Abstract
The interferon (IFN)-stimulated genes (ISGs) ISG-49, ISG-54, and ISG-56 are highly responsive to viral infection, yet the regulation and function of these genes in vivo are unknown. We examined the simultaneous regulation of these ISGs in the brains of mice during infection with either lymphocytic choriomeningitis virus (LCMV) or West Nile virus (WNV). Expression of the ISG-49 and ISG-56 genes increased significantly during LCMV infection, being widespread and localized predominantly to common as well as distinct neuronal populations. Expression of the ISG-54 gene also increased but to lower levels and with a more restricted distribution. Although expression of the ISG-49, ISG-54, and ISG-56 genes was increased in the brains of LCMV-infected STAT1 and STAT2 knockout (KO) mice, this was blunted, delayed, and restricted to the choroid plexus, meninges, and endothelium. ISG-56 protein was regulated in parallel with the corresponding RNA transcript in the brain during LCMV infection in wild-type and STAT KO mice. Similar changes in ISG-49, ISG-54, and ISG-56 RNA levels and ISG-56 protein levels were observed in the brains of wild-type mice following infection with WNV. Thus, the ISG-49, ISG-54, and ISG-56 genes are coordinately upregulated in the brain during LCMV and WNV infection; this upregulation, in the case of LCMV, was totally (neurons) or partially (non-neurons) dependent on the IFN-signaling molecules STAT1 and STAT2. These findings suggest a dominant role for the ISG-49, ISG-54, and ISG-56 genes in the host response to different viruses in the central nervous system, where, particularly in neurons, these genes may have nonredundant functions.
The interferon (IFN) family of cytokines, in addition to their involvement in cell growth regulation and antitumor protection, are pivotal mediators in the innate and adaptive immune responses to microbial pathogens such as viruses and bacteria (reviewed in references 6 and 14). These cytokines fall into two distinct subgroups, type I (containing many members, including IFN-α and IFN-β) and type II (containing a single member, IFN-γ). The type I IFNs are produced by a wide variety of cells in direct response to infection by viruses and bacteria, while type II IFN production is largely restricted to activated CD4-positive Th1 and CD8-positive T lymphocytes and natural killer cells.
Type I and type II IFNs signal through separate unique cognate receptors, IFNAR (type I IFNs) and IFNGR (type II IFN), and activate signaling cascades primarily involving the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway (reviewed in references 29 and 36). The binding of the IFNs triggers receptor subunit heterodimerization, and the consequent activation of receptor-associated JAKs (type I IFN receptors, Jak1 and Tyk2; type II IFN receptors, Jak1 and Jak2). This leads to the recruitment of specific STAT molecules to the receptor and their subsequent phosphorylation by the JAKs. For the type I IFN-stimulated pathway, STAT1 and STAT2 molecules are recruited and activated by JAK-mediated tyrosine phosphorylation, before dissociating from the receptor complex and forming a heterodimer. This STAT1/STAT2 heterodimer then translocates to the nucleus and associates with a third molecule, IFN regulatory factor 9 (IRF-9). The heterotrimeric complex formed, IFN-stimulated gene factor 3 (ISGF3), binds to the cis-acting IFN-stimulated response element (ISRE), located upstream of most type I IFN-regulated genes. The type II IFN signaling pathway differs from the type I IFN pathway in that STAT1 molecules are recruited to the IFNGR and, after tyrosine phosphorylation, dissociate to form homodimers (gamma-activated factor [GAF]) that migrate to the nucleus and bind to a DNA recognition motif called the gamma activation sequence (GAS). In each case, the binding of ISGF3 to the ISRE or GAF to the GAS results in the altered transcription of a large number of overlapping as well as distinct genes (7).
Gene expression profiling studies have found three genes within the extensive category of IFN-stimulated genes (ISGs) to be among the most responsive in a variety of both human and rodent cells following exposure to IFN (7), as well as to viruses (12, 18, 24, 30, 42) and double-stranded RNA (dsRNA) (13). Encoding members of a highly conserved family of proteins containing multiple tetratricopeptide repeat (TPR) domains, these three genes in the mouse are termed ISG-56 (known also as IFIT-1 or GARG-16), ISG-54 (known also as IFIT-2 or GARG-39), and ISG-49 (known also as IFIT-3 or GARG-49) and are homologous to the human HuISG-56, HuISG-54, and HuISG-60 genes, respectively (8, 33). The regulation of this family of genes has been studied extensively in vitro with specific cell lines. In murine cells, ISG-49, ISG-54, and ISG-56 are rapidly induced by both type I (35, 37) and type II (35) IFNs, as well as by lipopolysaccharide (21, 22, 35). In addition to IFN and lipopolysaccharide, the expression of ISG-54 and ISG-56 is also induced directly by the Sendai virus (SeV) as well as by dsRNA (37). The structurally similar human orthologues HuISG-54 and HuISG-56 are also induced by IFN (17), dsRNA, and a range of viruses, including SeV, encephalomyocarditis virus, and cytomegalovirus (17, 31).
While there is considerable information about how the ISG-54 and ISG-56 genes are regulated in vitro, much less is known about the functional consequences of the induction of these genes, a subject that has only recently received experimental attention (33, 37). It is now clear that ISG-54 and ISG-56, along with their human counterparts, HuISG-54 and ISG-56, are intracellular proteins that are capable of inhibiting the initiation of protein synthesis. This is accomplished by the binding of these molecules to either the “c” (murine) or the “e” (human) subunit of the translation initiation factor eIF3, one of at least 11 factors involved in the initiation of protein synthesis in eukaryotes (16, 37). Consistent with this molecular action, cells transfected with and expressing constitutively the ISG-56 gene have a decrease in overall protein synthesis and exhibit reduced cell proliferation (17). Collectively, the foregoing observations suggest an important role for the ISG-49, ISG-54, and ISG-56 genes in antiviral and possibly antibacterial defense.
To date, the vast majority of information concerning this family of IFN-inducible genes is derived almost exclusively from studies performed in vitro. Little if anything is known about the coordinate regulation of expression of the ISG-49, ISG-54, and ISG-56 genes in vivo. Viral infection of the central nervous system (CNS) presents a dilemma for the host, in that an appropriate antiviral response must be generated that clears infectious agents without causing significant neuronal injury and loss, which otherwise would have catastrophic consequences (28). In this context, we hypothesized that the ISG-49, ISG-54, and ISG-56 genes might play a crucial role in antiviral responses in the CNS. As a first step toward examining this hypothesis, as well as to begin to clarify the nature of the regulation of these genes in vivo, we examined the simultaneous expression and localization of the ISG-49, ISG-54, and ISG-56 genes in the CNS of wild-type (WT), STAT1, or STAT2 knockout (KO) mice during infection with the arenavirus lymphocytic choriomeningitis virus (LCMV). In this widely studied model, intracranial infection of immunocompetent adult mice with LCMV leads to viral infection of mostly superficial areas of the brain, including the meninges, choroid plexus, and ependymal membranes, and results in the development of a fatal lymphocytic choriomeningitis which is mediated by antiviral CD8+ T cells (4, 9). Furthermore, to determine how general was the involvement of the ISG-49, ISG-54, and ISG-56 genes in the host response to viral infection in the CNS, we also examined the outcome of infection of the murine CNS with the flavivirus West Nile virus (WNV). In contrast to LCMV, WNV infects many areas of the brain, including many neuronal populations in the cortex, hippocampus, and ventral forebrain (20, 40). However, like LCMV, WNV infection is fatal, accompanied by demonstrable immunopathology in the brain involving antiviral CD4+ and CD8+ T cells.
