Abstract
Human metapneumovirus (hMPV) is a recently described paramyxovirus that is a major cause of upper and lower respiratory infection in children and adults worldwide. A safe and effective vaccine could decrease the burden of disease associated with this novel pathogen. We previously reported the development of the cotton rat model of hMPV infection and pathogenesis (J. V. Williams et al., J. Virol. 79:10944-10951, 2005). We report here the immunogenicity of an hMPV fusion (F) protein in this model. We constructed DNA plasmids that exhibited high levels of expression of hMPV F in mammalian cells (DNA-F). These constructs were used to develop a novel strategy to produce highly pure, soluble hMPV F protein lacking the transmembrane domain (FΔTM). We then immunized cotton rats at 0 and 14 days with either control vector, DNA-F alone, DNA-F followed by FΔTM protein, or FΔTM alone. All groups were challenged intranasally at 28 days with live hMPV. All three groups that received some form of hMPV F immunization mounted neutralizing antibody responses and exhibited partial protection against virus shedding in the lungs compared to controls. The FΔTM-immunized animals showed the greatest degree of protection (>1,500-fold reduction in lung virus titer). All three immunized groups showed a modest reduction of nasal virus shedding. Neither evidence of a Th2-type response nor increased lung pathology were present in the immunized animals. We conclude that sequence-optimized hMPV F protein protects against hMPV infection when delivered as either a DNA or a protein vaccine in cotton rats.
Human metapneumovirus (hMPV) is a recently described respiratory pathogen that is a major cause of upper and lower respiratory infection in children and adults worldwide (5, 13, 14, 45, 50, 52). hMPV is related to respiratory syncytial virus (RSV), which is the most significant respiratory pathogen of infancy and early childhood. Epidemiologic studies showed that hMPV is associated with significant morbidity in infants and other high-risk populations, such as immunocompromised patients and individuals with underlying conditions, including prematurity, asthma, and cardiopulmonary disease (6, 34, 49, 51). Hospitalization rates due to hMPV infection in previously healthy infants or these high-risk groups are comparable to those caused by other common respiratory viruses such as RSV, parainfluenza virus (PIV), or influenza virus. Thus, hMPV has a major impact on human health, and safe, effective vaccines could decrease the burden of disease associated with this novel pathogen.
A variety of vaccine strategies have been investigated for the related virus RSV, including formalin-inactivated virus, live-attenuated, and vectored or protein subunit vaccines. Original trials of formalin-inactivated vaccine in the 1960s led to enhanced disease in vaccinees when they subsequently were infected naturally with wild-type RSV, thus halting further attempts to utilize this as a vaccine approach (20, 21). Animal studies have suggested that this phenomenon was due to an imbalanced Th2-type T-cell response to virus antigens (10). Fusion (F) protein subunit vaccines for RSV have been shown to be immunogenic and safe in seropositive children and adults (15, 18, 35). However, in rodent studies, Th2-type responses have been observed after inoculation with purified F protein, leading to concern that this could occur in seronegative infants and produce enhanced disease with natural infection (30, 31). An immature or non-native conformation of F protein in these preparations may account for the phenomenon of immune-mediated enhanced disease (41). hMPV shares substantial homology with RSV in its surface glycoproteins, leading to concern that many of these issues will hinder the development of vaccines for hMPV. Furthermore, the inability to produce large quantities of pure, conformationally intact fusion protein has been a major obstacle to subunit vaccine efforts and the paramyxovirus field in general.
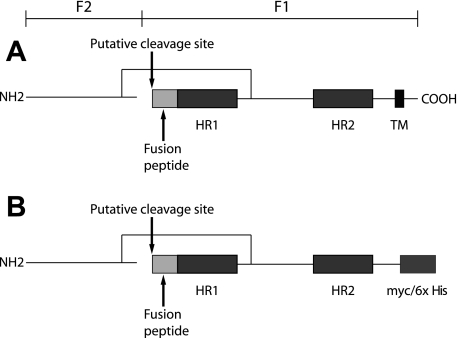
hMPV F protein clearly is related to other paramyxovirus fusion proteins and appears to have homologous regions that may have similar functions. Paramyxovirus fusion proteins are synthesized as inactive precursors (F0) that are cleaved by host cell proteases into the biologically fusion-active F1 and F2 domains (Fig. 1). hMPV has one putative cleavage site, in contrast to the two sites established for RSV F, and only shares 34% amino acid sequence identity with RSV F. F2 is extracellular and disulfide linked to F1. Fusion proteins are type I glycoproteins existing as trimers, with two 4-3 heptad repeat domains at the N- and C-terminal regions of the protein (HR1 and HR2), which form coiled-coil α-helices. These coiled coils become apposed in an antiparallel fashion when the protein undergoes a conformational change into the fusogenic state. There is a hydrophobic fusion peptide N-proximal to the N-terminal heptad repeat, which is thought to insert into the target cell membrane, while the association of the heptad repeats brings the transmembrane domain into close proximity, inducing membrane fusion (2). This mechanism has been proposed for a number of different viruses, including RSV, influenza virus, and human immunodeficiency virus (7, 16, 23). Fusion proteins are major antigenic determinants for all known paramyxoviruses and for other viruses that possess similar fusion proteins such as human immunodeficiency virus, influenza virus, and Ebola virus. Two groups have shown that hMPV F expressed in a chimeric, live-attenuated PIV vaccine was immunogenic and protective in rodents (42, 43). Consequently, we hypothesized that F expressed alone as a subunit protein or DNA vaccine would be immunogenic and protective.
FIG. 1.
(A) Schematic of hMPV fusion protein-based amino acid sequence and analogy to other paramyxoviruses. (B) F ectodomain construct FΔTM. HR, heptad repeat; TM, transmembrane domain.
The technique of DNA immunization against viruses has been studied extensively in animal and human studies (1, 3, 4, 8, 9, 12, 24, 25, 44, 47). Ulmer et al. first showed that mice could be protected against a lethal influenza virus challenge by immunization with a DNA plasmid containing an influenza virus gene (44). Numerous studies since in animal models, including mice, ferrets, and nonhuman primates, have shown plasmid DNA inoculation to be a safe and efficacious method of vaccination against a specific pathogen. Trials have been conducted in humans with malaria DNA vaccines (12, 47). However, DNA immunization alone has been poorly immunogenic in nonhuman primates and humans, leading to the development of the “prime-boost” strategy in which nonhuman primates are immunized with DNA followed by boosting either with a protein or with a recombinant virus vector. This strategy has been highly successful in immunizing chimpanzees against SIV (1). This method of immunization offers promise to overcome many of the obstacles of safety and imbalanced immune responses that have hindered the development of vaccines for viruses closely related to hMPV.
To test the hypothesis that F as a soluble protein or as a DNA construct is a major protective antigen for hMPV, we generated a soluble, epitope-tagged construct of hMPV F protein lacking the transmembrane region (FΔTM) and evaluated its immunogenicity in a cotton rat model. Our data indicate that hMPV FΔTM is expressed in mammalian cells and secreted in monomeric, dimeric, and trimeric forms. FΔTM is recognized by convalescent anti-hMPV sera from animals and humans. We demonstrate that immunization of cotton rats with FΔTM emulsified in adjuvant induces robust neutralizing antibody responses and protects animals against virus shedding in the lungs after wild-type virus challenge. Our findings indicate that FΔTM retains several important characteristics of native hMPV F protein, suggesting that it is a promising vaccine candidate and a valuable reagent for functional and structural studies of this important hMPV protein.
MATERIALS AND METHODS
Cloning of hMPV F full-length and hMPV F ectodomain.
