Abstract
The mature virion of the alphaherpesvirus pseudorabies virus (PrV) contains a minimum of 31 structural proteins which are recruited into the virus particle by a network of protein-protein interactions which is only incompletely understood. We show here that deletion of the tegument protein pUL21 resulted in a drastic decrease in the incorporation of the pUL46, pUL49, and pUS3 tegument components into mature virions. Moreover, the attenuated PrV strain Bartha (PrV-Ba), which, among other defects, carries mutations in pUL21, also fails to package pUL46, pUL49, and pUS3 efficiently. By the reconstitution of wild-type pUL21 expression to PrV-Ba and the transfer of mutated PrV-Ba pUL21 into wild-type PrV, we demonstrate that this phenotype is due to the mutated pUL21.
Alphaherpesvirus virions contain in excess of 30 structural proteins which form the nucleocapsid, the tegument, and constituents of the viral envelope. The tegument, which links the viral envelope and nucleocapsid, is the least understood structure of the herpesvirus virion. It consists of numerous viral and several cellular proteins which are recruited into the virus particle by a network of protein-protein interactions which is only incompletely understood (22, 23). Generally, the importance of viral proteins in virus replication has been analyzed by mutating single viral gene products and ascribing the observed phenotypes directly to the mutated protein. However, both functional redundancy, which precludes a direct correlation of a phenotype to the absence of one viral protein (2, 6, 15), and trans effects of mutations in a specific protein on other viral components have been observed (4, 25). Thus, the assignment of a particular phenotype to a specific mutation requires an in-depth analysis of possible effects on other viral or cellular factors.
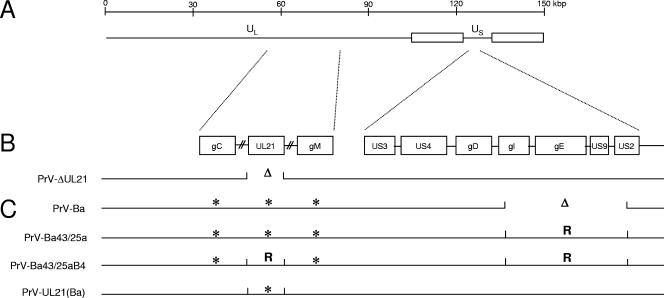
The pUL21 protein of the alphaherpesvirus pseudorabies virus (PrV) has been described as a capsid-associated protein involved in capsid maturation during cleavage and encapsidation of the viral genome (3, 33). However, a UL21 mutant of PrV is only slightly impaired in replication in cell culture (13, 14) and in a murine intranasal infection model (12) but is strongly attenuated in PrV's natural host, the pig (13). This indicates that pUL21 is important for PrV replication in vivo. Although little is known about pUL21 function, interestingly the live attenuated PrV strain Bartha (PrV-Ba) (1), besides containing a large deletion in the unique short (US) region eliminating the genes for glycoprotein I (gI) (pUS7), gE (pUS8), pUS9, and pUS2 (17, 24, 29), contains point mutations in the signal sequence of gC (30), in gM (5), in pUS3 (20), and in pUL21 (13). Marker rescue experiments showed that the deleted genes as well as UL21 are important for attenuation since their repair with the corresponding genes from PrV strain Kaplan (PrV-Ka) also restored to PrV-Ba the virulent phenotype of PrV-Ka (13, 17, 18). However, the molecular basis for this phenotype and which role pUL21 plays in PrV replication are still unclear.
Lyman and colleagues demonstrated by Western blot analysis of purified virions that PrV-Ba fails to package the tegument proteins pUS3 and pUL49 (VP22) (20). However, this has been challenged by Granzow et al. (9), who detected pUS3 and pUL49 in highly purified PrV-Ba virion preparations. Restoration of the large deletion in the US region by the corresponding gene from the PrV strain Becker did not restore pUS3 packaging into virions (20), and restoration of gE expression did not restore pUL49 incorporation despite the known interaction between the two (7). Thus, a discrepancy still exists about the question of whether PrV-Ba is indeed unable to package pUS3 and pUL49 and, if it is, which PrV-Ba component is responsible for this phenotype.