MATERIALS AND METHODS
Animals.
WT mice (strain background C57BL/6 or 129/Sv) were purchased from the Animal Resources Centre, Canning Vale, WA, Australia. STAT1 KO mice (strain background 129/Sv/Ev [23] or C57BL/6 [11]) were kindly provided by Robert D. Schreiber, Washington University School of Medicine, St. Louis, MO, or Joan Durbin, Children's Hospital, Columbus, OH, respectively, and STAT2 KO (129/Sv/Ev background [27]) mice were provided by Chris Schindler, Columbia University, New York, NY. The various genotypes of homozygous mutant mice were maintained by interbreeding, and their identities were verified by PCR analysis of tail DNA. Approval for the use of all mice in this study was obtained from the University of Sydney Animal Care and Ethics Committee.
LCMV infection.
Adult WT, STAT1 KO, and STAT2 KO mice, approximately 3 months of age, were injected intracranially in the frontal cortex with 250 PFU of the Armstrong 53b strain of LCMV, as described previously (5). Aged-matched control animals were injected intracranially with vehicle buffer alone. At 2, 4, or 6 days following infection, the animals were euthanized and their brains were removed pending analysis by either RNase protection assay (RPA) or in situ hybridization histochemistry and immunohistochemistry. For RPA, poly(A)+ RNA was isolated from snap-frozen hemibrain by an oligo(dT) cellulose (Ambion, Austin, TX) method (2).
WNV infection.
Eight- to 10-week-old female C57BL/6 mice were obtained from the Blackburn Animal House, University of Sydney, Sydney, Australia. Mice were anesthetized with Avertin (Aldrich Chemical Company, Milwaukee, WI) and inoculated intranasally with 10 μl WNV (Sarafend strain, lineage II; 6 × 104 PFU) in sterile phosphate-buffered saline (PBS) (Gibco BRL). Sham infections were conducted by inoculation with sterile PBS only. Mice were euthanized 7 days postinfection, and the brains were prepared and analyzed as described above.
Construction of ISG probes.
For RPA and in situ hybridization, the specific target sequences used to generate probes against the ISG-49, ISG-54, and ISG-56 RNA transcripts are listed in Table 1. The cDNA fragment corresponding to each of these ISGs was synthesized by reverse transcription-PCR using total RNA prepared from the brain of a GFAP-IFN-α transgenic mouse and amplified with specific oligonucleotide primers flanked by HindIII (antisense primer) and EcoRI (sense primer) restriction enzyme sites. The primers were used to generate fragments of the desired lengths (Table 1) that could be conveniently separated on a standard polyacrylamide sequencing gel. After PCR, the amplified fragments were incubated with polynucleotide kinase (Promega, Madison, WI), ligated with T4 ligase (Promega), subsequently digested with HindIII/EcoRI (Promega), and then ligated into pGEM-4Z (Promega). The specific identity of each ISG clone was subsequently verified by sequence analysis. The orientation of the fragment allowed antisense and sense RNA synthesis from the flanking T7 and SP6 RNA polymerase promoters, respectively. A cDNA fragment, RPL32-4A (10) (kindly provided by M. Hobbs, The Scripps Research Institute), also cloned in pGEM-4, served as a probe for the ribosomal protein L32 and was included as an internal control for RNA loading.
TABLE 1.
Target ISG cDNA sequences used to derive the ISG RPA probe set
RPA.
RPAs were performed as described previously (5) with the ISG probe set, consisting of an equimolar pool of EcoRI-linearized templates (15 ng each). RNA levels were quantified from scanned autoradiograms by densitometry with NIH Image software (version 1.63) as previously described (25).
Dual-label in situ hybridization and immunohistochemistry.
Brains were removed from virus-infected and sham-infected control mice and fixed overnight in 4% paraformaldehyde in PBS (pH 7.4) at 4°C. Paraffin-embedded sections (8 μm) were incubated with 33P-labeled cRNA probes transcribed from linearized ISG-49, ISG-54, ISG-56, or LCMV nucleoprotein (NP) RPA plasmids and processed for in situ hybridization as described previously (1, 5). Sections were then processed for immunohistochemistry to detect astrocytes (rabbit anti-glial fibrillary acidic protein antibody; DAKO Cytomation, Botany, NSW, Australia), microglia (biotinylated lectin from Lycopersicon esculentum; Sigma-Aldrich, St. Louis, MO), and neurons (mouse anti-neuron-specific nuclear protein; Chemicon, Boronia, VIC, Australia). Bound antibody or lectin was detected with Vectastain ABC kits (Vector Laboratories, Burlingame, CA). Diaminobenzidine/H2O2 reagent (Vector Laboratories) was used as the immunoperoxidase substrate.
To detect WNV, immunohistochemistry was employed as described above, with the modification that paraffin-embedded sections were pretreated for antigen retrieval with Proteinase K (Sigma-Aldrich) at 37°C for 15 min. Tissue sections were then incubated with a monoclonal antibody directed against the flavivirus nonstructural protein 1 (NS1) (biotin-conjugated clone 4G4, a gift from Roy Hall, University of Queensland, Queensland, Australia). As a control, similarly processed adjacent tissue was incubated with mouse immunoglobulin G1 (BD Biosciences).
Immunohistochemistry for the ISG-56 protein was performed as described above, with the modification that paraffin-embedded sections were pretreated for antigen retrieval by boiling for 15 min in 10 mM citrate buffer, pH 6.0, prior to incubation with rabbit polyclonal antisera to murine ISG-56 (37). As a control, similarly processed adjacent sections were incubated with a rabbit preimmune serum.
RESULTS
ISG-49, ISG-54, and ISG-56 mRNA levels are increased in the brain in response to LCMV infection.
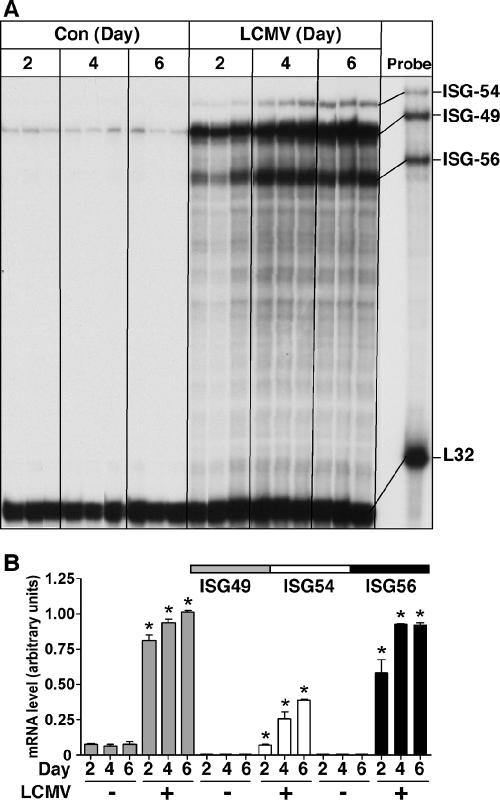
To assess the response of the ISG-49, ISG-54, and ISG-56 genes to infection in the CNS with the arenavirus LCMV, WT (C57BL/6) mice were injected intracranially (i.c.) with LCMV. LCMV infection caused a delayed recruitment (around day 5) of antiviral T cells to the CNS, the site of infection. WT mice died from cerebral seizures 6 to 8 days after intracranial injection of LCMV. In brains from WT (sham-injected) control mice, ISG-49 mRNA transcripts were detectable at low levels, while ISG-54 and ISG-56 mRNAs were undetectable (Fig. 1A and B). At day 2 postinfection, the levels of all three mRNA transcripts were significantly upregulated, with ISG-49 being the highest, followed by ISG-56 and ISG-54. Over the course of LCMV infection, ISG-49 and ISG-54 mRNA levels continued to increase at day 4 and further still at day 6 postinfection. ISG-56 mRNA levels increased at day 4 postinfection and remained unchanged from this level at day 6 postinfection. These results indicate that LCMV infection was accompanied by a significant upregulation of ISG-49, ISG-54, and ISG-56 mRNA levels from as early as day 2 postinfection.