We used reverse transcription-PCR (RT-PCR) to amplify a full-length F sequence from a pathogenic clinical isolate TN/92-4, a prototype A2 lineage strain according to the proposed nomenclature (46). The primers were 5′-CAAGAACGGGACAAATAAAAATG-3′ and 5′-CTAATTATGTGGTATGAAGCC-3′. The TN/92-4 PCR product was cloned into a commercial vector (pGEM; Promega), and the sequence was confirmed. Subsequently, EcoRI digestion was used to restriction clone TN/92-4 F into the mammalian expression vector pcDNA3.1 (Invitrogen), and the sequence of both strands was confirmed. The full TN/92-4 F sequence was sequence optimized by a commercial source (Aptagen) to alter suboptimal codon usage for mammalian tRNA bias, improve secondary mRNA structure, and remove AT-rich regions, increasing mRNA stability. The optimized full-length F sequence was cloned into pcDNA3.1 to generate the construct pcDNA3.1-F (DNA-F). To generate the hMPV F ectodomain construct (pcDNA3.1-FΔTM), the optimized full-length cDNA of the F gene was PCR amplified with the primers 5′-GGAGGTACCATGAGCTGGAAG-3′ and 5′-GAAGCGGCCGCTGCCCTTCTC-3′, and PCR product was digested and ligated into the KpnI/NotI sites (restriction sites underlined in primer sequences) of vector pcDNA3.1/myc-HisB (Invitrogen). Ligations were transformed into Escherichia coli strain DH5α-competent cells, and plasmids were purified with the QIAprep miniprep kit (QIAGEN). All plasmid constructs were sequenced on an ABI 3730xl DNA analyzer in the Vanderbilt DNA Sequencing Core Facility to confirm in-frame cloning with the C-terminal c-myc epitope and polyhistidine (His6) tag of the expression vector.
HMPV F expression in mammalian cells.
Both the native F gene cloned into pcDNA3.1 and the optimized DNA-F construct were transfected into LLC-MK2 cells in monolayer culture by using Effectene (QIAGEN) according to the manufacturer's instructions. The pcDNA3.1-FΔTM recombinant plasmid was transfected into suspension 293-F cells as recommended by the manufacturer (Freestyle 293 Expression System; Invitrogen). Both the cell fraction and the supernatant were assayed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting for FΔTM protein at 24, 48, 72, and 96 h. At the indicated time points posttransfection, the cells were centrifuged for 5 min at 100 × g at room temperature, and the supernatant and cells were harvested separately. Supernatant was filtered through 0.2-μm-pore-size filters before purification. Cells were lysed in buffer consisting of 50 mM Tris-Cl, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, and phenylmethylsulfonyl fluoride protease inhibitor cocktail (Sigma).
Purification of His6-tagged F ectodomain.
Protein purification was performed on an ÁKTA FPLC system controlled by UNICORN 4.12 software (GE Healthcare). The His-tagged F ectodomain FΔTM was purified by immobilized metal ion affinity chromatography using prepacked HisTrap Ni-Sepharose columns (GE Healthcare). The sample was diluted with concentrated binding buffer stock to adjust pH and the salt and imidazole concentration before purification. The protein was loaded onto a 5-ml HisTrap column with a loading flow rate of 5.0 ml/min, and the binding buffer contained 20 mM sodium phosphate, 0.5 M NaCl, and 30 mM imidazole (pH 7.4). Wash and elution protocols were optimized extensively for imidazole concentration and wash-elution column volumes (data not shown). Unrelated proteins were eluted in elution step 1 using 4 column volumes of 8% elution buffer, and the His6-tagged F protein was eluted in elution step 2 with 4 column volumes of 25% elution buffer. The elution buffer contained 20 mM sodium phosphate, 0.5 M NaCl, and 500 mM imidazole (pH 7.4). Purified protein was concentrated and dialyzed against phosphate-buffered saline (PBS; Invitrogen) through Amicon Ultra centrifugal filters with molecular weight cutoffs of 30,000 and 100,000 (Millipore).
Gel electrophoresis, Coomassie staining, and Western blotting.
Purified protein fractions were loaded onto NuPAGE 4 to 12% Bis-Tris gels (Invitrogen) and run at 200 V constant voltage in MES-SDS running buffer (Invitrogen). Gels were stained with Simply Blue SafeStain (Invitrogen) or Silver Stain Plus (Bio-Rad) to visualize protein bands. For Western blot analysis, separated protein bands were transferred to Invitrolon polyvinylidene difluoride membrane (Invitrogen) at 30 V for 1 h. After blocking with 5% milk in Tris-buffered saline with 0.05% Tween 20 (TBS-T), membranes were incubated with anti-polyhistidine mouse monoclonal antibody (Sigma) or anti-hMPV polyclonal guinea pig serum at a 1:500 dilution. Anti-hMPV guinea pig serum was generated in our laboratory as previously described (52). Membranes were washed with TBS-T and incubated with horseradish peroxidase-conjugated goat anti-mouse or goat anti-guinea pig secondary antibodies diluted 1:1,000 (Southern Biotech). Membranes were washed again with TBS-T and developed with TMB membrane peroxidase substrate (KPL).
Immunofluorescence detection of expressed F protein.
LLC-MK2 cell culture monolayers were transfected with pcDNA3.1-F recombinant plasmid by using TransFectin lipid reagent (Bio-Rad). At 24 h after transfection, the cells were fixed with 80% methanol, washed with PBS-T, and then incubated with anti-hMPV polyclonal guinea pig serum diluted 1:500 in PBS-T-milk for 1 h at 37°C. After being washed with PBS-T, the cells were stained with Alexa Fluor 568-conjugated goat anti-guinea pig immunoglobulin or Alexa Fluor 568-conjugated goat anti-rat immunoglobulin antibody diluted 1:1,000 (Molecular Probes) in PBS-T-milk for 1 h at 37°C. Cell monolayers were examined on an inverted Nikon Diaphot microscope, and the images were captured with a Nikon D100 digital camera. The images were cropped and the figures were constructed by using Adobe Photoshop and Illustrator without digital adjusting or reprocessing of the images.
Immunization of animals.
Animals were purchased at 5 to 6 weeks of age from a commercial breeder (Harlan, Indianapolis, IN), fed standard diet and water ad libitum, and kept in microisolator cages. Animals were anesthetized by methoxyflurane (Metofane) inhalation prior to immunization, blood sampling, or virus inoculation. Cotton rats in groups of four were immunized with 100 μg of DNA by the intramuscular route of either control vector pcDNA3.1 or pcDNA3.1-F (DNA-F) or received protein immunization with 25 μg of FΔTM adjuvanted 1:1 with TiterMax Gold (Sigma). TiterMax Gold was chosen as an adjuvant for these initial experiments based on our previous experience with its use and immunogenicity in rodents. Serum was collected from cotton rats by retro-orbital bleed. hMPV-neutralizing titers in serum were determined by a plaque reduction assay as previously described (53).
hMPV challenge.
The virus strain used was a pathogenic clinical isolate designated hMPV strain TN/96-12, a genotype group A1 virus, according the proposed nomenclature (46). The stock used had been passaged seven times since the primary isolation. Virus was grown in LLC-MK2 cells and purified over a 20%-60% discontinuous sucrose gradient as previously described (53). This virus stock was determined to have a titer of 106 PFU/ml by plaque titration in LLC-MK2 cell monolayer cultures. Cotton rats were inoculated on day 28 intranasally with 105 PFU in a volume of 100 μl. Four days later, the animals were sacrificed by CO2 asphyxiation and exsanguinated. Nasal and right lung tissues were harvested separately, weighed individually for each animal, and homogenized immediately, whereas the left lungs were inflation fixed with 4% paraformaldehyde. The right lungs were pulverized in ice-cold glass homogenizers, and nasal turbinates were ground with sterile sand in a cold porcelain mortar and pestle in 3 ml of ice-cold Hanks balanced salt solution. Tissue homogenates were centrifuged at 4°C for 10 min at 300 × g, and the supernatants were collected, divided into aliquots in cryovials, and snap-frozen in liquid nitrogen. Virus yields were measured by plaque titration as previously described (53). The Vanderbilt Institutional Animal Care and Use Committee approved the study.
Determination of gene expression levels and cytokine responses in cotton rats.
