221G-224A) are shown in Fig. 4D. While not likely to be involved in Watson-Crick pairing, the internal loop sequences do, however, present the intriguing possibility for loop E motifs (34). The loop E motif uses a series of noncanonical base pairs to form a distorted helical region containing an S turn (9). The sequence signature for loop E is as follows: (i) an A-G pair (which can be A-A), (ii) an absolutely conserved U-A pair (noncanonical H bonding), (iii) a bulged nucleotide, and (iv) an A-A pair (which can vary to R-R, Y-Y, A-Y, and Y-A but rarely G-Y or Y-G). Figure 4C shows the loop E motif with the elements numbered accordingly. The noncanonical pairing in the loop E motif produces a characteristic pattern of chemical modification. Both purines of the A-G pair are accessible, the U in the U-A pair is protected while the A is accessible, the bulged nucleotide is protected, and the A-A pair is accessible (35). Two consecutive loop E sequence patterns are present in the proposed internal loops of domain III from CVB3, an upper loop utilizing bases 197-200:217-219 and a lower loop utilizing bases 191-194:202-204. These are labeled E1 and E2, respectively, in Fig. 4B. Our chemical modification results lend support to the upper loop E sequence (E1) but do not support the lower loop E sequence (E2). In the upper loop E sequence, 197A, 199U, 200A, 217G, 218A, and 219A are modified. Of these, all but 199U are predicted to be accessible in the loop E structure (34). An H bond to the backbone by the bulged nucleotide in the loop E structure should provide protection from chemical modification. Our results show such protection for 198A. Sequence comparison also provides solid evidence for the upper loop E motif. Among the 105 enterovirus sequences, none show deviation from the signature loop E sequence. Element 1 is A-G in 96 sequences, A-A in 8 sequences, and G-A in one sequence. Element 2 is conserved as a U-A pair, occurring in 104 of 105 sequences, with the only exception a C-U. The bulged nucleotide (element 3) is an A 104 times and a G once. Element 4 is variable, being A-A in 64 cases, A-G in 2, U-A in 22, and C-A in 17, but never G-Y or Y-G. Evidence for the lower loop E is lacking, both in the modification pattern and in the sequence comparison. For example, element 1, which should be A-A or A-G, is C-G 9 times, U-A 11 times and U-G 19 times; element 2, which should be U-A only, shows examples of A-A (11 times), A-G (1 time), C-A (4 times), G-A (14 times), G-G (7 times), and G-U (1 time).
In contrast to the lower part of domain III, the apical stem-loop is solidly supported by experimental evidence. The stem region is protected, and the proposed loop nucleotides are all modified. Sequence comparison also adds to this support. In the six proposed pairs in the stem, four never show examples of mismatches and the other two show a total of only four mismatches. Even these mismatches are balanced by a substantial number of compensatory base changes at these positions. The hairpin loop capping domain III is always 4 nucleotides, which is suggestive of a tetraloop. However, the sequence of the loop is highly variable among enteroviruses and does not show any clear tetraloop consensus pattern. All four of these bases are accessible to chemical probes. The connecting region between domain III and domain IV is very highly conserved. In fact, 234U, 236C, 238C, and 239C do not vary among 105 sequences, and 235A and 240G change only one time. All of the connecting-region bases are susceptible to modification.
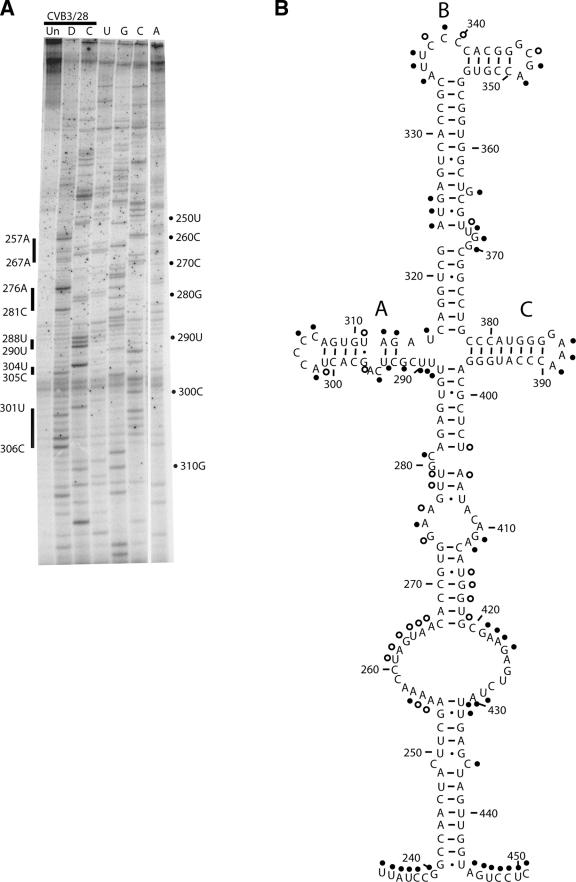
(iv) Domain IV.