FIG. 1.
ISG-49, ISG-54, and ISG-56 mRNA levels in brains of sham- or LCMV-infected WT mice. Mice (C57BL/6) were injected i.c. with vehicle or vehicle plus 250 PFU of LCMV (ARM). At the times shown, mice were euthanized, poly(A)+ RNA was isolated from the brains, and 2 μg was analyzed by RPA as described in Materials and Methods. (A) Low constitutive levels of ISG-49 mRNA transcripts were present in normal, uninfected brain, while ISG-54 and ISG-56 mRNA transcripts were undetectable. The levels of ISG-49, ISG-54, and ISG-56 mRNA transcripts increased significantly (P < 0.05) by day 2 postinfection and continued to increase to day 4 postinfection. At day 6 postinfection, the levels of ISG-49 and ISG-54 mRNA continued to increase significantly from those seen at day 4, while levels of ISG-56 mRNA transcripts remained unchanged between days 4 and 6 postinfection. (B) Quantification of ISG mRNA levels in LCMV-infected WT mice. Densitometric analysis of each lane was performed on scanned autoradiographs with NIH Image software (version 1.63), with each individual mRNA density normalized to that of the corresponding L32 loading control. Statistical analysis was performed with Student's t test. *, significant increase compared to uninfected mice (P < 0.05).
Increased ISG-49, ISG-54, and ISG-56 mRNA levels in response to CNS infection with LCMV are variably dependent on STAT1 and STAT2.
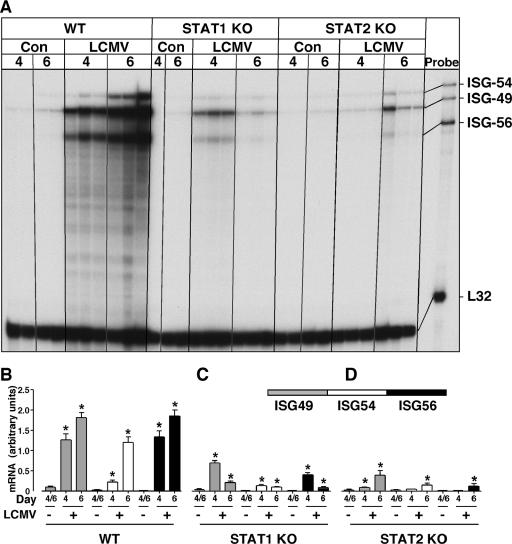
To determine whether the ISGs were regulated through the classical ISGF3/ISRE type I IFN pathway that requires STAT1 and STAT2, we next investigated ISG-49, ISG-54, and ISG-56 gene expression in LCMV-infected mice that were deficient in either STAT1 or STAT2. The levels of ISG-49, ISG-54, and ISG-56 mRNA in uninfected mice were similar between the different genotypes and similar to those seen in the previous experiment (Fig. 1). Following LCMV infection in WT (129/Sv) mice, the ISG-49, ISG-54, and ISG-56 mRNA levels increased significantly at day 4 and increased further by day 6 (Fig. 2A and B).
FIG. 2.
ISG-49, ISG-54, and ISG-56 mRNA levels in brains of sham- or LCMV-infected WT, STAT1 KO, and STAT2 KO mice. Mice (129/Sv) were manipulated as described in the legend to Fig. 1. (A) In WT mice, ISG-49, ISG-54, and ISG-56 mRNA levels were significantly (P < 0.05) increased by day 4 post-LCMV infection and continued to increase to day 6. There was a significant increase in the level of ISG-49, ISG-54, and ISG-56 mRNAs at day 4 postinfection in the STAT1 KO mice, which decreased but remained significantly above control levels by day 6. A minor but significant increase occurred in the level of ISG-49 mRNA only at day 4 postinfection in the STAT2 KO brain. At day 6 postinfection in STAT2 KO mice, ISG-49, ISG-54, and ISG-56 mRNA transcripts all increased slightly. (B, C, and D) Quantification of ISG expression in WT, STAT1 KO, and STAT2 KO mice, respectively. Densitometric analysis of each lane was performed on scanned autoradiographs with NIH Image software (version 1.63), with each individual mRNA density normalized to that of the corresponding L32 loading control. Statistical analysis was performed with Student's t test. *, significant increase compared to uninfected mice (P < 0.05).
Although considerably blunted compared with WT animals, LCMV infection of STAT1 KO (129/Sv) mice produced a significant increase in ISG-49 and ISG-56 mRNA levels at day 4 postinfection (Fig. 2A and C). However, the levels of these mRNA transcripts decreased at day 6 postinfection, although this was to levels still significantly above those seen in control STAT1 KO brains. An even smaller but significant increase in ISG-54 mRNA levels was seen at day 4 postinfection in STAT1 KO mice, and this did not change at 6 day postinfection. These findings indicate that in the brain the expression of the ISG-49, ISG-54, and ISG-56 mRNAs in response to LCMV infection required STAT1 for optimal induction.
STAT2 KO (129/Sv) mice differed from WT and STAT1 KO mice in their cerebral ISG response to LCMV infection. At day 4 postinfection in STAT2 KO mice, there was no significant change in either the ISG-54 or ISG-56 mRNA level, while there was a small but significant increase in the ISG-49 mRNA level (3.4-fold). By day 6 postinfection in STAT2 KO mice, small but significant increases were seen in cerebral ISG-49, ISG-54, and ISG-56 mRNA levels, with ISG-49 being the highest, followed by ISG-54 and ISG-56 (Fig. 2A and D). These results highlight a dominant but not total dependence on STAT2 signaling for the upregulation of the ISG-49, ISG-54, and ISG-56 genes in the brain following LCMV infection.
Differential anatomic localizations of ISG-49, ISG-54, and ISG-56 RNA transcripts in the brain following LCMV infection.
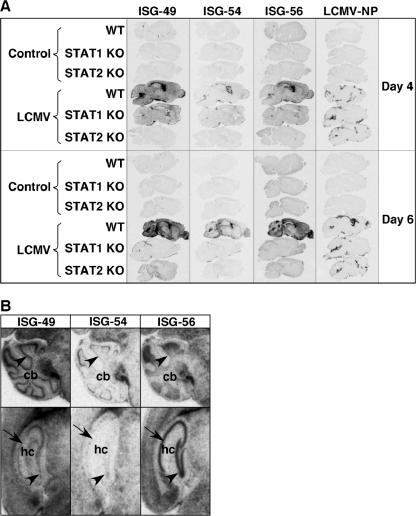
The gross anatomic localizations of the ISG-49, ISG-54, and ISG-56 RNA transcripts following LCMV infection were examined in the brain by in situ hybridization and compared for WT, STAT1 KO, and STAT2 KO mice. No ISG hybridization was detectable above the sense probe negative controls (not shown) in uninfected WT (Fig. 3A), STAT1 KO (not shown), or STAT2 KO (not shown) brain with ISG-49, ISG-54, or ISG-56 probes.
FIG. 3.