Lung tissue was harvested from infected rats at the time of sacrifice, and a 3-by-3-mm piece was placed into MagNApure mRNA lysis buffer (Roche). The tissue was homogenized by using a hand-held motorized homogenizer (VWR). mRNA was extracted from the lung homogenates by using a MagNApure LC automated nucleic acid extractor (including DNase digestion) and stored at −80°C until testing. Extracted mRNA was subjected to quantitative RT-PCR for cotton rat genes using the Quantitect Probe RT-PCR kit (QIAGEN) and a Smart Cycler II (Cepheid). Primers and probes for cotton rat genes were designed by using Primer Express (ABI) based on GenBank sequences for GAPDH (glyceraldehyde-3-phosphate dehydrogenase), interleukin-4 (IL-4), IL-5, and gamma interferon (IFN-γ) (accession numbers AF512009, AF421390, AF148211, and AF167349). The development of these assays is described elsewhere (H. S. Mok, A. Herrygers, S. Tollefson, and J. V. Williams, unpublished data). Briefly, probes were labeled at the 5′ end with 6-carboxyfluorescein (FAM) and at the 3′ end with the nonfluorescent quencher Blackhole Quencher 1 (BHQ1; Operon Biotechnologies, Inc.). Primers and probes were tested and optimized against mRNA extracted from cotton rat splenocytes. Extensive optimization was carried out to determine the optimal annealing temperatures, cycle times, and primer and probe concentrations (Mok et al., unpublished). Amplified products were sequenced to confirm the specificity of the primers. All reactions were performed in single-tube assays, with appropriate positive and negative controls. Cycle threshold (CT) data for each run were normalized to GAPDH and uninfected rats were used as the calibrator data set for the comparative 2−ΔΔCt method of relative quantitation of gene expression (26). The data were expressed as the mean fold change from the calibrator group of uninfected cotton rats.
Supernatants from lung homogenates obtained at the time of harvest were snap-frozen in a dry-ice ethanol slurry and stored at −80°C until assay. IFN-γ and IL-4 levels were measured by a sandwich capture enzyme-linked immunosorbent assay specific for cotton rat IFN-γ and IL-4, according to the manufacturer's instructions (R&D Systems). The enzyme-linked immunosorbent assay was optimized for coating conditions, blocking, and secondary antibody concentrations using recombinant cotton rat IFN-γ and IL-4 (R&D Systems). Lung supernatants were tested in duplicate, and each run included a dilutional series of recombinant cytokine standards. Ultra-TMB substrate (Pierce) was used to develop the plates, 1 N H2SO4 was used to stop the reaction after 20 min, and the optical density at 450 nm was read on a Spectramax Pro plate reader (Molecular Devices).
Pathological examination.
Specimens for histological examination were embedded in paraffin and processed, and slides were prepared in the Vanderbilt Immunohistochemistry Core, as previously described (53). All lobes of the lung were examined, including two sections per lung. Sections were reviewed by a pathologist experienced in small animal studies of RSV and hMPV (J.E.J.) without knowledge of the immunization status of the specimens. Slides were examined in their entirety in a group-blinded fashion. The following compartments of the lung were assessed: alveolar spaces, airways at all levels, the interstitium, and vessels (both arteries and veins). Inflammatory infiltrates were evaluated for location, severity, and composition (the cell types included small mononuclear cells, transformed lymphocytes, histiocytes, neutrophils, and eosinophils). The perivascular “cuff thickness” was assessed semiquantitatively as a measure of the severity of inflammation and was evaluated at the point of minimal diameter of the structure. The degrees of inflammation were graded as follows: 0, no infiltrate; 1+, most vessels had an infiltrate up to four cells thick; 2+, most vessels had an infiltrate five to seven cells thick; and 3+, most vessels had an infiltrate greater than seven cells thick. Interstitial alveolar cellularity was graded as follows: 0, no infiltrate; 1+, minimal increased cellularity without widening of septa; 2+, obvious increased cellularity with widening of septa; and 3+, markedly increased cellularity with thickened septa. This score also included blood or edema fluid in the tissue space. Lung sections also were stained with periodic acid-Schiff (PAS) to quantitate the mucus.
Statistical analysis.
Tissue virus titers were log10 transformed to adjust for non-Gaussian distribution. Mean virus titers, serum neutralizing titers, and gene expression levels were compared between groups with a two-tailed t test assuming unequal variance. Cytokine levels were compared by using the Mann-Whitney U test.
RESULTS
Sequence-optimized hMPV F is expressed in mammalian cells.
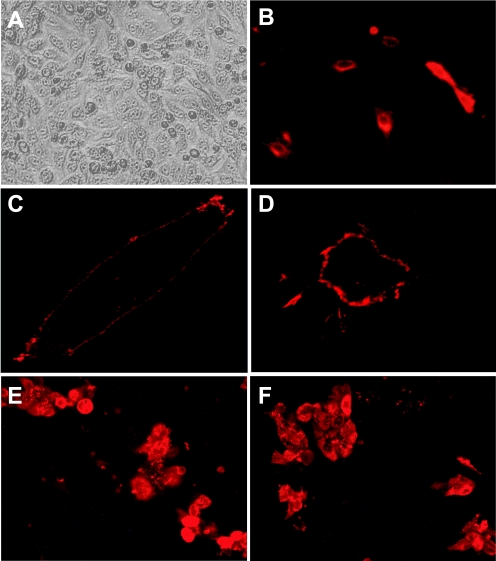
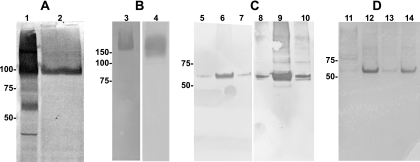
The native viral sequence was cloned into a mammalian expression plasmid and transfected into LLC-MK2 and 293T cells. This construct did not appear to express, since hMPV F protein could not be detected on cell membranes or in the cytoplasm by either immunofluorescent or Western blot assays, using anti-hMPV serum that reacted strongly with hMPV-infected cells in both assays (data not shown). Therefore, a sequence optimization strategy was undertaken to enhance expression of hMPV F in mammalian cells. Analysis of the native sequence revealed numerous rare mammalian codons, AT-rich sequences, mRNA instability motifs including poly(A) regions and cryptic splice sites, and several potential RNA secondary structure elements. In contrast to the native sequence, the sequence-optimized construct DNA-F was expressed and readily detectable by immunofluorescence when transfected into LLC-MK2 cells (Fig. 2A and B). Furthermore, DNA-F protein was detected by immunofluorescence in a membrane distribution similar to that observed in hMPV-infected cells (Fig. 2C and D). The predicted molecular mass of hMPV F protein monomer based on the nucleotide sequence is 58 kDa. Denaturing, nonreducing SDS-PAGE and immunoblot analysis of DNA-F-transfected cells showed that the majority of the hMPV F protein was detected as a dimer, whereas native F protein in hMPV-infected cell lysates was detected in monomeric, dimeric, and trimeric forms at the appropriate predicted molecular masses (Fig. 3A, lanes 1 and 2). Analysis of both hMPV-infected and DNA-F-transfected cells showed that under native, nondenaturing conditions, F protein was detected only as a trimer (Fig. 3B, lane 3, and data not shown). Conversely, under reducing conditions, F protein in both DNA-F-transfected cells and hMPV-infected cells was detected only as a monomer (data not shown). The antiserum we used to detect recombinant DNA-F in both immunofluorescent and immunoblot assays was generated previously by infecting guinea pigs with live hMPV (53). Recombinant DNA-F protein also was detected in both immunofluorescence and immunoblot assays by human serum with an hMPV-neutralizing titer of 1:640 (data not shown).
FIG. 2.
(A) DNA-F transfected LLC-MK2 cells by light microscopy. (B) Same field viewed by immunofluorescence, stained with guinea pig anti-hMPV serum and Alexa Fluor 568-goat anti-guinea pig immunoglobulin. (C) Confocal microscopy image of a single DNA-F transfected cell, showing the membrane distribution of the F protein, stained as in panel B. (D) Confocal microscopy image of a single hMPV-infected cell, showing the membrane distribution of the F protein, stained as in panel B. (E) hMPV-infected LLC-MK2 cells, stained with DNA-F immunized cotton rat serum at a 1:160 dilution and Alexa Fluor 568-goat anti-rat immunoglobulin. (F) hMPV-infected LLC-MK2 cells, stained with FΔTM protein-immunized cotton rat serum at A 1:640 dilution and secondary antibody as in panel E.