The predicted structure for domain IV is very complicated and contains a long complex helical region topped by a junction loop, from which three stem-loop regions radiate (labeled A to C in Fig. 5B). Primer extension results for domain IV are shown in Fig. 5A, and a modification map is shown in Fig. 5B. The lower stem region of domain IV has 14 proposed base pairs interrupted only by a C-C mismatch. All of the proposed pairs are protected from modification, and most of the paired positions are amply supported by compensatory base changes. It is fascinating that the 249C-426C mismatch is conserved in all 105 enterovirus sequences. Only the 3′ partner (436C) shows exposure to chemical modification. The lower stem is followed by an extremely large internal loop, whose bases are largely protected. On the 5′ side of the loop only 1 base of the 12, 257A, is strongly modified, while 255A-256A and 261U-266A are lightly modified. On the 3′ side of the loop, 421G-424G and 429U-430A are strongly modified, but five bases are protected. These results suggest noncanonical pairing or other types of interaction for this region of the molecule. Above the internal loop, a long helical region is interrupted by a small internal loop and a bulge loop involving nt 275-281:404-412. This proposed structure is supported by modification data showing that most of the single-stranded nucleotides are accessible. The exception is the trio 408A, 409C, and 410A, which show protection. Again, comparative sequence analysis provides excellent support for the vast majority of the complex stem region.
FIG. 5.
Chemical probing results for domain IV. A. A 12% sequencing gel showing primer extension analysis of modified and unmodified CVB3 RNA. Labels on the right indicate nucleotide positions according to the sequencing tracks (lanes U, G, C, and A), and labels on the left identify positions that are modified. Lane Un, unmodified; lane D, DMS; lane C, CMCT. B. Predicted secondary structure map of domain IV, showing modified positions. Filled circles identify strongly modified positions; open circles identify moderately modified positions.
The junction-loop region at the top of domain IV displays a modification pattern that is largely as predicted. Stem-loop A contains two interesting structural features. The region that emerges from the junction is extremely accessible, with modifications at most positions from 288U to 297G and 311U-315U, including three proposed base pairs. Sequence analysis shows that 290U covaries with 314A to maintain a pair in all cases (U-A 66 times and G-C 39 times). Thus, it seems likely that this pair would form, but the chemical modification results are only partly supportive. Whereas 314A is protected, 290U is exposed. Together the results suggest that the bases in stem-loop A near the junction are structurally dynamic. The apical region of stem-loop A is protected in the stem region as expected. In the C-rich hairpin loop region, two of the four highly conserved cytidines are protected from chemical modification. This is the only example in the entire 5′NTR where hairpin loop nucleotides are shown to be protected. This loop is known to be functionally important as a recognition feature for poly(rC) binding protein (PCBP) in poliovirus (19) and contains a core 5′-ACCCC-3′ that is present in all 105 enterovirus sequences. According to our probing results, the C-rich loop nucleotides are adopting a complicated structure that protects some of the positions.
In stem-loop B the modification pattern is as expected from the model. The long stem region is protected, aside from the bulge-loop nucleotides near the center of the helix. The pyrimidine-rich bulge loop from position 335 to 340 is exposed, as is the tetraloop from position 347 to 349. The initial base of the tetraloop, 347G, is not marked on the diagram in Fig. 5 because a strong stop in primer extension did not allow analysis of this base. Stem-loop C displayed a pattern that matches the proposed structure, with only the tetraloop nucleotides in the hairpin showing modification.
The connecting region between domain IV and domain V is seven nucleotides, four of which are completely conserved (448C to 451C). All seven positions are accessible to chemical probes.
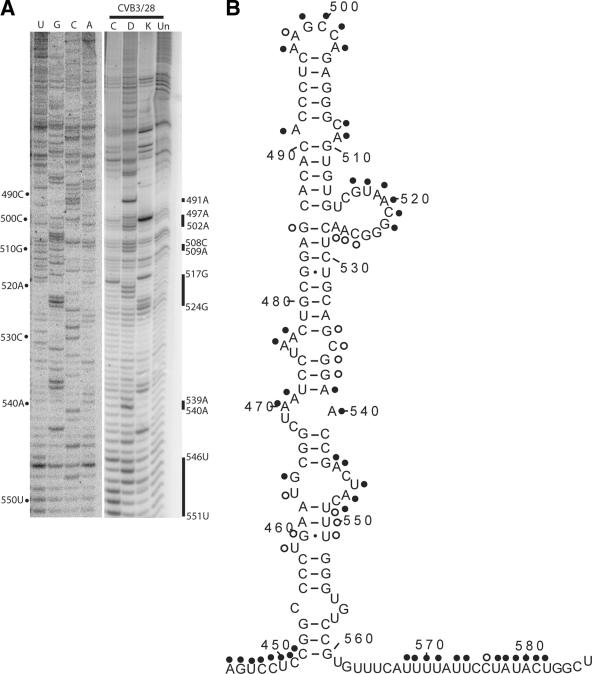
(v) Domain V.
Attenuating mutations for the Sabin vaccine strains of poliovirus are located in domain V. Consequently, this domain has received a great deal of experimental attention. As shown in Fig. 6A and B, our probing results for domain V of CVB3 reveal that the predicted secondary structure is very well supported experimentally. Our modification results support the current model suggesting that the molecule folds into a long complex stem-loop containing several bulge loops and internal loops. The majority of the basal stem region is protected from modification, including a proposed internal loop composed of 455C, 555U, 556G, and 557U. A bulged U at position 459 is partially modified, as are the bases in the adjacent pair involving 460U-551G. Overall, the protection of this section of domain V indicates that it is involved in a structure beyond the simple base-pairing interactions. Every base pair between 452C-560G and 474C-537G is extremely conserved. Of the 15 proposed pairs, 11 are completely conserved, 2 have a single compensatory change, and 2 have two compensatory changes.
