Abstract
The varicella-zoster virus (VZV) ORF9 protein is a member of the herpesvirus UL49 gene family but shares limited identity and similarity with the UL49 prototype, herpes simplex virus type 1 VP22. ORF9 mRNA is the most abundantly expressed message during VZV infection; however, little is known concerning the functions of the ORF9 protein. We have found that the VZV major transactivator IE62 and the ORF9 protein can be coprecipitated from infected cells. Yeast two-hybrid analysis localized the region of the ORF9 protein required for interaction with IE62 to the middle third of the protein encompassing amino acids 117 to 186. Protein pull-down assays with GST-IE62 fusion proteins containing N-terminal IE62 sequences showed that amino acids 1 to 43 of the acidic transcriptional activation domain of IE62 can bind recombinant ORF9 protein. Confocal microscopy of transiently transfected cells showed that in the absence of other viral proteins, the ORF9 protein was localized in the cytoplasm while IE62 was localized in the nucleus. In VZV-infected cells, the ORF9 protein was localized to the cytoplasm whereas IE62 exhibited both nuclear and cytoplasmic localization. Cotransfection of plasmids expressing ORF9, IE62, and the viral ORF66 kinase resulted in significant colocalization of ORF9 and IE62 in the cytoplasm. Coimmunoprecipitation experiments with antitubulin antibodies indicate the presence of ORF9-IE62-tubulin complexes in infected cells. Colocalization of ORF9 and tubulin in transfected cells was visualized by confocal microscopy. These data suggest a model for ORF9 protein function involving complex formation with IE62 and possibly other tegument proteins in the cytoplasm at late times in infection.
Varicella-zoster virus (VZV) is the etiologic agent of varicella (chicken pox) during primary infection of the human host and herpes zoster (shingles) upon reactivation from latency. The vital genome is approximately 125 kb in size and encodes at least 71 open reading frames (ORFs). Among the first viral proteins encountering the host cell after herpesvirus infection are those of the tegument, a proteinaceous layer lying between the viral capsid and envelope (7). VZV tegument proteins thus far identified experimentally include those encoded by ORFs 4, 62, 63, 10, and 47 (26, 29, 47).
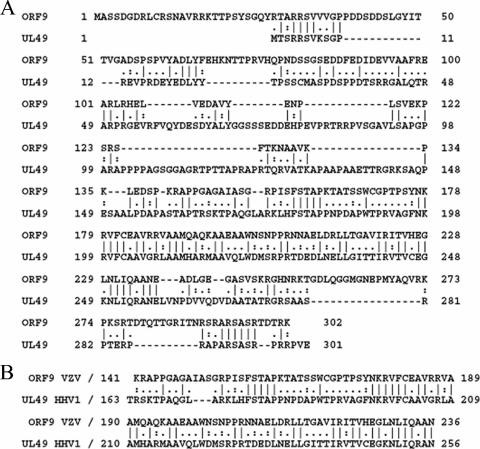
The product of ORF9, the VZV ORF9 protein, is predicted to be a 302-amino-acid (aa) polypeptide expressed at late postinfection times. The ORF9 protein is a member of the alphaherpesvirus UL49 gene family and, as the orthologue of herpes simplex virus type 1 (HSV-1) VP22, the prototype of that family, is believed to be a major component of the VZV virion tegument. The VZV ORF9 protein has not been well characterized, despite the fact that its transcript is the most abundant viral message expressed during lytic infection (11, 23). VZV ORF9 displays 25% identity and 34% similarity to the HSV VP22 protein, with the most divergent regions occurring at the amino and carboxyl termini of the two proteins (Fig. 1A). Further, the core UL49 homology region in the ORF9 protein is only 43% identical and 56% similar to the corresponding region in VP22 (Fig. 1B). Thus, while it is likely that the ORF9 protein may show some functions similar to those of VP22, it is equally likely that it will be found to display different properties as well, in keeping with several other VZV proteins that have been found to differ to various extents from their HSV orthologues (9, 26, 29). Further, orthologues of VP22 from other alphaherpesviruses have also been shown to exhibit properties and functions different from those of VP22. As an example, while VP22 is not required for cell growth in tissue culture, its Marek's disease virus orthologue has been shown to be essential for growth (13). Similarly, bovine herpesvirus VP22 (BVP22) has been reported to display distinct qualitative differences in microtubule, nuclear, and chromatin associations (20).
FIG. 1.
Sequence comparison of the VZV ORF9 protein and HSV-1 UL49 (VP22). (A) Alignment of the complete ORF9 and VP22 amino acid sequences. (B) Alignment of the UL49 homology domains of ORF9 and VP22.
VP22 has been reported to interact with the potent C-terminal acidic activation domain of the HSV-1 VP16 protein (15). Both of these proteins are present in the HSV-1 virion tegument, and VP16 is absolutely required for viral growth (1, 41, 50). The relatively weak activation domain of the VZV orthologue of VP16, the ORF10 protein, is situated near its N terminus, and the ORF10 protein is dispensable for growth in tissue culture (8, 10, 35). In contrast, IE62, the major VZV transactivator, contains a potent N-terminal acidic activation domain and is present in significant levels in the tegument of viral particles (9, 29, 39).
The mechanism by which IE62 is incorporated into the viral tegument is largely unknown. The viral kinases encoded by VZV ORF47 and ORF66 have been shown to be capable of phosphorylating IE62. The ORF47 kinase, which is also present in the viral tegument, has been shown to form a stable complex with IE62. The region of the kinase involved in this complex is distinct from the enzyme's catalytic domain (5). The specific sites within IE62 that are phosphorylated by the ORF47 kinase are unknown, but at least nine have been predicted based on their similarity to the consensus ORF47 phosphorylation motif (24). The ORF66 kinase phosphorylates IE62 at late times during infection at serine residues in close proximity to the IE62 nuclear localization signal (14). This phosphorylation event is required for the redistribution/exclusion of IE62 from the nucleus to the cytoplasm and subsequent incorporation into the viral particle (28).
In this study, we show that the VZV ORF9 protein interacts with the major VZV transactivator IE62. We show that the two proteins can be coimmunoprecipitated from infected cell extracts. We have mapped the regions required for this interaction to the central portion of the ORF9 protein (aa 117 to 186) via yeast two-hybrid assays. The region of IE62 involved in this interaction, based on glutathione S-transferase (GST) pull-down assays, is the N-terminal half of the IE62 acidic activation domain. Confocal microscopy of cells transfected with plasmids expressing the ORF9 protein and IE62 showed that the ORF9 protein, in contrast to VP22, localized almost exclusively to the cytoplasm while IE62 is nuclear. Cotransfection with a third plasmid expressing the ORF66 kinase resulted in the redistribution of IE62 from the nucleus to the cytoplasm and colocalization of the ORF9 protein and IE62 in the cytoplasm in filamentous structures. The colocalization of the two proteins under these conditions was similar to that observed in infected cells. Confocal microscopy of cells cotransfected with plasmids expressing the ORF9 protein and the VZV ORF47 kinase showed that the two polypeptides colocalized in the cytoplasm. Luciferase reporter assays showed that the ORF9 protein does not affect the transcriptional activity of IE62 under our experimental conditions in the presence or absence of either of the viral kinases. Taken together, these data suggest a role for the VZV ORF9 protein in the recruitment of IE62 and possibly the ORF47 kinase to complexes within the cytoplasm. The complexes may be involved in viral tegument formation and cell-to-cell spread of the virus during late times in infection.
MATERIALS AND METHODS
Cells and viruses.
Melanoma cells (MeWo) and HeLa cells (ATCC CCL2) were grown in Eagle's minimal essential medium supplemented with 10% fetal bovine serum as previously described (25, 48). VZV strain MSP was propagated in MeWo cell monolayers as described by Lynch et al. (32) and Peng et al. (38).
Plasmids.