FIG. 3.
(A) Denaturing, nonreducing SDS-PAGE, immunoblotted with anti-hMPV serum. Lane 1, hMPV-infected cell lysate; lane 2, DNA-F-transfected cell lysate. (B) Native PAGE. Lane 3, hMPV-infected cell lysate; lane 4, FΔTM protein (both immunoblotted with anti-hMPV serum). (C) Reducing SDS-PAGE immunoblots of eluted fractions 2 to 4 of purified FΔTM protein. Lanes 5 to 7, blotted with anti-hMPV serum; lanes 8 to 10, blotted with anti-His monoclonal antibody (Sigma). (D) Reducing SDS-PAGE, silver-stained gel. Lane 11, original supernatant from pcDNA3.1-FΔTM-transfected 293F cells; lane 12, Ni-nitrilotriacetic acid column-purified fraction; lane 13, slight amount of FΔTM lost in flowthrough; lane 14, final purified FΔTM. Molecular mass markers in kilodaltons are shown to the left of each image. The predicted molecular mass of native F protein is 58 kDa and that of FΔTM is 56 kDa.
hMPV F ectodomain is expressed as a soluble trimer.
In order to generate a soluble form of hMPV F lacking the transmembrane domain but possessing an artificial affinity purification epitope, we cloned the truncated hMPV F gene into the plasmid pcDNA3.1 to generate the construct pcDNA3.1-FΔTM (Fig. 1). SDS-PAGE and immunoblot analysis of pcDNA3.1-FΔTM-transfected 293-F cells showed that the hMPV F ectodomain (FΔTM) was present in the medium rather than the cell lysate and thus was secreted (data not shown). The peak of FΔTM production was at 96 h posttransfection. FΔTM protein was highly pure, as determined by Coomassie blue and silver staining of SDS-PAGE gels (Fig. 3D, lanes 11 to 14). The typical yield of purified FΔTM protein was 0.5 to 1 mg/30 ml (16 to 33 mg/liter) of culture. FΔTM protein was detected in multimeric form consistent with trimers when analyzed by native PAGE and Western blotting (Fig. 3B, lane 4). The difficulty of determining the exact molecular mass under native gel conditions makes it impossible to precisely determine the multimeric state of FΔTM in this assay. However, the migration of the FΔTM multimer (Fig. 3B, lane 4) was slightly more rapid than native F in hMPV-infected cells (Fig. 3B, lane 3), which is consistent with the lower predicted molecular mass of FΔTM (56 kDa). SDS-PAGE analysis of successive eluted 2-ml fractions (fractions 2 to 4 of six fractions) of FΔTM under reducing conditions showed monomeric forms at the predicted molecular mass either by anti-hMPV immunoblotting (Fig. 3C, lanes 5 to 7) or by anti-His immunoblotting (Fig. 3C, lanes 8 to 10). FΔTM protein also was detected in immunoblot assays by human serum with an hMPV-neutralizing titer of 1:640 (data not shown). Notably, polyclonal guinea pig antiserum generated against live virus detected the FΔTM construct in Western blot (Fig. 3C, lanes 5 to 7). Both full-length F protein and FΔTM had apparent molecular masses consistent with uncleaved F0, and we did not see evidence of separate F1 and F2 subunits in the absence of trypsin treatment (data not shown).
Immunization with hMPV F DNA or protein is immunogenic and protective in cotton rats.
We immunized cotton rats intramuscularly twice, at 2-week intervals, with pcDNA3.1 alone (vector control), DNA-F, FΔTM protein adjuvanted with Titermax Gold (Sigma), or DNA-F followed by FΔTM, as described in Materials and Methods. Animals were bled on day 27 for measurement of serum antibodies to hMPV. All groups were challenged subsequently intranasally with 105 PFU of hMPV on day 28. At 4 days postinfection, the animals were sacrificed and nasal and lung tissue titers of virus were measured by plaque assay.
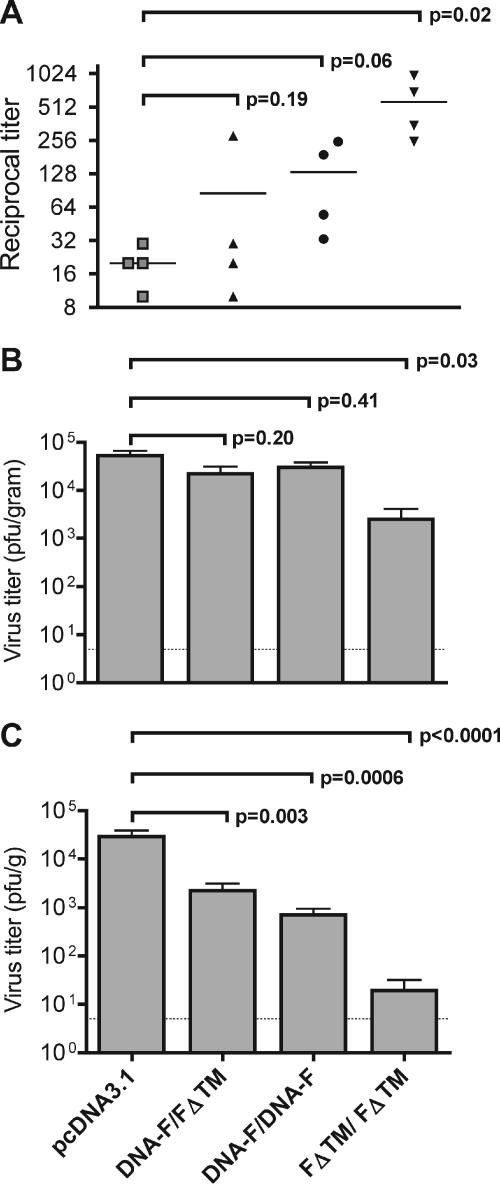
All groups except the vector control group had elevated immunofluorescent antibody titers to hMPV-infected LLC-MK2 cells (range, 1:320 to 1:1,280; Fig. 2E and F). We measured in vitro serum neutralizing titers in all groups prior to challenge. The FΔTM/FΔTM immunized animals developed a statistically significant rise in hMPV-neutralizing titers (mean, 1:570; range, 1:250 to 1:984; Fig. 4A). This titer was threefold higher than the mean serum neutralizing titer of 1:180 we previously observed in cotton rats after primary hMPV infection (53). The DNA-F/DNA-F group exhibited a rise in serum neutralizing titer that approached significance (mean, 1:132; range, 1:33 to 1:250; Fig. 4C), whereas only one animal in the DNA-F/FΔTM group developed a neutralizing antibody titer (mean, 1:86; range, 1:10 to 1:282; Fig. 4A), despite this group exhibiting a statistically significant reduction in lung virus shedding (Fig. 4C). Animals immunized twice with FΔTM showed a modest but statistically significant level of protection against shedding in nasal tissues compared to control animals (mean, 2.5 × 103 PFU/g versus 5.3 × 104 PFU/g, P = 0.03; Fig. 4B), whereas DNA-F/FΔTM and DNA-F/DNA-F groups showed nonsignificant reductions in nasal virus shedding (mean, 2.2 × 104 PFU/g [P = 0.20] and 3 × 104 PFU/g [P = 0.41], respectively; Fig. 4A). Conversely, two doses of FΔTM protein alone provided substantial protection against lung virus shedding, giving a >1,500-fold reduction in mean lung hMPV titer compared to controls (mean, 1.9 × 101 PFU/g versus 2.9 × 104 PFU/g, P < 0.0001; Fig. 4C). DNA-F/FΔTM and DNA-F/DNA-F groups showed modest but statistically significant reduction of virus replication in the lungs (mean, 2.2 × 103 PFU/g [P = 0.003] and 7 × 102 PFU/g [P = 0.0006], respectively; Fig. 4C). Cotton rats immunized with the vector control exhibited nasal and lung tissue virus replication similar to naive cotton rats during primary hMPV infection (53).
FIG. 4.