FIG. 6.
Chemical probing results for domain V. A. A 12% sequencing gel showing primer extension analysis of modified and unmodified CVB3 RNA. Labels on the left indicate nucleotide positions according to the sequencing tracks (lanes A, C, G, and U), and labels on the right identify positions that are modified. Lane Un, unmodified; lane K, kethoxal; lane D, DMS; lane C, CMCT. B. Predicted secondary structure map of domain V, showing modified positions. Filled circles identify strongly modified positions; open circles identify moderately modified positions.
Above the initial stem, a series of three internal loops is proposed by the model. Modification results are largely as expected for these loops, with the proposed single-stranded bases showing modification and the proposed double-stranded bases remaining protected. The apical portion of the domain is dominated by a long stem region, and all of the proposed pairs, aside from the G at position 485, are unreactive. A large bulged loop from position 515 to 527 is modified between 517G and 522G. The first two bases leading into the loop and the last five bases ending the loop show full or partial protection. An interesting possibility, first investigated by Malnou et al. (41) for poliovirus, that is consistent with our probing results and sequence analysis is the formation of a single pair between 523G and 528C. Sequence comparison shows that 523G-528C, which is present 65 times, covaries to 523C-528G 36 times and 523A-528U 4 times. A pair between 523C and 528G would break the 485G-528C pair suggested in the current model, which is consistent with the modification results and supported by the presence of noncanonical substitutions at that pair 37 times among the 105 sequences. The 523C-528G pair also frames a canonical GNRA tetraloop sequence, a sequence motif that is completely conserved among all 105 enterovirus sequences. At the apex of domain V, the three-base internal loop involving 491A, 508C, and 509A is accessible, as are the proposed single-stranded nucleotides in the hairpin-loop, 497A, 498A, 499G, 500C, 501C, and 502A.
(vi) Domain VI.
Two features in domain VI are critical for translation initiation directed by the IRES element of picornaviruses: a pyrimidine-rich region (known as box A) followed by an AUG (box B) (Fig. 7B). These features are separated by a spacer element of 15 to 25 nucleotides (57). In enteroviruses and rhinoviruses, the pyrimidine-rich region is located in a long connecting region between domain V and domain VI and the AUG is located in a stem region of domain VI. Our probing results for these regions are shown in Fig. 7A and B. Most of the nucleotides in the pyrimidine-rich connecting region are accessible for modification. However, within this region we encountered a large number of strong stops; we are unable to determine the exposure of these bases. Strong stops occurred at 561U, 562G, 566C, 567A, and 572A. Interestingly, we observed protection of positions 563U, 564U, and 565U. These bases are part of the conserved 5′-UGUUUC-3′ sequence that is complementary to domain II and that we propose to be involved in a long-range pairing interaction. The long stretch of nucleotides from 568U to 582U showed accessibility; this stretch contains the putative Shine-Dalgarno-like sequence proposed to serve as the initial interaction site for the ribosome during translation initiation (64). In the hairpin-loop of domain VI, all of the proposed base pairs were protected from modification, including the pairs housing the AUG of box B and the mismatch 599A-610C. The GAGA tetraloop capping the domain was accessible, as was the bulged U at position 620. Sequence analysis reveals that this tetraloop changes to a triloop 34 times and often varies from the GNRA motif.
DISCUSSION
Theoretical approaches to RNA structure, such as energy minimization (43, 67) and comparative sequence analysis (65), are extremely well developed and generally reliable, particularly for secondary structure interactions. However, experimental determination of structure is critically important for verification of predicted models and for identification of novel higher-order interactions. To experimentally analyze the structure of this RNA molecule, we have employed chemical modification analysis using DMS, CMCT, and kethoxal to establish structural interactions in the 5′NTR of CVB3. Such analysis monitors nucleotides whose Watson-Crick pairing positions are available to solvent; most commonly as part of an accessible single-stranded region but also as a pair found at a helix end, a G-U pair, or a pair adjacent to a G-U pair (16). Our results verified many of the predicted structures in the enterovirus 5′NTR (65) (Fig. 1), but interestingly, several functionally important regions showed results that are indicative of more complex and dynamic structures. Using a combination of chemical probing results as a constraint for structure prediction in the RNASTRUCTURE algorithm (43) and comparative sequence analysis, we have generated a model that proposes novel additions to the current model for the 5′NTR from CVB3 (Fig. 8). We have used Mfold (42, 67) to compare the overall free energy of the new model (−206.6 kcal/mol) to that of the previous model (−213.9 kcal/mol), and we find the difference to be less than 4%.
FIG. 8.
Proposed structure model for the CVB3 5′NTR. For each domain, chemical probing results were entered into the RNASTRUCTURE algorithm (43) to generate a proposed structure. This structure was refined using results from the comparative sequence analysis. Strongly modified positions are indicated in red. Weakly modified positions are indicated in green.