We recently adapted a highly quantitative method for the analysis of proteins in purified herpesvirus virions (25). The original procedure has been designated stable isotope labeling by amino acids in cell culture (SILAC) (27). Here, the samples to be compared were differentially labeled metabolically with either unmodified leucine or leucine carrying three atoms of the stable heavy hydrogen isotope deuterium. After gel electrophoresis and mass spectrometry, the ratio of each protein in the virus particle was determined with high accuracy, using the constant amount of 960 copies per capsid of the pUL19 major capsid protein (32) as the standard. Thus, we reanalyzed the protein composition of PrV-Ba in comparison to that of wild-type PrV-Ka (10), to that of a UL21 deletion mutant of PrV-Ka (PrV-ΔUL21) (14) as well as to that of rescuants of PrV-Ba isolated after marker rescue with the US region of PrV-Ka (PrV-Ba43/25a) (17), and, in addition, with BamHI fragment 4 of PrV-Ka, which includes the pUL21 mutations (18). Moreover, the PrV-Ba UL21 gene was used to restore UL21 expression to PrV-ΔUL21, which carries a complete deletion of the UL21 gene (Fig. 1). Viruses were propagated on porcine kidney cells and purified by sucrose density gradient centrifugation (11). Purified PrV-Ka was propagated in cells cultured in Dulbecco's modified Eagle's medium-F-12 medium (catalog no. D-9785; Sigma-Aldrich) containing exclusively deuterated leucine (l-leucine-5,5,5-d3, 99 atom% D, catalog no. 486825; Sigma-Aldrich), which resulted in the insertion of a mass tag of 3 Da per leucine residue. PrV-Ka virions served as a global internal standard for the SILAC procedure. All other viruses were propagated on cells maintained in medium supplemented with conventional leucine. To compare the relative amounts of proteins in the different viruses in relation to those in PrV-Ka, purified virion preparations were mixed at a 1:1 protein ratio and mixtures were separated by standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (16). Thereafter, bands of interest were cut from the Coomassie-stained gels (26). After tryptic digestion (31), peptide mass fingerprint spectra were obtained on an Ultraflex mass spectrometer (Bruker, Bremen, Germany), and the data were further processed by FlexAnalysis and BioTools software (Bruker). Database queries were performed with MASCOT software (Matrixscience, London, United Kingdom) (28), using an in-house database representing the PrV proteome as compiled from the Swiss-Prot database (8). Quantitative evaluation of the spectra was carried out using self-developed software. Mass lists were screened for peak pairs representing leucine-containing peptides of the respective protein in the conventional and the mass-tagged variant, and an average intensity ratio reflecting the relative amounts of protein originating from PrV-Ka, PrV-Ba, or the recombinant viruses was calculated from all qualified peaks of the spectrum. Intensity ratios of the tegument proteins were normalized to the intensity ratios of the major capsid protein, which is present in every intact virus particle in the capsid in 960 copies (32), to correct for any variations in protein input (25).
FIG. 1.
Diagram of viruses used in this study. (A) A schematic map of the PrV genome with location of the long (UL) and short (US) unique regions. (B) Localization of the genes of interest in wild-type PrV-Ka. In panel C, the defects in the analyzed virus mutants are indicated. Δ indicates deletion of the corresponding portion of the viral genome. * indicates the presence of mutations within the respective open reading frames. R, repair with PrV-Ka sequences.