(A) Reciprocal serum neutralizing antibody titers. Bars indicate the means. (B) Nasal titer of hMPV. (C) Lung titer of hMPV. The groups are as defined in the text. Comparisons between groups were made by using the Student t test (two tailed), assuming unequal variance. Tissue virus titers were log transformed for statistical analysis. The dotted lines in panels B and C indicate the limits of detection (5 PFU/g); bars represent mean ± the standard error of the mean (SEM).
Assessment of lung histopathology after hMPV infection in immunized cotton rats.
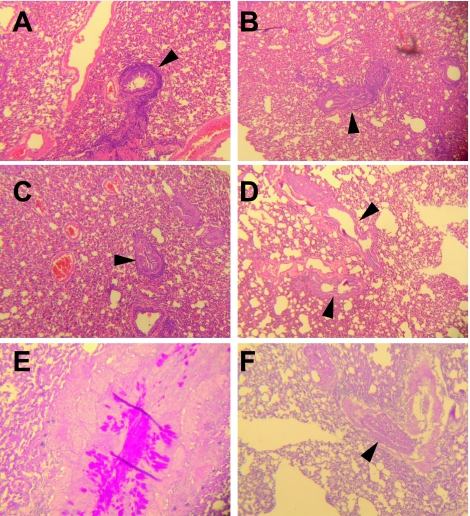
None of the challenged animals exhibited respiratory symptoms (data not shown). Lung sections from all animals were examined and scored in a blinded fashion. There were no major differences in pathological changes seen in the lungs of any groups. However, subtle differences between groups were noted (Table 1 and Fig. 5). Animals that were immunized with vector control (and thus hMPV-naive) had mild (1+) mononuclear cell interstitial infiltrates, with pronounced (1 to 2+) peribronchiolar lymphocytic cuffing (Fig. 5A), similar to changes we previously described in cotton rats with primary hMPV infection (53). All animals in the control group had copious (3+) mucus plugs present in the large airways (Fig. 5E). Animals immunized with DNA-F/FΔTM exhibited mild to moderate (1 to 2+) interstitial and circumferential peribronchiolar mononuclear cell infiltrates (Fig. 5B), with mucus plugs present in only one of four animals. Lungs from cotton rats immunized with DNA-F/DNA-F exhibited mild to moderate (1 to 2+) interstitial infiltrates, with thickening of the interstitium, and moderate (2+) circumferential peribronchiolar lymphoid cuffs (Fig. 5C). Cotton rats immunized with FΔTM/FΔTM showed mild (1+) interstitial mononuclear infiltrates and an absence of peribronchiolar mononuclear cuffs (Fig. 5D). Mild (1+) airway mucus was detected in two of four animals (Fig. 5F). Cellular infiltrates in all animals were mononuclear and appeared to be predominantly lymphocytic. Neither perivascular infiltrates nor alveolitis were present in any lung sections.
TABLE 1.
Histopathological scoring of lung sections from immunized groups of animals after hMPV challengea
| Lung section source | Score in immunized groups
|
|||
|---|---|---|---|---|
| 3.1/3.1 | DNA-F/FΔTM | DNA-F/DNA-F | FΔTM/FΔTM | |
| Interstitial infiltrates | 1+ | 1-2+ | 1-2+ | 1+ |
| Peribronchiolar infiltrates | 1-2+ | 1+ | 2+ | 0 |
| PAS+ (mucus plugs) | 3+ (all animals) | 1+ (1 of 4 animals) | 0 | 1+ (2 of 4 animals) |
Lung sections were viewed and scored by a pathologist (J. E. Johnson) without prior knowledge of the treated and control group assignment of the animals. Scoring ranged from 0 (absent) to 3+ (severe) as described in the text. All cellular infiltrates were mononuclear. There were no perivascular infiltrates in any group.
FIG. 5.
Representative sections demonstrating the histopathology of hMPV infection in the lungs of immunized cotton rats. (A) The lung of a control vector 3.1/3.1-immunized animal exhibits mild to moderate interstitial and significant peribronchiolar mononuclear infiltrates (arrowhead). (B) The lung of a DNA-F/ FΔTM-immunized animal exhibits moderate interstitial and moderate peribronchiolar mononuclear infiltrates (arrowhead). (C) The lung of a DNA-F/DNA-F-immunized rat exhibits moderate interstitial and mild peribronchiolar mononuclear cell infiltrates (arrowhead). (D) In contrast, the lung of a FΔTM/FΔTM-immunized animal exhibits minimal interstitial mononuclear infiltrates, no peribronchiolar infiltrates, and no alveolitis (arrowheads). (E) PAS stain of a control vector 3.1/3.1-immunized animal's lung reveals copious mucus in large airways. (F) PAS stain of a FΔTM/FΔTM-immunized animal's lung shows minimal mucus in airway lumen (arrow). A to D, hematoxylin and eosin staining. Original magnification (all sections), ×25.
Gene expression levels and cytokine responses in cotton rat lungs.
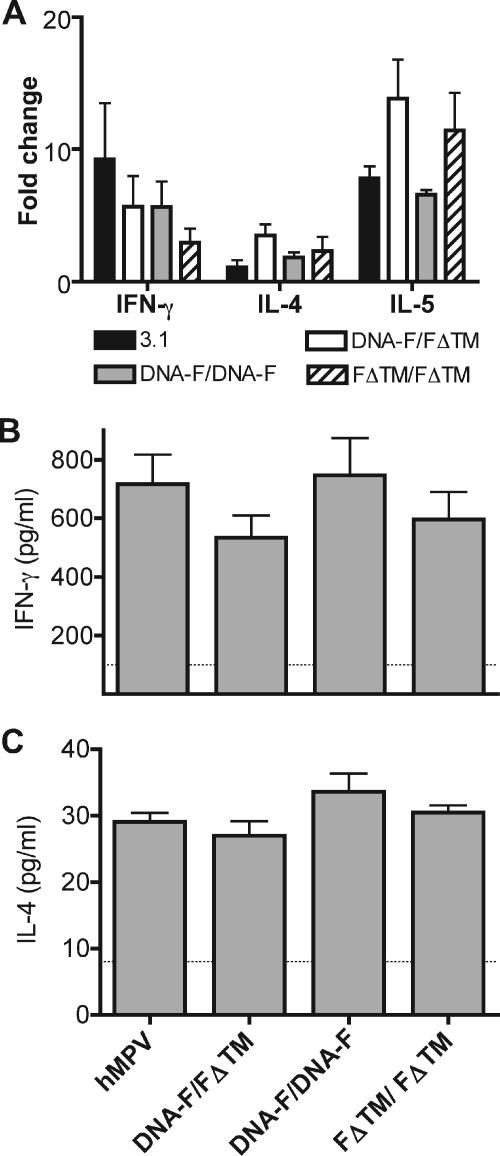
Cytokine gene expression levels were substantially upregulated in all infected groups compared to uninfected animals (Fig. 6A). However, there were no statistically significant differences between untreated hMPV-infected animals and immunized hMPV-infected animals. The only cytokine gene upregulation that approached significance was the increase in IL-4 transcription in the DNA-F/FΔTM group compared to the 3.1 control animals (P = 0.06). Animals previously immunized with FΔTM/FΔTM had less upregulation of IFN-γ and more upregulation of IL-4 than control animals, but neither of these were significant (P = 0.27 and P = 0.34, respectively). Similarly, both Th1 and Th2 soluble cytokines were present in the lungs of all infected groups, but there were no significant differences between groups (Fig. 6B and C).
FIG. 6.
(A) Cytokine gene expression levels expressed as the mean fold change ± the SEM compared to hMPV-uninfected animals. (B) IFN-γ levels in lung homogenates of hMPV-infected naive and immunized cotton rats. (C) IL-4 levels in lung homogenates of hMPV-infected naive and immunized cotton rats. Comparisons between groups were made by using the Student t test (two tailed), assuming unequal variance. The dotted line in panels B and C indicates the limit of detection; bars represent the mean ± the SEM.