In domain I, the overriding result is the stability of the proposed cloverleaf structure. For the vast majority of the domain, the chemical modifications follow what is expected by the established cloverleaf arrangement. The single major exception occurs at a proposed internal loop involving bases 54 to 56 and 71 to 73, all of which we show to be inaccessible to chemical modification. In the most direct interpretation, this is indicative of a continuation of the cloverleaf helix d through the internal loop, using non-Watson-Crick base pairs. We have included these pyrimidine-pyrimidine pairs in our structure model (Fig. 8). Indeed, recent NMR analysis of a molecule containing only the 30-nucleotide apical portion of stem-loop d from CVB3 shows three pyrimidine-pyrimidine pairs, consistent with the chemical protection results (47). Our results also support the idea that the 30-nucleotide region of stem-loop d used for NMR adopts a similar structure whether alone or part of the entire 5′NTR. The stability and independence of stem-loop d were proven by our earlier studies on naturally occurring CVB3 genomes containing 5′-terminal deletions of up to 49 nucleotides (29).
The primary role of the cloverleaf structure is replication. Ample evidence points to its function in assembling the protein factors of PCBP, viral protein 3CD and 3Cpro, and poly(A) binding protein to initiate and regulate replication (1, 2, 18, 19, 22, 47, 49, 66). Our probing results confirm that the cloverleaf region consists of a four-stem structure emanating from a single junction loop where the exposure of nucleotides in the loop regions of stem-loop b, stem-loop c, and the tetraloop on stem-loop d provide available bases for recognition proteins that have been shown to interact with this region of the molecule, as has been shown in poliovirus, by PCBP, viral protein 3CD and 3Cpro, and the complex formed between poly(A) binding protein, PCBP and 3CD.
A much more complex interpretation is needed for the long, pyrimidine-rich connecting region between domains I and II and the basal region of domain II. In the connecting region, probing results show an extended string of protection in bases predicted to be single stranded. It is likely that this region is involved in some type of interaction, perhaps folding into the interior of a globular portion of the molecule or participating in a triple-stranded structure. Since this pyrimidine-rich region of the 5′NTR is very highly conserved in class I IRES elements, this complex structural role is likely to be critical for the function of the molecule. Protection of the connecting region by an alternative secondary structure would be expected if this region serves as a buttress for the adjacent long-range pairing interaction that we have proposed for domain II. Together, these elements would control the structural relationship between the cloverleaf and the IRES element downstream.
Several studies with poliovirus, particularly those conducted in vitro, have established that the cloverleaf and IRES function independently, with the cloverleaf directing replication and the IRES directing translation (39, 45, 46). However, solid evidence also exists for cloverleaf-mediated effects on translation and IRES-mediated effects on replication (5, 24, 25, 45, 53, 55). Our model for a structural organization in the enterovirus 5′NTR that brings the cloverleaf and the IRES together provides a foundation for the mechanism underlying these synergistic effects. The model also accentuates the importance of the connecting region between the cloverleaf and domain II. The cloverleaf is generally reported as nt 1 to 88 and the IRES as nt 127 to 608 (CVB3 numbering) (10, 45, 63), which leaves nt 89 to 126 unassigned. It is clear from the recent mutagenesis study by De Jesus et al. (10) that this region is critical for poliovirus virulence. In addition, deletion studies designed to prove the role of the cloverleaf in replication used constructs that included nt 89 to 110 (38, 39). Our results call attention to the important structural role of the region between the cloverleaf and the IRES and emphasize the need to focus studies in that area.
Based on the experimental results, major changes are proposed for domain II. The helix originally suggested at the base of the domain shows the expected protections on the 5′ side, but many of the partnering bases on the 3′ side are modified. This behavior continues through the large internal loop above the helix, where the 5′ side is protected but the 3′ side is exposed. These results suggest that the base of the domain is not consistently paired. This is supported by comparative sequence analysis showing that some class I IRES elements do not have the potential to pair in this region (65). Indeed, our own analysis of 105 enterovirus sequences shows that several positions in the basal helix change to mismatches in a substantial number of genomes. We have proposed a long-range pairing interaction that involves the 5′ strand of domain II (Fig. 8). This pairing would bring domain II together with domain V, which links the 5′ end of the IRES with the critically important polypyrimidine track (box A) and AUG (box B) at the 3′ end of the IRES. A functional interaction between domain II and domain V is also amply supported by evidence showing that both are critical for virulence in enteroviruses (6, 14, 17, 27, 33, 40). As alluded to earlier for the connecting region between the cloverleaf and domain II, the long-range interactions may account for the protection of bases in domain II on either side of the interaction if these bases are buried into a globular domain of the molecule. The apical stem-loop of domain II is present in all class I IRES elements, and our sequence analysis shows numerous examples of compensatory base changes. Modification results provide strong support for this predicted structure.