The pCDNA-ORF9 vector was constructed by inserting the ORF9 tegument protein, generated by PCR amplification, into the pCDNA3.1(+) plasmid (Invitrogen, Carlsbad, CA). In order to generate a plasmid expressing the VZV ORF66 serine threonine kinase, the ORF66 coding region was amplified from the pBACgus-2cp/ORF66 vector by PCR with the following primers: 5′-GCTAGCATGAACGACGTTGATGCAACAG, including an NheI restriction site, and 3′-GGTACCTTAATCTCCAACTTCCATTGG, including a KpnI restriction site. The resulting PCR product was inserted as an NheI-KpnI fragment into plasmid pCDNA 3.1(+) (Invitrogen, Carlsbad, CA). In-frame positioning and correctness of the inserted sequence were verified by gene sequencing. The p61GL2 reporter vector was constructed by inserting the ORF61 promoter, generated by PCR amplification using the following primers: 5′-CCCGGGGCTGTATACCCGGCCCAAGGTTATAC-3′, which generated an SmaI site, and 3′-GCTAGCATGTCCGGGCATCCAAACACGTAGC-5′, which generated an NheI site. The resulting PCR product was inserted as an SmaI-NheI fragment into the pGL2B plasmid (Promega, Madison, WI) upstream of the firefly luciferase reporter gene. The ORF29 reporter plasmid with the ORF29 promoter driving expression of the firefly luciferase gene was the gift of Min Yang. The expression plasmids pCAGGS-ORF47.12 and pCMV62 have previously been described (25, 32).
Expression and purification of ORF9 from recombinant baculovirus.
The VZV tegument protein ORF9 was expressed using an Ek/LIC cloning kit and a BacVector transfection kit (Novagen, Madison, WI) according to the manufacturer's instructions. The ORF9 coding region was amplified from the pGADT7-ORF9 plasmid by PCR. The primers (Integrated DNA Technologies, Coralville, IA) were as follows: 5′-GACGACGACAAGATGGCATCTTCCGACGGTGAC and 3′-GAGGAGAAGCCCGGTCTATTTTCGCGTATCAGT. The purified PCR product was annealed to the pBACgus-2cp vector (Novagen). The pBACgus-2cp vector encodes an S tag and a His tag that were incorporated at the N terminus of the ORF9 protein. The expression of the ORF9 protein from plaque-purified stocks of recombinant baculovirus was verified by immunoblotting using an anti-ORF9 protein antibody. The recombinant protein exhibits an increase of 8.5 kDa in apparent molecular mass compared to the wild type due to the presence of the tags. The recombinant ORF9 protein was purified from Sf21 insect cells infected with the recombinant baculovirus using His-binding resin (Novagen, Madison, WI) following the manufacturer's instructions.
Antibodies.
The mouse monoclonal (H6) and rabbit polyclonal antibodies against IE62 used in these experiments have been described previously (45). Rabbit polyclonal antiserum and affinity-purified antibodies against ORF9 were generated by Proteintech Group Inc. (Chicago, IL), using the purified baculovirus-expressed ORF9 protein (BAC-ORF9) generated in our laboratory.
Coimmunoprecipitation of ORF9 protein and IE62.
Protein G-Sepharose 4 fast-flow beads (Amersham Biosciences, Piscataway, NJ) were blocked with 4% milk-phosphate-buffered saline (PBS) for 1 hour at 4°C. Beads (100 μl) were conjugated with 50 μg of anti-IE62 monoclonal antibody for 2 hours. The antibody-conjugated beads were washed three times with PBS-1% Tween 80. The beads were then incubated with MeWo cell nuclear extract (500 μg of protein) for 3 hours at 4°C. Beads were washed with PBS-1% Tween 80. Bound proteins were eluted by boiling in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer, separated by a 10% SDS-PAGE gel, and transferred to nitrocellulose membranes. Polyclonal rabbit antibody generated against purified baculovirus-expressed recombinant ORF9 protein was used as the primary antibody for detection of the ORF9 protein, and polyclonal rabbit anti-IE62 antibody (48) was used as the primary antibody for detection of IE62. Both primary antibodies were generated in our laboratory. Reactive bands were visualized using goat anti-rabbit immunoglobulin G (IgG) conjugated with horseradish peroxidase (Chemicon, Temecula, CA) in conjunction with SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL).
Yeast two-hybrid analysis.
The full-length ORF9 coding region was PCR amplified and then cloned into the NdeI/EcoRI sites of pGADT7 (Clontech, Palo Alto, CA) in frame with the GAL4 activation domain to create pGADT7-ORF9. The pGADT7 plasmid has the phenotype LEU2 Ampr. An N-terminal fragment of the IE62 coding region encompassing amino acids 1 to 201 was also PCR amplified and cloned into the NdeI/EcoRI sites of pGBKT7 (Clontech, Palo Alto, CA) in frame with the GAL4 DNA binding domain to create pGBKT7-IE62. The pGBKT7 plasmid has the phenotype TRP1 Ampr. Three smaller ORF9 fragments were generated encompassing the coding sequences for amino acids 1 to 117, 93 to 210, and 187 to 302, creating the plasmids pGADT7-ORF9 1/3F, pGADT7-ORF9 1/3S, and pGADT7-ORF9 1/3T, respectively.
Yeast two-hybrid analysis to test for protein-protein interaction was performed using Matchmaker Two-Hybrid System 3 (Clontech, Palo Alto, CA) with yeast strain Y190. Saccharomyces cerevisiae strain Y190 was sequentially cotransformed with pGBKT7-IE62 and pGADT7-ORF9 (or with the smaller ORF9 fragments). Transformed yeast cells were plated on selective dropout (SD) medium lacking tryptophan and leucine to determine the efficiency of transformation. Medium also lacking histidine (to prevent false positives) in the presence of 35 mM 3-amino-1,2,4 triazole selected for two-hybrid interactions between the ORF9 and IE62 proteins. In Y190, the tryptophan and leucine biosynthesis genes select for the maintenance of activation and binding domain plasmids on SD medium lacking tryptophan and leucine, while GAL4-activated histidine biosynthesis genes select for yeast two-hybrid interactions between activation and DNA binding domain fusions. Our positive control for expression of β-galactosidase was the pGAL4 control plasmid, which expresses the entire coding sequence of the wild-type GAL4 protein with the phenotype LEU2 Ampr. As a positive control for interaction, we used the plasmids p53, which expresses the murine p53 protein fused to the GAL4 DNA binding domain with the phenotype TRP1 Ampr, and pSV40, which expresses the simian virus 40 (SV40) large T antigen fused to the GAL4 activation domain with the phenotype LEU2 Ampr. As a negative control, we used pLaminC, which expresses the human lamin C protein fused to the GAL4 binding domain with the phenotype TRP1 Ampr, in conjunction with the pSV40 plasmid.
Verification of positive protein interaction was determined by a colony lift filter assay in order to detect β-galactosidase activity. Briefly, Whatman no. 5 filter paper was placed over the surface of a plate with fresh colonies growing in the respective selective medium. When the filter was evenly wetted, it was lifted off the agar plate, frozen in liquid nitrogen for 10 seconds, and thawed at room temperature three times. Then, the filter was placed colony side up on a new plate containing a presoaked filter in Z buffer (16.1 g Na2HPO4 · 7H2O, 5.5 g NaH2PO4 · H2O, 0.75 g KCl, 0.246 g MgSO4 · 7H2O)-X-Gal solution (100 ml Z buffer, 0.27 ml β mercaptoethanol, 1.67 ml X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside], 20 mg/ml in N,N′-dimethylformamide) and incubated at room temperature. The appearance of blue colonies indicated a protein-protein interaction.
Protein affinity pull-down assays.
The ORF9 protein was expressed as a His-tagged fusion protein in recombinant baculovirus and purified from infected cell cultures. IE62 fragments were fused to GST and expressed in E. coli DH5α as fusion proteins following induction with IPTG (isopropyl-β-d-thiogalactopyranoside), and crude lysates were prepared and clarified as previously described (38, 48). Aliquots (200 μl) of the bacterial cell lysates were then added to 100 μl of glutathione-Sepharose beads, which were washed twice with PBS-1% Triton X-100 (PBST). A mixture of 40 μg of recombinant ORF9 protein and 200 μg of bovine serum albumin in 350 μl of PBST was added to the beads. The resulting slurries were incubated for 2 hours at 4°C with gentle rocking. The beads were then collected by low-speed centrifugation and washed three times with 500 μl of PBST. Bound proteins were eluted by boiling in SDS-PAGE loading buffer, separated by a 10% SDS-PAGE gel, and transferred to nitrocellulose membranes. To detect the ORF9 protein, rabbit polyclonal anti-ORF9 antibody was used. Reactive bands were visualized using goat anti-rabbit IgG conjugated with horseradish peroxidase (Chemicon, Temecula, CA) in conjunction with SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL).