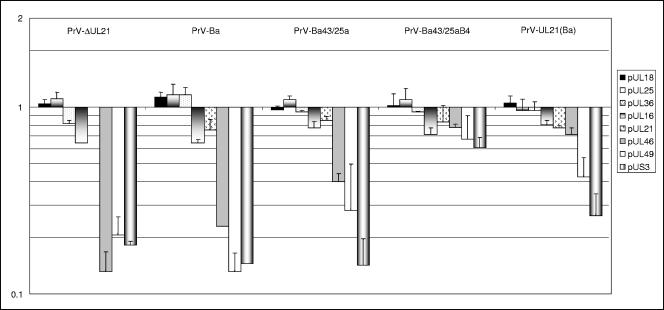
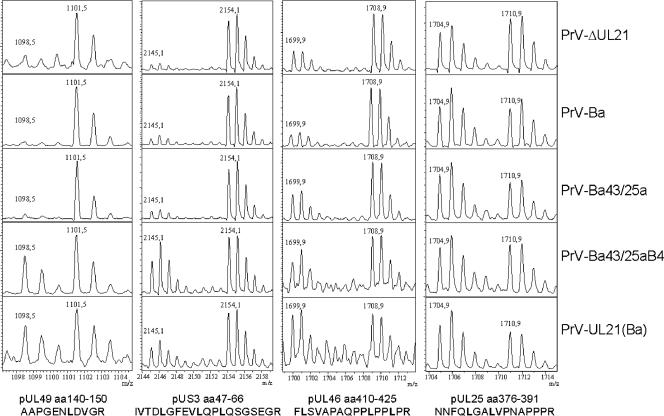
Deletion of UL21 from PrV-Ka resulted in a decrease in the incorporation of the pUL46, pUL49, and pUS3 proteins into virions (Fig. 2) by 80 to 90%. A similar reduction in the incorporation of these proteins compared to PrV-Ka was observed with PrV-Ba, indicating that PrV-Ba still packages pUL46, pUL49, and pUS3 but at strikingly lower levels. Thus, the differences observed in the assessment of whether PrV-Ba packages pUS3 and pUL49 (9, 20) can be explained by the nonquantitative nature of the immunoblot analysis and differences in the sensitivities of the immunological reagents used. Restoration of expression of gI, gE, pUS9, and pUS2 in PrV-Ba43/25a did not correct the pUS3 incorporation defect (Fig. 2), which is in line with previous experiments (20), but increased the incorporation of pUL46 and pUL49. The latter finding may be due to the interaction of gE with pUL49 (7). Interestingly, the incorporation of pUL46, pUL49, and pUS3 was restored to nearly wild-type levels when, in addition to the repair of the US deletion, an intact wild-type UL21 gene derived from PrV-Ka was present in the PrV-Ba genome. Thus, in a PrV-Ba background, PrV-Ka pUL21 aids in the incorporation of pUL46 and pUL49 and is required for the efficient virion localization of pUS3. This is further corroborated after transfer of the mutated UL21 gene of PrV-Ba into PrV-Ka. In PrV-UL21(Ba), pUS3 incorporation is decreased by more than 70% and that of pUL49 by ca. 60%, whereas incorporation of pUL46 was only slightly affected. To allow a direct comparison of the obtained spectra, examples of relevant regions of the peptide mass fingerprints are compiled in Fig. 3. In all virus preparations, the amounts of the capsid protein pUL18 and the capsid-associated pUL25 did not differ significantly from that of wild-type PrV-Ka, as indicated by their values around 1. The same was observed for the inner tegument protein pUL36 and tegument proteins pUL16 and pUL21 (Fig. 2). PrV-Ka pUL21 has been shown to form a complex with pUL16 (14), which, in turn, may interact with pUL11 (19). Complex formation with pUL16 has also been observed for PrV-Ba pUL21 (data not shown), so the mutations apparently do not interfere with interaction between pUL16 and pUL21. The intracellular levels of these proteins did not differ significantly between the different virus mutants, which correlated with similar RNA expression profiles as detected by Northern blot analysis (data not shown).
FIG. 2.
Quantitation of capsid (pUL18), capsid-associated (pUL25), and tegument (pUL36, pUL16, pUL21, pUL46, pUL49, and pUS3) proteins in purified virions. Results are expressed on a logarithmic scale in multiples of the amount of the respective protein present in wild-type PrV-Ka. Values below 1 indicate a decrease in incorporation. Error bars indicate the standard deviations for at least three different independent virus preparations.
FIG. 3.