Studies with poliovirus prove that domain III is dispensable for IRES activity and for viral infection (21). However, comparative sequence analysis shows that this domain is quite variable among class I IRES elements (65), raising the possibility that it distinguishes the biological differences between one type of virus and another. For example, bovine enterovirus contains only the apical stem-loop region, while other enteroviruses and rhinoviruses have a lower complex helix in addition to the apical stem-loop (Fig. 4). For all of these viruses, the apical loop is 4 nucleotides, but the sequence motif is highly variable and does not follow a tetraloop pattern. When a lower helix is present, there is at least one loop E sequence signature, and for most there are two such sequence motifs. The loop E motif is a helical element composed of three noncanonical base pairs surrounding a bulged purine (34). The motif was first discovered in the loop E region of 5S RNA and has since been found in numerous RNA molecules (35), where it provides a recognition feature for interactions with protein and RNA. The non-Watson-Crick base pairs in the motif form a characteristic tertiary structure that includes an important recognition feature called an S turn (9). The tertiary structure also gives a signature pattern of chemical modifications (34). In rRNAs, loop E motifs provide the key structures for complex protein interactions by translation factors (9) and zinc fingers (32). A loop E in domain III of the enterovirus 5′NTR may play a similar role. In CVB3 there are two potential loop E motifs in the complex helix of domain III (shown in Fig. 4B). Modification results are consistent with the formation of an upper loop E motif but not with the lower loop E motif. This is strongly supported by sequence comparison, where the loop E signature is present in the upper portion of the stem in every enterovirus genome, while the signature is not found in many cases in the lower portion of the stem. The upper loop E has been incorporated into our proposed 5′NTR structure (Fig. 8).
With two exceptions, modifications in domain IV follow the predicted pattern. One exception occurs in the extremely large internal loop involving bases 254 to 266 and 420 to 430. Within these supposed single-stranded regions there are long stretches of protected nucleotides. Such behavior suggests that the bases are involved in structural interactions, likely at the tertiary level. The other exception is in the stem that leads to stem-loop A of the junction loop. Modification results suggest that this stem is very flexible near the junction loop. Three elements in the junction loop region of domain IV, i.e., the C-rich loop of stem-loop A, the bulge loop in stem-loop B, and the bulge loop at position 370, are recognized by the protein PCBP in poliovirus (19). This is a key interaction for IRES function and for the regulatory step controlling translation and replication. Presentation of the PCBP recognition features is sure to depend upon the three-dimensional folding of the junction loop, which our results show to be dynamic. Indeed, a recent NMR study by Du et al. (13) showed that a single-base heterogeneity in enteroviral sequences at position 337 produces drastically different structures in stem-loop B. Whereas the loop containing 337C adopts a flexible L shape, the loop containing 337U adopts a rigid U shape.
Domain V has received a great deal of experimental attention because the attenuation mutations for the vaccine strains of poliovirus are located in this domain (17, 40). In our chemical probing experiments, the predicted structure of domain V is well supported. The large bulge loop around position 520 is of particular interest because it is conserved and is critical for poliovirus virulence (41). The mutagenesis study by Malnou et al. (41) specifically addressed the presence and role of two conserved GNRA motifs in this bulge loop. Their results showed that mutation of both sequences had the most dramatic effect, but when assayed separately, mutation of 524G-525C had the most impact. Our chemical probing is also consistent with their biochemical results, which showed the bulge loop to be somewhat protected from RNase attack and lead probes (41). However, the majority of the loop (nt 517 to 522) is exposed to solvent, as shown by Stewart and Semler (58) for poliovirus. Based upon probing results and a convincing sequence covariation pattern, we suggest that an intraloop pairing involving 523G-528C, which frames a GCAA tetraloop, is present in the bulge loop. This has been incorporated into our model of the 5′NTR (Fig. 8).
The connecting region between domains V and VI, as well as domain VI itself, contains sequences important for translation initiation. One such region is an extended pyrimidine-rich sequence called box A, and the other is a properly spaced AUG called box B (57). The box A sequence is complementary to 18S rRNA and has been implicated in a Shine-Dalgarno-like interaction (37, 64). The box B sequence is predicted to be part of a stem structure in domain VI. We show that the long connecting region containing box A is accessible to modification in the region complementary to 18S rRNA, making it quite available for a potential base-pairing interaction. We also propose a long-range pairing interaction that brings box A into proximity with domain II. In a study by Yang et al. (64), designed to test the participation of the polypyrimidine box A in ribosome binding by enhancing the pairing potential to 18S rRNA, bases 561 to 566 were mutated to nucleotides that would no longer participate in the pairing interaction proposed here. In bicistronic assays, the mutant IRES did not significantly decrease (or increase) in vitro translation. These results suggest that the pairing interaction structure is not required for the in vitro bicistronic translation assays conducted in the study, but they do not rule out the existence or importance of the interaction. Since the assays in the study by Yang et al. (64) were conducted using HeLa cell extracts, the effects of disrupting the long-range pairing interaction may have been masked. HeLa cells are extremely permissive for CVB3 and other enteroviruses. In fact, CVB3 mutants that show dramatic growth and virulence phenotypes in mouse fetal heart fibroblasts show no altered phenotype in HeLa cells (14, 33). Similarly, the Sabin strains of poliovirus show no growth phenotype in HeLa cells but are severely attenuated in cells of neuronal origin (31).