DISCUSSION
We generated a sequence-optimized clone of the hMPV F gene that was expressed in mammalian cells, with a plasma membrane distribution. This recombinant protein was detected by immunofluorescence on the surface of transfected cells and by immunoblotting transfected cell lysates, using antiserum produced by infecting animals with live virus or convalescent human serum. These findings show that the recombinant F protein displayed at least some epitopes present in the native F protein and exhibited the expected plasma membrane trafficking pattern. The majority of the recombinant F protein appeared as a dimer when analyzed by immunoblot (Fig. 3A, lane 2). The majority of native F in hMPV-infected cells also appeared to be dimeric by immunoblotting (Fig. 3A, lane 1). This phenomenon also has been described for RSV F in immunoblots of RSV-infected cell lysates (40) and is likely an artifact, since paramyxovirus fusion proteins are thought to exist primarily in trimeric forms on viral and infected cell membranes.
The hMPV F ectodomain construct is also expressed in mammalian cells, and FΔTM was secreted in a soluble state and formed trimers under nondenaturing conditions. The yield of purified FΔTM protein was 16 to 33 mg/liter, comparable to reported yields from baculovirus expression systems of 5 to 25 mg/liter (27, 48). It is likely that the truncated ectodomain does not form a completely stable trimer, and the results of our purification optimization strategy support this hypothesis. The predicted molecular mass of an FΔTM trimer is 168 kDa, and yet the majority of the purified protein could be passed through a 100,000-molecular-mass cutoff membrane with little retention (data not shown). These results suggest that while FΔTM is capable of forming trimers, trimerization may be concentration and solution dependent. Similar to full-length DNA-F, the FΔTM construct was detected in immunoblots by guinea pig antiserum generated by infection with live virus and by human convalescent-phase serum. These results suggest that FΔTM retains some epitopes that are present in native protein.
Both DNA-F and FΔTM, alone or in combination, were immunogenic in cotton rats and induced partial protection against virus shedding. hMPV F-specific antibody responses evaluated by immunofluorescence against hMPV-infected cells were higher in the FΔTM/FΔTM group at >1:640, but immunofluorescence titers were detected in all vaccinated groups after two immunizations. Animals immunized with FΔTM twice showed a modest (>20-fold) but highly significant reduction in nasal virus titer. DNA-F/DNA-F- and DNA-F/FΔTM-immunized animals showed a nonsignificant reduction in nasal virus titers. In contrast to the limited protection observed in the upper respiratory tract, we found more significant reductions in virus titer in the lungs. Remarkably, two doses of FΔTM were highly protective in the lungs with a >1,500-fold reduction in lung virus titer. DNA-F/DNA-F- and DNA-F/FΔTM-immunized animals also exhibited significant reductions in lung virus titer compared to control animals, although the reductions were more modest in these groups. The reduced protection of the upper compared to the lower respiratory tract is similar to the discrepancy between upper and lower respiratory tract protection induced by primary hMPV infection in cotton rats (53). This finding also has been reported in both animal and human studies of RSV, where passively acquired or actively induced serum neutralizing antibodies protect the lower much more efficiently than the upper airways (37, 38). Epidemiologic studies have shown that humans can experience recurrent upper respiratory tract infection with hMPV and that lower respiratory tract infection early in life may be followed by upper respiratory tract infection alone with subsequent hMPV infection (54).
Serum neutralizing antibody responses were robust in the FΔTM/FΔTM group, correlating with the greater degree of lung protection in these animals. In fact, the serum titers in this group were markedly higher than the mean titer of 1:180 we previously reported in cotton rats after primary infection with hMPV (53). The antibody titers of FΔTM/FΔTM-immunized animals approximate those induced in cotton rats after multiple experimental reinfections with hMPV (G. Cseke, S. J. Tollefson, and J. V. Williams, unpublished data). These data show that FΔTM is immunogenic and capable of inducing protection. It is important to note that the present study does not establish the optimum adjuvant for FΔTM. TiterMax Gold is not approved for use in humans. Other groups have shown that a recombinant chimeric PIV strain expressing hMPV F induces protection against challenge (42, 43). Our findings suggest that F protein immunization using a nonreplicating antigen also is capable of inducing protection.
The two groups immunized with DNA-F mounted less-robust serum neutralizing titers and showed less protection against virus shedding. We hypothesize that the lower level of protection we observed with DNA immunization may be related to a dose-response relationship, since others have reported that the immunogenicity of DNA vaccination correlates with the DNA dose-body weight ratio (19, 28, 33). We did not attempt to determine an optimum dose or schedule of immunizations in these experiments. Further experiments will be needed to elucidate the relationship between the dose and route of administration and the character and quality of the induced immune response. These issues are of major importance for pursuing DNA immunization as a valid vaccine strategy in human trials (19, 33). Interestingly, the DNA-F/FΔTM group showed a >10-fold reduction in lung virus titers that was significant, despite having relatively low serum antibody titers. The mechanism underlying this apparent discrepancy is not clear. We speculate that the animals may have been partially protected by antigen-specific cytotoxic T lymphocytes, since intramuscular administration of DNA vaccines often is associated with improved cell-mediated immune responses. However, the serum neutralizing titer that correlates with lung protection is not known, and the degree of protection in this group was modest. Further studies will be needed to clarify mechanisms of immunity to hMPV.
A concern frequently raised with protein subunit vaccination for the related virus RSV is the potential for enhanced disease upon challenge, similar to the phenomenon observed in human children who received the formalin-inactivated RSV vaccine in the 1960s (20, 22). We considered this possibility in the present experiments. Evidence of enhanced lung histopathology after challenge was not seen in the lungs of cotton rats immunized with FΔTM/FΔTM in comparison to other immunized groups or control animals. In fact, the lung histopathologic scores were lower in the FΔTM/FΔTM-immunized group than in control animals, with no peribronchiolar infiltrates and diminished airway mucus. All immunized groups had less-prominent peribronchiolar infiltrates, but the absence of these infiltrates was most marked in the animals immunized with FΔTM/FΔTM alone. The absence of peribronchial lymphocytic infiltrates likely reflects decreased viral replication, given the extremely high serum neutralizing titer in this group. Notably, the FΔTM/FΔTM group did not exhibit alveolitis or increased eosinophilic or polymorphonuclear infiltrates, all characteristics of enhanced disease associated with RSV challenge after FI-RSV immunization in previous studies (11, 17, 20, 21, 32). Furthermore, determination of both lung gene expression and cytokine secretion did not reveal evidence of a Th2-type response in the FΔTM-immunized rats. The phenomenon of enhanced RSV disease has been experimentally induced in cotton rats after FI-RSV or purified RSV F protein (31, 36, 39), but increased lung pathology was not observed until virus challenge several months after vaccination. Thus, we cannot exclude this possibility in the present experiments. One recent study suggested that formalin inactivation mediates the formation of reactive carbonyl groups that are responsible for inducing Th2-type responses (29). Of note, it has not yet been established whether inactivated hMPV vaccines are associated with such a response, and thus there is currently no adequate positive control for these experiments.
In summary, the FΔTM protein construct offers potential as a subunit vaccine, and we demonstrate here that both DNA-F and FΔTM are immunogenic alone and in combination and induce a protective immune response in cotton rats. These data support the hypothesis that nonreplicating F alone is a protective antigen for hMPV. These findings have important implications for the development of vaccines and prophylactic antibodies. The soluble FΔTM protein appears to retain some epitopes that are present on the native protein and is capable of inducing virus-neutralizing antibodies. Future studies will be needed to determine whether late or waning immunity induced by FΔTM will be associated with Th2-type immune responses. This construct will facilitate structural studies of this important hMPV protein. Development of this soluble antigen as a vaccine candidate could have broader applicability to other paramyxoviruses.
Acknowledgments
This study was supported by grants from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (K08 AI-56170 [J.V.W.] and R01 AI-57933 [J.E.C.]); a Vanderbilt Department of Pediatrics Hazinski-Turner Award (J.V.W.); and a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research (J.E.C.).
We thank Thomas Utley for assistance with confocal microscopy, Bryan Shepherd for assistance with the biostatistics, Kelly Parman and the Vanderbilt Immunohistochemistry Core for assistance with specimen processing, Maggie McTighe for animal care, and Deb Macheca for administrative assistance.