Peptide mass spectra of selected peak pairs representing pUL49, pUS3, pUL46, and pUL25. The x axis indicates molecular masses (m/z values), and the y axis is given in arbitrary relative intensity units. The lighter peak of every diagram (left) represents the tryptic peptide whose sequence is given at the bottom and which originates from the virus indicated on the right. Heavy peaks (right) represent the corresponding peptides of the deuterium-labeled protein originating from wild-type PrV-Ka, which had been added as an internal standard. Note that the number of leucine residues (in bold) corresponds to the mass distance of 3 Da (pUL49), 9 Da (pUS3 and pUL46), or 6 Da (pUL25) of the respective light and heavy peaks in the spectra. Only minute amounts of light peaks (1,098.5 Da) were detected for the pUL49-specific peptide of PrV-ΔUL21, PrV-Ba, and PrV-Ba43/25a. aa, amino acids.
In summary, we demonstrate that PrV pUL21 is a virion component which is necessary for the efficient incorporation of pUL46, pUL49, and pUS3 into mature PrV virions. The serine/threonine kinase pUS3 and the major tegument protein pUL49 are not packaged efficiently into mature virions in the absence of pUL21 or in the presence of mutated PrV-Ba pUL21, which indicates that the point mutations in pUL21 of PrV-Ba interfere with the incorporation of pUS3 and pUL49 but not pUL46. Thus, different domains of the protein may be important for this differential trans effect of pUL21 on virion localization of these PrV tegument proteins.
It has been described that the absence of pUS3 results in inefficient virion localization of pUL46 in herpes simplex virus type 2 (21) and PrV-Ka (25). Thus, the defects observed in pUL46 incorporation in PrV-ΔUL21, PrV-Ba, or PrV-Ba43/25a may be an indirect effect of the deficient pUS3 incorporation. However, PrV-UL21(Ba) packages nearly wild-type amounts of pUL46 while incorporation of pUS3 is strikingly decreased, indicating that there is no direct interaction between the two.
Our data demonstrate another hitherto unknown direct or indirect interaction between herpesvirus tegument proteins yielding an additional piece for the herpesvirus assembly puzzle (22). pUL21 is a virion protein conserved throughout the Herpesviridae, whereas pUS3, pUL46, and pUL49 are found only in the alphaherpesviruses. Thus, it is conceivable that pUL21 of beta- and gammaherpesviruses interacts with other nonconserved tegument proteins during virion morphogenesis. Quantitation of virion components by SILAC and mass spectrometry should help to analyze whether this is indeed the case.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (DFG Me 854/5-2).
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Bartha, A. 1961. Experimental reduction of virulence of Aujeszky's disease virus. Magy. Allatorv. Lapja 16:42-45. [Google Scholar]
- 2.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 43:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Wind, N., F. Wagenaar, J. Pol, T. Kimman, and A. Berns. 1992. The pseudorabies virus homolog of the herpes simplex virus UL21 gene product is a capsid protein which is involved in capsid maturation. J. Virol. 66:7096-7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dietz, P., B. G. Klupp, W. Fuchs, B. Köllner, E. Weiland, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J. Virol. 74:5083-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dijkstra, J. M., T. C. Mettenleiter, and B. G. Klupp. 1997. Intracellular processing of pseudorabies virus glycoprotein M (gM): gM of strain Bartha lacks N-glycosylation. Virology 237:113-122. [DOI] [PubMed] [Google Scholar]
- 6.Farnsworth, A., K. Goldsmith, and K. G. Kousoulas. 2003. Herpes simplex virus glycoproteins gD and gE/gI serve essential but redundant functions during acquisition of the virion envelope in the cytoplasm. J. Virol. 77:8481-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2004. The pseudorabies virus US3 protein is a component of primary and of mature virions. J. Virol. 78:1314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7:394-407. [DOI] [PubMed] [Google Scholar]
- 11.Karger, A., B. Bettin, H. Granzow, and T. C. Mettenleiter. 1998. Simple and rapid purification of alphaherpesviruses by chromatography on a cation exchange membrane. J. Virol. Methods 70:219-224. [DOI] [PubMed] [Google Scholar]