Overall the predicted model shown in Fig. 1 for the secondary structure of the CVB3 5′NTR is supported by our chemical modification and comparative sequence analysis. In our working model of the CVB3 5′NTR, we have identified regions of the model that need revision, including novel interactions not previously characterized between both local and distant nucleotides and localized sites likely to participate in a long-range pairing interaction that could mediate the functions of the cloverleaf and IRES. We have pinpointed a loop E motif likely to serve as a protein recognition feature, and we have provided evidence for a tetraloop closed by a lone pair. Although the probing results were generated with CVB3, the proposals for novel interactions can be expanded to all enteroviruses, as they were all supported by sequence comparison. The structural insights presented will be critical references to better guide mechanistic studies designed to gain a more complete understanding of the picornaviral 5′NTR.
Acknowledgments
We thank Jill Banaszak for excellent technical assistance and Nora Chapman and Steve Tracy for plasmid constructs as well as helpful discussions and critical reviews of the manuscript.
This work was supported by NIH grant P20 RR16469 from the INBRE Program of the National Center for Research.
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369-380. [DOI] [PubMed] [Google Scholar]
- 2.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand RNA synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belsham, G. J., and N. Sonenberg. 1996. RNA-protein interactions in regulation of picornavirus translation. Microbiol. Rev. 60:499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belsham, G. J., and N. Sonenberg. 2000. Picornavirus RNA translation: roles for cellular proteins. Trends Microbiol. 8:330-335. [DOI] [PubMed] [Google Scholar]
- 5.Borman, A. M., F. G. Deliat, and K. M. Kean. 1994. Sequences within the poliovirus internal ribosome entry segment control viral RNA synthesis. EMBO J. 13:3149-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradrick, S. S., E. A. Lieben, B. M. Carden, and J. R. Romero. 2001. A predicted secondary structure domain within the internal ribosome entry site of echovirus 12 mediates a cell-type specific block to viral replication. J. Virol. 75:6472-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman, N., Z. Tu, S. Tracy, and C. Gauntt. 1994. An infectious cDNA copy of the genome of a non-cardiovirulent coxsackievirus B3: its complete sequence analysis and comparison to the genomes of cardiovirulent coxsackieviruses. Arch. Virol. 135:115-130. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C., and P. Sarnow. 1995. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268:415-417. [DOI] [PubMed] [Google Scholar]
- 9.Correll, C. C., I. G. Wool, and A. Munishkin. 1999. The two faces of the Escherichia coli 23 S rRNA sarcin/ricin domain: the structure at 1.11 Å resolution. J. Mol. Biol. 292:275-287. [DOI] [PubMed] [Google Scholar]
- 10.De Jesus, N., D. Franco, A. Paul, E. Wimmer, and J. Cello. 2005. Mutation of a single conserved nucleotide between the cloverleaf and internal ribosome entry site attenuates poliovirus neurovirulnece. J. Virol. 79:14235-14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dildine, S. L., and B. L. Semler. 1989. The deletion of 41 proximal nucleotides reverts a poliovirus mutant containing a temperature-sensitive lesion in the 5′ noncoding region of the genomic RNA. J. Virol. 63:847-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du, Z., J. Yu, R. Andino, and T. L. James. 2003. Extending the family of UNCG-like tetraloop motifs: NMR structure of a CACG tetraloop from coxsackievirus B3. Biochemistry 42:4373-4383. [DOI] [PubMed] [Google Scholar]
- 13.Du, Z., N. B. Ulyanov, J. Yu, R. Andino, and T. L. James. 2004. NMR structures of loop B RNAs from the stem-loop IV domain of the Enterovirus internal ribosome entry site: a single C to U substitution drastically changes the shape and flexibility of RNA. Biochemistry 43:5757-5771. [DOI] [PubMed] [Google Scholar]
- 14.Dunn, J. J., N. M. Chapman, S. Tracy, and J. R. Romero. 2000. Natural genetics of cardiovirulence in coxsackievirus B3 clinical isolates: Localization to the 5′ nontranslated region. J. Virol. 74:4787-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn, J. J., S. S. Bradrick, N. M. Chapman, S. M. Tracy, and J. R. Romero. 2003. The stem loop II within the 5′ nontranslated region of clinical coxsackievirus B3 genomes determines cariovirulence phenotype in a murine model. J. Infect. Dis. 187:1552-1561. [DOI] [PubMed] [Google Scholar]
- 16.Ehresmann, C., F. Baudin, M. Mougel, P. Romby, J. Ebel, and B. Ehresmann. 1987. Probing the structure of RNAs in solution. Nucleic Acids Res. 15:9109-9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans, D., G. Dunn, P. D. Minor, G. C. Schild, A. J. Cann, G. Stanway, J. W. Almond, K. Currey, and J. V. Maizel. 1985. Increased neurovirulence associated with a single nucleotide change in a noncoding region of the Sabin type 3 poliovaccine genome. Nature 314:548-550. [DOI] [PubMed] [Google Scholar]
- 18.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodwin, G. H., and A. E. Dahlberg. 1982. Electrophoresis of ribosomes and polyribosomes, p. 213-225 In D. Rickwood and B. D. Hames (ed.) Polyacrlamide gel electrophoresis of nucleic acids—a practical approach. IRL Press Ltd., Oxford, United Kingdom.