Transfections and reporter assays.
MeWo cells were grown in Eagle's minimal essential medium supplemented with 10% fetal bovine serum as previously described (48). For transfections, MeWo cells were grown to approximately 90% confluence in 6-well or 12-well cell culture plates. Plasmids were cotransfected with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. All transfections were performed in triplicate. The plasmid pCDNA3.1(+) (without any insert) was used to normalize the total amount of the cytomegalovirus (CMV) promoter transfected into cells. The plasmid pEFRL (Promega, Madison, WI) was used to normalize the luciferase activity of the assay by using the Renilla luciferase reporter gene. Cells were collected at 48 h posttransfection and lysed in the appropriate buffers by alternating three freeze-thaw cycles. Dual-luciferase assays were performed using the Dual-Luciferase reporter assay system, allowing the measurement of both firefly luciferase and Renilla luciferase in the same lysate (Promega, Madison, WI) as per the manufacturer's instructions. In experiments with titrations of the ORF47 and ORF66 viral kinases, the concentrations of plasmids expressing these proteins ranged from 0.1 to 2 μg.
Immunofluorescence and confocal microscopy.
MeWo cells were seeded onto coverslips in six-well dishes, grown in Eagle's minimal essential medium with 10% fetal bovine serum to confluence, and infected with VZV MSP (44, 45). At 24 h postinfection, the infected cells were fixed and permeabilized with 2% paraformaldehyde in 0.2 M Na2HPO4 with 0.05% Triton X-100 for 1 hour and then washed five times with PBS (pH 7.4). The samples were blocked with 5% dry milk in PBS (wt/vol) for 30 min. Primary antibodies were diluted in PBS containing 1% milk: rabbit polyclonal anti-ORF9 was diluted 1:1,000, and monoclonal antibody 5C6, which recognizes IE62, was diluted 1:1,000. After being incubated overnight at 4°C and washed with PBS, the samples were incubated with secondary antibodies and TOTO-3, a DNA marker (Molecular Probes, Inc.). Secondary antibodies, including Texas Red-conjugated goat anti-mouse and Alexa 488-conjugated goat anti-rabbit antibodies, were diluted 1:1,000 in PBS. Samples were analyzed by laser scanning confocal microscopy with the accompanying proprietary software (LSM 510; Zeiss, Germany). For transfections, HeLa and MeWo cells were grown in six-well dishes and plasmids expressing the ORF9 protein, IE62, the ORF47.12 (ORF47) kinase, and the ORF66 kinase were cotransfected with Fugene 6 transfection reagent (Roche, Indianapolis, IN) following the manufacturer's instructions. All transfections were performed in triplicate. Cells were fixed at 24 or 48 h posttransfection. The ORF9 protein and IE62 were visualized as describe above. The ORF47.12 kinase was visualized using antibody directed against the 3B3 epitope of VZV gE present in the ORF47.12 construct (25). Subsequent reactions with secondary antibodies and examination by confocal microscopy were carried out as described above.
For tubulin labeling, transfected MeWo cells were fixed in 100% methanol for 10 min and washed three times for 5 min each in PBS, followed by permeabilization in PBS with 0.1% Triton X-100 for 5 min. Samples were washed three times for 5 min each in PBS and blocked for 30 min in PBS with 10% fetal bovine serum. Sheep anti-tubulin antibody diluted 1:500 in block solution was added for 1 h at room temperature. Samples were washed three times for 10 min each in PBS and incubated for 1 h at room temperature in donkey anti-sheep secondary antibody diluted 1:1,250 in block solution. After three 10-min washes in PBS, coverslips were mounted on slides with Vectashield. Images were acquired on a Zeiss LSM 510 confocal microscope and processed with Zeiss LSM Image Browser version 3.5.0.376 software. Sheep anti-tubulin polyclonal antibody was purchased from Cytoskeleton, Inc. (Denver, CO). Alexa Fluor 546 donkey anti-sheep IgG (heavy plus light chains) was purchased from Molecular Probes, Eugene, OR. Vectashield was purchased from Vector Laboratories, Inc., Burlingame, CA.
RESULTS
Coimmunoprecipitation of the ORF9 protein and IE62.
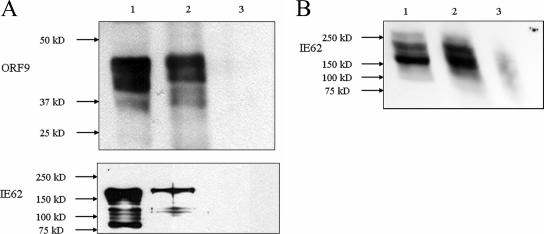
HSV-1 VP22 has been shown to interact with HSV VP16 through that protein's acidic transactivating domain, and this interaction has been postulated to act as a recruitment mechanism for VP16 into the tegument of HSV virions (15, 17, 19, 42). The VZV orthologue of VP16, the ORF10 protein, is a component of the virion tegument but lacks an acidic C-terminal activation domain. An internal activation domain has been identified in the ORF10 protein but is weaker functionally than the VP16 activation domain (35). Further, unlike VP16, the VZV ORF10 protein is dispensable for growth in tissue culture (8, 10). The VZV major transactivator IE62, however, contains a potent N-terminal acidic transactivation domain (aa 1 to 86) and, unlike its HSV orthologue, ICP4, has been demonstrated to be present in significant quantities in the virion tegument (29). Based on these facts, in conjunction with preliminary protein-protein blot experiments (47) suggesting a potential interaction between the ORF9 protein and IE62 coimmunoprecipitation, experiments were performed using VZV-infected MeWo cell extracts. The results are presented in Fig. 2 and show that monoclonal antibody directed against IE62 was capable of coimmunoprecipitating the ORF9 protein present in infected cell extracts and that polyclonal antibody directed against ORF9 coprecipitated IE62. Thus, these results indicate that the ORF9 protein and IE62 are present in a complex or complexes within infected cells.
FIG. 2.
Coimmunoprecipitation of the ORF9 protein and IE62. (A) Coimmunoprecipitation of the ORF9 protein and IE62 using the monoclonal H6 anti-IE62 antibody. Lane 1, infected cell extract; lane 2, material bound to protein G-Sepharose beads coupled with anti-IE62 antibody; lane 3, beads alone. Upper panel, ORF9 protein; lower panel, IE62. (B) Coimmunoprecipitation of IE62 using polyclonal anti-ORF9 protein antibody. The lane designations are identical to those in panel A.
Direct physical interaction between VZV IE62 and the ORF9 protein.
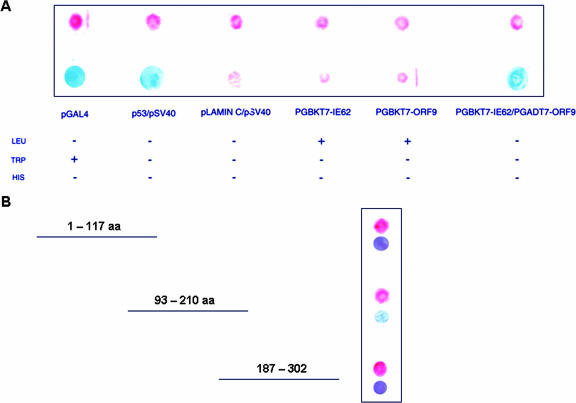
The coimmunoprecipitation data presented above do not differentiate between the direct interaction of the ORF9 protein and IE62 and the possibility that the putative interaction is indirect and mediated by one or more bridging molecules. The possibility of a direct physical interaction was examined by yeast two-hybrid analysis. In these experiments, cDNA segments encoding the full-length ORF9 protein and an IE62 fragment containing the entire acidic activation domain (9, 39) plus an additional 115 amino acids (IE62-201) were generated by PCR and subcloned into the yeast expression vectors pGADT7 and pGBKT7, resulting in fusion to the GAL4 activation domain and the GAL4 DNA binding domain, respectively. The results from these yeast two-hybrid experiments are presented in Fig. 3A. They show that the ORF9 protein interacted with the IE62 fragment, thus establishing that a direct interaction can occur between these proteins in the context of the yeast two-hybrid system in the absence of other VZV-encoded polypeptides. The negative controls showed that neither fusion protein alone had the ability to activate the reporter genes.
FIG. 3.