Anatomic localization of ISG-49, ISG-54, ISG-56, and LCMV NP RNA in the brain. WT, STAT1 KO, and STAT2 KO mice were injected i.c. as described in the legend to Fig. 1, and brains were removed at days 4 and 6 for in situ hybridization. Paraffin-embedded sagittal sections (8 μm) were hybridized with 33P-labeled cRNA probes transcribed from linearized ISG-49, ISG-54, ISG-56, or LCMV NP RNA plasmids and exposed to Kodak MR film for 3 days as outlined in Materials and Methods. (A) No hybridization above background of the ISG-49, ISG-54, ISG-56, or LCMV NP probes was evident in uninfected (control) brains. Following LCMV infection, in WT mice high levels of ISG-49 and ISG-56 hybridization were evident throughout the brain at day 4 and increased further by day 6. In contrast, there was a lower and more restricted level of ISG-54 hybridization. Increased ISG-49 and ISG-56 hybridization was also evident in the brain of STAT1 KO mice at day 4 but not day 6 postinfection, particularly in the meninges and ventricles. A small increase in ISG-49 hybridization was evident in the meninges and choroid plexus in the brain of STAT2 KO mice at day 6 postinfection. (B) Differential patterns of ISG-49, ISG-54, and ISG-56 hybridization at day 6 postinfection. In the Purkinje cell layer (arrowhead) of the cerebellum (cb), high levels of ISG-49 but low levels of ISG-54 and ISG-56 hybridization were seen. In the CA1 pyramidal neurons (arrow) and dentate gyrus (arrowhead) of the hippocampus (hc), high levels of ISG-56 but low levels of ISG-49 and ISG-54 hybridization were seen.
Following LCMV infection in WT brain, the overall changes in the levels of ISG-49, ISG-54, and ISG-56 RNA detected by in situ hybridization correlated with the RPA findings and increased progressively from day 4 to day 6, with the levels of ISG-49 and ISG-56 mRNAs being clearly higher than ISG-54 mRNA (Fig. 3A). ISG-49 RNA was distributed diffusely throughout the parenchyma, as well as in the meninges, with particularly high levels in the Purkinje cell layer of the cerebellum, corpus callosum, and choroid plexus (Fig. 3A). In contrast with ISG-49, ISG-54 RNA showed a much more restricted distribution, being found predominantly in the choroid plexus and paraventricular regions. Similar to ISG-49, ISG-56 RNA was distributed widely throughout the brain parenchyma and meninges, with highest levels found in olfactory bulb, dentate gyrus, and CA1 to CA4 pyramidal neurons. Clear differences were observed in the anatomic distributions of the three ISG RNAs, as highlighted in Fig. 3B. For example, while ISG-49 RNA was present at high levels in the Purkinje cell (Fig. 3B, upper panels, arrowhead), molecular, and granule neuron layers of the cerebellum, the ISG-54 and ISG-56 RNAs were seen largely in the Purkinje cell layer and granule neuron layer, respectively. Striking differences were also seen in the hippocampus, with high levels of ISG-56 RNA in the dentate gyrus (Fig. 3B, lower panels, arrowhead) and CA1 to CA2 pyramidal neuron layers (Fig. 3B, lower panels, arrows), while the ISG-49 and ISG-54 RNAs were little expressed by these structures.
Comparison of the distribution of ISG gene expression with LCMV NP gene expression in brains from wild-type mice revealed that the expression of the ISG-49 and ISG-56 RNA transcripts was far more widespread throughout the brain than was LCMV NP RNA (Fig. 3A). However, the restricted pattern of LCMV NP RNA expression in the meninges, ventricles, and choroid plexus overlapped more closely the expression of ISG-54 RNA.
Changes in the levels of the ISG-49, ISG-54, and ISG-56 RNAs detectable in the brain by in situ hybridization following LCMV infection of STAT1 or STAT2 KO mice also reflected those found by RPA (Fig. 3A). In LCMV-infected STAT1 KO mice, increased levels of the ISG-49 and ISG-56 RNAs were evident in the brain at day 4 but decreased by day 6 postinfection. RNA transcripts for both of these genes were present in the meninges and ventricles, as well as a diffuse pattern of expression throughout the parenchyma. In contrast, ISG-54 RNA was present in these brains at low levels only, at both day 4 and day 6 postinfection, and was localized predominantly to the meningeal areas. The only alteration in ISG RNA levels apparent in brains from LCMV-infected STAT2 KO mice compared with uninfected controls was for the ISG-49 gene, which showed a modest increase throughout the brain at day 6 postinfection.
ISG-49, ISG-54, and ISG-56 RNA transcripts show overlapping as well as differential cellular localizations following LCMV infection.
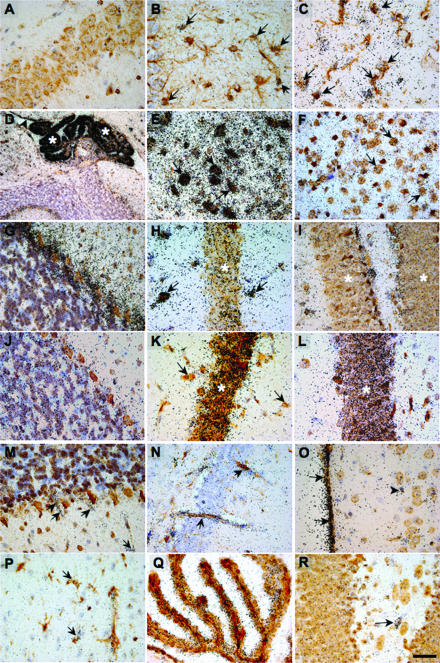
The cellular localizations of the ISG-49, ISG-54, and ISG-56 RNA transcripts were identified by dual-label in situ hybridization immunohistochemistry. An overall summary of the findings is given in Table 2, with selected images depicted in Fig. 4. For reference purposes, the hybridization signal for the ISG-49 RNA in the brain of a WT, noninfected mouse is given in Fig. 4A. This level of hybridization was representative of all three ISG RNA transcripts in noninfected WT brains and was similar to the hybridization signal observed for the sense (negative) control probe for each ISG. In WT brains at day 6 following LCMV infection, a high level of ISG-49 RNA (Fig. 4B to I) was seen in various neuronal populations, including neurons in the deep cerebellar gray matter (Fig. 4E, arrows), cortex (Fig. 4F, arrows) Purkinje cells (Fig. 4G), and CA2 pyramidal neurons (Fig. 4H). In addition, cells of the choroid plexus (Fig. 4D, asterisks) and ependymal layer (Fig. 4D, arrowhead), as well as astrocytes (Fig. 4B, arrows) and microglial cells (Fig. 4C, arrows), were also seen to be positive for ISG-49 RNA. Neurons were found to differentially express ISG-49 RNA, with neuronal subgroups in the brainstem (Fig. 4E, arrows) and hippocampal hilus (Fig. 4H, arrows) exhibiting very high levels, while the expression levels in hippocampal CA1 pyramidal neurons were lower (Fig. 4H, asterisk) and negligible in neurons of the dentate gyrus (Fig. 4I). In the brain of STAT1 KO mice at day 4 postinfection, ISG-49 RNA was not detectable in neurons (Fig. 4M to O); however, nonneuronal cell populations, including the meninges (Fig. 4O, arrows) and blood vessels (Fig. 4 M and N, arrows), were positive. In the brain of STAT2 KO mice at day 6 postinfection, expression of ISG-49 RNA was limited to meninges, blood vessels, and infiltrating immune cells compared to that seen in the LCMV-infected WT brain (not shown).
TABLE 2.