- 21.Haller, A. A., J. H. Nguyen, and B. L. Semler. 1993. Minimum internal ribosome entry site required for poliovirus infectivity. J. Virol. 67:7441-7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ihle, Y., O. Ohlenschläger, S. Häfner, E. Duchardt, M. Zacharias, S. Seitz, R. Zell, R. Ramachandran, and M. Gorlach. 2005. A novel cGUUAg tetraloop structure with a conserved yYNMGg-type backbone conformation from cloverleaf 1 of bovine enterovirus 1 RNA. Nucleic Acids Res. 33:2003-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishii, T., K. Shiroki, D.-H. Hong, T. Aoki, Y. Ohta, S. Abe, S. Hashizume, and A. Nomoto. 1998. A new internal ribosome entry site 5′ boundary is required for poliovirus translation in a mouse system. J. Virol. 72:2398-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishii, T., K. Shiroki, A. Iwai, and A. Nomoto. 1999. Identification of a new element for RNA replication within the internal ribosome entry site of poliovirus RNA. J. Gen. Virol. 80:917-920. [DOI] [PubMed] [Google Scholar]
- 26.Jang, S., H.-G. Krausslich, M. J. Nicklin, G. Duke, A. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawamura, N. M., M. Kohara, S. Abe, T. Komatsu, K. Tago, M. Arita, and A. Nomoto. 1989. Determinants in the 5′ noncoding region of poliovirus Sabin 1 RNA that influence the attenuation phenotype. J. Virol. 63:1302-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kieft, J. S., K. Zhou, R. Jubin, M. G. Murray, J. Y. N. Lau, and J. Doudna. 1999. The hepatitis C internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 292:513-529. [DOI] [PubMed] [Google Scholar]
- 29.Kim, K. S., S. Tracy, W. E. Tapprich, J. Bailey, C. G. Lee, K. Kim. W. H. Barry, and N. M. Chapman. 2005. 5′ terminal deletions occur in coxsackievirus B3 during replication in murine hearts and cardiac myocyte cultures and correlate with encapsidation of negative-strand viral DNA. J. Virol. 79:7024-7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klump, W. M., I. Bergmann, B. C. Muller, D. Ameis, and R. Kandolf. 1990. Complete nucleotide sequence of infectious coxsackievirus B3 cDNA: two initial 5′ uridine residues are regained during plus-strand RNA synthesis. J. Virol. 64:1573-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.La Monica, N., and V. R. Racaniello. 1989. Differences in replication of attenuated and neurovirulent polioviruses in human neuroblastoma cell line SH-SY5Y. J. Virol. 63:2357-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, B. M., J. Xu, B. K. Clarkson, M. A. Martinez-Yamount, H. J. Dyson, D. A. Case, J. M. Gottesfeld, and P. E. Wright. 2006. Induced fit and “lock and key” recognition of 5S RNA by zinc fingers of transcription factor IIIA. J. Mol. Biol. 357:275-291. [DOI] [PubMed] [Google Scholar]
- 33.Lee, C.-K., K. Kono, E. Haas, K.-S. Kim, K. M. Drescher, N. M. Chapman, and S. Tracy. 2005. Characterization of an infectious cDNA copy of the genome of a naturally occurring, avirulent coxsackievirus B3 clinical isolate. J. Gen. Virol. 86:197-210. [DOI] [PubMed] [Google Scholar]
- 34.Leontis, N., and E. Westhof. 1998a. A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J. Mol. Biol. 283:571-583. [DOI] [PubMed] [Google Scholar]
- 35.Leontis, N., and E. Westhof. 1998b. The 5S RNA loop E: chemical probing and phylogenetic data versus crystal structure. RNA 4:1134-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindberg, A. M., P. O. Stalhandske, and U. Pettersson. 1987. Genome of coxsackievirus B3. Virology 156:50-63. [DOI] [PubMed] [Google Scholar]
- 37.Liu, Z., C. M. Carthy, P. Cheung, L. Bohunek, J. E. Wilson, B. M. McManus, and D. C. Yang. 1999. Structural and functional analysis of the 5′ untranslated region of coxsackievirus B3 RNA: in vivo translational and infectivity studies of full-length mutants. Virology 265:206-217. [DOI] [PubMed] [Google Scholar]
- 38.Lu, H. H., and E. Wimmer. 1996. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosome entry site of hepatitis C virus. Proc. Natl. Acad. Sci. USA 93:1412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyons, T., K. E. Murray, A. W. Roberts, and D. J. Barton. 2001. Poliovirus 5′ terminal cloverleaf RNA is required in cis for VPg uridylation and the initiation of negative-strand RNA synthesis. J. Virol. 75:10696-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macadam, A. J., S. R. Pollard, G. Ferguson, G. Dunn, R. Skuce, J. Almond, and P. D. Minor. 1991. The 5′ noncoding region of the type 2 poliovirus vaccine strain contains determinants of attenuation and temperature sensitivity. Virology 181:451-458. [DOI] [PubMed] [Google Scholar]
- 41.Malnou, C. E., T. A. A. Pöyry, R. J. Jackson, and K. M. Kean. 2002. Poliovirus internal ribosome entry segment structure alterations that specifically affect function in neuronal cell: molecular genetic analysis. J. Virol. 76:10617-10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 43.Mathews, D. H., M. D. Disney, J. L. Childs, S. J. Schroeder, M. Zuker, and D. H. Turner. 2004. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA 101:7287-7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moine, H., B. Ehresmann, C. Ehresmann, and P. Romby. 1998. Probing RNA structure and function in solution, p. 77-116 In R. W. Simons and M. Grunberg-Manago (ed.) RNA structure and function. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 45.Murray, K. E., B. P. Steil, A. W. Roberts, and D. J. Barton. 2004. Replication of poliovirus RNA with complete internal ribosome entry site deletions. J. Virol. 78:1393-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicholson, R., J. Pelletier, S. Y. Le, and N. Sonenberg. 1991. Structural and functional analysis of the ribosome landing pad of poliovirus type 2: in vivo translation studies. J. Virol. 65:5886-5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohlenschläger, O., J. Wohnert, E. Bucci, S. Seitz, S. Hafner, R. Ramachandran, R. Zell, and M. Görlach. 2004. The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure 12:237-248. [DOI] [PubMed] [Google Scholar]
- 48.Pallansch, M., and R. P. Roos. 2001. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p. 723-776 In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lipincott Williams and Wilkins, Philadelphia, PA.