Results of yeast two-hybrid analysis of the potential ORF9 protein/IE62 interaction. (A) Filter colony lift β-galactosidase assay results using the intact ORF9 protein and an IE62 fragment containing the IE62 acidic activation domain (aa 1 to 201). The upper line shows yeast growth; the lower line shows β-galactosidase expression. The table indicates the selective growth conditions using the indicated SD medium to select and test for specific phenotypes (described in Materials and Methods). As a positive control for interaction, the plasmids p53, which expresses the murine p53 protein fused to the GAL4 DNA binding domain, and pSV40, which expresses the SV40 large T antigen fused to the GAL4 activation domain, were used. As a negative control, pLaminC, which expresses the human lamin C protein fused to the GAL4 binding domain, was used in conjunction with the pSV40 plasmid. (B) Yeast two-hybrid analysis using fragments of the ORF9 protein-coding sequences and the N-terminal IE62 fragment.
The next series of experiments was aimed at determining the region of the ORF9 protein required for interaction with IE62 (aa 1 to 201). Three ORF9 gene fragments covering the N-terminal third, middle third, and C-terminal third of the coding region (Fig. 3B) were generated and cloned into the yeast vector pGADT7 and tested in the yeast two-hybrid assay. The ORF9 fragment designated ORF9 1/3S, encompassing amino acids 93 to 210, interacted with IE62 as demonstrated by the positive β-galactosidase activity of this fragment in the colony filter assay. These results, accounting for the overlaps between the ORF9 fragments, narrow the region of the ORF9 protein required for direct interaction with IE62 to amino acids 117 to 186.
Mapping of the region of IE62 required for interaction with the ORF9 protein.
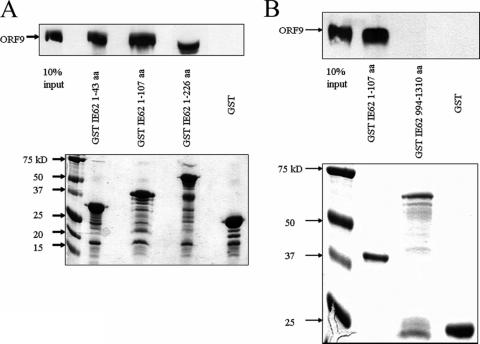
To complement our results with the yeast two-hybrid assay and to fine map the region of the IE62 N terminus required for interaction with the ORF9 protein, we performed GST pull-down assays using purified baculovirus-expressed ORF9 protein and N-terminal IE62 fragments fused to GST. As shown in Fig. 4A, purified ORF9 protein interacted with all of the GST-IE62 fusions, including the shortest, containing only amino acids 1 to 43 of IE62, but not with GST alone. The IE62 fusion protein present in Fig. 4A, lane 4, following coelution from the beads migrates at a position similar to that of the ORF9 protein and is present at a high concentration, resulting in a distortion of the gel at this position. In contrast to the results with the N-terminal IE62 fragments, binding to a GST fusion protein containing the C-terminal 316 amino acids of IE62 was not observed (Fig. 4B). These data confirm the existence of a direct physical association between the ORF9 protein and IE62. They also indicate that this interaction does not directly require phosphorylation of either the IE62 fragments or the ORF9 protein by viral kinases or kinases present in mammalian cells. The overall level of binding was roughly the same with all three of the IE62 N-terminal fragments used, suggesting that the complete ORF9 protein binding region lies within amino acids 1 to 43. This N-terminal region of the acidic activation domain of IE62 has previously been shown to be the most critical portion of the domain for transactivation (9).
FIG. 4.
Mapping of the minimal region of IE62 that interacts with the ORF9 protein. Results from protein pull-down assays using GST-IE62 fusions showing the presence or absence of the ORF9 protein in samples eluted from glutathione-Sepharose beads. (A) N-terminal IE62 fragments. (B) C-terminal IE62 fragments. The lower portion of each panel is a Coomassie gel showing the levels of the GST-IE62 fusions and GST bound to the beads in these assays. Arrows in upper panels indicate the position of the intact ORF9 protein.
ORF9 has no effect on IE62 transactivation in the absence of other viral proteins.
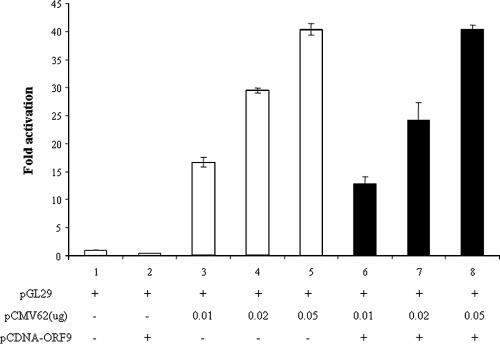
Based on the above results concerning the interaction of the ORF9 protein with the IE62 activation domain, we wished to determine whether the presence of the ORF9 protein had any effect on the ability of IE62 to transactivate representative VZV promoters. This seemed to be a reasonable possibility since HSV-1 VP22 has been reported to be present in both the nucleus and the cytoplasm of infected cells (16, 18, 20, 22). In these experiments, various amounts of the pCMV62 expression plasmid were cotransfected with a reporter plasmid containing the VZV ORF29 promoter driving expression of firefly luciferase in the presence and absence of a constant high level of the pCDNA-ORF9 protein expression plasmid. The amounts of the pCMV62 plasmid chosen corresponded to the linear response range of this assay. The results are presented in Fig. 5 and show that the presence of the ORF9 protein had no significant effect, either positive or negative, on the ability of IE62 to activate the VZV ORF29 promoter. Similar results were also obtained with the VZV ORF61 and ORF67 promoters (data not shown).
FIG. 5.
The ORF9 protein does not affect IE62 transactivation activity in the absence of other viral proteins. The results of luciferase reporter assays showing IE62 activation of the VZV ORF29 promoter in the presence and absence of the ORF9 protein are shown.
Intracellular localization of the ORF9 protein and IE62.
Our hypothesis regarding the role of the ORF9 protein during VZV infection included the possibility of it being part of the mechanism of recruitment of IE62 to the tegument. If this were the case, interaction between these two proteins could occur in the cytoplasm rather than the nucleus, and it was possible that the ORF9 protein showed no effect on IE62 transactivation due to its absence from the nucleus. If this were true, it would present a contrast to VP22, which (as documented above), while predominantly cytoplasmic, is also found in the nuclei of infected and transfected cells (16, 18, 22).
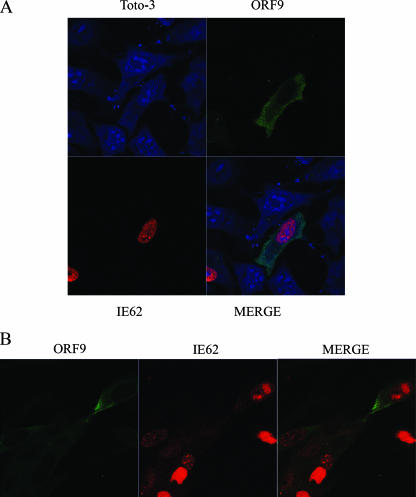
The intracellular localization of the ORF9 protein and IE62 was examined by confocal microscopy. In the first series of experiments, plasmids expressing the ORF9 protein and IE62 were transfected into HeLa cells and the expressed proteins were detected by immunofluorescence and confocal microscopy. In the transfected cells, IE62 was detected only in the nuclei. In contrast, the ORF9 protein was cytoplasmic, although low levels of signal were detected over nuclei (Fig. 6A). Similar results were obtained with transfections in MeWo cells (data not shown).
FIG. 6.
Cellular localization of the ORF9 protein and IE62. (A) Confocal microscopy of HeLa cells transfected with ORF9- and IE62-expressing plasmids at 24 h posttransfection. TOTO-3 was used as a nuclear marker. (B) Confocal microscopy of MeWo cells at 24 h post-VZV infection.
The intracellular locations of the ORF9 protein and IE62 were also examined in VZV-infected MeWo cells. At 24 hours postinfection, the ORF9 protein exhibited a predominantly cytoplasmic localization. IE62 was present in both the nuclei and cytoplasm of infected cells (Fig. 6B). The presence of IE62 in the cytoplasm and the nucleus is in agreement with previous findings which show that at later stages of infection, IE62 is phosphorylated by the viral ORF66 kinase, resulting in a redistribution of IE62 from the nucleus to the cytoplasm (14, 28).