Footnotes
Published ahead of print on 18 October 2006.
REFERENCES
- 1.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 2.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 3.Bembridge, G. P., N. Rodriguez, R. Garcia-Beato, C. Nicolson, J. A. Melero, and G. Taylor. 2000. DNA encoding the attachment (G) or fusion (F) protein of respiratory syncytial virus induces protection in the absence of pulmonary inflammation. J. Gen. Virol. 81:2519-2523. [DOI] [PubMed] [Google Scholar]
- 4.Bhardwaj, D., A. Kushwaha, S. K. Puri, A. Herrera, N. Singh, and V. S. Chauhan. 2003. DNA prime-protein boost immunization in monkeys: efficacy of a novel construct containing functional domains of Plasmodium cynomolgi CS and TRAP. FEMS Immunol. Med. Microbiol. 39:241-250. [DOI] [PubMed] [Google Scholar]
- 5.Boivin, G., Y. Abed, G. Pelletier, L. Ruel, D. Moisan, S. Cote, T. C. Peret, D. D. Erdman, and L. J. Anderson. 2002. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J. Infect. Dis. 186:1330-1334. [DOI] [PubMed] [Google Scholar]
- 6.Cane, P. A., B. G. van den Hoogen, S. Chakrabarti, C. D. Fegan, and A. D. Osterhaus. 2003. Human metapneumovirus in a hematopoietic stem cell transplant recipient with fatal lower respiratory tract disease. Bone Marrow Transplant. 31:309-310. [DOI] [PubMed] [Google Scholar]
- 7.Carr, C. M., and P. S. Kim. 1993. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73:823-832. [DOI] [PubMed] [Google Scholar]
- 8.Cherpelis, S., I. Shrivastava, A. Gettie, X. Jin, D. D. Ho, S. W. Barnett, and L. Stamatatos. 2001. DNA vaccination with the human immunodeficiency virus type 1 SF162ΔV2 envelope elicits immune responses that offer partial protection from simian/human immunodeficiency virus infection to CD8+ T-cell-depleted rhesus macaques. J. Virol. 75:1547-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coban, C., M. T. Philipp, J. E. Purcell, D. B. Keister, M. Okulate, D. S. Martin, and N. Kumar. 2004. Induction of Plasmodium falciparum transmission-blocking antibodies in nonhuman primates by a combination of DNA and protein immunizations. Infect. Immun. 72:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crowe, J. E., Jr. 2001. Respiratory syncytial virus vaccine development. Vaccine 20(Suppl. 1):S32-S37. [DOI] [PubMed] [Google Scholar]
- 11.Durbin, J. E., and R. K. Durbin. 2004. Respiratory syncytial virus-induced immunoprotection and immunopathology. Viral Immunol. 17:370-380. [DOI] [PubMed] [Google Scholar]
- 12.Epstein, J. E., Y. Charoenvit, K. E. Kester, R. Wang, R. Newcomer, S. Fitzpatrick, T. L. Richie, N. Tornieporth, D. G. Heppner, C. Ockenhouse, V. Majam, C. Holland, E. Abot, H. Ganeshan, M. Berzins, T. Jones, C. N. Freydberg, J. Ng, J. Norman, D. J. Carucci, J. Cohen, and S. L. Hoffman. 2004. Safety, tolerability, and antibody responses in humans after sequential immunization with a PfCSP DNA vaccine followed by the recombinant protein vaccine RTS,S/AS02A. Vaccine 22:1592-1603. [DOI] [PubMed] [Google Scholar]
- 13.Esper, F., R. A. Martinello, D. Boucher, C. Weibel, D. Ferguson, M. L. Landry, and J. S. Kahn. 2004. A 1-year experience with human metapneumovirus in children aged <5 years. J. Infect. Dis. 189:1388-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falsey, A. R., D. Erdman, L. J. Anderson, and E. E. Walsh. 2003. Human metapneumovirus infections in young and elderly adults. J. Infect. Dis. 187:785-790. [DOI] [PubMed] [Google Scholar]
- 15.Falsey, A. R., and E. E. Walsh. 1997. Safety and immunogenicity of a respiratory syncytial virus subunit vaccine (PFP-2) in the institutionalized elderly. Vaccine 15:1130-1132. [DOI] [PubMed] [Google Scholar]
- 16.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276-279. [DOI] [PubMed] [Google Scholar]
- 17.Graham, B. S. 1995. Pathogenesis of respiratory syncytial virus vaccine-augmented pathology. Am. J. Respir. Crit. Care Med. 152:S63-S66. [DOI] [PubMed] [Google Scholar]
- 18.Groothuis, J. R., S. J. King, D. A. Hogerman, P. R. Paradiso, and E. A. Simoes. 1998. Safety and immunogenicity of a purified F protein respiratory syncytial virus (PFP-2) vaccine in seropositive children with bronchopulmonary dysplasia. J. Infect. Dis. 177:467-469. [DOI] [PubMed] [Google Scholar]
- 19.Gurunathan, S., D. M. Klinman, and R. A. Seder. 2000. DNA vaccines: immunology, application, and optimization. Annu. Rev. Immunol. 18:927-974. [DOI] [PubMed] [Google Scholar]
- 20.Kapikian, A. Z., R. H. Mitchell, R. M. Chanock, R. A. Shvedoff, and C. E. Stewart. 1969. An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am. J. Epidemiol. 89:405-421. [DOI] [PubMed] [Google Scholar]
- 21.Kim, H. W., J. G. Canchola, C. D. Brandt, G. Pyles, R. M. Chanock, K. Jensen, and R. H. Parrott. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 89:422-434. [DOI] [PubMed] [Google Scholar]
- 22.Kim, H. W., J. G. Canchola, A. J. Vargosko, J. O. Arrobio, J. L. De Meio, and R. H. Parrott. 1966. Immunogenicity of inactivated parainfluenza type 1, type 2, and type 3 vaccines in infants. JAMA 196:819-824. [PubMed] [Google Scholar]
- 23.Lawless-Delmedico, M. K., P. Sista, R. Sen, N. C. Moore, J. B. Antczak, J. M. White, R. J. Greene, K. C. Leanza, T. J. Matthews, and D. M. Lambert. 2000. Heptad-repeat regions of respiratory syncytial virus F1 protein form a six-membered coiled-coil complex. Biochemistry 39:11684-11695. [DOI] [PubMed] [Google Scholar]
- 24.Letvin, N. L., D. C. Montefiori, Y. Yasutomi, H. C. Perry, M. E. Davies, C. Lekutis, M. Alroy, D. C. Freed, C. I. Lord, L. K. Handt, M. A. Liu, and J. W. Shiver. 1997. Potent, protective anti-HIV immune responses generated by bimodal HIV envelope DNA plus protein vaccination. Proc. Natl. Acad. Sci. USA 94:9378-9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, X., S. Sambhara, C. X. Li, M. Ewasyshyn, M. Parrington, J. Caterini, O. James, G. Cates, R. P. Du, and M. Klein. 1998. Protection against respiratory syncytial virus infection by DNA immunization. J. Exp. Med. 188:681-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 27.McCall, E. J., A. Danielsson, I. M. Hardern, C. Dartsch, R. Hicks, J. M. Wahlberg, and W. M. Abbott. 2005. Improvements to the throughput of recombinant protein expression in the baculovirus/insect cell system. Protein Expr. Purif. 42:29-36. [DOI] [PubMed] [Google Scholar]
- 28.McCluskie, M. J., C. L. Brazolot Millan, R. A. Gramzinski, H. L. Robinson, J. C. Santoro, J. T. Fuller, G. Widera, J. R. Haynes, R. H. Purcell, and H. L. Davis. 1999. Route and method of delivery of DNA vaccine influence immune responses in mice and non-human primates. Mol. Med. 5:287-300. [PMC free article] [PubMed] [Google Scholar]
- 29.Moghaddam, A., W. Olszewska, B. Wang, J. S. Tregoning, R. Helson, Q. J. Sattentau, and P. J. Openshaw. 2006. A potential molecular mechanism for hypersensitivity caused by formalin-inactivated vaccines. Nat. Med. 12:905-907. [DOI] [PubMed] [Google Scholar]