- 49.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 50.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 51.Rivera, V., J. Welsh, and J. V. Maizel, Jr. 1988. Comparative sequence analysis of the 5′ noncoding region of the enteroviruses and rhinoviruses. Virology 165:42-50. [DOI] [PubMed] [Google Scholar]
- 52.Rohll, J. B., N. Percy, R. Ley, D. J. Evans, J. W. Almond, and W. S. Barclay. 1994. The 5′-untranslated regions of picornavirus RNAs contain independent functional domains essential for RNA replication and translation. J. Virol. 68:4384-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shiroki, K., T. Ishii, T. Aoki, M. Kobashi, S. Ohka, and A. Nomoto. 1995. A new cis-acting element for RNA replication within the 5′ noncoding region of poliovirus type I RNA. J. Virol. 69:6825-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiroki, K., T. Ishii, T. Aoki, Y. Ota, W.-X. Yang, T. Komatsu, Y. Ami, M. Arita, S. Abe, S. Hashizume, and A. Nomoto. 1997. Host range phenotype induced by mutations in the internal ribosomal entry site of poliovirus RNA. J. Virol. 71:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simoes, E. A., and P. Sarnow. 1991. An RNA hairpin at the extreme 5′ end of the poliovirus RNA genome modulates viral translation in human cells. J. Virol. 65:467-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skinner, M., V. Racaniello, G. Dunn, J. Cooper, P. D. Minor, and J. W. Almond. 1989. New model for the secondary structure of the 5′ non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J. Mol. Biol. 207:379-392. [DOI] [PubMed] [Google Scholar]
- 57.Stewart, S. R., and B. L. Semler. 1997. RNA determinants of picornavirus cap-independent translation initiation. Semin. Virol. 8:242-255. [Google Scholar]
- 58.Stewart, S. R., and B. L. Semler. 1998. RNA structure adjacent to the attenuation determinant in the 5′-non-coding region influences poliovirus viability. Nucleic Acids Res. 26:5318-5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tracy, S., K. M. Drescher, N. M. Chapman, K. S. Kim, S. D. Carson, S. Pirruccello, P. H. Lane, J. Romero, and J. S. Leser. 2002. Toward testing the hypothesis that group B coxsackieviruses (CVB) trigger insulin-dependent diabetes: inoculating nonobese diabetic mice with CVB markedly lowers diabetes incidence. J. Virol. 76:12097-12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trono, D., J. Pelletier, N. Sonnenberg, and D. Baltimore. 1988. Translation in mammalian cells of a gene linked to the poliovirus 5′ noncoding region. Science 241:445-448. [DOI] [PubMed] [Google Scholar]
- 61.Tu, Z., N. Chapman, G. Hufnagel, S. Tracy, J. R. Romero, W. H. Barry, L. Zhao, K. Currey, and B. Shapiro. 1995. The cardiovirulent phenotype of coxsackievirus B3 is determined at a single site in the genomic 5′ nontranslated region. J. Virol. 69:4607-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willian, S., N. M. Chapman, J. S. Leser, J. R. Romero, and S. Tracy. 2000. Mutations in a conserved enteroviral RNA sequence: correlation between predicted RNA structural alteration and diminished viability. Arch. Virol. 145:2061-2086. [DOI] [PubMed] [Google Scholar]
- 63.Wimmer, E., C. U. T. Hellen, and X. M. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]
- 64.Yang, D., P. Cheung, Y. Sun, J. Yuan, H. Zhang, C. M. Carthy, D. M. Anderson, L. Bohunek, J. E. Wilson, and B. McManus. 2003. A Shine-Dalgarno-like sequence mediates in vitro entry and subsequent scanning for translation initiation of coxsackievirus B3 RNA. Virology 305:31-43. [DOI] [PubMed] [Google Scholar]
- 65.Zell, R., and A. Stelzner. 1997. Application of genome sequence information to the classification of bovine enteroviruses: the importance of 5′- and 3′-nontranslated regions. Virus Res. 51:213-229. [DOI] [PubMed] [Google Scholar]
- 66.Zell, R., K. Sidigi, E. Bucci, A. Stelzner, and M. Görlach. 2002. Determinants of the recognition of enteroviral cloverleaf RNA by coxsackie B3 proteinase 3C. RNA 8:188-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]