Effect of the viral ORF66 kinase on the intracellular localization of the ORF9 protein and IE62.
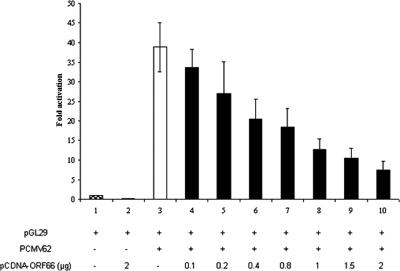
The ORF66 kinase was expressed from the pCDNA-ORF66 plasmid generated in this study. Coexpression of the ORF66 kinase and IE62 resulted in the loss of IE62 transcriptional activation (Fig. 7), as would be expected if the kinase were active (14, 27, 28). Confocal microscopy experiments were then performed to determine the intracellular localization of the ORF9 protein and IE62 in the presence of the ORF66 kinase. As shown in Fig. 8, coexpression of the ORF66 kinase in addition to IE62 and the ORF9 protein resulted in a redistribution of IE62 from the nucleus (observed in the absence of the kinase) to the cytoplasm whereas the ORF9 protein remained cytoplasmic. These results suggested that significant cytoplasmic colocalization of the ORF9 protein and IE62 occurs in the presence of ORF66.
FIG. 7.
Effect of the ORF66 viral kinase on IE62 transactivation and cellular distribution of the ORF9 protein and IE62. (A) Luciferase reporter assay results from transient transfections of MeWo cells using constant amounts of the pCMV62 expression plasmid and increasing amounts of a plasmid expressing the VZV ORF66 kinase.
FIG. 8.
Effect of the ORF66 viral kinase on the intracellular distribution of the ORF9 protein and IE62. Confocal microscopy results of HeLa cells transfected with plasmids expressing the ORF9 protein, IE62, and the ORF66 kinase at 24 h posttransfection are shown. The presence of the kinase results in a redistribution of IE62 from the nucleus to the cytoplasm.
Effect of the viral ORF47 kinase on the intracellular localization of the ORF9 protein and IE62.
In addition to being a target of the VZV ORF66 kinase, the IE62 protein is phosphorylated by the VZV ORF47 kinase and has been shown to form a stable complex with that enzyme (5, 25). Computer analysis (6) of the primary structure of the ORF9 protein revealed the presence of numerous potential sites of phosphorylation by cellular kinases throughout the ORF9 protein sequence. In addition, three consensus phosphorylation sites for the ORF47 kinase, S/T-X-D/E-D/E (24), are present in the N-terminal region of the ORF9 protein at amino acid positions 81, 82, and 84 (Fig. 1A). Besser et al. (5), however, showed that mutants of the ORF47 kinase lacking enzymatic activity and/or mutants containing a C-terminal truncation of the ORF47 kinase exhibited significant nuclear colocalization with IE62, whereas the wild-type ORF47 kinase was found in the cytoplasm.
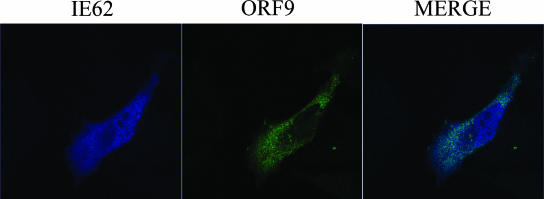
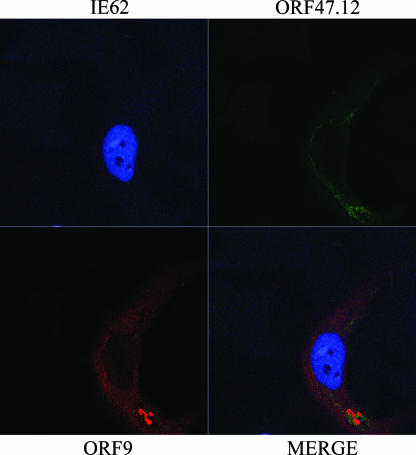
Based on the above information, we wished to assess the effect of the presence of the ORF47 kinase on the intracellular localization of the ORF9 protein and IE62. Cotransfection of HeLa cells with a plasmid expressing the VZV ORF47.12 kinase did not alter the nuclear and cytoplasmic localization of the ORF9 protein and IE62 in the absence of other viral proteins (Fig. 9). Both ORF9 and the ORF47 kinase were localized in the cytoplasm, with the signals showing some, but not complete, overlap. Similar results were observed with MeWo cells, and the cytoplasmic localization of ORF47 is consistent with data from other laboratories (4, 5).
FIG. 9.
Effect of the VZV ORF47 kinase on the cellular localization of the ORF9 protein and IE62. Confocal microscopy results of MeWo cells transfected with plasmids expressing the ORF9 protein, IE62, and the ORF47.12 kinase are shown. IE62 is confined to the nucleus. The ORF9 protein and the ORF47 kinase are both cytoplasmic but show different distributions.
Effects of the ORF47 and ORF66 kinases on IE62 transactivation.
A series of transient transfection experiments was performed to assess the effects of the presence of the two viral kinases on IE62 transactivation in the absence and presence of the ORF9 protein. These assays examined the formal possibility that phosphorylation of the ORF9 protein by one or both of the viral kinases could lead to rapid shuttling of the ORF9 protein into and out of the nucleus, resulting in alteration of IE62 transcriptional activation. The existence of such shuttling events is strongly suggested by the data of Besser et al. (5) in the context of the ORF47 kinase mutants. Cotransfection with the ORF9-expressing plasmid along with the IE62-expressing plasmid and either or both of the ORF47 and ORF66 kinases yielded no effects beyond those already observed with the kinases alone (data not shown). Thus, the presence of the two VZV kinases appears to have no effect on the ability of the ORF9 protein to influence the transcriptional activity of IE62.
Interaction of ORF9 with tubulin.
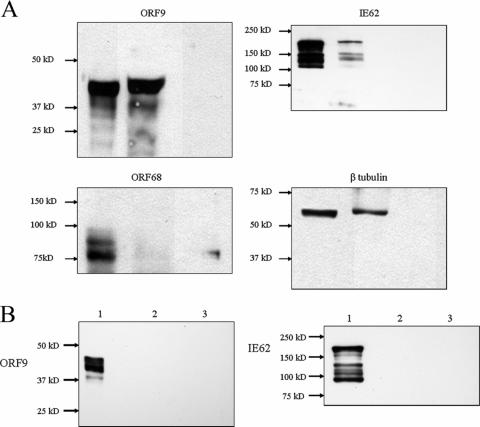
HSV-1 VP22 has been shown to associate with microtubules, and the region of VP22 required for this interaction lies within the amino acid stretch showing the highest homology with the ORF9 protein (19). Based on this information, coimmunoprecipitation experiments were performed using infected cell extracts and monoclonal anti-β-tubulin antibody. The resulting precipitates were probed for the presence of the ORF9 protein, IE62, β-tubulin (positive control), and VZV glycoprotein E. Antibodies of the same isotype directed against the Xpress tag peptide (Invitrogen, Carlsbad, CA) were used in control coimmunoprecipitations. The results are presented in Fig. 10 and show that the ORF9 protein, IE62, and tubulin but not gE were coprecipitated by the antitubulin antibody. Neither the ORF9 protein nor IE62 was precipitated by the anti-Xpress tag antibody.
FIG. 10.
ORF9 and IE62 are coprecipitated by anti-β-tubulin antibodies. (A) Immunoblot analysis of coimmunoprecipitation experiments using anti-β-tubulin antibodies. Lane 1, input from whole-cell extract; lane 2, eluate from antibody-bound beads; lane 3, eluate from beads alone without antibody. Arrows indicate the positions of the ORF9 protein, IE62, gE, and β-tubulin. (B) Results of control experiments using monoclonal IgG directed against the Xpress peptide and analyzed for the presence of the ORF9 protein and IE62. The lane designations are as described for panel A.