Summary of the cellular localizations of ISG-49, ISG-54, and ISG-56 RNA in the brains of mice following infection with LCMV
| Mouse genotype | Hybridization signal increase at indicated locationa
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glia
|
Neurons
|
Other
|
Infiltrating immune cells | ||||||||||||||
| Astrocytes | Microglia | Olfactory bulb | Neocortex | Purkinje cells | Granule cells | Deep cerebellar neurons | CA1 | CA2 | CA3 & CA4 | Hippocampal hilus | Dentate gyrus | Choroid plexus | Ependymal layer | Meninges | Blood vessels | ||
| ISG-49 | |||||||||||||||||
| WT LCMV (day 6) | + | + | ++ | ++ | ++ | +++ | +++ | + | + | ++ | +++ | + | +++ | ++ | +++ | ++ | + |
| STAT1 KO LCMV (day 4) | − | + | − | − | − | − | − | − | − | − | − | − | + | + | ++ | ++ | + |
| STAT2 KO LCMV (day 6) | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | + | + |
| ISG-54 | |||||||||||||||||
| WT LCMV (day 6) | ++ | + | +++ | + | +++ | + | ++ | + | + | + | + | − | +++ | + | + | ++ | + |
| STAT1 KO LCMV (day 4) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + |
| STAT2 KO LCMV (day 6) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| ISG-56 | |||||||||||||||||
| WT LCMV (day 6) | +++ | + | − | ++ | − | + | +++ | +++ | +++ | ++ | − | +++ | +++ | ++ | +++ | ++ | + |
| STAT1 KO LCMV (day 4) | − | + | − | − | − | − | − | − | − | − | − | − | ++ | − | ++ | + | + |
| STAT2 KO LCMV (day 6) | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | + | + |
For scoring, hybridization signals were compared with those from uninfected control brains of the same genotype. Symbols: −, no increase; +, small increase; ++, moderate increase; +++, large increase.
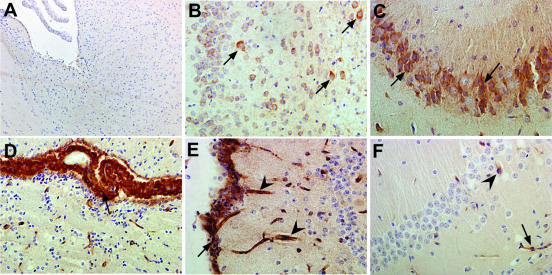
FIG. 4.
Cellular localization of ISG-49, ISG-54, and ISG-56 RNA in the brain. Mice were injected i.c. with saline or 250 PFU of LCMV (ARM), and the brain was removed at day 4 and day 6 postinfection for in situ hybridization and immunohistochemistry. Eight-micrometer-thick paraffin-embedded sections were hybridized with 33P-labeled cRNA probes transcribed from linearized ISG-49 or ISG-56 RPA plasmids and then processed for immunohistochemistry for cell-specific markers to identify astrocytes, microglia, and neurons as outlined in Materials and Methods. Slides were coated with photographic emulsion, developed after 2 weeks, and visualized with bright-field microscopy. Photomicrographs are of uninfected (A) and day 6 LCMV-infected (B to L) brain sections from WT mice hybridized for ISG-49 (A to I) or ISG-56 (J to L). ISG-49 hybridization was localized to astrocytes (B, arrows, GFAP stained) and microglia (C, arrows, lectin stained), gigantocellular neurons (E, arrows, NeuN stained), cortical neurons (F, arrows, NeuN stained), Purkinje cells (G, arrows NeuN stained), and hippocampal hilus (H, arrows, NeuN stained). Lower magnification revealed high-level expression of ISG-49 (D, NeuN stained) in the choroid plexus (asterisk) and ependymal layer (arrowhead). In the hippocampus, there was low ISG-49 hybridization in neurons of the CA1 region (H, asterisk, NeuN stained) and little hybridization to dentate gyrus (I, asterisk, NeuN stained). By contrast, ISG-56 hybridization was very high in both CA1 (K, asterisk, NeuN stained) and dentate gyrus (L, asterisk, NeuN stained) neurons of the hippocampus but low in Purkinje cells (J, NeuN stained) and neurons of the hippocampal hilus (K, arrows, NeuN stained). In LCMV infection of STAT1 KO mice at day 4 postinfection, ISG-49 hybridization was localized to vascular endothelium (M and N, arrows) and meninges (O, arrows, NeuN stained). A similar cellular localization was found for ISG-56 hybridization, which was seen predominantly in vascular endothelium and microglia (P, arrows, lectin stained) and choroid plexus (Q, lectin stained). Note the lack of hybridization for both ISG-49 (M to O) and ISG-56 (R) to neurons. Bars, 50 μm (A-C, E-G, J, M, O, and P), 100 μm (H, I, K, L, N, and R), 150 μm (Q), and 300 μm (D).
In the brain of WT mice at day 6 postinfection, ISG-54 RNA was localized to cells of the choroid plexus, astrocytes, endothelial cells, and neurons, particularly those adjacent to the ventricles, as well as the Purkinje cells (not shown). In the brain of STAT1 KO mice at day 4 postinfection, ISG-54 RNA was localized to the infiltrating cells only (not shown).
Like that of ISG-49, ISG-56 RNA was localized to a number of different cell types and was particularly high in various neuronal populations in the brains of LCMV-infected WT mice. However, there were also notable differences in the expression of the ISG-56 gene compared with the ISG-49 gene. In particular, ISG-56 RNA showed little change in the Purkinje cell layer (Fig. 4J) and neurons of the hippocampal hilus, while very high levels of this RNA were observed in both the CA1 pyramidal neurons (Fig. 4K, asterisk) and dentate gyrus (Fig. 4L, asterisk) of the hippocampus. Like ISG-49 at day 4 postinfection, in the STAT1 KO mouse brain no neuronal expression of ISG-56 RNA was detectable (Fig. 4P to R), while nonneuronal cell populations, including blood vessels and microglia (Fig. 4R, arrows), the choroid plexus (Fig. 4Q), and meninges (not shown), were positive. In the brain of STAT2 KO mice at day 6 postinfection, expression of ISG-56 RNA was limited to meninges, blood vessels, and infiltrating immune cells compared to that seen in the LCMV-infected WT brain (not shown).
Cerebral ISG-56 protein levels are increased in parallel with RNA transcripts following LCMV infection.
Although the expression levels of all three ISG RNA transcripts were increased significantly in the brain following LCMV infection, whether this was reflected by changes in the corresponding proteins remained unknown. A rabbit polyclonal antiserum to murine ISG-56, used previously for immunoblotting analysis of ISG-56 protein in cultured murine cells (37), was employed in the present study to determine the level and cellular localization of ISG-56 protein in the brain following LCMV infection. Although this rabbit antiserum proved to be unsuitable for immunoblotting analysis of whole brain tissue lysates, we were successful in using this reagent for immunohistochemistry (Fig. 5). On sections of brain tissue from sham-inoculated WT, STAT1 KO, or STAT2 KO mice, there was very little detectable staining with either a rabbit polyclonal preimmune serum (not shown) or the ISG-56 antiserum (Fig. 5A). By contrast, sections of brain tissue from LCMV-infected WT mice at day 4 and day 6 postinfection exhibited strong immunostaining that was localized to various cell populations throughout the brain, including cortical neurons (Fig. 5B, arrows), CA2 pyramidal neurons (Fig. 5C, arrows), the choroid plexus (Fig. 5D, arrow), and the meninges and vascular endothelium (Fig. 5E, arrow and arrowhead, respectively). Similar to the RNA transcript, ISG-56 protein was increased to much lower levels in the brain of LCMV-infected STAT1 KO mice and was associated mainly with the vascular endothelium (Fig. 5F, arrows), meninges, and choroid plexus (not shown). There was little detectable protein above control levels in the brain of LCMV-infected STAT2 KO mice. Overall, these findings show that there was good concordance between the ISG-56 protein and corresponding RNA transcripts, both in relative levels and cellular localizations in the brain following LCMV infection.