- 12.Klopfleisch, R., B. G. Klupp, W. Fuchs, M. Kopp, J. P. Teifke, and T. C. Mettenleiter. 2006. Influence of pseudorabies virus proteins on neuroinvasion and neurovirulence in mice. J. Virol. 80:5571-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klupp, B. G., B. Lomniczi, N. Visser, W. Fuchs, and T. C. Mettenleiter. 1995. Mutations affecting the UL21 gene contribute to avirulence of pseudorabies virus vaccine strain Bartha. Virology 212:466-473. [DOI] [PubMed] [Google Scholar]
- 14.Klupp, B. G., S. Boettcher, H. Granzow, M. Kopp, and T. C. Mettenleiter. 2005. Complex formation between the UL16 and UL21 tegument proteins of pseudorabies virus. J. Virol. 79:1510-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, and T. C. Mettenleiter. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 78:3024-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of the structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Lomniczi, B., S. Watanabe, T. Ben-Porat, and A. S. Kaplan. 1984. Genetic basis of the neurovirulence of pseudorabies virus. J. Virol. 52:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lomniczi, B., S. Watanabe, T. Ben-Porat, and A. S. Kaplan. 1987. Genome location and identification of functions defective in the Bartha vaccine strain of pseudorabies virus. J. Virol. 61:769-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loomis, J. S., R. J. Courtney, and J. W. Wills. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 77:11417-11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyman, M. G., G. L. Demmin, and B. W. Banfield. 2003. The attenuated pseudorabies virus strain Bartha fails to package the tegument proteins Us3 and VP22. J. Virol. 77:1403-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuzaki, A., Y. Yamauchi, A. Kato, F. Goshima, Y. Kawaguchi, T. Yoshikawa, and Y. Nishiyama. 2005. US3 protein kinase of herpes simplex virus type 1 is required for the stability of the UL46-encoded tegument protein and its association with virus particles. J. Gen. Virol. 86:1979-1985. [DOI] [PubMed] [Google Scholar]
- 22.Mettenleiter, T. C. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet. Microbiol. 113:163-169. [DOI] [PubMed] [Google Scholar]
- 23.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Mol. Biol. 9:423-429. [DOI] [PubMed] [Google Scholar]
- 24.Mettenleiter, T. C., N. Lukacs, and H.-J. Rziha. 1985. Pseudorabies virus avirulent strains fail to express a major glycoprotein. J. Virol. 56:307-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michael, K., B. G. Klupp, T. C. Mettenleiter, and A. Karger. 2006. Composition of pseudorabies virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J. Virol. 80:1332-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neuhoff, V., N. Arold, D. Taube, and W. Ehrhardt. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9:255-262. [DOI] [PubMed] [Google Scholar]
- 27.Ong, S. E., B. Blagoev, I. Kratchmarova, D. B. Kristensen, H. Steen, A. Pandey, and M. Mann. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1:376-386. [DOI] [PubMed] [Google Scholar]
- 28.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 29.Petrovskis, E. A., J. G. Timmins, T. M. Gierman, and L. E. Post. 1986. Deletions in vaccine strains of pseudorabies virus and their effect on synthesis of glycoprotein gp63. J. Virol. 60:1166-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robbins, A. K., J. P. Ryan, M. E. Whealy, and L. W. Enquist. 1989. The gene encoding the gIII envelope protein of pseudorabies virus vaccine strain Bartha contains a mutation affecting protein localization. J. Virol. 63:250-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenfeld, J., J. Capdevielle, J. C. Guillemont, and P. Ferrara. 1992. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203:173-179. [DOI] [PubMed] [Google Scholar]
- 32.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, W. M. Burnett, and R. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, NY.
- 33.Wagenaar, F., J. M. A. Pol, N. de Wind, and T. G. Kimman. 2001. Deletion of the UL21 gene in pseudorabies virus results in the formation of DNA-deprived capsids: an electron microscopy study. Vet. Res. 32:47-54. [DOI] [PubMed] [Google Scholar]