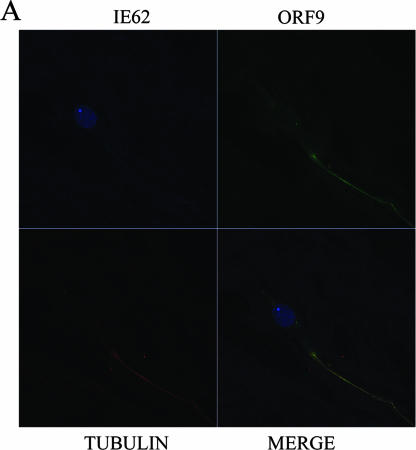
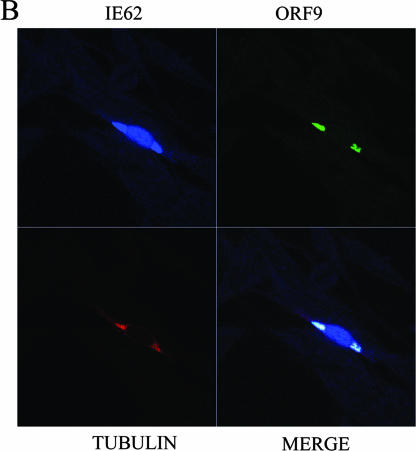
Confocal microcopy experiments were then performed using MeWo cells transfected with combinations of plasmids expressing ORF9, IE62, and the ORF47 and ORF66 kinases in order to assess the extent of colocalization of ORF9 and IE62 with tubulin. Figure 11A shows a cell transfected with plasmids expressing ORF9 and IE62 and examined for the intracellular locations of those two proteins as well as tubulin. The relatively weak IE62 signal is confined to the nucleus as anticipated due to the lack of the ORF66 kinase, whereas the ORF9 and tubulin signals colocalize to an extended filamentous structure in the cytoplasm. Three-dimensional projection of this cell confirmed the colocalization of ORF9 and tubulin (data not shown). Figure 11B shows a cell transfected with plasmids expressing ORF9, IE62, and the ORF47 and ORF66 kinases. In this case, IE62 is present in both the nucleus and the cytoplasm and the IE62 signal in the cytoplasm colocalizes with the strongest signals from both ORF9 and tubulin. Thus, the confocal imaging data support the existence of a complex between the ORF9 protein and tubulin that can also include other ORF9 binding partners such as IE62 under specific circumstances.
FIG.11.
Results of confocal microscopy visualizing intracellular tubulin, the ORF9 protein, and IE62. (A) MeWo cell transfected with plasmids expressing the ORF9 protein and IE62. The IE62 signal is confined to the nucleus. The ORF9 protein and tubulin colocalize in a long filamentous structure. (B) MeWo cell transfected with plasmids expressing the ORF9 protein, IE62, and the ORF66 and ORF47 kinases. IE62 is present in both the nucleus and the cytoplasm. ORF9 and tubulin signals overlap with the IE62 signal in the cytoplasm.
DISCUSSION
The VZV ORF9 protein is a member of the herpesvirus UL49 gene family. Members of this protein family are components of the virion tegument (17, 20, 34). The prototype of this family, HSV VP22, has been shown to be capable of interaction with viral and cellular proteins, including HSV VP16, tubulin, histones, and template-activating factor I, with this last interaction resulting in inhibition of nucleosome assembly (17, 19, 43, 49). Confocal microscopy of HSV-infected cells and cells transfected with plasmids expressing VP22 shows both nuclear and cytoplasmic localization. However, work with the BVP22 homologue has demonstrated differences as well as similarities regarding intracellular localization and microtubule interaction between the two (20). Specifically, while both proteins associated with chromatin, BVP22 showed a haloed or marbled distribution within the nucleus whereas VP22 presented a speckled or nucleolus-bound pattern. Further, BVP22 exhibited a much less pronounced association with microtubules. In the work presented here, we have shown that the VZV ORF9 protein can interact directly with the major VZV transactivator IE62 and that unlike VP22, the ORF9 protein exhibits a predominantly, if not exclusively, cytoplasmic localization in cells.
One of the unique aspects of the VZV IE62 protein lies in the fact that, in contrast to its HSV homologue, ICP4, it is present in significant amounts in the virion tegument (29). The details of the mechanism by which the IE62 protein becomes incorporated into the tegument of the VZV particle are currently unknown. The data presented here show that the ORF9 protein is capable of interacting directly with the N-terminal half of the acidic activation domain of IE62. The region of the ORF9 protein involved in this interaction maps to the central third of the molecule (aa 117 to 186), overlapping the region of major homology found in all members of the UL49 gene family (Fig. 1B). The majority of this region coincides with that recently mapped in the HSV-1 VP22 protein as being required for its interaction with the VP16 acidic activation domain (aa 160 to 212) (19). These sequences within the two proteins constitute the N-terminal half of the UL49 gene family homology domain. Thus, the two proteins appear to share similar mechanisms for interaction with the two viral transactivators. Finer mapping of the interaction region within the ORF9 protein was not possible due to the instability of smaller fragments, both in yeast and in bacteria, derived from the central portion of the molecule.
Several possibilities were then examined as to the potential function of this interaction. While the confocal microscopic evaluation of the intracellular location of the ectopically expressed ORF9 protein indicated that it was almost exclusively cytoplasmic, low levels of signal were sometimes observed over nuclei and the existence of a rapid shuttling of the ORF9 protein between the cytoplasm and the nucleus remained a formal possibility. Based on the region within the IE62 protein involved in the interaction, we examined the possibility that the ORF9 protein influenced the transactivating function of IE62. No reproducible effect was observed using representative promoters from all three putative kinetic classes of VZV genes (Fig. 5; data not shown). Thus, even if the two proteins do interact within nuclei, alteration of IE62 transactivation does not appear to occur. In contrast to the overall lack of colocalization seen in cells transfected only with plasmids expressing IE62 and the ORF9 protein, cells in the later stages of infection and cells transfected with plasmids expressing the VZV ORF66 kinase in addition to those expressing IE62 and the ORF9 protein exhibited colocalization of the two proteins within the cytoplasm. The redistribution of IE62 is due to its well-documented phosphorylation by the ORF66 kinase near the IE62 nuclear localization signal and subsequent exclusion from the nucleus (14, 28).
HSV-1 VP22 is known to associate with microtubules (17, 19). Coimmunoprecipitation experiments with antitubulin antibody showed that both ORF9 and IE62 were coprecipitated in a complex with tubulin. The examination of confocal images of cells transfected with plasmids expressing the ORF9 protein and IE62 showed that while IE62 remained nuclear, the ORF9 protein colocalized with tubulin. Further, in cells cotransfected with the ORF66 kinase, the IE62 signal present in the cytoplasm localized in the regions with the highest tubulin and ORF9 protein signals. These findings represent the first description of IE62 bound either directly or indirectly to tubulin. Since the vast majority of β-tubulin is present in microtubules (37), our results indicate that a fraction of the ORF9 protein and IE62 can associate with microtubules. HSV-1 VP22 has been reported to be capable of intercellular trafficking and delivering nucleic acids and proteins to uninfected and untransfected cells (7, 12, 18, 36, 40, 46). The bovine herpesvirus type 1 VP22 homologue has also been reported to shuttle between the nuclei of expressing and nonexpressing cells. We explored these possibilities by examining whether the ORF9 protein expressed from plasmids either alone or fused to enhanced green fluorescent protein could shuttle between cells. We also attempted to shuttle IE62 and plasmids expressing enhanced green fluorescent protein between cells by using the ORF9 protein as an intermediary. All of these experiments proved to be negative (data not shown). Thus, we currently have no evidence that the ORF9 protein shares these properties with HSV-1 VP22 and other alphaherpesvirus orthologues. This may be a reflection of the fact that we rarely observed the ORF9 protein within nuclei. Two regions of VP22 have been shown to be involved in nuclear targeting (2), encompassing amino acids 81 to 121 and 267 to 301 of the VP22 molecule. These regions, particularly aa 81 to 121, exhibit low homology with the ORF9 protein primary sequence (Fig. 1A). These differences are manifested as significant gaps as well as lack of direct sequence homology, suggesting that the regions in question may have significantly different structures and functions.