FIG. 5.
Localization and levels of ISG-56 protein in the brains of sham- or LCMV-infected mice. Mice were injected i.c. with saline or 250 PFU of LCMV (ARM), and the brains were removed at day 4 and day 6 postinfection and processed for immunohistochemistry for ISG-56 protein as outlined in Materials and Methods. (A) Sham-infected WT control specimen. ISG-56 protein immunostaining was not detectable and was comparable to adjacent sections treated with a preimmune rabbit sera. (B to E) LCMV-infected WT specimens (day 6). Prominent neuronal staining was evident in the cortex (B, arrows) and CA2 region of the hippocampus (C, arrows). Strong immunostaining for ISG-56 was also seen in the choroid plexus (D, arrow), meninges (E, arrow), and vascular endothelium (E, arrowhead). (F) LCMV-infected STAT1 KO specimen (day 6). Compared with LCMV-infected WT mice, ISG-56 immunostaining was reduced considerably. In this representative section, positive immunostaining can be seen in the vascular endothelium (arrow) and some scattered cells (arrowhead) likely to be microglia. Total magnification, ×100 (A and D) and ×200 (B, C, E, and F).
Cerebral ISG-49, ISG-54, and ISG-56 RNA levels and ISG-56 protein levels are increased significantly throughout the brain following WNV infection.
Intranasal infection of immunocompetent adult C57BL/6 mice with WNV resulted in fatal encephalitis and death of the animals by day 7 postinfection. RPA analysis of brain RNA at day 7 postinfection with WNV revealed that there were significant increases in the levels of the ISG-49, ISG-54, and ISG-56 transcripts, with ISG-49 > ISG-56 ≥ ISG-54 compared with sham-infected controls (Fig. 6A). In situ hybridization analysis revealed widespread and diffuse expression of the ISG transcripts throughout the brain of WNV-infected mice compared with controls (Fig. 6B). On the whole, the regional distributions of the three ISG transcripts in the WNV-infected brain overlapped and were particularly high in various neuronal populations throughout the brain, most prominently in the granule neuron layer of the cerebellum (Fig. 6B). Thus, as with LCMV infection, WNV infection of the brain was associated with a robust and widespread increase in the expression of the ISG-49, ISG-54, and ISG-56 genes that was very prominent in neurons.
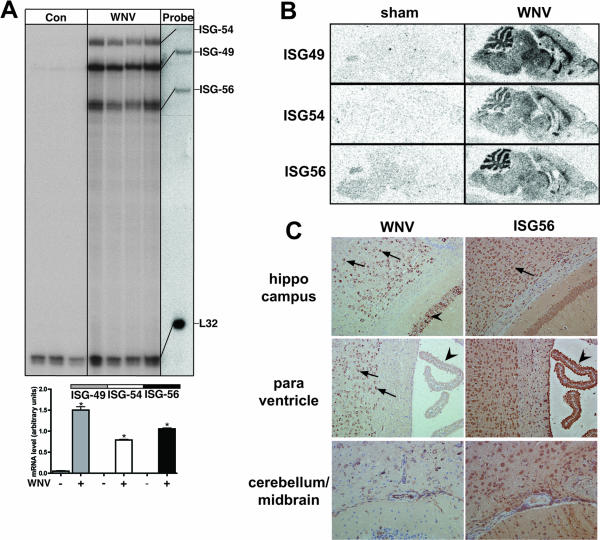
FIG. 6.
Regulation of ISG-49, ISG-54, and ISG-56 mRNA and ISG-56 protein in the brains of WNV-infected WT mice. Mice were inoculated intranasally with vehicle or vehicle plus 6 × 104 PFU of WNV (Sarafend strain). At day 7 postinfection, mice were euthanized and the brains were removed and analyzed as described in Materials and Methods. (A) RNase protection analysis revealed that compared with sham-inoculated controls the levels of ISG-49, ISG-54, and ISG-56 mRNA transcripts increased significantly (*, P < 0.05) in WNV infection. (B) Anatomic localization of ISG RNA transcripts in the brain by in situ hybridization. Compared with sham-inoculated controls, during WNV infection the levels of all three ISG RNAs increased markedly and were distributed widely throughout the brain. (C) Immunohistochemical localization of the WNV nonstructural protein NS1 compared with the ISG-56 protein. Infection of many neurons by WNV is evident in most regions of the cerebrum but was largely absent from the choroid plexus (left middle panel, arrows) and cerebellar neurons. ISG-56 protein was found to be more widespread in the majority of neurons in both cerebrum and cerebellum, as well as in the choroid plexus (right middle panel, arrowhead). Total magnification, ×100.
Immunohistochemical analysis was performed to identify regions and cells in the brain that were infected with WNV, as well as on adjacent sections producing the ISG-56 protein. WNV NS1 protein immunostaining was detectable at high levels in the forebrain and midbrain regions but not in the cerebellum. At the cellular level, WNV NS1 protein was located predominantly in various neuronal populations in these regions, including cortical (Fig. 6C, upper and middle left panels, arrows) and hippocampal pyramidal (Fig. 6C, upper left panel, arrowheads) neurons. The lower left panel of Fig. 6C shows an area of midbrain and cerebellar cortex that, in contrast to the cerebral cortex and hippocampus, contained few WNV NS1-positive cells. Similarly, the choroid plexus (Fig. 6C, middle left panel, arrowhead) was largely devoid of WNV NS1 protein. Analysis of adjacent sections for the ISG-56 protein revealed that while there was general overlap of this protein with WNV NS1 protein in specific neurons, such as cortical neurons (Fig. 6C, upper right panel, arrows), the distribution of the ISG-56 protein was far more widespread and included various regions such as the cerebellum (Fig. 6C, bottom right panel) and cells such as the choroid plexus (Fig. 6, middle right panel, arrowhead). These findings indicate that the production of ISG-56 protein is increased dramatically during CNS infection with WNV and localizes with, as well as extends beyond, regions of viral infection.
DISCUSSION
The ability of type I (IFN-α/β) and type II (IFN-γ) IFNs to confer host resistance to microbial infection is accomplished by the regulation of a large number of cellular genes, from many of which the proteins produced have direct antimicrobial actions (6, 34). Previous DNA microarray studies showed that during infection of the CNS or neuronal cultures with different classes of viruses, some of the most highly upregulated genes belonged to the structurally related, TPR-containing ISG family represented by the ISG-49, ISG-54, and ISG-56 genes (18, 30). Similar findings were observed in cultured murine neurons following treatment with IFN-α (38). However, the regulation and role of the ISG-49, ISG-54, and ISG-56 genes in viral infection of the CNS or other tissues are not well defined. Here we analyzed the simultaneous temporal and spatial expression of these genes in the brains of mice following infection with two different neurotropic viruses, LCMV and WNV. Our findings highlighted the dynamic and coordinate regulation of these genes during infection of the CNS with both viruses. Also notable, particularly in the case of LCMV infection, were differences in the level and cellular localization of expression of the individual ISG genes. In all, these findings provide novel insights into the regulation and spatial expression patterns of the ISG-49, ISG-54, and ISG-56 gene family during viral infection in vivo and suggest that these genes may have nonredundant functions as gatekeepers of antiviral host defense in the CNS.