- 30.Murphy, B. R., A. Sotnikov, P. R. Paradiso, S. W. Hildreth, A. B. Jenson, R. B. Baggs, L. Lawrence, J. J. Zubak, R. M. Chanock, J. A. Beeler, et al. 1989. Immunization of cotton rats with the fusion (F) and large (G) glycoproteins of respiratory syncytial virus (RSV) protects against RSV challenge without potentiating RSV disease. Vaccine 7:533-540. [DOI] [PubMed] [Google Scholar]
- 31.Murphy, B. R., A. V. Sotnikov, L. A. Lawrence, S. M. Banks, and G. A. Prince. 1990. Enhanced pulmonary histopathology is observed in cotton rats immunized with formalin-inactivated respiratory syncytial virus (RSV) or purified F glycoprotein and challenged with RSV 3-6 months after immunization. Vaccine 8:497-502. [DOI] [PubMed] [Google Scholar]
- 32.Openshaw, P. J., F. J. Culley, and W. Olszewska. 2001. Immunopathogenesis of vaccine-enhanced RSV disease. Vaccine 20(Suppl. 1):S27-S31. [DOI] [PubMed] [Google Scholar]
- 33.Oran, A. E., and H. L. Robinson. 2003. DNA vaccines, combining form of antigen, and method of delivery to raise a spectrum of IFN-gamma and IL-4-producing CD4+ and CD8+ T cells. J. Immunol. 171:1999-2005. [DOI] [PubMed] [Google Scholar]
- 34.Pelletier, G., P. Dery, Y. Abed, and G. Boivin. 2002. Respiratory tract reinfections by the new human metapneumovirus in an immunocompromised child. Emerg. Infect. Dis. 8:976-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piedra, P. A., S. Grace, A. Jewell, S. Spinelli, D. Bunting, D. A. Hogerman, F. Malinoski, and P. W. Hiatt. 1996. Purified fusion protein vaccine protects against lower respiratory tract illness during respiratory syncytial virus season in children with cystic fibrosis. Pediatr. Infect. Dis. J. 15:23-31. [DOI] [PubMed] [Google Scholar]
- 36.Prince, G. A., S. J. Curtis, K. C. Yim, and D. D. Porter. 2001. Vaccine-enhanced respiratory syncytial virus disease in cotton rats following immunization with Lot 100 or a newly prepared reference vaccine. J. Gen. Virol. 82:2881-2888. [DOI] [PubMed] [Google Scholar]
- 37.Prince, G. A., V. G. Hemming, R. L. Horswood, and R. M. Chanock. 1985. Immunoprophylaxis and immunotherapy of respiratory syncytial virus infection in the cotton rat. Virus Res. 3:193-206. [DOI] [PubMed] [Google Scholar]
- 38.Prince, G. A., R. L. Horswood, and R. M. Chanock. 1985. Quantitative aspects of passive immunity to respiratory syncytial virus infection in infant cotton rats. J. Virol. 55:517-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prince, G. A., A. B. Jenson, V. G. Hemming, B. R. Murphy, E. E. Walsh, R. L. Horswood, and R. M. Chanock. 1986. Enhancement of respiratory syncytial virus pulmonary pathology in cotton rats by prior intramuscular inoculation of formalin-inactiva ted virus. J. Virol. 57:721-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roder, C., T. Krusat, K. Reimers, and H. Werchau. 2000. Purification of respiratory syncytial virus F and G proteins. J. Chromatogr. B Biomed. Sci. Appl. 737:97-106. [DOI] [PubMed] [Google Scholar]
- 41.Sakurai, H., R. A. Williamson, J. E. Crowe, J. A. Beeler, P. Poignard, R. B. Bastidas, R. M. Chanock, and D. R. Burton. 1999. Human antibody responses to mature and immature forms of viral envelope in respiratory syncytial virus infection: significance for subunit vaccines. J. Virol. 73:2956-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skiadopoulos, M. H., S. Biacchesi, U. J. Buchholz, J. M. Riggs, S. R. Surman, E. Amaro-Carambot, J. M. McAuliffe, W. R. Elkins, M. St. Claire, P. L. Collins, and B. R. Murphy. 2004. The two major human metapneumovirus genetic lineages are highly related antigenically, and the fusion (F) protein is a major contributor to this antigenic relatedness. J. Virol. 78:6927-6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang, R. S., K. Mahmood, M. Macphail, J. M. Guzzetta, A. A. Haller, H. Liu, J. Kaur, H. A. Lawlor, E. A. Stillman, J. H. Schickli, R. A. Fouchier, A. D. Osterhaus, and R. R. Spaete. 2005. A host-range restricted parainfluenza virus type 3 (PIV3) expressing the human metapneumovirus (hMPV) fusion protein elicits protective immunity in African green monkeys. Vaccine 23:1657-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ulmer, J. B., J. J. Donnelly, S. E. Parker, G. H. Rhodes, P. L. Felgner, V. J. Dwarki, S. H. Gromkowski, R. R. Deck, C. M. DeWitt, A. Friedman, et al. 1993. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science 259:1745-1749. [DOI] [PubMed] [Google Scholar]
- 45.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van den Hoogen, B. G., S. Herfst, L. Sprong, P. A. Cane, E. Forleo-Neto, R. L. de Swart, A. D. Osterhaus, and R. A. Fouchier. 2004. Antigenic and genetic variability of human metapneumoviruses. Emerg. Infect. Dis. 10:658-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vuola, J. M., S. Keating, D. P. Webster, T. Berthoud, S. Dunachie, S. C. Gilbert, and A. V. Hill. 2005. Differential immunogenicity of various heterologous prime-boost vaccine regimens using DNA and viral vectors in healthy volunteers. J. Immunol. 174:449-455. [DOI] [PubMed] [Google Scholar]
- 48.Wang, K., K. M. Holtz, K. Anderson, R. Chubet, W. Mahmoud, and M. M. Cox. 2006. Expression and purification of an influenza hemagglutinin; one step closer to a recombinant protein-based influenza vaccine. Vaccine 24:2176-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams, J. V., J. E. Crowe, Jr., R. Enriquez, P. Minton, R. S. Peebles, Jr., R. G. Hamilton, S. Higgins, M. Griffin, and T. V. Hartert. 2005. Human metapneumovirus infection plays an etiologic role in acute asthma exacerbations requiring hospitalization in adults. J. Infect. Dis. 192:1149-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams, J. V., P. A. Harris, S. J. Tollefson, L. L. Halburnt-Rush, J. M. Pingsterhaus, K. M. Edwards, P. F. Wright, and J. E. Crowe, Jr. 2004. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N. Engl. J. Med. 350:443-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams, J. V., R. Martino, N. Rabella, M. Otegui, R. Parody, J. M. Heck, and J. E. Crowe, Jr. 2005. A prospective study comparing human metapneumovirus with other respiratory viruses in adults with hematologic malignancies and respiratory tract infections. J. Infect. Dis. 192:1061-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams, J. V., S. J. Tollefson, P. W. Heymann, H. T. Carper, J. Patrie, and J. E. Crowe. 2005. Human metapneumovirus infection in children hospitalized for wheezing. J. Allergy Clin. Immunol. 115:1311-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams, J. V., S. J. Tollefson, J. E. Johnson, and J. E. Crowe, Jr. 2005. The cotton rat (Sigmodon hispidus) is a permissive small animal model of human metapneumovirus infection, pathogenesis, and protective immunity. J. Virol. 79:10944-10951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams, J. V., C. K. Wang, C. F. Yang, S. J. Tollefson, F. S. House, J. M. Heck, M. Chu, J. B. Brown, L. D. Lintao, J. D. Quinto, D. Chu, R. R. Spaete, K. M. Edwards, P. F. Wright, and J. E. Crowe, Jr. 2006. The role of human metapneumovirus in upper respiratory tract infections in children: a 20-year experience. J. Infect. Dis. 193:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]