Based on the above results, we hypothesized that the major function of the IE62/ORF9 protein interaction would most likely be in the recruitment/incorporation of IE62 into the viral tegument. Based on this assumption, we examined the intracellular localization of the ORF9 protein in the presence of the VZV ORF47 kinase. The ORF47 kinase is known to be a component of the VZV virion and has been shown to be capable of autophosphorylation in addition to phosphorylation of the IE62 protein both in vitro and in vivo (24, 25). Besser et al. (5) showed that mutations of the ORF47 protein that ablated kinase activity resulted in nuclear retention of both the ORF47 kinase and IE62, indicating a role for the ORF47 kinase as well as the ORF66 kinase in the cytoplasmic localization of IE62 in infected cells. Confocal microscopy of cells transfected with plasmids expressing the ORF47 kinase, the ORF9 protein, and/or IE62 showed that the intact ORF47 protein exhibited a cytoplasmic localization and had no effect on the intracellular localization of IE62 (nuclear). Further, we found no evidence of influence of the presence of the ORF47 kinase on the transcriptional activation properties of IE62 (data not shown). These findings are congruent with the results of Besser et al. (5), who showed that direct physical interaction of the ORF47 protein with IE62 rather than the kinase activity constitutes the essential contribution of ORF47 to VZV replication in vivo in a SCID-hu mouse model. Visualization of the ORF9 protein and the ORF47 kinase in transfected cells showed that the two proteins had cytoplasmic distributions, with partial overlap of the ORF9 protein and ORF47 kinase signals.
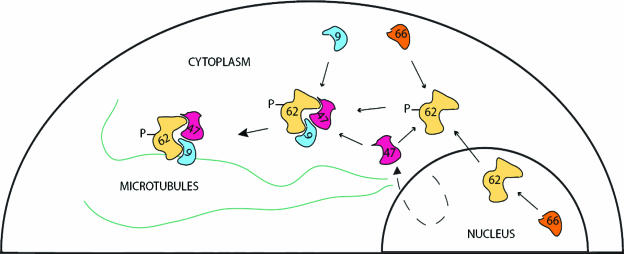
Taken together, these results suggest a model in which the VZV ORF9 protein plays a central role in the nucleation and/or recruitment of complexes of VZV tegument proteins. In this model (Fig. 12), the concerted action of the VZV ORF66 (either in the nucleus or in the cytoplasm) and ORF47 kinases as well as cellular kinases results in the relocalization of IE62 from the nucleus to the cytoplasm. The ORF9 protein would bind to IE62 and be capable of delivering it to the cellular microtubule network. Based on our data for transfected cells, this association does not require the ORF47 kinase. However, it is possible that the ORF9 protein could recruit the IE62/ORF47 protein complex as well as other known IE62 complexes, including those involving the VZV IE63 and ORF4 proteins (3, 32, 48), both of which are known to be present within the viral tegument (26). Thus, the assembly of such complexes may be involved in virion formation, as has been postulated for the association of VP22 and VP16 (15, 17, 19, 42). While this remains a possibility, the ORF47 kinase and the ORF66 kinase are not required for growth in tissue culture and virus particles lacking these proteins along with IE62 remain infectious (5, 21, 27). Thus, the recruitment of these proteins for incorporation into the virus tegument may not be the sole, or even the most important, function of the ORF9 protein.
FIG. 12.
Model of ORF9 protein/IE62 interactions. In this model, IE62 is phosphorylated by the ORF66 kinase either in the nucleus or in the cytoplasm late in infection, resulting in its exclusion from the nucleus. IE62 can then interact with the ORF9 protein and potentially, either simultaneously or sequentially, with the ORF47 kinase. This complex then binds to the microtubules via the ORF9 protein. An alternative mechanism could involve initial binding of ORF9 to microtubules, followed by recruitment of the other proteins.
At least one alternative possibility for the function of the ORF9 protein-orchestrated assembly of tegument proteins, including IE62, on the cellular microtubule network may be that proposed for the axonal transport of nonenveloped virus particles and tegument structures for HSV-1 in neurons (30, 31, 33). This is also of relevance in the case of VZV based on its cell-to-cell spread in tissue culture and the fact that Besser et al. (4) showed that in skin, virion formation is not necessary for infectivity. The most straightforward method for assessing the role or roles of the ORF9 protein in viral infection would be the generation of viral mutants. This being said, attempts to propagate mutant viruses with a point mutation in the start codon, which are incapable of expressing ORF9, have thus far proven to be unsuccessful. The generated viral mutants exhibit extremely slow growth, followed by a high rate of reversion after a few passages in tissue culture (N. Osterrieder, personal communication). Thus, future efforts to examine ORF9 protein function will require the identification of specific amino acids whose mutation disrupts the interaction of the ORF9 protein with specific viral and cellular proteins, the generation of viruses carrying these mutations, and the evaluation of the effects of these mutations by utilizing in situ and in vivo models of VZV infection.
Acknowledgments
This work was supported by NIH grants AI18449 and AI053846.
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Ace, C. I., T. A. McKee, J. M. Ryan, J. M. Cameron, and C. M. Preston. 1989. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J. Virol. 63:2260-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aints, A., H. Guven, G. Gahrton, C. I. Smith, and M. S. Dilber. 2001. Mapping of herpes simplex virus-1 VP22 functional domains for inter- and subcellular protein targeting. Gene Ther. 8:1051-1056. [DOI] [PubMed] [Google Scholar]
- 3.Baiker, A., C. Bagowski, H. Ito, M. Sommer, L. Zerboni, K. Fabel, J. Hay, W. Ruyechan, and A. M. Arvin. 2004. The immediate-early 63 protein of varicella-zoster virus: analysis of functional domains required for replication in vitro and for T-cell and skin tropism in the SCIDhu model in vivo. J. Virol. 78:1181-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Besser, J., M. Ikoma, K. Fabel, M. H. Sommer, L. Zerboni, C. Grose, and A. M. Arvin. 2004. Differential requirement for cell fusion and virion formation in the pathogenesis of varicella-zoster virus infection in skin and T cells. J. Virol. 78:13293-13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besser, J., M. H. Sommer, L. Zerboni, C. P. Bagowski, H. Ito, J. Moffat, C. C. Ku, and A. M. Arvin. 2003. Differentiation of varicella-zoster virus ORF47 protein kinase and IE62 protein binding domains and their contributions to replication in human skin xenografts in the SCID-hu mouse. J. Virol. 77:5964-5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blom, N., S. Gammeltoft, and S. Brunak. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294:1351-1362. [DOI] [PubMed] [Google Scholar]
- 7.Brewis, N., A. Phelan, J. Webb, J. Drew, G. Elliott, and P. O'Hare. 2000. Evaluation of VP22 spread in tissue culture. J. Virol. 74:1051-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Che, X., L. Zerboni, M. H. Sommer, and A. M. Arvin. 2006. Varicella-zoster virus open reading frame 10 is a virulence determinant in skin cells but not in T cells in vivo. J. Virol. 80:3238-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen, J. I., D. Heffel, and K. Seidel. 1993. The transcriptional activation domain of varicella-zoster virus open reading frame 62 protein is not conserved with its herpes simplex virus homolog. J. Virol. 67:4246-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen, J. I., and K. Seidel. 1994. Varicella-zoster virus (VZV) open reading frame 10 protein, the homolog of the essential herpes simplex virus protein VP16, is dispensable for VZV replication in vitro. J. Virol. 68:7850-7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohrs, R. J., M. P. Hurley, and D. H. Gilden. 2003. Array analysis of viral gene transcription during lytic infection of cells in tissue culture with varicella-zoster virus. J. Virol. 77:11718-11732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dilber, M. S., A. Phelan, A. Aints, A. J. Mohamed, G. Elliott, C. I. Smith, and P. O'Hare. 1999. Intercellular delivery of thymidine kinase prodrug activating enzyme by the herpes simplex virus protein, VP22. Gene Ther. 6:12-21. [DOI] [PubMed] [Google Scholar]
- 13.Dorange, F., B. K. Tischer, J.-F. Vautherot, and N. Osterrieder. 2002. Characterization of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 76:1959-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisfeld, A. J., S. E. Turse, S. A. Jackson, E. C. Lerner, and P. R. Kinchington. 2006. Phosphorylation of the varicella-zoster virus (VZV) major transcriptional regulatory protein IE62 by the VZV open reading frame 66 protein kinase. J. Virol. 80:1710-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott, G., and P. O'Hare. 2000. Cytoplasm-to-nucleus translocation of a herpesvirus tegument protein during cell division. J. Virol. 74:2131-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott, G., and P. O'Hare. 1998. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J. Virol. 72:6448-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliott, G., and P. O'Hare. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223-233. [DOI] [PubMed] [Google Scholar]