Despite the restricted localization of LCMV to the meninges, choroid plexus, and ependymal regions of the brain, the induction of the ISG-49 and ISG-56 genes, in particular, occurred not only in these regions but throughout the brain, while expression of ISG-54 exhibited much more restricted anatomic localization. The early and widespread expression of the ISG-49 and ISG-56 genes constitutes part of a more generalized response of the brain to LCMV infection that, as we have previously noted, encompasses a number of other genes involved in the host response, including STAT1 (I. L. Campbell, unpublished data), IRF-9, IRF-7 (26), and ISG-15 (38). This suggests that the expression of the ISG-49 and ISG-56 genes, in particular, may contribute to a common defensive response in the brain against viral infection.
It was notable from our findings that neurons showed some of the most prominent cellular responses to viral infection of the brain, with substantial induction of the ISG genes and the ISG-56 protein. Since neurons are essentially nonrenewable, this raises the possibility that the high-level expression of these genes could represent an antiviral response to protect these crucial cells. Although in the week following intracranial inoculation, LCMV infection remained limited largely to the brain surfaces in WT mice, a similar distribution of virus was also seen in STAT1 and STAT2 KO mice that did not exhibit a significant host response with upregulation of the ISG genes. Thus, factors other than those associated with the neuronal ISG response limit LCMV infection in the acute phase of infection prior to the death of the host. However, STAT2 KO mice, which survive intracranial inoculation with LCMV and lack an ISG response, develop a persistent infection of the CNS in which the virus spreads to most of the brain, infecting neurons (M. J. Hofer and I. L. Campbell, unpublished observations).
While in general there was considerable coexpression of the ISG-49 and ISG-56 genes by neurons, notable differences were also seen in the expression of these, as well as the ISG-54 gene, in specific neuronal populations. Thus, for a subset of specialized neurons there was a clear compartmentalization in the neuronal response to LCMV infection, suggesting that there may be cell-specific functions linked to the expression of the ISG-49, ISG-54, and ISG-56 genes. A variety of factors are known to regulate the expression of the murine ISG-49, ISG-54, and ISG-56 genes in cells in vitro, including type I and type II IFNs (35, 37), dsRNA, and viruses such as SeV (37). We have noted previously that, following intracranial infection of mice with LCMV, there are low levels of local expression of the type I IFN genes while higher expression levels of the type II IFN gene, IFN-γ, occur late in infection (1, 26). Despite the apparent low levels of type I IFN production in the LCMV-infected brain, our findings point to these cytokines as having a key role as the dominant stimulatory factors mediating ISG-49, ISG-54, and ISG-56 gene expression. First, we observed that optimal induction of the expression of these genes was dependent on the key type I IFN signal transduction molecules STAT1 and STAT2. Second, both the relative levels and anatomic and cellular distributions of the ISG-49, ISG-54, and ISG-56 genes found in the brain following LCMV infection were similar to those in transgenic mice with chronic astrocyte production of the type I IFN, IFN-α (unpublished observations). Finally, previous in vitro studies show that neurons are very sensitive to IFN-α and markedly upregulate their ISG-49, ISG-54, and ISG-56 gene expression levels (38). In the case of WNV infection, IFN-α/β production is found in the CNS of infected mice (32) and is therefore likely to have a dominant role in upregulating ISG gene expression.
The finding that there are neuron-specific differences in the expression of the ISG-49, ISG-54, and ISG-56 genes raises the question of how these ISGs show such cellular specificity in response to an identical stimulating factor. One possible explanation could be the presence of differences in the promoter sequences of these genes. The promoter regions of ISG-54 and ISG-56 contain one and two functional ISRE sites, respectively (3), and additional differences cannot be excluded, since these gene regions have not yet been thoroughly investigated. Differences in the promoter elements of these genes may also help to explain the overall lower levels of induction of the ISG-54 gene that were observed in the brain following LCMV infection and in the GFAP-IFN-α transgenic mouse. Characterization of the promoter region of the murine ISG-49 gene has not been reported. However, it will be of interest to learn whether the murine ISG-49 gene, like its human orthologue, ISG-60 (8), contains two functional ISRE sites.
Previous in vitro cell culture studies by our group established that neurons (38) and astrocytes (C. Wacher and I. L. Campbell, unpublished observations) have an obligate requirement for STAT1 for type I IFN induction of the ISG-49, ISG-54, and ISG-56 genes; this was confirmed for induction of the ISG-49, ISG-54, and ISG-56 genes following LCMV infection in the present study. However, numerous nonneuronal cells in the brain, including the vascular endothelium, microglia, meningeal cells, and choroid plexus cells, clearly exhibited STAT1-independent induction of these genes. It is unclear presently as to the identity of the factor(s) responsible for the STAT1-independent upregulation of the ISG-49, ISG-54, and ISG-56 genes in these cells in the brain following LCMV infection. STAT1-independent signaling leading to functional responses can be mediated by the type I IFNs in the CNS (39). Therefore, it is possible that these cytokines constitute such factors. Activated IRF-3 can also mediate the direct IFN-α/β- and STAT1-independent induction of ISG-56 via the ISREs of the ISG-56 promoter (15). IRF-3 is normally latent within cells but is activated in direct response to viral infection (19). In the present study, there was a lack of concordance between the sites of LCMV infection and the expression of the ISG-56 gene, as well as little upregulation of ISG-56 despite the presence of high levels of LCMV in STAT2 KO mice. These observations suggest that IRF-3 may not be the primary mediator of the STAT-independent stimulation of ISG-56 gene expression found in the CNS during LCMV infection.
Overall, the present studies implicate ISG-49, ISG-54, and ISG-56 gene expression in antiviral host defense in the CNS and, in particular, in neurons and lay the foundations for ongoing studies examining further the regulation of these genes in vivo. Of course, a central question concerns the precise mode of action of the products of these genes in vivo. From in vitro biochemical and cellular studies, it is known that the human counterparts of ISG-54 and ISG-56 are located intracellularly and that both the murine and the human proteins are capable of inhibiting cellular protein synthesis by binding to either the “c” or the “e” subunit of the translation initiation factor eIF3 (16, 37). ISG-56 has been identified as a key antiviral effector suppressing the hepatitis C virus internal ribosome entry site and inhibiting viral RNA translation during the IFN response (41). On the other hand, cellular expression of the HuISG-56 gene by transfection failed to inhibit replication of either vesicular stomatitis virus or encephalomyocarditis virus, despite evidence that cellular protein synthesis was reduced (17). Therefore, at present the function of ISG-49, ISG-54, and ISG-56 in the antiviral host response in vivo is uncertain and may be limited to specific classes of viruses. Clarification of these outstanding issues will no doubt come in the future from studies aimed at manipulating the expression of these genes in mice to accomplish their tissue-specific overexpression or deletion.
Acknowledgments
This work was supported by U.S. Public Health Service NIH grant MH62231 and by a startup grant from the University of Sydney to I.L.C. and by NIH grant CA 68782 to G.C.S. M.J.H. is a postdoctoral fellow of the Deutsche Forschungsgemeinschaft (HO3298/1-1). M.M. is a postdoctoral fellow of the Deutsche Forschungsgemeinschaft (Mu17-07/3-1) and was also supported by the “Innovative Medical Research” fund of the University of Münster Medical School, Münster, Germany. D.R.G. is supported by an Australian Postgraduate Award.
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Asensio, V. C., C. Kincaid, and I. L. Campbell. 1999. Chemokines and the inflammatory response to viral infection in the central nervous system with a focus on lymphocytic choriomeningitis virus. J. Neurovirol. 5:65-75. [DOI] [PubMed] [Google Scholar]
- 2.Badley, J. E., G. A. Bishop, T. St. John, and J. A. Frelinger. 1988. A simple, rapid method for the purification of poly A+ RNA. BioTechniques 6:114-116. [PubMed] [Google Scholar]
- 3.Bluyssen, H. A., R. J. Vlietstra, P. W. Faber, E. M. Smit, A. Hagemeijer, and J. Trapman. 1994. Structure, chromosome localization, and regulation of expression of the interferon-regulated mouse Ifi54/Ifi56 gene family. Genomics 24:137-148. [DOI] [PubMed] [Google Scholar]
- 4.Buchmeier, M. J., R. M. Welsh, F. J. Dutko, and M. B. Oldstone. 1980. The virology and immunobiology of lymphocytic choriomeningitis virus infection. Adv. Immunol. 30:275-331. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, I. L., M. V. Hobbs, P. Kemper, and M. B. A. Oldstone. 1994. Cerebral expression of multiple cytokine genes in mice with lymphocytic choriomeningitis. J. Immunol. 152:716-723. [PubMed] [Google Scholar]
- 6.Decker, T., S. Stockinger, M. Karaghiosoff, M. Muller, and P. Kovarik. 2002. IFNs and STATs in innate immunity to microorganisms. J. Clin. Investig. 109:1271-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Der, S. D., A. Zhou, B. R. G. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon a, b, or g using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Veer, M. J., H. Sim, J. C. Whisstock, R. J. Devenish, and S. J. Ralph. 1998. IFI60/ISG60/IFIT4, a new member of the human IFI54/IFIT2 family of interferon-stimulated genes. Genomics 54:267-277. [DOI] [PubMed] [Google Scholar]
- 9.Doherty, P. C., J. E. Allan, F. Lynch, and R. Ceredig. 1990. Dissection of an inflammatory process induced by CD8+ T cells. Immunol. Today 11:55-59. [DOI] [PubMed] [Google Scholar]
- 10.Dudov, K. P., and R. P. Perry. 1984. The gene family encoding the mouse ribosomal protein L32 contains a uniquely expressed intron-containing gene and an unmutated processed gene. Cell 37:457-468. [DOI] [PubMed] [Google Scholar]
- 11.Durbin, J. E., R. Hackenmiller, M. C. Simon, and D. E. Levy. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84:443-450. [DOI] [PubMed] [Google Scholar]
- 12.Elco, C. P., J. M. Guenther, B. R. Williams, and G. C. Sen. 2005. Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-κB, and interferon but not Toll-like receptor 3. J. Virol. 79:3920-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geiss, G., G. Jin, J. Guo, R. Bumgarner, M. G. Katze, and G. C. Sen. 2001. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J. Biol. Chem. 276:30178-30182. [DOI] [PubMed] [Google Scholar]
- 14.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral responses and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 15.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo, J., D. J. Hui, W. C. Merrick, and G. C. Sen. 2000. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 19:6891-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo, J., K. L. Peters, and G. C. Sen. 2000. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology 267:209-219. [DOI] [PubMed] [Google Scholar]
- 18.Johnston, C., W. Jiang, T. Chu, and B. Levine. 2001. Identification of genes involved in the host response to neurovirulent alphavirus infection. J. Virol. 75:10431-10445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131-137. [DOI] [PubMed] [Google Scholar]
- 20.King, N. J., and A. M. Kesson. 2003. Interaction of flaviviruses with cells of the vertebrate host and decoy of the immune response. Immunol. Cell Biol. 81:207-216. [DOI] [PubMed] [Google Scholar]
- 21.Kitamura, Y., O. Spleiss, H. Li, T. Taniguchi, H. Kimura, Y. Nomura, and P. J. Gebicke-Haerter. 2001. Lipopolysaccharide-induced switch between retinoid receptor (RXR) alpha and glucocorticoid attenuated response gene (GARG)-16 messenger RNAs in cultured rat microglia. J. Neurosci. Res. 64:553-563. [DOI] [PubMed] [Google Scholar]
- 22.Lee, C. G., J. Demarquoy, M. J. Jackson, and W. E. O'Brien. 1994. Molecular cloning and characterization of a murine LPS-inducible cDNA. J Immunol. 152:5758-5767. [PubMed] [Google Scholar]
- 23.Meraz, M. A., J. M. White, K. C. F. Sheehan, E. A. Bach, S. J. Rodig, A. S. Dighe, D. H. Kaplan, J. K. Riley, A. C. Greenlund, D. Campbell, K. Carver-Moore, R. N. DuBois, R. Clark, M. Aguet, and R. D. Schreiber. 1996. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84:431-442. [DOI] [PubMed] [Google Scholar]
- 24.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ousman, S. S., and I. L. Campbell. 2005. Regulation of murine interferon regulatory factor (IRF) gene expression in the central nervous system determined by multiprobe RNase protection assay. Methods Mol. Med. 116:115-134. [DOI] [PubMed] [Google Scholar]
- 26.Ousman, S. S., J. Wang, and I. L. Campbell. 2005. Differential regulation of interferon regulatory factor (IRF)-7 and IRF-9 gene expression in the central nervous system during viral infection. J. Virol. 79:7514-7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park, C., S. Li, E. Cha, and C. Schindler. 2000. Immune response in Stat2 knockout mice. Immunity 13:795-804. [DOI] [PubMed] [Google Scholar]
- 28.Patterson, C. E., J. K. Daley, and G. F. Rall. 2002. Neuronal survival strategies in the face of RNA viral infection. J. Infect. Dis. 186(Suppl. 2):S215-S219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Platanias, L. C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5:375-386. [DOI] [PubMed] [Google Scholar]
- 30.Prehaud, C., F. Megret, M. Lafage, and M. Lafon. 2005. Virus infection switches TLR-3-positive human neurons to become strong producers of beta interferon. J. Virol. 79:12893-12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Preston, C. M., A. N. Harman, and M. J. Nicholl. 2001. Activation of interferon response factor 3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75:8909-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samuel, M. A., and M. S. Diamond. 2005. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. J. Virol. 79:13350-13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarkar, S. N., and G. C. Sen. 2004. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol. Ther. 103:245-259. [DOI] [PubMed] [Google Scholar]
- 34.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 35.Smith, J. B., and H. R. Herschman. 1996. The glucocorticoid attenuated response genes GARG-16, GARG-39, and GARG-49/IRG2 encode inducible proteins containing multiple tetratricopeptide repeat domains. Arch. Biochem. Biophys. 330:290-300. [DOI] [PubMed] [Google Scholar]
- 36.Stark, G. R., I. M. Kerr, B. R. G. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 37.Terenzi, F., S. Pal, and G. C. Sen. 2005. Induction and mode of action of the viral stress-inducible murine proteins, P56 and P54. Virology 340:116-124. [DOI] [PubMed] [Google Scholar]
- 38.Wang, J., and I. L. Campbell. 2005. Innate STAT1-dependent genomic response of neurons to the antiviral cytokine alpha interferon. J. Virol. 79:8295-8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, J., R. D. Schreiber, and I. L. Campbell. 2002. STAT1 deficiency unexpectedly and markedly exacerbates the pathophysiological actions of IFN-α in the central nervous system. Proc. Natl. Acad. Sci. USA 99:16209-16214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, T., and E. Fikrig. 2004. Immunity to West Nile virus. Curr. Opin. Immunol. 16:519-523. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Y., M. Lobigs, E. Lee, and A. Mullbacher. 2003. CD8+ T cells mediate recovery and immunopathology in West Nile virus encephalitis. J. Virol. 77:13323-13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]