- 19.Hafezi, W., E. Bernard, R. Cook, and G. Elliott. 2005. Herpes simplex virus tegument protein VP22 contains an internal VP16 interaction domain and a C-terminal domain that are both required for VP22 assembly into the virus particle. J. Virol. 79:13082-13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harms, J. S., X. Ren, S. C. Oliveira, and G. A. Splitter. 2000. Distinctions between bovine herpesvirus 1 and herpes simplex virus type 1 VP22 tegument protein subcellular associations. J. Virol. 74:3301-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heineman, T. C., K. Seidel, and J. I. Cohen. 1996. The varicella-zoster virus ORF66 protein induces kinase activity and is dispensable for viral replication. J. Virol. 70:7312-7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutchinson, I., A. Whiteley, H. Browne, and G. Elliott. 2002. Sequential localization of two herpes simplex virus tegument proteins to punctate nuclear dots adjacent to ICP0 domains. J. Virol. 76:10365-10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kennedy, P. G., E. Grinfeld, M. Craigon, K. Vierlinger, D. Roy, T. Forster, and P. Ghazal. 2005. Transcriptomal analysis of varicella-zoster virus infection using long oligonucleotide-based microarrays. J. Gen. Virol. 86:2673-2684. [DOI] [PubMed] [Google Scholar]
- 24.Kenyon, T. K., E. Homan, J. Storlie, M. Ikoma, and C. Grose. 2003. Comparison of varicella-zoster virus ORF47 protein kinase and casein kinase II and their substrates. J. Med. Virol. 70:(Suppl. 1)S95-S102. [DOI] [PubMed] [Google Scholar]
- 25.Kenyon, T. K., J. Lynch, J. Hay, W. Ruyechan, and C. Grose. 2001. Varicella-zoster virus ORF47 protein serine kinase: characterization of a cloned, biologically active phosphotransferase and two viral substrates, ORF62 and ORF63. J. Virol. 75:8854-8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinchington, P. R., D. Bookey, and S. E. Turse. 1995. The transcriptional regulatory proteins encoded by varicella-zoster virus open reading frames (ORFs) 4 and 63, but not ORF 61, are associated with purified virus particles. J. Virol. 69:4274-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinchington, P. R., K. Fite, A. Seman, and S. E. Turse. 2001. Virion association of IE62, the varicella-zoster virus (VZV) major transcriptional regulatory protein, requires expression of the VZV open reading frame 66 protein kinase. J. Virol. 75:9106-9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinchington, P. R., K. Fite, and S. E. Turse. 2000. Nuclear accumulation of IE62, the varicella-zoster virus (VZV) major transcriptional regulatory protein, is inhibited by phosphorylation mediated by the VZV open reading frame 66 protein kinase. J. Virol. 74:2265-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinchington, P. R., J. K. Hougland, A. M. Arvin, W. T. Ruyechan, and J. Hay. 1992. The varicella-zoster virus immediate-early protein IE62 is a major component of virus particles. J. Virol. 66:359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klopfleisch, R., J. P. Teifke, W. Fuchs, M. Kopp, B. G. Klupp, and T. C. Mettenleiter. 2004. Influence of tegument proteins of pseudorabies virus on neuroinvasion and transneuronal spread in the nervous system of adult mice after intranasal inoculation. J. Virol. 78:2956-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaVail, J. H., A. N. Tauscher, E. Aghaian, O. Harrabi, and S. S. Sidhu. 2003. Axonal transport and sorting of herpes simplex virus components in a mature mouse visual system. J. Virol. 77:6117-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch, J. M., T. K. Kenyon, C. Grose, J. Hay, and W. T. Ruyechan. 2002. Physical and functional interaction between the varicella zoster virus IE63 and IE62 proteins. Virology 302:71-82. [DOI] [PubMed] [Google Scholar]
- 33.Mettenleiter, T. C., T. Minson, and P. Wild. 2006. Egress of alphaherpesviruses. J. Virol. 80:1610-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michael, K., B. G. Klupp, T. C. Mettenleiter, and A. Karger. 2006. Composition of pseudorabies virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J. Virol. 80:1332-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moriuchi, H., M. Moriuchi, R. Pichyangkura, S. J. Triezenberg, S. E. Straus, and J. I. Cohen. 1995. Hydrophobic cluster analysis predicts an amino-terminal domain of varicella-zoster virus open reading frame 10 required for transcriptional activation. Proc. Natl. Acad. Sci. USA 92:9333-9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Normand, N., H. van Leeuwen, and P. O'Hare. 2001. Particle formation by a conserved domain of the herpes simplex virus protein VP22 facilitating protein and nucleic acid delivery. J. Biol. Chem. 276:15042-15050. [DOI] [PubMed] [Google Scholar]
- 37.Pellegrini, F., and D. R. Budman. 2005. Tubulin function, action of antitubulin drugs, and new drug development. Cancer Investig. 23:264-273. [DOI] [PubMed] [Google Scholar]
- 38.Peng, H., H. He, J. Hay, and W. T. Ruyechan. 2003. Interaction between the varicella zoster virus IE62 major transactivator and cellular transcription factor Sp1. J. Biol. Chem. 278:38068-38075. [DOI] [PubMed] [Google Scholar]
- 39.Perera, L. P., J. D. Mosca, W. T. Ruyechan, G. S. Hayward, S. E. Straus, and J. Hay. 1993. A major transactivator of varicella-zoster virus, the immediate-early protein IE62, contains a potent N-terminal activation domain. J. Virol. 67:4474-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phelan, A., G. Elliott, and P. O'Hare. 1998. Intercellular delivery of functional p53 by the herpesvirus protein VP22. Nat. Biotechnol. 16:440-443. [DOI] [PubMed] [Google Scholar]
- 41.Poon, A. P. W., and B. Roizman. 1995. The phenotype in vitro and in infected cells of herpes simplex virus 1 α trans-inducing factor (VP16) carrying temperature-sensitive mutations introduced by substitution of cysteines. J. Virol. 69:7658-7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Potel, C., and G. Elliott. 2005. Phosphorylation of the herpes simplex virus tegument protein VP22 has no effect on incorporation of VP22 into the virus but is involved in optimal expression and virion packaging of ICP0. J. Virol. 79:14057-14068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren, X., J. S. Harms, and G. A. Splitter. 2001. Bovine herpesvirus 1 tegument protein VP22 interacts with histones, and the carboxyl terminus of VP22 is required for nuclear localization. J. Virol. 75:8251-8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santos, R. A., C. C. Hatfield, N. L. Cole, J. A. Padilla, J. F. Moffat, A. M. Arvin, W. T. Ruyechan, J. Hay, and C. Grose. 2000. Varicella-zoster virus gE escape mutant VZV-MSP exhibits an accelerated cell-to-cell spread phenotype in both infected cell cultures and SCID-hu mice. Virology 275:306-317. [DOI] [PubMed] [Google Scholar]
- 45.Santos, R. A., J. A. Padilla, C. Hatfield, and C. Grose. 1998. Antigenic variation of varicella zoster virus Fc receptor gE: loss of a major B cell epitope in the ectodomain. Virology 249:21-31. [DOI] [PubMed] [Google Scholar]
- 46.Sciortino, M. T., B. Taddeo, A. P. Poon, A. Mastino, and B. Roizman. 2002. Of the three tegument proteins that package mRNA in herpes simplex virions, one (VP22) transports the mRNA to uninfected cells for expression prior to viral infection. Proc. Natl. Acad. Sci. USA 99:8318-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spengler, M., N. Niesen, C. Grose, W. T. Ruyechan, and J. Hay. 2001. Interactions among structural proteins of varicella zoster virus. Arch. Virol. Suppl. 17:71-79. [DOI] [PubMed] [Google Scholar]
- 48.Spengler, M. L., W. T. Ruyechan, and J. Hay. 2000. Physical interaction between two varicella zoster virus gene regulatory proteins, IE4 and IE62. Virology 272:375-381. [DOI] [PubMed] [Google Scholar]
- 49.van Leeuwen, H., M. Okuwaki, R. Hong, D. Chakravarti, K. Nagata, and P. O'Hare. 2003. Herpes simplex virus type 1 tegument protein VP22 interacts with TAF-I proteins and inhibits nucleosome assembly but not regulation of histone acetylation by INHAT. J. Gen. Virol. 84:2501-2510. [DOI] [PubMed] [Google Scholar]
- 50.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O'Boyle. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]